SUMMARY
Trimethylation on H3K27 (H3K27me3) mediated by the Polycomb repressive complex 2 (PRC2) has been linked to embryonic stem (ES) cell identity and pluripotency. EZH2, the catalytic subunit of PRC2, has been reported as the sole histone methyltransferase which methylates H3K27 and mediates transcriptional silencing. Analysis of Ezh2−/− ES cells suggests existence of additional enzyme(s) catalyzing H3K27 methylation. We have identified EZH1, a homolog of EZH2 that is physically present in a non-canonical PRC2 complex, as an H3K27 methyltransferase in vivo and in vitro. EZH1 colocalizes with the H3K27me3 mark on chromatin, and preferentially preserves this mark on development-related genes in Ezh2−/− ES cells. Depletion of Ezh1 in cells lacking Ezh2 abolishes residual methylation on H3K27 and derepresses H3K27me3 target genes, demonstrating a role of EZH1 in safeguarding ES cell identity. Ezh1 partially complements Ezh2 in executing pluripotency during ES cell differentiation, suggesting that cell fate transitions require epigenetic specificity.
INTRODUCTION
Polycomb group (PcG) proteins serve in transcriptional control to preserve cellular identity and maintain epigenetic memory during metazoan development. Three PcG proteins, enhancer of zeste 2 (EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 homolog (SUZ12), comprise the core of the Polycomb repressive complex 2 (PRC2), which mediates methylation on histone H3 lysine 27 (H3K27) (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002). EZH2, a SET domain containing histone methyltransferase (HMTase), functions as the catalytic subunit of PRC2. EED contains five repeats of WD40 domains and functions as a scaffold protein by physically linking EZH2 and histone H3 substrates (Tie et al., 2007). SUZ12 is required for nucleosome recognition and stability of EZH2 (Nekrasov et al., 2005; Pasini et al., 2004). In addition to the essential role of EED and SUZ12 in PRC2, histone-binding protein RBBP4 (or RbAp48/46) and the zinc finger protein AEBP2 bind to PRC2 and optimize its enzymatic activity in vitro (Cao and Zhang, 2004).
Mouse embryos lacking Eed, Ezh2 or Suz12 show defects in gastrulation around 7 – 9 days post coitus (Faust et al., 1995; O’Carroll et al., 2001; Pasini et al., 2004). In addition to essential roles in early development, PRC2 is required for diverse physiological functions including B-lymphoid development, myogenic differentiation, imprinted X-chromosome inactivation, and reprogramming of migrating primordial germ cells (Caretti et al., 2004; Kalantry et al., 2006; Seki et al., 2007; Su et al., 2003). Moreover, PRC2 members are associated with cancer (Valk-Lingbeek et al., 2004).
Three states of methylation, mono-, di-, and tri-, are observed on H3K27. H3K27me3 is characteristic of PcG target genes and functional in repression (Schwartz and Pirrotta, 2007). Genome-wide location analysis in embryonic stem (ES) cells has revealed that many PcG target genes encode transcription factors important in development (Boyer et al., 2006). Derepression of PcG target genes in Eed−/− ES cells suggests a requirement of PRC2 in maintaining ES cell pluripotency and plasticity. Suz12 is implicated in regulating ES cell differentiation (Pasini et al., 2007). However, Ezh2−/− ES cells have not been isolated and Ezh2-deficient blastocysts have been reported to be impaired in outgrowth (O’Carroll et al., 2001). It has been speculated that EZH2 may have independent functions that do not involve H3K27 methylation (Martin and Zhang, 2005; Pasini et al., 2007; Rajasekhar and Begemann, 2007; Schwartz and Pirrotta, 2007).
Higher eukaryotes often have multiple HMTases with similar specificity, for example, six HMTases acting on H3K4 and seven H3K9 HMTases (Glaser et al., 2006; Martin and Zhang, 2005). There are two sequence homologs of EZH, including EZH2 and EZH1. Human Ezh1 has been reported to interact with Eed by yeast-2-hybrid and coIP assays (Jones et al., 1998; van Lohuizen et al., 1998). Expression of murine Ezh1 restores gene repression in S. cerevisiae mutants impaired in telomeric silencing (Laible et al., 1997). However, whether EZH1 is part of PRC2 or contains enzymatic activity is unknown.
As EZH2 has been reported as the sole HMTase acting on H3K27, we have investigated the role of Ezh2 in mouse ES cells in an effort to determine whether Ezh2 is directly required for establishment and maintenance of ES cell identity and pluripotency. We generated a conditional knockout allele of Ezh2 and established Ezh2−/− ES cells. Despite global loss of di- and trimethyaltion on H3K27, Ezh2−/− ES cells preserve H3K27me3 at a subset of developmental genes and show robust H3K27me1, indicating that the EZH2-containing PRC2 complex is not the sole HMTase responsible for methylation on H3K27. Importantly, we have demonstrated that EZH1 is part of a non-canonical PRC2 complex that catalyzes addition of methyl groups on H3K27 and prevents the derepression of PRC2 target genes.
RESULTS
Residual H3K27me3 and robust H3K27me1 in Ezh2−/− ES cells
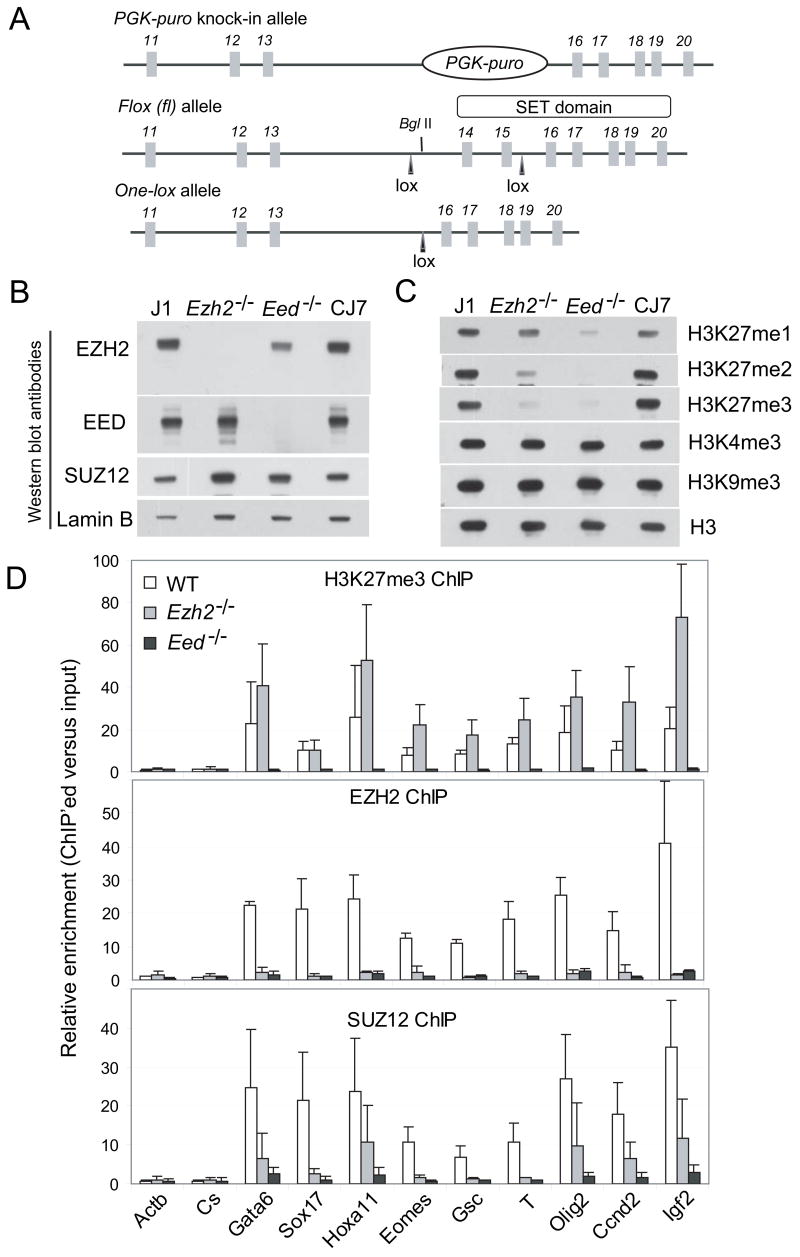
Previously, O’Carroll et al. failed to establish Ezh2−/− ES cells through direct outgrowth of the inner cell mass, and hypothesized an essential role in ES cell derivation (O’Carroll et al., 2001). To investigate the role of Ezh2 in ES cell pluripotency, we generated a conditional knockout allele of Ezh2 by inserting loxP (fl) sites flanking exons 14 and 15, which encode part of the catalytic SET domain (Figure 1A). Deletion of exon 14 – 15 introduces a frameshift and subsequent termination mutation, which effectively deletes the SET domain.
