Summary
The C-C chemokine receptor 5 (CCR5), a member of G-protein-coupled receptors, serves as a co-receptor for human immunodeficiency virus type 1 (HIV-1). In the present study, we examined the interactions between CCR5 and novel CCR5 inhibitors containing the spirodiketopiperazine (SDP) scaffold, AK530 and AK317, both of which were lodged in the hydrophobic cavity located between the upper transmembrane domain and the second extracellular loop (ECL2) of CCR5. Although substantial differences existed between the two inhibitors: AK530 had 10-fold greater CCR5-binding affinity (KD: 1.4 nM) than AK317 (16.7 nM), their antiviral potency was virtually identical (IC50: 2.1 and 1.5 nM, respectively). Molecular dynamics simulations for inhibitor-unbound CCR5 showed hydrogen bond interactions among transmembrane residues Y108, E283, and Y251, which were crucial for HIV-1-gp120/sCD4 complex binding and HIV-1 fusion. Indeed, AK530 and AK317, when bound to CCR5, disrupted these inter-helix hydrogen bond interactions, a salient molecular mechanism enabling allosteric inhibition. Mutagenesis and structural analysis showed that ECL2 consists of a part of the hydrophobic cavity for both inhibitors, although AK317 is more tightly engaged with ECL2 than AK530, explaining their similar anti-HIV-1 potency despite the difference in KD values. We also found that amino acid residues in the β-hairpin structural motif of ECL2 are critical for HIV-1-elicited fusion and the binding of the SDP-based inhibitors to CCR5. The direct ECL2-engaging property of the inhibitors likely produces an ECL2 conformation, which HIV-1 gp120 cannot bind to, but also prohibits HIV-1 from utilizing the "inhibitor-bound" CCR5 for cellular entry, a mechanism of HIV-1’s resistance to CCR5 inhibitors. The data should not only help delineate the dynamics of CCR5 following inhibitor binding but also aid in designing CCR5 inhibitors that are more potent against HIV-1 and prevent or delay the emergence of resistant HIV-1 variants.
Keywords: HIV-1, CCR5 inhibitor, GPCR structure, allosteric inhibition, extracellular loop
Introduction
C-C chemokine receptor 5 (CCR5) is a member of G protein-coupled, seven-transmembrane segment receptors (GPCRs), which comprise the largest superfamily of proteins in the body.1 In 1996, it was revealed that CCR5 represents one of the two essential co-receptors for HIV-1 entry to human CD4+ cells, thereby serving as an attractive target for possible intervention of infection by HIV-1 that uses CCR5 as a co-receptor (R5-HIV-1).2–5 The second extracellular loop (ECL2) of GPCRs is known to play a critical role in ligand binding and ensuing signal transduction. The ECL2 of CCR5 is also thought to play an important role in CCR5 interactions with HIV-1 envelope. To date, scores of newly designed and synthesized CCR5 inhibitors have been reported to be potent against R5-HIV-1,6–15 and one such inhibitor, maraviroc,11; 15 has recently been approved by the US Food and Drug Administration (FDA) for treatment of HIV-1-infected individuals who do not respond to any existing antiretroviral regimens.
HIV-1-gp120 interacts with CCR5 following its binding to CD4, and such an interaction is thought to involve the V3 region of gp120 and the N-terminus and extracellular loops (ECLs) of CCR5.16;17 Recent reports 18–21 have determined the orientation and location of CCR5 inhibitors within CCR5 and shown that those inhibitors are all located in a hydrophobic cavity formed by the transmembrane domains of CCR5. In fact, earlier reports demonstrated that mutations in the extracellular loops did not have any effect on the binding of CCR5 inhibitors SCH-C and TAK-779.19; 22; 23 Taking these observations together, the binding sites in CCR5 for CCR5 inhibitors distinctly differ from the binding sites in CCR5 for HIV-1 gp120, strongly suggesting that CCR5 inhibitors block the interactions of CCR5 with HIV-1 gp120 through eliciting allosteric changes in ECL structures. 9; 22; 23