Figure 1. Altered methylation on H3K27 in Ezh2−/− cells.
(A) Targeting strategy to generate Ezh2 conditional and knockout alleles. Exon number is indicated above each exon shown in boxes. Lines represent introns.
(B–C) Western blot analysis of nuclear extracts (B) and histones (C) isolated from wild-type (J1 and CJ7) and mutant (Ezh2−/− and Eed−/−) ES cells.
(D) ChIP-qPCR analysis of H3K27me3, EZH2 and SUZ12 binding at promoters of PcG target genes in wild-type (WT), Ezh2−/− and Eed−/− ES cells. The y-axis shows relative enrichment of ChIP’ed DNA versus the inputs. Actb promoter and an intergenetic region (Cs) serve as negative controls.
By either sequentially targeting both alleles of Ezh2 locus or in vitro Cre excision of the fl allele of Ezh2fl/fl (Supplemental notes), we were able to establish Ezh2−/− ES cells, showing a non-essential role of Ezh2 in ES cell maintenance. Western blot analysis confirmed the absence of EZH2 (or a shorter form) in Ezh2−/− cells (Figure 1B), suggesting that the deletion of the SET domain in vivo may adversely affect protein folding and result in degradation of truncated EZH2. Levels of EED and SUZ12 are normal in Ezh2−/− cells. SUZ12 coimmunoprecipitates with EZH2 in Eed−/− but not in Ezh2−/− cells (Figure S1A).
Ezh2−/− mouse embryos are deformed and underdeveloped as compared to wild-type littermates and die at ~ E8.5 (Figure S1B). To reassess whether Ezh2 is required for de novo establishment of ES cells, we sought to isolate null ES cells directly from knockout embryos. Out of 50 blastocysts obtained from mating of Ezh2−/+ mice, we established 28 ES lines, including 13 (46.4%) heterozygous lines, 8 (28.6%) wild-type and 7 lines (25%) that are Ezh2−/−(Figure S1C). Thus, contrary to expectations based on prior work, Ezh2 is required neither for establishment nor maintenance of ES cells. Ezh2−/− ES cells generated by the above methods are indistinguishable. The discrepancy between O’Carroll et al. and our work may reflect improved contemporary methods for derivation of ES cell lines from blastocysts. It is also possible that in the O’Carroll et al. allele an in-frame fusion of LacZ with the first 200 amino acids of EZH2 (which contain EED-interacting domain) generated a dominant negative protein or perturbed functions beyond PRC2.
In contrast to Eed−/− cells that lack di- and tri-methylation on H3K27 and show dramatic reduction of H3K27me1, Ezh2−/− cells show decreased H3K27me2 and H3K27me3, yet retain robust H3K27me1 (Figure 1C), indicating additional HMTase(s) catalyzing H3K27me1 in an EED-dependent manner. To confirm the loss of the H3K27me3 mark on individual PcG targets (Boyer et al., 2006), we performed chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR) (Figure 1D). Surprisingly, the H3K27me3 mark is still significantly enriched at promoters of PcG target genes in Ezh2−/− cells, but entirely lost in Eed−/− cells. The loss of EZH2 binding at these loci rules out possible residual activity of EZH2 in Ezh2−/− cells, indicating that additional HMTase(s) catalyze H3K27me3. In addition, SUZ12 binding at PcG sites is reduced in Ezh2−/− cells, but abolished in Eed−/−cells. It was reported that G9a, the HMTase catalyzing methylation on H3K9, might mediate H3K27 methylation (Tachibana et al., 2001). However, we observed no binding of G9a at PcG target loci in either wild-type or Ezh2−/− ES cells (data not shown). Therefore, robust H3K27me1 and residual H3K27me3 imply the presence of additional HMTase(s) beyond EZH2 acting on H3K27.
EZH1 interacts with core components of PRC2
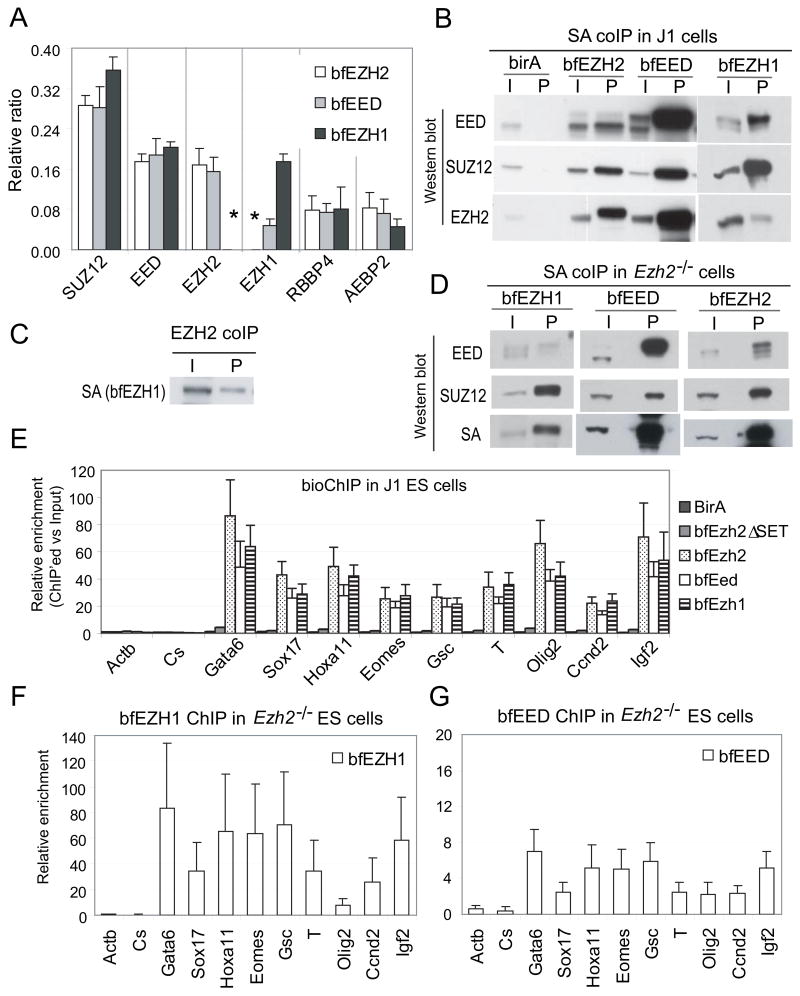
To identify other HMTase(s) associated with EED, we generated ES cell lines stably expressing sub-endogenous levels of EED containing a biotin tag and a FLAG epitope at its amino terminus (herein referred to as bfEED, Figure S2A–S2C). Five components of PRC2 are pulled down by FLAG-antibody and streptavidin-mediated tandem coimmunoprecipitation (coIP) and identified by Mass Spec (MS) sequencing (Figure 2A). Interestingly, in addition to EZH2 (represented by a total of 224 peptides), bfEED also pulls down EZH1 (72 peptides), a sequence homolog of EZH2 (Table S1). More peptides corresponding to EZH2 than EZH1 are recovered. Although the numbers of peptides identified by MS analysis provide only a rough estimate of protein abundance, consistent differences in peptide numbers in multiple pull-down experiments suggest a greater abundance of EZH2 than EZH1 polypeptides in association with EED.
Figure 2. EZH1 interacts and co-localizes with EZH2 and EED at PcG target loci.
(A) Proteins interacting with bfEZH2, bfEED or bfEZH1 were pulled down by FLAG and biotin-streptavidin (SA) mediated tandem coIP and subsequently identified by Mass Spec (MS) sequencing. Y-axis shows relative ratio of peptides representing an identified protein (x–axis) in total number of peptides identified by MS. Error bars represent standard deviations of ratios of peptides identified by MS in independent pull-down experiments. Neither bfEZH2 nor bfEZH1 pulls down each other (indicated by asterisks).
(B) One-step SA coIP and Western blotting of J1 ES cells expressing birA alone or with any of bfEZH2, bfEZH1 and bfEED. “I” represents inputs and “P” represents coIP’ed fractions.
(C) bfEZH1 is coIP’ed with endogenous EZH2.
(D) SA coIP and Western blot analysis of Ezh2−/− ES cells expressing bfEZH1, bfEED or bfEZH2.
(E) Colocalization of bfEZH1 with bfEZH2 and bfEED at promoters of PcG targets shown by bioChIP-qPCR analysis in J1 ES cells.
(F–G) The binding of bfEZH1 (F) and bfEED (G) to PcG target loci in Ezh2−/− ES cells.
By using a similar strategy, we constructed ES cell lines stably expressing sub-endogenous levels of bfEZH2 or bfEZH1 (Figure S2B–S2C). Core components of PRC2 including SUZ12, EED, RBBP4 and AEBP2 are pulled down with both EZH2 and EZH1 (Figure 2A and Table S1). However, neither bfEZH2 nor bfEZH1 tandem pull-down identifies any peptides representing each other, suggesting the presence of EZH2 and EZH1 in alternative protein complexes. In addition, five core components of the EZH1-containing complex have similar stoichiometry as found in the canonical EZH2-containing PRC2 complex (Figure 2A).
To confirm these data, we performed one-step streptavidin-mediated coIP of bfEZH1. EZH2 is detected in the coIP’ed fraction as a weak protein band with about half of the intensity as the input. However, both EED and SUZ12 show ~ 5-fold higher intensity in the coIP’ed fractions than in the inputs (Figure 2B). In addition, bfEZH1 is pulled down with endogenous EZH2 (Figure 2C). Therefore, the interaction of EZH2 and EZH1 is evident in a less stringent one-step coIP but not in tandem purification, suggesting that EZH1 and EZH2 may indirectly interact through their binding to EED and SUZ12. Thus, we have identified an alternative PRC2 complex containing a different subunit, EZH1, which might provide catalytic activity.
To examine the integrity of PRC2 complexes in absence of EZH2, we generated Ezh2−/−ES cells stably expressing sub-endogenous bfEZH1 (Figure S2B). Although bfEZH1 efficiently pulls down SUZ12 in absence of EZH2, the amount of EED pulled down by bfEZH1 in Ezh2−/−cells is about 10-fold less than that in wild-type cells (Figure 2B and 2D). Thus, the interaction between EED and bfEZH1 appears dramatically reduced, though detectable in Ezh2−/− cells. This result suggests a possible role of EZH2 in regulating EZH1-containing PRC2 by promoting EZH1-EED protein interactions.
EZH1 colocalizes with H3K27me3 in wild-type and Ezh2−/− ES cells
To investigate the role of EZH1 within the alternative PRC2 complex, we performed biotin-streptavidin mediated ChIP (bioChIP) and qPCR to analyze EZH1 association with chromatin. bfEZH2 and bfEED, but not birA control and SET domain-deleted EZH2 (bfEZH2δSET), show enrichment on PcG target genes (Figure 2E), demonstrating the specificity of the bioChIP procedure. Interestingly, bfEZH1 shows significant enrichment at PcG target loci not only in wild-type but also in Ezh2−/− cells (Figure 2E–2F). H3K27me3 modification on PcG target loci is unaltered irrespective bfEzh1 expression (Figure S2D–S2E), suggesting that exogenous bfEZH1 does not disturb the balance of methylation. In addition, Ezh2−/− cells do not up-regulate Ezh1 transcripts to compensate the loss of Ezh2 (Figure S2B). Moreover, bfEED enrichment at PcG loci in Ezh2−/− cells is dramatically decreased compared to that in wild-type cells (Figure 2G), a finding consistent with an attenuated interaction of EED and bfEZH1 in the absence of EZH2.
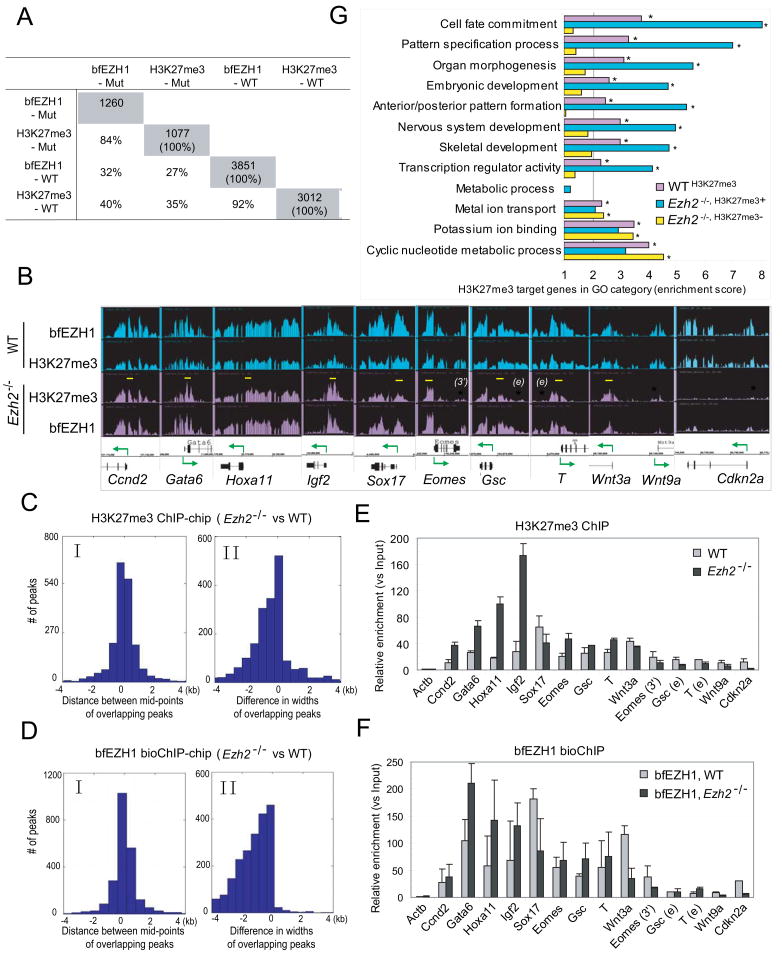
We then determined whether EZH1 colocalizes with H3K27me3 in a genome-wide manner and where the residual H3K27me3 mark is located in Ezh2−/− ES cells. Whole-genome ChIP-chip analysis of H3K27me3 detects 7045 peaks (p < 1e-7) which are associated with 3012 genes in wild-type ES cells, but 1997 peaks associated with 1077 genes in Ezh2−/− cells suggesting a 65% or 72% loss of H3K27me3 at peak or gene level, respectively (Figure 3A and Table S2–S6). BioChIP-chip analysis of bfEZH1 reveals 7478 peaks (p < 1e-7) associated with 3851 genes, overlapping with 92% of H3K27me3 targets in wild-type cells. The average probe signals of H3K27me3 and bfEZH1 are dramatically reduced in Ezh2−/− cells (Figure S3B). Ezh2−/−cells display 70% or 68% loss of bfEZH1 binding at peak or gene level, respectively. Importantly, target genes of H3K27me3 and bfEZH1 in Ezh2−/− cells are largely overlapping (79% – 84%).
Figure 3. bfEZH1 colocalizes with H3K27me3 on chromatin.
(A) The overlapping percentages of target genes identified by H3K27me3 and bfEZH1 ChIP-chip in wild-type (WT) and Ezh2−/− mutant (Mut) ES cells. The total numbers of target genes (p < 1e-7) is highlighted in grey.
(B) Representative view of bfEZH1 and H3K27me3 occupancy at various loci in WT (shown in blue) and Ezh2−/− ES cells (in purple) by Affymetrix Integrated Genome Browser. Arrows indicate transcription starts and directions. Yellow bars indicate regions retaining strong binding in Ezh2−/− cells. Asterisks in red indicate regions losing ChIP peaks in Ezh2−/− cells. qPCR primers used in Panel E and F were designed in the above marked regions.
(C–D) Comparisons of peak positions and widths of H3K27me3 (C) or bfEZH1 (D) ChIP-chip in Ezh2−/− and wild-type ES cells. Panel I shows deviations of mid-points of corresponding ChIP peaks in Ezh2−/− and wild-type cells. The distribution is centered at 0 and the deviations, by large part, are within 1 kb on both sides. Panel II shows the distribution of width differences of corresponding ChIP peaks in Ezh2−/− and wild-type cells. The distribution is heavily skewed to the left, indicating less broad peaks in Ezh2−/− cells than in wild-type cells in general.