We previously reported a small molecule CCR5 inhibitor, aplaviroc (APL), which has a high affinity to CCR5 (KD values of 3 nM) and exerts potent activity against a wide spectrum of R5-HIV-1 isolates, including multi-drug resistant R5-HIV-1 strains.14;24 APL significantly reduced viremia in patients with HIV-1 infection as examined in a phase 2a clinical trial in the United States. However, in phase 2b clinical trials enrolling about 300 patients, four individuals receiving APL, developed Grade 3 or greater treatment-emergent elevations in ALT, and in late 2005, the clinical development of APL was terminated. However, using APL as a specific probe, we further conducted structural analyses of CCR5 inhibitor interactions with CCR5, employing homology modeling, robust structure refinement, and molecular docking based on the site-directed mutagenesis-based saturation binding assay data of CCR5 inhibitors.22
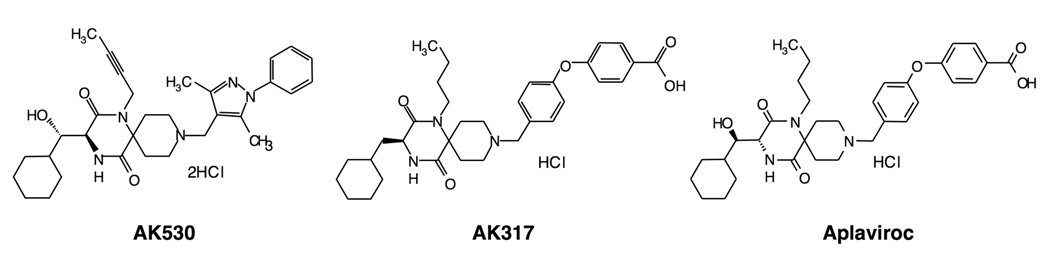
In the current study, we determined the structural and molecular interactions of two novel CCR5 inhibitors, AK530 and AK317 (Fig. 1), both of which contain a novel spirodiketopiperazine (SDP) scaffold. We found that these two inhibitors lodge in a hydrophobic cavity located between the upper transmembrane domain and ECL2 of CCR5. We found substantial differences between the two molecules: AK530 had 10-fold greater CCR5-binding affinity (KD: 1.4 nM) than AK317 (16.7 nM), while their antiviral potency was virtually identical (IC50: AK530, 2.1 nM; AK317, 1.5 nM). Modeling analysis showed that AK530 has least interactions with S180 and K191 of ECL2, with which AK317 has a close association, suggesting that the interaction profile of the inhibitors with ECL2 residues is one of the important determinants of antiviral potency. We also found that the hairpin motif in the N-terminal half of ECL2 is critical for HIV-1 envelope-elicited fusion event. The direct ECL2-engaging property of the inhibitors likely produces an ECL2 conformation, which HIV-1 gp120 cannot bind to, but also prohibits or substantially delays the emergence of HIV-1 that utilizes the "inhibitor-bound" CCR5 for cellular entry, a mechanism of HIV-1’s resistance to CCR5 inhibitors. We also carried out molecular dynamics simulations of inhibitor-unbound CCR5 and compared the conformation with inhibitor-bound CCR5. Critical inter-helix hydrogen bond interactions and interactions between the helices and ECL2 seen in the unbound CCR5 were lost when transmembrane helix residues rearranged to accommodate AK530 and AK317 in the binding pocket. These observations add considerable insights to the mechanism of allosteric inhibition of CCR5-gp120 interaction by CCR5 inhibitors.
Fig. 1.
Structures of small molecule CCR5 inhibitors, AK530, AK317, and aplaviroc (APL).
Results
Structural modeling of unliganded human CCR5
It is thought that the second extracellular loop (ECL2) of human CCR5 (Fig. 2), plays an important role in the binding of CC-chemokines to CCR5 as well as the binding of HIV-1-gp120/CD4 complex to CCR5 in the cellular entry of HIV-1.25; 26 On the other hand, certain amino acid residue substitutions such as Y108A, Y251A, and E283A, all of which are located in the transmembrane domain (Fig. 2a) significantly reduce both HIV-1-gp120/CD4 complex binding to CCR5 and HIV-1 susceptibility of CCR5-expressing cells, as previously described. 22
Fig. 2. The trans-membrane helices and extra-cellular loop regions of CCR5.