(E–F) qPCR verification of H3K27me3 ChIP (E) in WT and Ezh2−/− ES cells without exogenous bfEzh1 and bfEZH1 bioChIP (F) in cells expressing bfEzh1.
(G) Gene Ontology (GO) analysis of H3K27me3 target genes. GO terms with an enrichment score larger than 2 (i.e. one-fold above genome background) and a p-value less than 1e-6 are considered significantly enriched and indicated by asterisks. The WTH3K27me3 gene set comprises of all H3K27me3 target genes. Ezh2−/−, H3K27me3+ or Ezh2−/−, H3K27me3− genes represent a subset of H3K27me3 target genes which retain or lose this mark in Ezh2−/− cells, respectively.
We observed that H3K27me3 and bfEZH1 binding on DNA was often broad in wild-type ES cells covering distal, proximal promoters and gene bodies, but became focused at proximal promoters near transcription start sites in Ezh2−/− ES cells (Figure 3B and S3A). Indeed, the widths of corresponding H3K27me3 or bfEZH1 peaks in Ezh2−/− cells are narrower than those in wild-type cells despite similar positioning on chromatin (Figure 3C–3D). Therefore, loss of H3K27me3 or bfEZH2 binding is underestimated if the narrowed binding pattern of these marks in mutant cells is not taken into account.
Consistent with ChIP-chip, enrichment patterns of bfEZH1 and H3K27me3 in a few target regions shown by ChIP-qPCR are very similar (Figure 3B–3F). For example, in Ezh2−/−cells, genes such as Cdkn2a and Wnt9a lose the association of both H3K27me3 and bfEZH1 marks, while enhancer or 3′UTR regions of Eomes, Gsc and T show reduced binding. H3K27me3 and bfEZH1 signals appear higher at promoters of Ccnd2, Gata6, Hoxa11 and Igf2 in Ezh2−/− cells than in wild-type cells. However, only 91 out of 1077 genes that retain the H3K27me3 mark in Ezh2−/− cells show higher ChIP-chip signals (Table S7), suggesting that greater enrichment of H3K27me3 in selected regions is not general to all targets.
Thus, colocalization of EZH1 and H3K27me3 on chromatin in both wild-type and Ezh2−/−ES cells implies a direct role of EZH1 in formation of the H3K27me3 mark. Loss of Ezh2 negatively affects EZH1 association with chromatin, indicating that DNA binding ability of the EZH1 interacting complex is at least partially dependent on EZH2. Furthermore, we found that over 94% target genes revealed by bfEZH2 bioChIP-chip analysis were co-occupied by bfEZH1 and H3K27me3 (data not shown), suggesting that EZH2 and EZH1 bind overlapping sets of target genes, perhaps in a coordinated manner.
To gain insights into the functions of genes that preserve or lose the H3K27me3 mark in Ezh2−/− ES cells, we performed gene ontology (GO) analysis. The overlapping target genes of H3K27me3 and bfEZH1 in wild-type (WT) ES cells are referred to WTH3K27me3 genes, representing overall target genes marked by H3K27me3 (Table S8). Consistent with previous reports (Boyer et al., 2006; Lee et al., 2006), WTH3K27me3 genes are enriched in development-related GO terms including cell fate commitment, embryonic development and transcription regulator activity etc (Figure 3G). Interestingly, of WTH3K27me3 genes, the subset of genes (referred to Ezh2−/−, H3K27me3+) that retain both H3K27me3 and bfEZH1 marks in Ezh2−/− cells show even higher enrichment scores on developmental terms. On the contrary, the subset (referred to Ezh2−/−, H3K27me3−) of genes that lose both marks in Ezh2−/− cells shows no enrichment on development-related GO terms, but is enriched in cyclic nucleotide metabolic process, potassium ion binding and metal ion transport. In comparison, none of the three sets shows significant enrichment in metabolic processes. Therefore, Ezh2−/− ES cells preferentially preserve EZH1 binding and the H3K27me3 mark on genes related to development, suggesting a possible role of EZH1 in ES cell function.
Ezh2 is dispensable for the maintenance of ES cell identity
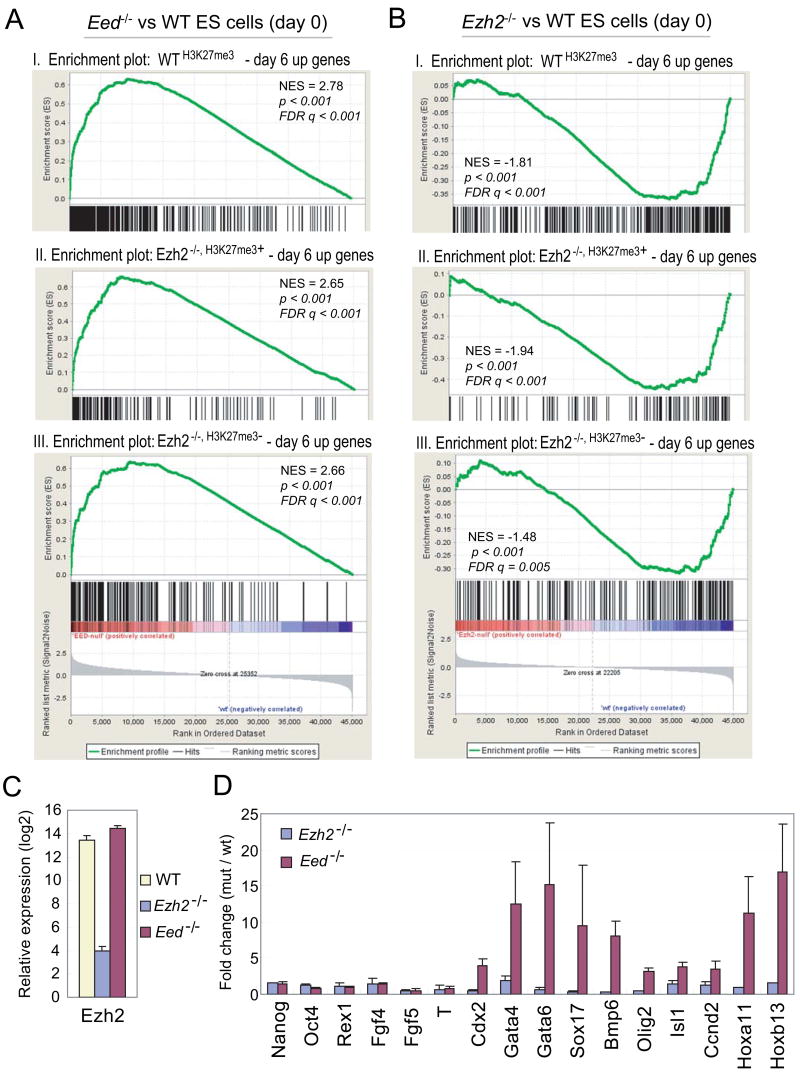
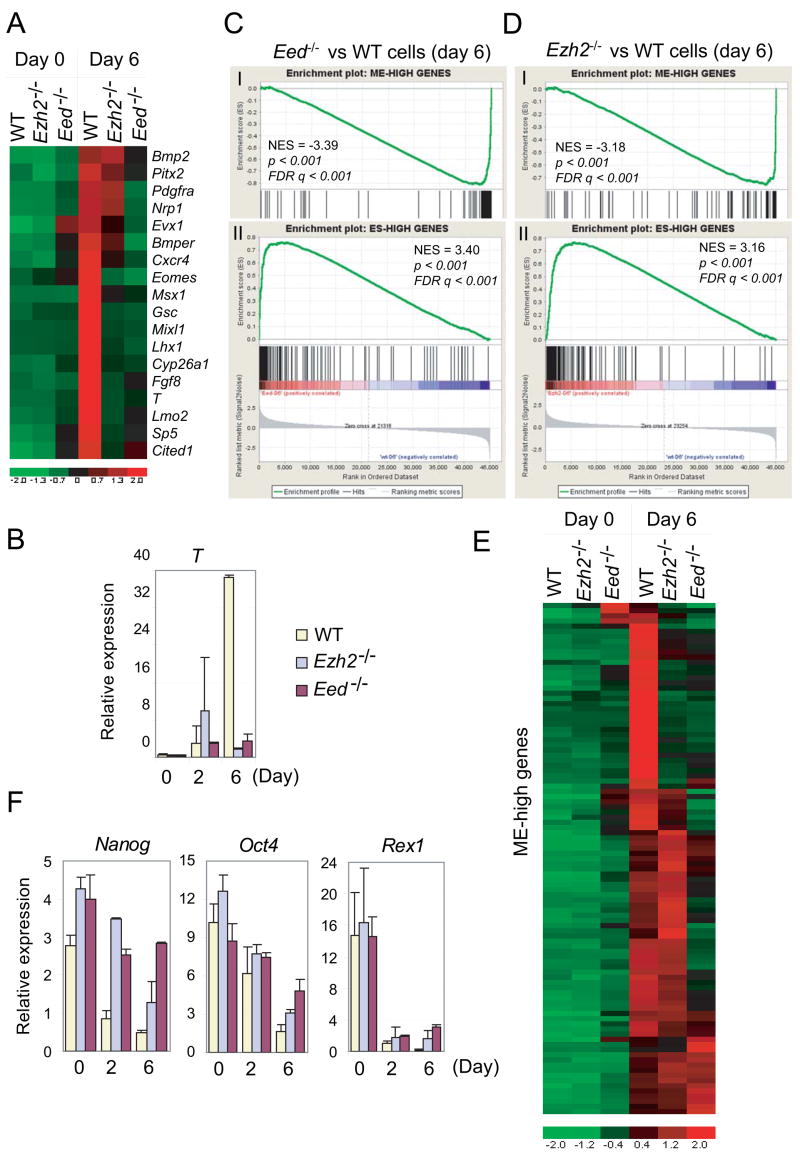
To determine functional consequences of residual H3K27me3 possibly mediated by EZH1 in maintaining ES cell identity, we compared microarray expression profiles of wild-type and mutant ES cells and performed gene set enrichment analysis (GSEA) on three predefined subsets of H3K27me3 target genes that are activated at day 6 differentiation of wild-type ES cells including WTH3K27me3 – day 6 up, Ezh2−/−, H3K27me3− – day 6 up and Ezh2−/−, H3K27me3+ – day 6 up genes (Table S9). All three gene sets remain suppressed in Ezh2−/− ES cells despite substantial reduction in the H3K27me3 mark, but are up-regulated in Eed−/− ES cells which lose all H3K27me3 (Figure 4A–4B and S4).
Figure 4. Ezh2 is dispensable for the maintenance of ES cell identity.
(A–B) GSEA profiles of three subsets of H3K27me3 targets by comparing undifferentiated Eed−/−(A) or Ezh2−/− ES cells (B) to wild-type (WT) ES cells. Panel I, WTH3K27me3 – day 6 up genes; Panel II, Ezh2−/−, H3K27me3+ – day 6 up genes; Panel III, Ezh2−/−, H3K27me3− – day 6 up genes. All three subsets show positive normalized enrichment scores (NES) in Eed−/− ES cells but negative NES in Ezh2−/− ES cells, suggesting derepression in Eed−/− ES cells but repression in Ezh2−/− ES cells. (C) Ezh2 transcript is reduced ~ 1000 fold in Ezh2−/− cells compared to that in WT and Eed−/− cells. (D) Fold changes of gene expression in mutants versus wild-type cells.
Reverse transcription and qPCR analysis confirms the loss of Ezh2 transcripts in Ezh2−/−cells and dramatic up-regulation of differentiation-induced, H3K27me3 genes in Eed−/− cells but not in Ezh2−/− ES cells, despite normal expression of pluripotency markers in both mutant cells (Figure 4C–4D). To rule out the possibility of derepression as a consequence of partial differentiation due to the unstable nature of Eed−/− ES cells, we analyzed a number of single colonies grown on feeders that showed undifferentiated ES cell morphology. Of 52 H3K27me3 and PRC2 target genes analyzed, 28 genes are up-regulated in Eed−/− cells as compared to wild-type ES cells (Figure S5), suggesting that derepression is intrinsic to the loss of Eed. Thus, complete loss of the H3K27me3 mark in absence of Eed leads to global derepression of polycomb genes, whereas Ezh2−/− ES cells appear to be normal in this respect. These data suggest that the repression of developmental genes by residual H3K27me3 mediated by EZH1 may contribute to the maintenance of ES cell identity in absence of Ezh2.
EZH1 complements EZH2 in mediating H3K27 methylation and gene repression
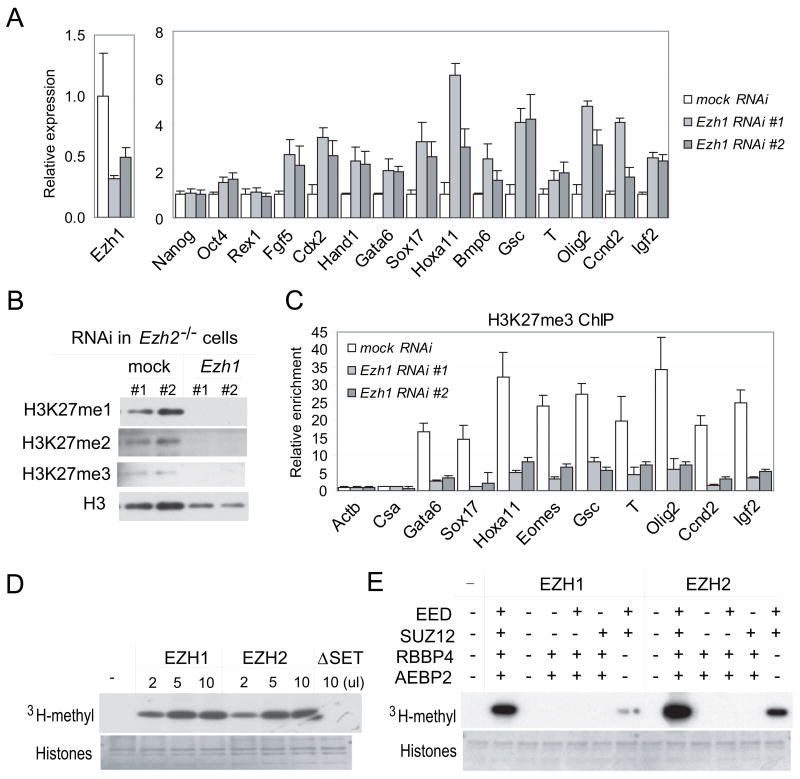
To assess whether EZH1 is responsible for methylation on H3K27 and required for repression of developmentally regulated PcG target genes in Ezh2−/− cells, we used RNA interference (RNAi) to knock down Ezh1 expression. Compared to cells treated with mock RNAi, Ezh1 RNA transcripts decrease 50% – 70% in Ezh1 RNAi (Figure 5A, S6A–S6B). In wild-type cells depleted of Ezh1, no change in global methylation on H3K27 is observed despite modest reduction of the H3K27me3 mark at PcG target loci (Figure S6C and S6E). In addition, Ezh1 RNAi did not affect the expression of PcG target genes (Figure S6A), supporting EZH2 as the major histone methyltransferase acting on H3K27.
Figure 5. Ezh1 mediates methylation on H3K27 in vivo and in vitro.
(A–C) Ezh1 RNAi in Ezh2−/− ES cells. (A) RT-qPCR analysis of Ezh1 and marker genes. Relative expression was normalized to that in untreated Ezh2−/− ES cells. (B) Histone analysis. (C) Reduced H3K27me3 binding at PRC2 targets in Ezh1 RNAi cells shown by ChIP-qPCR.
(D–E) HMTase assays. (D) FLAG-tagged EZH1, EZH2 or EZH2ΔSET was co-expressed with EED, SUZ12, RBBP4 and AEBP2 in Sf21 cells. (E) FLAG-tagged EZH1 or EZH2 was co-expressed with indicated PRC2 components in Sf21 cells. Protein complexes were pulled down by anti-FLAG agarose and assayed for HMTase activity. The upper panels show the autoradiography and the lower panels show equal addition of core histone substrates.
Interestingly, all three forms of methylation on H3K27 are barely detected in Ezh2−/− cells treated with Ezh1 RNAi (Figure 5B), demonstrating a role of EZH1 in catalyzing mono-, di- and trimethylation on H3K27 in absence of Ezh2. Moreover, in Ezh2−/− cells depleted of Ezh1, the H3K27me3 mark is dramatically reduced at PcG target loci (Figure 5C and S6D), and PcG targets are up-regulated, despite normal expression of pluripotency markers (Figure 5A and S6B). Therefore, cells lacking Ezh2 and depleted of Ezh1 resemble Eed−/− ES cells. These findings demonstrate an Eed-dependent, complementary role of Ezh1 in suppressing differentiation and maintaining the identity of Ezh2−/− ES cells.