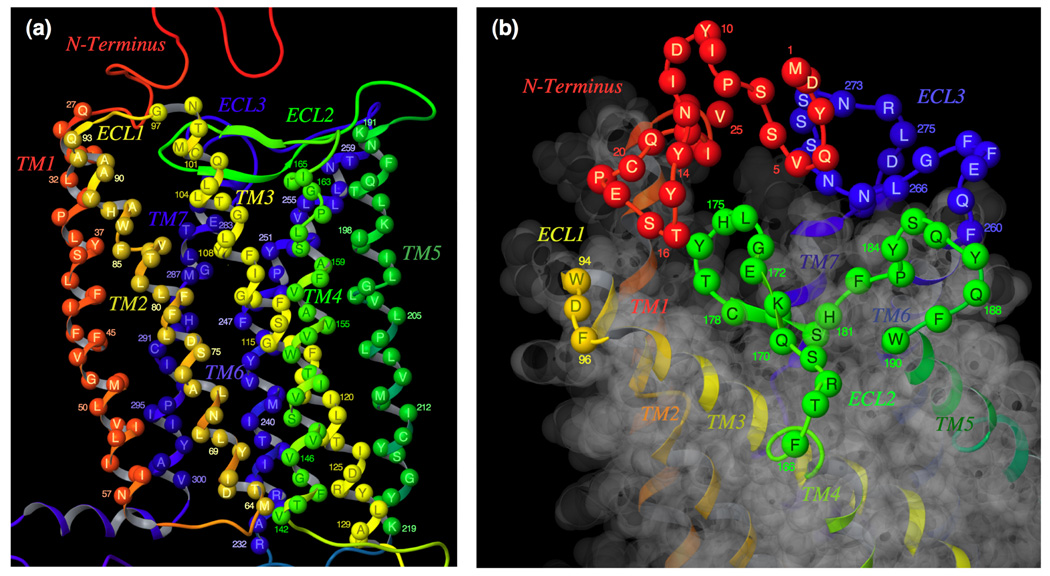
(a) A side-view of the TM domains. TM2, TM3, and TM4 are above the plane of the paper, and TM6 and TM7 are below the plane. The following assignments have been made for TM helices: TM1, residues 27 to 57; TM2, residues 64 to 93; TM3, residues 97 to 130; TM4, residues 142 to 165; TM5, residues 191 to 219; TM6, residues 232 to 259; TM7, residues 279 to 300. (b) A top-view of the extra-cellular loop regions. The following assignments have been made for loops: N-terminus, 1 to 26; ECL1, residues 94 to 96; ECL2, residues 166 to 190; ECL3, residues 260 to 278.
In the present study, in an attempt to examine the interplays of ECL2 and selected amino acid residues that consist of the largest hydrophobic cavity within CCR5, which accommodates small molecule CCR5 inhibitors, we generated a homology model of CCR5, without any ligands bound, using the crystal structure of bovine rhodopsin as a template.27 In generating the model, the following structural assignment to CCR5 was made: residues 1 through 26 to the N-terminus region; residues 27 through 57 to transmembrane region-1 (TM1); residues 58 through 63 to the first cytoplasmic region (C1); residues 64 through 93 to TM2; residues 94 through 96 to first extracellular loop (ECL1); residues 97 through 130 to TM3; residues 131 through 141 to C2; residues 142 through 165 to TM4; residues 166 through 190 to ECL2; residues 191 through 219 to TM5; residues 220 through 231 to C3; residues 232 through 259 to TM6; residues 260 through 278 to ECL3; residues 279 through 300 to TM7; residues 303 through 312 to the helix region (H8) in the cytoplasmic domain; and residues from 313 as forming the C-terminus (Fig. 2).
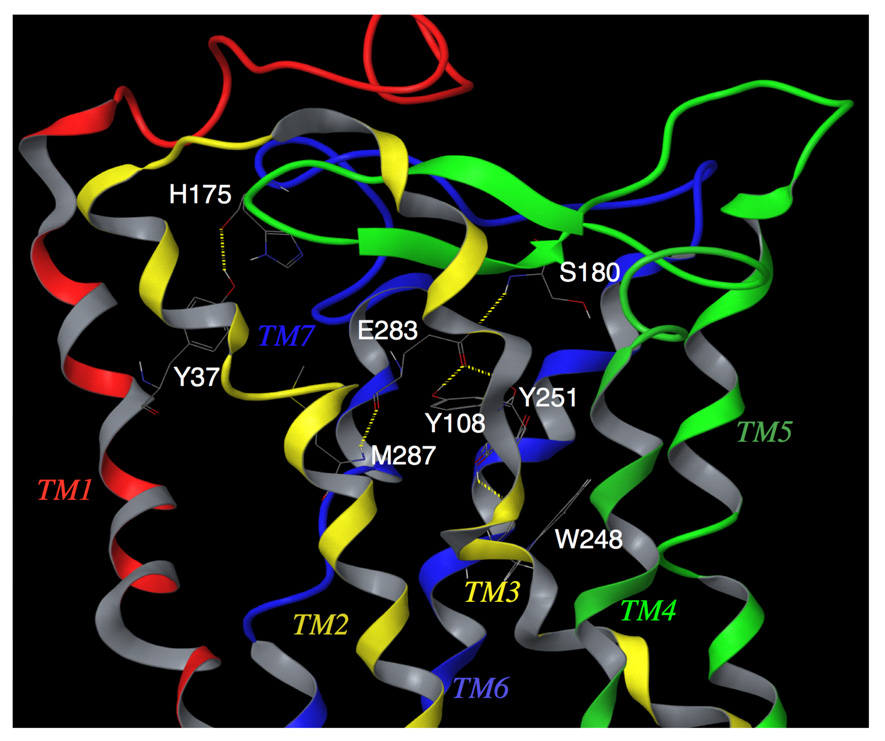
To more effectively sample the conformational space occupied by CCR5, we carried out molecular dynamics simulation for 4800 pico-seconds (ps) to efficiently explore the conformational space of CCR5. The simulation was carried out in implicit water with a time step of 1-femto-second, and without any distance cut-offs for non-bonded van der Waals and electrostatic interactions. These stringent conditions added considerably to the overall computation time, but are thought to provide robust results. The conformations at intervals of 50 ps were analyzed. As expected, there are several hydrogen bonds between nearby residues in the same transmembrane helix that is important for maintaining the helical structure. Examples of such interactions are between W248 and Y251 in TM6, and between E283 and M287 in TM7 (Fig. 3).
Fig. 3.
Transmembrane residues Y108(TM3) / E283(TM7) / Y251(TM6) / M287(TM7) / S180(ECL2) of unliganded CCR5, forming a hydrogen bond network. Y37 has hydrogen bond interaction with H175. This shows that transmembrane residues, implicated in CCR5 inhibitor binding, have direct interactions with ECL2 residues in the unliganded receptor. It is assumed that these intramolecular interactions are responsible for maintaining a conformation of ECL2 that is favorable for binding with HIV-1 gp120/CD4 complex. AK530 interacts with Y37, Y108, E283, and Y251 in the binding cavity and disrupts the intramolecular interactions of Y108 and Y251 with E283, and those of E283 with the ECL2 (Fig. 5). It is assumed that these structural changes, after inhibitor binding, alter the conformation of the ECL2, resulting in loss of association with gp120. The analysis strongly suggests that the loss of hydrogen bonds between helices may cause allosteric conformational changes in ECL2 leading to the inhibition of HIV-1 gp120 binding to CCR5.
A hydrogen bond network was identified, involving multiple helices and ECL2: Y108 (TM3), E283 (TM7), Y251 (TM6), and S180 (ECL2) (Fig. 3). These hydrogen bond interactions were considered particularly important for the following reason. Inter-helical hydrogen bonds have been shown to be critical in maintaining inactive conformations of G-protein coupled receptors.28 For rhodopsin, whose structure-function relationships have been studied widely using biochemical and spectroscopic methods, movement of TM3 and TM6 helices are known to produce changes in the loop conformations.29 This inter-helix movement takes place due to the loss of hydrogen bond interactions between TM3 and TM6 residues. The loss of hydrogen bond network between transmembrane helices, mediated through water molecules, have also been thought to be responsible for carrying out changes in loop conformations with functional implications.30 When AK530 and AK317 were bound to CCR5, Y108 rotated away from Y251, and the hydrogen bonds between Y108 and E283 and between E283 and S180 were lost (see below). The change in this polar interaction might be responsible for changes in CCR5 loop conformation for inhibiting gp120 interaction.
Activity against R5-HIV-1 and the CCR5 binding affinity of CCR5 inhibitors
We determined the activity of two novel spiro-diketopiperazine (SDP) - based CCR5 inhibitors, AK530 and AK317 (Fig. 1), against R5-HIV-1 in a cell-based acute R5-HIV-1 exposure assay using the HeLa-CD4-LTR-β-gal indicator cell line expressing human CCR5 [CD4+, CCR5+ MAGI cells] and PHA-PBMCs as target cells with two different R5-HIV-1 species, HIV-1JRFL and HIV-1Ba-L. We also conducted a saturation binding assay using 3H-labeled AK530 and AK317 and CCR5WT-expressing CHO cells and determined their binding affinity to CCR5WT (KD; dissociation constant values) as previously described.14 As shown in Table 1, AK530 exerted potent antiviral activity against both HIV-1JRFL and HIV-1Ba-L in two different target cells with IC50 values ranging 2.1 – 32 nM and proved to have a high binding affinity to CCR5 with a KD value of 1.4 ± 0.9 nM. AK317 was comparably potent against the virus (IC50: 1.5 – 25 nM), but had a lower CCR5-binding affinity by about a factor of 10 (KD: 16.7 ± 7.5 nM) than AK530. Neither of these CCR5 inhibitors had activity against X4-HIV-1 (data not shown). For comparison, the anti-viral potency and binding affinity of APL 14 is illustrated in Table 1. APL had a greater anti-HIV-1 activity (IC50: 0.2 – 0.7 nM) than AK530 and AK317, but its binding affinity (KD value: 3.6 ± 1.3 nM) was slightly less than that of AK530.
TABLE 1.