EZH1 is a histone methyltransferase
To directly evaluate the enzymatic activity of EZH1, we coexpressed FLAG-tagged EZH1 with EED, SUZ12, RBBP4 and AEBP2 in Sf21 cells. These four proteins are coIP’ed with FLAG-tagged EZH1, indicating their existence in a core complex (Figure S7A–S7B). The EZH1 complex is capable of transferring 3H-methyl to core histones and shows enzymatic activity at a level roughly comparable to the EZH2 complex (Figure 5D).
We next determined how the other four components of PRC2 affect catalytic activity of EZH1. Both EZH1 and EZH2 require the presence of EED and SUZ12 for enzymatic activity (Figure 5E). The presence of RBBP4 and AEBP2 significantly enhances the activity of both enzymes. Similar to EZH2 protein, EZH1 is unstable and does not interact with RBBP4 and AEBP2 outside of PRC2 (Figure S6C). Co-expression of EED or SUZ12 greatly enhances the stability of EZH1 and EZH2. SUZ12, but not EED, is required for the interaction of the either EZH protein with RBBP4 and AEBP2. Therefore, EZH1 and EZH2 show similar dependence on other components of PRC2 complexes for their activities.
Differentiation of Ezh2−/− ES cells is impaired but not to the extent seen in Eed−/− cells
Withdrawal of LIF induces differentiation of mouse ES cells into various embryonic and extraembryonic lineages, a phenomena mimicking in vivo gastrulation that requires the establishment of specific gene expression programs temporally and spatially (Keller, 2005). We determined whether and how Ezh2 and Eed are involved in lineage specification. Among the top differentially expressed genes, we found 18 genes that are known to be involved in mesodermal commitment, gastrulation and embryonic patterning (Figure 6A). While 14 of them fail to be activated in Ezh2−/− cells, all 18 genes remain inactive in Eed−/− cells. In addition, T, an early marker for gastrulation and mesoendoderm (ME)-committed cells that can further differentiate into mesoderm and/or endoderm (Fehling et al., 2003), is activated ~ 91 fold at day-6 differentiation in wild-type cells, but only 7–10 fold in Ezh2−/− and Eed−/− cells (Figure 6B).
Figure 6. Ezh2 and Eed are required for mesoendodermal (ME) differentiation.
(A) Heat map showing impaired activation of known mesodermal markers in Ezh2−/− and Eed−/−cells as compared to wild-type (WT) cells.
(B) RT-qPCR analysis of T expression along ES cell differentiation by LIF withdrawal.
(C–D) GSEA profiles of ME-high (panel I) and ES-high (panel II) genes by comparing day-6 differentiated Eed−/− (C) or Ezh2−/− (D) cells to wild-type cells.
(E) Heat map of ME-high genes in wild-type and mutant ES cells.
(F) RT-qPCR analysis of pluripotency markers along ES cell differentiation.
To ask whether this defect is limited to a select set of genes or is more general for the mesoendodermal lineage, we performed GSEA to assess ME-specific genes in a global fashion. About 97 genes are activated > 7-fold in mesoendoderm-committed cells compared to undifferentiated ES cells and herein referred to as ME-high genes (Table S9 and Supplemental notes). However, this set of genes is negatively enriched in both mutant cells (Figure 6C–6D). Of 97 ME-high genes, 40 in Ezh2−/− cells and 65 in Eed−/− cells are down-regulated at least 2-fold compared to their expression in wild-type cells at day-6 differentiation (Table S9). In addition, clustering reveals a more severe defect in ME differentiation observed in Eed−/− cells than in Ezh2−/− cells (Figure 6E).
Moreover, we analyzed a set of 143 genes that are highly expressed in neuronal progenitor (NP) cells (Table S9 and Supplemental notes). Clustering reveals that a majority of NP-high genes are activated in day-6 differentiated Ezh2−/− cells to a level comparable to, or even higher than, that in differentiated wild-type cells (Figure S8A), However, about one third of NP-high genes fail to be fully activated in differentiated Eed−/− cells, suggesting a role of Eed in neuronal lineage. In addition, abnormal activation of genes (for example, Cdkn2a) is observed in both mutant cells (Figure S9A). De-regulated activation of genes beyond ME- and NP lineages is more evident in Eed−/− cells (Figure S8B and Table S9).
Interestingly, a set of ES cell specific genes remains at higher levels of expression in both day-6 differentiated Ezh2−/− and Eed−/− cells despite overall decreased expression as compared to undifferentiated ES cells (Figure 6C–6F, S9B and Table S10). Thus, mutant cells fail to extinguish ES-specific genes. Pluripotency regulators NANOG and DAX1 are found bound to promoters of T, Gsc and Evx1, key transcription regulators of mesoendodermal development (Figure S10). Thus, it is possible that constitutively high levels of pluripotency-related genes may inhibit lineage development.
The less severe differentiation defect seen in Ezh2−/− than Eed−/− ES cells is consistent with a complementary role of Ezh1 in ES cell function. However, Ezh1 cannot fully compensate for the loss of Ezh2 during lineage commitment.
DISCUSSION
Complementary roles of Ezh1 and Ezh2 in H3K27 methylation
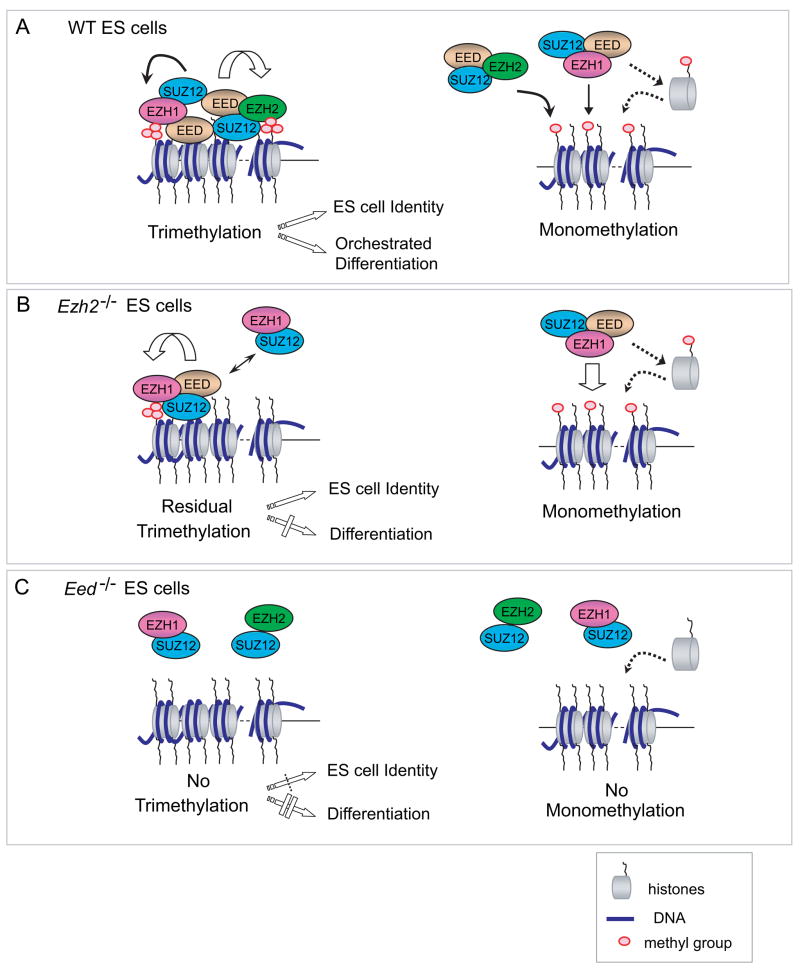
Mammalian Ezh1 has been an “orphan” since its discovery a decade ago. EZH1 has not previously been identified within a Polycomb repressive complex by biochemical purifications or coIP in Hela or 293T cells (Cao et al., 2002; Kuzmichev et al., 2002). A trivial explanation is possibly low-level expression of Ezh1 in these cells. Here, we have demonstrated in vivo association of EZH1 with PRC2. The non-canonical EZH1-mediated PRC2 coexists with the classic EZH2-mediated PRC2 in catalyzing H3K27me3 at overlapping genes (Figure 7).
Figure 7. Complementary but non-redundant roles of EZH1 and EZH2 in mediating methylation on H3K27 and ES cell function.