Anti-HIV activity against wt-R5-HIV-1 and binding affinity to wt-CCR5 of CCR5 inhibitors
| Antiviral assay (Virus) [IC50a and IC 90, nM] |
Binding Assay (KD, nM) |
||||
|---|---|---|---|---|---|
| Compound | MAGI with HIV-1JRFL | MAGI with HIV-1Ba-L | PHA-PBM with HIV-1Ba-L | ||
| AK530 | IC50 | 2.8 ± 1.5b | 2.1 ± 1.1 | 32 ± 27 | 1.4 ± 0.9 |
| IC90 | 51 ± 2.5 | 79 ± 34 | 430 ± 113 | ||
| AK317 | IC50 | 2.0 ± 0.2 | 1.5 ± 1.9 | 25 ± 8 | 16.7 ± 7.5 |
| IC90 | 19 ± 14 | 25 ± 3 | 171 ± 45 | ||
| APL | IC50 | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.7 ± 0.4 | 3.6 ± 1.4 |
| IC90 | 1.8 ± 0.7 | 2.9 ± 1.6 | 12 ± 10 | ||
The IC50 values were determined with the MAGI assay and the PHA-PBM (p24) assay (see "Materials and Methods").
The numbers denote the IC50, IC90 or KD values (mean ± 1 S.D.).
Binding affinity profiles of 3H-AK530 and 3H-AK317 to a panel of mutant CCR5-expressing cells
In order to explain the difference in the anti-HIV-1 activity and CCR5-binding profiles described above, we determined the binding affinity of 3H-AK530 and 3H-AK317 employing a panel of mutant CCR5-overexpressing CHO cells. As shown in Table 2, we found that the KD values of AK530 drastically increased as examined in CHO cells expressing such CCR5 mutations as Y37A, C101A, F109A, G163R, C178A, T195P, T195S, and Y251A with all KD values of >100 nM (>71-fold greater than that with CCR5WT). AK317, whose affinity to wild-type CCR5 was nearly ten-fold lower than AK530, also had a significant decrease in its binding affinity with these mutations with all KD values greater than 100 nM (>5.9-fold greater than that with CCR5WT). As for G163, a substitution from Gly to Arg was examined in this study, because the G163R substitution has been reported to reduce the susceptibility to HIV.31 In addition, AK317 had a significantly decreased binding affinity to CCR5 when it contained P84H, Y108A, K191A/N, I198A, W248A, or E283A (all KD values > 100 nM). It was seen that APL significantly decreased its binding affinity (KD values of >100 nM; >27-fold lower than that with CCR5WT) to CCR5 when it contained a P84H, C101A, F109A, G163R, C178A, K191A, T195P/S or E283A substitution. Thus, AK317, whose CCR5 binding affinity to CCR5WT was > 4-fold lower than that of APL, had virtually the same binding profile as that of APL, while two additional mutations (Y108A, and Y251A), which decreased the CCR5 binding affinity of APL (KD values < 100 nM), also nullified the CCR5 binding of AK317, consistent to the notion that the affinity of AK317 to CCR5WT is lower than that of APL. It was noted that the K191A substitution significantly reduced the CCR5 binding affinity of both AK317 and APL, it had moderately (4.2-fold) reduced the binding affinity of AK530. Moreover, the Y37A substitution, which caused only a less than 4-fold reduction in the CCR5 binding of APL (Table 2), produced a significant reduction in CCR5 binding of AK530 and AK317. Other dissociations in the KD profiles between APL and AK530 were seen when CCR5 had an amino acid substitution at Y108A (5.5-fold for APL vs. 43.3-fold for AK530), Y251A (10.1-fold for APL vs. >71-fold for AK530), or E283A (>55-fold for APL vs. 13.6-fold for AK530). Taken together, the results obtained with the saturation binding assay using 3H-labeled CCR5 inhibitors employed here suggest that the binding profile of AK317 is similar to that of APL; the binding profiles of AK530 and APL share some common features, and possibly have some important differences, while both AK530 and AK317 are comparably potent against R5-HIV-1.
TABLE 2.
Binding affinity of CCR5 inhibitors to mutant CCR5s
| KD value (nM) |
||||
|---|---|---|---|---|
| Mutant CCR5 on CHO cells | AK530 | AK317 | APL | |
| Wild type | 1.4 ± 0.9 | 16.7 ± 7.5 | 3.6 ± 1.4 | |
| D11A | NH2-terminus | 1.1 ± 0.5 | 14.6 ± 1.9 | 3.0 ± 0.6a |
| Y37A | TM1 | >100 | >100b | 12.8 ± 0.9 |
| P84H | TM2 | 37.3 ± 6.3 | >100 | >100 |
| C101A | TM3 | >100 | >100 | >100 |
| L104D | TM3 | 6.8 ± 0.7 | 93.3 ± 26.4 | 18.3 ± 3.6 |
| Y108A | TM3 | 60.7 ± 26.2 | >100 | 19.8 ± 4.4a |
| F109A | TM3 | >100 | >100 | >100 |
| F112L | TM3 | 5.4 ± 2.1 | 14.7 ± 2.5 | 4.0 ± 2.6 a |
| F112Y | TM3 | 5.2 ± 3.4 | 21.1 ± 8.9 | 6.8 ± 1.1 a |
| F113A | TM3 | 2.4 ± 0.3 | 43.3 ± 7.4 | 13.3 ± 2.3 a |
| F113Y | TM3 | 2.8 ± 0.5 | 48.2 ± 10.2 | 12.9 ± 3.1 |
| G163A | TM4 | 5.9 ± 2.6 | 14.7 ± 0.1 | 8.0 ± 4.2 a |
| G163R | TM4 | >100 | >100 | >200 a |
| R168A | ECL2 | 2.2 ± 0.7 | 24.6 ± 4.1 | 14.1 ± 9.4 |
| K171A/E172A | ECL2 | 3.5 ± 1.2 | 14.7 ± 0.1 | 2.8 ± 0.1 a |
| C178A | ECL2 | >100 | >100 | >200 a |
| S180A | ECL2 | 7.4 ± 1.4 | 34.5 ± 7.9 | 5.7 ± 1.2 a |
| S180T | ECL2 | 1.4 ± 0.6 | 14.9 ± 1.7 | 1.5 ± 0.6 a |
| S180E | ECL2 | 5.7 ± 2.3 | 61.8 ± 22.9 | 13.9 ± 1.7 a |
| Y184A/S185A | ECL2 | 2.2 ± 2.3 | 14.8 ± 0.2 | 2.0 ± 0.8 a |
| Y184A/S185A/Q186A/Y187A | ECL2 | 2.2 ± 0.4 | 22.0 ± 4.2 | 2.0 ± 0.6 a |
| Q186A/Y187A | ECL2 | 2.3 ± 0.1 | 14.4 ± 2.2 | 2.8 ± 0.5 a |
| Q188A | ECL2 | 1.9 ± 1.3 | 14.6 ± 3.6 | 6.6 ± 1.4 a |
| K191A | ECL2-TM5 | 6.0 ± 3.8 | >100 | >200 a |
| K191R | ECL2-TM5 | 5.2 ± 2.8 | 22.2 ± 7.7 | 9.0 ± 5.6 a |
| K191N | ECL2-TM5 | 12.5 ± 3.3 | >100 | 14.2 ± 1.1 a |
| T195A | TM5 | 5.6 ± 3.9 | 14.7 ± 0.1 | 48.1 ± 4.3 |
| T195P | TM5 | >100 | >100 | >100 |
| T195S | TM5 | >100 | >100 | >100 |
| K197A | TM5 | 7.6 ± 3.7 | 58.3 ± 18.7 | 10.7 ± 3.5 |
| I198A | TM5 | 9.8 ± 2.2 | >100 | 24.6 ± 4.8 a |
| W248A | TM6 | 43.4 ± 4.5 | >100 | 29.8 ± 4.6 |
| Y251A | TM6 | >100 | >100 | 36.5 ± 9.5 a |
| E283A | TM7 | 19.1 ± 2.5 | >100 | >200 a |
| M287A | TM7 | 2.4 ± 1.4 | 16.1 ± 2.1 | 6.8 ± 2.3 a |
| M287E | TM7 | 62.7 ± 17.8 | 87.1 ± 0.6 | 14.8 ± 1.7 a |
Data from Maeda et al. 22
KD values more than 5-fold compared to that with CCR5WT are shown in bold.
Amino acid residues of CCR5 crucial for interactions of CCR5 inhibitors with CCR5
We subsequently defined a three-dimensional model of human CCR5-CCR5 inhibitor complex by combining the results of site-directed mutagenesis-based analyses described above (Table 2) and molecular modeling that involved structural refinement and docking of inhibitors to an initial structure of CCR5 based on the crystal structure of bovine rhodopsin.30; 32