(A) A model in wild-type (WT) ES cells. Both EZH2 and EZH1 bind to EED and SUZ12, forming alternative but interacting PRC2 complexes containing HMTase activity. EZH2-mediated PRC2 is more abundant in ES cells and plays a major role in forming H3K27me3, which is required for ES cell identity and proper differentiation. Both EZH1 and EZH2 containing PRC2 complexes may mediate monomethylation on H3K27 in a non-targeted manner by either acting transiently on nucleosomes across the genome or on free H3 histones prior to nucleosomal assembly during DNA replication.
(B) In Ezh2−/− ES cells, EZH1-PRC2 mediates monomethylation and residual trimethylation on H3K27. EED may regulate EZH1-PRC2 complex in an EZH2-dependant manner. EZH1 mediates H3K27me3 on a set of developmental genes and prevents derepression of PRC2 target genes from globe loss of the H3K27me3 mark in Ezh2−/− ES cells. Hereby, EZH1 complements EZH2 in the maintenance of ES cell identity. However, EZH1-PRC2 cannot fully rescue defects in mesoendodermal development during Ezh2−/− cell differentiation, suggesting non-redundant functions of these two PRC2 complexes in lineage commitment.
(C) Eed−/− ES cells lack all levels of methylation on H3K27, indicating the essential role of EED in EZH1 or EZH2 mediated PRC2 complexes. Complete loss of H3K27me3 in the absence of Eed confounds ES cell identity by de-repressing a set of differentiation-regulated PRC2 target genes, and blocks the proper execution of pluripotent programs.
High correlation between EZH1 and H3K27me3 target genes suggests possible roles of EZH1 in both initiating and spreading the H3K27me3 mark. We cannot formally exclude, however, the possibility that exogenously introduced bfEZH1 may overrepresent EZH1 protein. Against this possibility however, we note that exogenously introduced bfEZH1 does not rescue the methylation defect shown in absence of endogenous EZH2, suggesting that we have not appreciably perturbed protein equilibrium in vivo. Extensive loss of H3K27me2 and me3 in Ezh2−/− cells and no global methylation change on H3K27 in wild-type cells depleted of Ezh1, point to EZH2 as the major H3K27 HMTase in ES cells (Figure 7).
Consistent with H3K27me1 as the primary product of EZH2-PRC2 reaction in vitro (Sarma et al., 2008), EZH2 is also involved in monomethylation in vivo. However, EZH1 can fully compensate the loss of EZH2 in forming H3K27me1 (Figure 7B). A recent genome-wide study conducted in human T cells revealed an inverse correlation between H3K27me1 and me3 (Barski et al., 2007). The H3K27me1 mark is associated with actively transcribed regions, whereas H3K27me3 is enriched at silenced promoters. As both EZH1 and EZH2 colocalize with H3K27me3 on chromatin, we suspect the EZH proteins may be regulated to leave the sites after addition of one methyl group to certain nucleosomes (Figure 7). Monomethylation may occur on free H3 histones prior to their assembly into nucleosomes during DNA replication. It is unclear whether H3K27me1 is simply an intermediate state or is functionally important.
Previous studies have shown that at limiting concentration EED may be preferentially assembled into a monomethylase complex and the removal of H3K27me1 may occur passively (Chamberlain et al., 2008; Montgomery et al., 2005). We show detectable, but significantly reduced, association between bfEZH1 and EED in Ezh2−/− cells. Consistent with the essential role of EED in all methylation on H3K27, our data support EED as the rate limiting factor regulating both EZH1- and EZH2-containing PRC2 complexes. It is also possible that post-translational modifications or conformation switches might affect the ability of EED to interact with the EZH proteins.
Robust H3K27me1 in Suz12−/− cells suggests a dispensable role of SUZ12 in mediating H3K27me1 (Pasini et al., 2004). Intriguingly, SUZ12 is required for the HMTase activity of both EZH1 and EZH2 in vitro. It is possible that other gene(s) or Suz12 homologue(s) in mammalian genome are yet to be identified and might compensate for the loss of Suz12 in vivo. Of potential relevance, in Caenorhabditis elegans EZH2 and EED homologs are present in a complex that retains H3K27 methyltransferase activity, yet lacks a SUZ12 homolog (Bender et al., 2004).
H3K27me3 and PRC2 complexes in stem cell maintenance
Our success in isolating Ezh2−/− ES cells directly from knockout blastocysts, and by inactivating Ezh2 in established ES cell lines, demonstrates that Ezh2 is dispensable for both establishment and maintenance of mouse ES cells. Our results are consistent with post-implantation lethality and the ability of pathenogenetic embryos lacking maternal and embryonic Ezh2 to develop to the blastocyst stage in vitro (Erhardt et al., 2003; O’Carroll et al., 2001).
Interestingly, deletion of Ezh2 does not result in transcriptional activation of ~ two thirds of H3K27me3 target genes that lose this mark. EZH1 in Ezh2−/− ES cells preferentially binds to and transfers the repressive H3K27me3 mark onto developmental genes whose expression promotes differentiation. Inactivation of EZH1 in Ezh2−/− ES cells resembles the loss of Eed, leading to the derepression of a set of PcG target genes. As complete loss of H3K27me3 leads to epigenetic instability where some genes are expressed and some are not, EZH1 thus provides an additional layer of protection to safeguard the pluripotent state by preventing aberrant gene expression.
We, and others, have observed that Eed−/− cells are prone to differentiate, even when maintained on feeders (Boyer et al., 2006; Chamberlain et al., 2008). In addition, we have observed that Eed−/− ES cells grow more slowly than Ezh2−/− cells. Therefore, the maintenance of H3K27me3 on a subset of development-related PcG genes is crucial to prevent the expression of conflicting epigenetic programs which may be detrimental to self-renewal and survival (Figure 7). Of note, the extent of derepression is insufficient to drive full differentiation. Eed−/− ES cells thus retain limited capacity to self-renew and differentiate upon stimulation. We speculate that Eed−/− ES cells may reflect an epigenetic state intermediate between undifferentiated and partially differentiated ES cells.
EZH2 is essential for the proliferation of fibroblasts and tumor cells (Valk-Lingbeek et al., 2004), but is dispensable for ES cell survival and proliferation, suggesting the lack of epigenetic restrictions in these cells. ES cells have a globally relaxed, hyperdynamic chromatin structure (Niwa, 2007). This permissive chromatin environment may be counter-balanced by repressive mechanisms, such as dynamic methylation at H3K9 and H3K27, repressive activities of NuRD, PRC2 and PRC1 complexes, and proteasomal degradation of cryptic transcripts in ES cells (Szutorisz et al., 2006; Wang et al., 2006). Together with transcriptional repressor activities of pluripotency regulators (Boyer et al., 2005), multiple repressive regulatory mechanisms in ES cells cooperate to keep developmental genes silenced but poised for eventual up-regulation as needed for cell lineage commitment.
EZH2-PRC2 in stem cell differentiation
Suz12−/− ES cells fail to up-regulate selected markers during embryoid body formation (Pasini et al., 2007). However, the roles of PRC2 in ES cell differentiation have not been investigated systematically. We show that Ezh2−/− and Eed−/− ES cells fail to fully activate the mesoendodermal transcription program, whereas neuronal gene activation is partially impaired in Eed−/− ES cells. These results indicate profound disturbances of differentiation in absence of Ezh2 and Eed.
Although Eed−/− ES cells are able to contribute to chimeric embryos, high contribution chimeras display some of the same defects as homozygous Eed−/− embryos showing allantois overgrowth, poor neurectoderm and scarce embryonic mesoderm (Chamberlain et al., 2008; Faust et al., 1995). Non-cell-autonomous functions for Eed in morphogenesis in the chimeric setting might compensate for some developmental blocks, thereby confounding a strict definition of pluripotency. Nevertheless, defects in lineage specification shown by in vitro differentiation of Ezh2−/− and Eed−/− ES cells are consistent with in vivo gastrulation defects observed both in high contribution chimeras of Eed−/− ES cells and in homozygous knockout embryos lacking Eed, Ezh2 or Suz12. Therefore, our study demonstrates that Ezh2 and Eed are required to orchestrate the sequence of events necessary to execute a pluripotency program.
The block in transcriptional activation observed in PRC2 mutants is initially paradoxical as the loss of the H3K27me3 repressive mark should promote gene activation. Faulty gene activation observed during differentiation of Ezh2−/− and Eed−/− cells may disrupt normal developmental programs. It is also possible that the failure to extinguish pluripotency genes lies at the basis of differentiation failure.