Of note, both amino acid substitutions, C101A (TM3) and C178A (ECL2), virtually nullified the binding of all three CCR5 inhibitors examined, AK530, AK317, and APL (Table 2). These findings confirmed the assumption that C101 and C178 form a disulfide bond that is crucial in maintaining the conformation of the second extracellular loop. These data also strongly suggest that either of the two amino acid substitutions disrupted the disulfide link, altered the conformation of the loop, and nullified the binding of the three CCR5 inhibitors to CCR5. This binding profile common to AK530, AK317, and APL indicates that their binding to CCR5 is sensitive to the ECL2 conformation and significantly differs from the binding profile of other CCR5 inhibitors such as SCH-C and TAK-779, which do not undergo drastic loss in their CCR5 binding with these mutations.19; 22; 23
The model of CCR5 complexed with a CCR5 inhibitor we generated in the present study was derived by taking the flexibility of both CCR5 and the inhibitor into account and by computationally designating a model that most suitably provided a rational explanation of the mutagenesis data. In the present study, we chose a few CCR5 residues, which were predicted to have significant interactions with the inhibitor based on our initial model.22 The residues chosen for mutation were, P84, L104, F109, T195, and W248. P84 was observed in close contact with APL in these models (Fig. 4c), and CCR5 containing a P84H substitution (CCR5P84H) was generated. The binding affinity of APL to CCR5P84H proved to decrease by nearly 30 times compared to wild-type CCR5 (CCR5WT) and confirmed that the binding of APL is indeed dependent on this residue (Table 2). L104 was also in the proximity of APL-binding cavity (Fig. 4c) and CCR5L104D was generated, which also proved to have a decrease in the binding affinity of APL by about 5-fold (Table 2). The binding affinity of SCH-C and TAK-779 for CCR5P84H and CCR5L104D were also determined, and there were minimal changes from the wild-type binding affinity (KD changed by only 2–3 fold). This indicates that P84 and L104 are likely in close contact with APL, and the decrease in binding affinity of CCR5P84H and CCR5L104D mutants is not predominantly due to any drastic conformational changes that might have accompanied these mutations. The models examined in this study showed that F109 forms a π-π interaction with APL (Fig. 4c). F109 is present in a cluster of aromatic residues involving other nearby phenylalanine and tyrosine residues in the binding pocket. CCR5F109A was generated, which proved to have a drastic loss of APL, AK317 and AK530 binding to CCR5 (Table 2). SCH-C and TAK-779 also had KD values higher than 100 nM, indicating that F109 is important for the binding of structurally diverse CCR5 inhibitors. T195 was chosen because it was observed that the inhibitors were shown to have different dependence on their binding affinity on this residue. The model for APL (Fig. 4c) showed that an intra-molecular hydrogen bond network involving S180/G163/K191/T195 is important for defining the binding cavity for APL, and APL per se also has a potential hydrogen bond interaction with T195 (Fig. 4c). The