Alternatively, ME induction may be initiated but cannot be sustained due to cell cycle arrest and apoptosis in mutant cells. It has been reported that decreased Ezh2 levels in senescent cells and aged stem cells lead to the activation of Ink4f-Arf locus encoding Cdkn2a, a negative regulator of cell cycle, and subsequent decreases in proliferation and stem cell self-renewal (Bracken et al., 2007). ES cells lacking Rb-E2F signaling are less sensitive to Cdkn2a activation (White et al., 2005). As somatic cells acquire Rb-E2F and G1 checkpoint regulations, high-level expression of Cdkn2a may be detrimental to cell proliferation. Indeed, Suz2−/− embryos show decreased proliferation and increased apoptosis (Pasini et al., 2004). We suspect Ezh2−/− embryos may die of the combined effects of differentiation failure and cell cycle related proliferation alterations.
Lastly, PRC2 binding to DNA regulatory elements could act positively to recruit specific transcription activators to initiate lineage commitment but repress transcription during terminal differentiation (Pasini et al., 2007). This hypothesis is intriguing as it suggests an instructive role of the H3K27me3 mark mediated by PRC2 complexes in cell fate switches beyond their well-known repressive functions in maintaining cellular memory during cell division.
Implications of EZH1 mediated non-canonical PRC2 in development and disease
Our work challenges the traditional view of EZH2 as the sole H3K27 HMTase in mammals. Coexisting canonical EZH2 and non-canonical EZH1 mediated PRC2 complexes are analogous to the SWI/SNF chromatin remodeling complex mediated by BRG1 or BRM, two mutually exclusive ATPase subunits (Roberts and Orkin, 2004). In addition, variants of the SWI/SNF complex exist, where tissue specific subunits provides distinct functions. For example, a switch in BAF45 and BAF53 subunit composition of the SWI/SNF complex is required for proliferating neural stem cells transiting into postmitotic neurons (Lessard et al., 2007). Similarly, Ezh1 is upregulated upon the transition of the presomitic mesoderm into the somites in E9.5 mouse embryos, suggesting a role of Ezh1 in somite patterning (Buttitta et al., 2003).
EZH1 cannot fully complement EZH2 during ES cell differentiation, suggesting their non-redundant roles during epigenetic establishment of cell fates. De novo recruitment of alternative PRC2 complexes to distinct chromatin loci may be required for lineage commitment. Similar redundancy of methyltransferases MLL and MLL2 acting on H3K4 has been reported. Despite regulation of different Hox complexes by MLL and MLL2 during differentiation, neither is essential for ES cell self-renewal (Glaser et al., 2006; Lubitz et al., 2007). Non-redundant functions of EZH1 and EZH2 in lineage commitment further support the notion that development entails networks of epigenetic specificities. Future studies of genome-wide location analysis of EZH1 and EZH2 in purified lineage-committed cells and the identification of interacting proteins which direct PRC2 complexes to DNA should provide insights on whether and how the two PRC2 complexes are coordinated to regulate development.
It is also possible that low-level expression of Ezh1 in embryonic cells, or temporal and spatial regulation of the EZH proteins (Laible et al., 1997; van Lohuizen et al., 1998), accounts for the failure of Ezh1 to fully complement Ezh2 in orchestrating proper differentiation and development in vivo and in vitro. For example, differential expression of Ezh1 and Ezh2 in B-lymphoid lineages may explain the specific block at the Pro-B cell stage upon deletion of Ezh2 in bone marrow cells (Su et al., 2003). We find that human Ezh1 is widely expressed by analysis of expressed sequence tag (EST) counts (Figure S11). Some tissues including brain, kidney, prostate and spleen express more Ezh1 than Ezh2, and tissues such as adipose, parathyroid and pituitary gland express Ezh1 exclusively. Therefore, Ezh1 may be physiologically more important in post-embryonic development. In addition, human cancers including kidney tumor, prostate cancer and skin tumor show similar Ezh1 EST counts as Ezh2, which serves as a diagnostic marker for various human cancers. Our study reveals EZH1 residency in an alternative PRC2 complex and suggests potential mechanisms by which inappropriate levels of EZH1 may cause epigenetic dysregulation and contribute to cancer. A more detailed knowledge of the recruitment and target genes of EZH1 and EZH2 in maintaining cellular identity and during cell fate switches will enable a better understanding of the role of PRC2 complexes during development and in disease.
EXPERIMENTAL PROCEDURES
ES cell lines and culture
Generation of Ezh2−/− ES cells, cells expressing biotin-FLAG tagged proteins and ES cell culture and differentiation methods were described in Supplemental notes.
ChIP and RNA analysis
ChIP and bioChIP was performed as described previously (Kim et al., 2008). DNA levels at various loci were normalized to an internal control region in the first intron of Actb. Relative enrichments were calculated by dividing normalized ChIP’ed DNA to input DNA. Error bars in ChIP-qPCR analysis represent standard deviations of relative enrichments from three biological replicates of ChIP. Total RNA was reversely transcribed by SuperScript III (Invitrogen). Gene expression was normalized to GADPH. Error bars in RNA analysis represent standard deviations of mean expression or fold changes based on at least three cell and RNA isolations. Primer sequences were listed in Table S11.
Microarrary hybridization and data processing
ChIP’ed DNA was amplified by ligation-mediated PCR (Kim et al., 2008), and hybridized to Affymetrix mouse tiling 2.0R array sets. Model-based Analysis of Tiling array (MAT) was used to predict target loci (p < 1e-7) (Johnson et al., 2006). RNA profilings were performed on Affymetrix mouse genome 430 2.0 arrays. GSEA were performed as described (Subramanian et al., 2005). Detailed procedures were described in Supplemental notes.
RNAi
RNAi was performed as described previously (Wang et al., 2006). Details were described in Supplementary notes.
Baculoviral expression, protein purification and HMTase assay
Bac-to-Bac expression system (Invitrogen) was used to generate baculoviruses expressing FLAG-tagged EZH1 (BC007135), FLAG-EZH2, FLAG-EZH2ΔSET, HA-tagged EED (isoform III), SUZ12, Myc-tagged RBBP4 and V5-tagged AEBP2 (BC031376). Sf21 cells were harvested in 70 hours post infection. Proteins purified by anti-FLAG M2-agarose (Sigma) were assayed for HMTase activity on core histones (Millipore, #17–330).
CoIP and Antibodies
Tandem purification, coIP and protein identification by LC-MS/MS were described previously (Wang et al., 2006). About 5% of nuclear extracts used for coIP were loaded as the input in Western analysis. Antibodies were described in Supplementary notes.
Supplementary Material
Supplemental Data
Supplemental methods and notes, 11 figures and 11 tables can be found at http://www.Raw data for Affymetrix microarray expression profiling were deposited in the Gene Expression Omnibus (GEO, accession #).
Acknowledgments
We are grateful to T. Magnuson for Eed−/− ES cells, G. Keller for T-GFP ES cells, A. Smith for 46C cells, and K. Helin for EED antibody. We thank R. Tomaino for performing LC-MS/MS, DFCI Microarray Core for processing microarray samples, and W. Li, T. Liu and X. S. Liu for assistance with MAT. We thank B. Wilson, C. Roberts, D. Levasseur and X. Xia for criticism, and members of Orkin lab for their assistance. G.C.Y. is support by the Claudia Adams Barr Program in Cancer Research. S.H.O. is an Investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta L, Tanaka TS, Chen AE, Ko MS, Fan CM. Microarray analysis of somitogenesis reveals novel targets of different WNT signaling pathways in the somitic mesoderm. Developmental biology. 2003;258:91–104. doi: 10.1016/s0012-1606(03)00116-7. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb Repressive Complex 2 is Dispensable for Maintenance of Embryonic Stem Cell Pluripotency. Stem cells (Dayton, Ohio) 2008 doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS. The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Mol Cell Biol. 1998;18:2825–2834. doi: 10.1128/mcb.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. Embo J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Molecular biology of the cell. 2007;18:2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Muller J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 2005;6:348–353. doi: 10.1038/sj.embor.7400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. Embo J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar VK, Begemann M. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem cells (Dayton, Ohio) 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nature reviews. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nature immunology. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tie F, Stratton CA, Kurzhals RL, Harte PJ. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol Cell Biol. 2007;27:2014–2026. doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Molecular biology of the cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data
Supplemental methods and notes, 11 figures and 11 tables can be found at http://www.Raw data for Affymetrix microarray expression profiling were deposited in the Gene Expression Omnibus (GEO, accession #).