intra-molecular hydrogen bond network of CCR5 was not observed in our models of CCR5 complexed with SCH-C or TAK-779. Moreover, SCH-C or TAK-779 do not have any groups or atoms capable of forming hydrogen bond interactions with T195. Indeed, the KD values for APL for CCR5T195P and CCR5T195S were greater than 100 nM (Table 2), while the binding profile of SCH-C and TAK-779 to CCR5 was not affected by T195P or T195S substitutions [KD values: SCH-C, 16.0 nM (WT), 28.9 nM (T195P), 21.7 nM (T195S); TAK-779, 30.2 nM (WT), 31.1 nM (T195P), 32.5 nM (T195S)]. Interestingly, in CCR5 complexed with AK530 and AK317, the hydrogen bond network with S180, G163, K191, and T195 was seen, suggesting that the network led to maintain a conformation of CCR5 suitable for binding of SDP-based inhibitors. Indeed, both AK530 and AK317 failed to bind to CCR5T195P and CCR5T195S, although AK530 did not have a direct interaction with the network (Fig. 5). Taken together, these results sustained our notion that the CCR5 inhibitor-CCR5 interactions generated based on the rhodopsin crystallographic data are reasonably reliable.
Fig. 4.
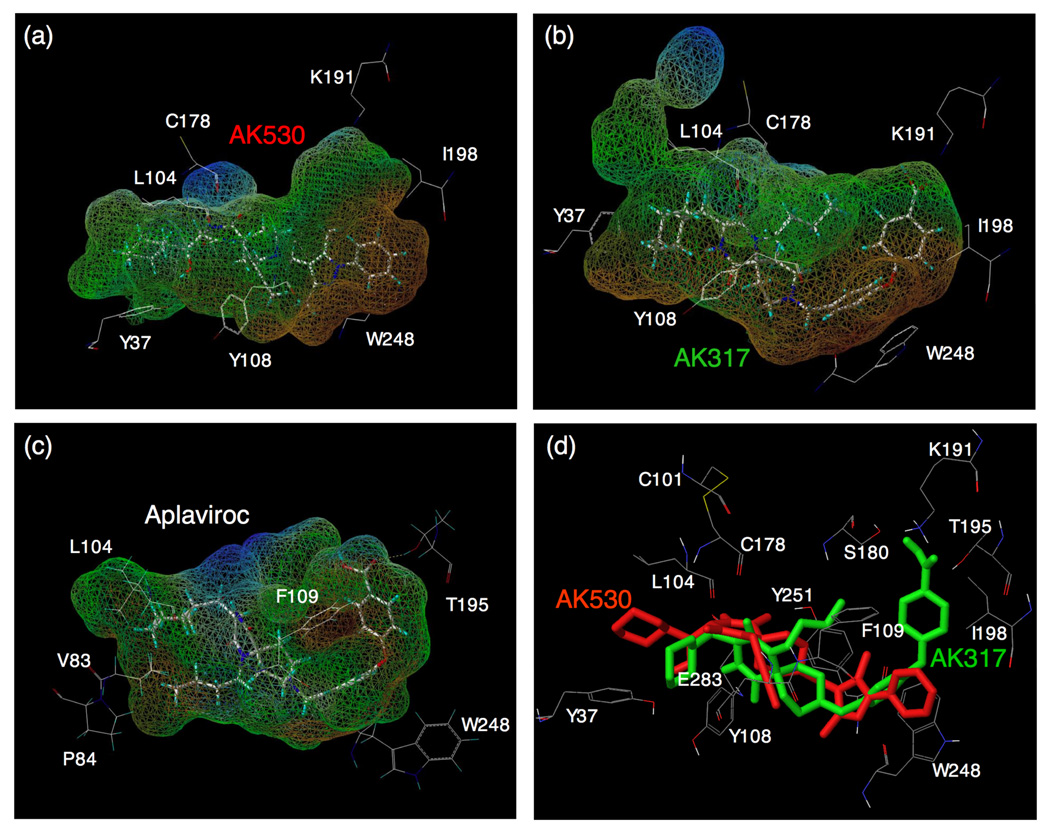
Lipophilic potential mapped on the binding cavity of CCR5 inhibitors. The lipophilic potential mapped onto the binding cavity when AK530, AK317, and APL bind to CCR5 are shown in (a), (b), and (c), respectively. The predominantly lipophilic region of the cavity is shown in brown (bottom region of the cavity which is towards the cytoplasmic region of CCR5). The blue regions are predominantly hydrophilic (present towards the extra-cellular region), and the green regions are moderately lipophilic. The figure was generated using MOLCAD. The shape of the binding cavity is slightly different for AK530 and AK317 as the receptor conformations are slightly different when these molecules bind to CCR5. The unoccupied volumes of the cavities suggest optimization ideas for improving the potency of these molecules. (d) The binding modes of AK530 and AK317 superimposed. AK530 is shown in red and AK317 in green. Note that the binding orientation in the vicinity of TM helices 5 and 6 differ. AK317 binds towards and around ECL2 residues, whereas AK530 bends towards the intracellular domain. AK530 has a high binding affinity probably because it binds “deeper” into the cavity. On the other hand, by being able to interact with ECL2 residues, AK317 maintains comparable anti-HIV-1 potency with AK530 even though its binding affinity is about ten fold lower.
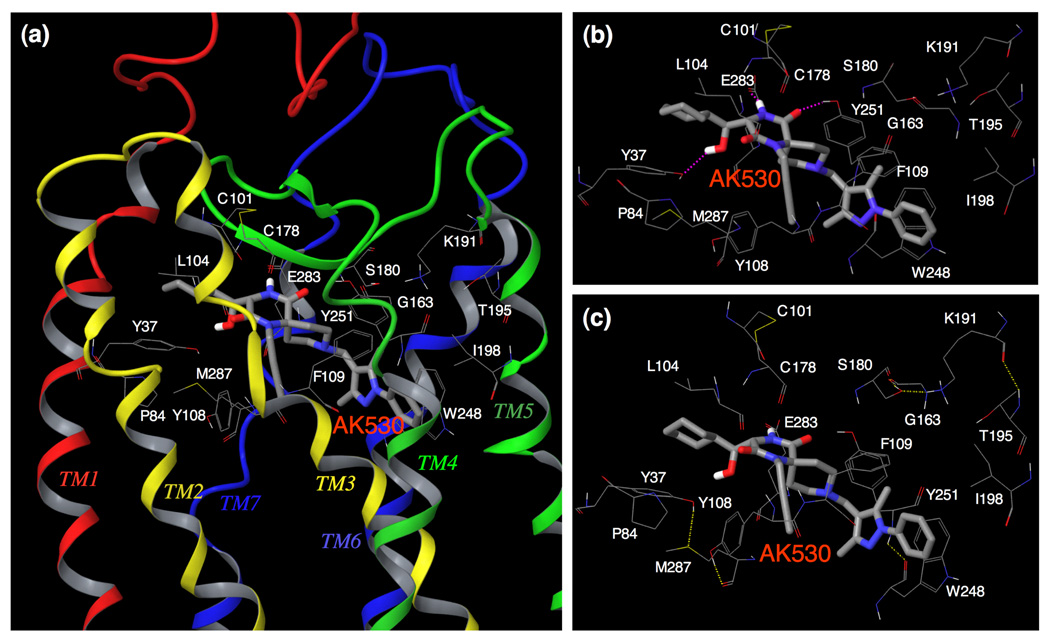
Fig. 5. Amino acid residues forming the binding cavity within CCR5 for AK530.
(a) The amino acid residues forming the binding cavity of CCR5 and the binding mode of AK530 is shown. The TM regions and the ECL2 enclose the cavity. Every TM helix has at least one residue that contributes towards forming the binding pocket for AK530. (b) AK530 is predicted to have hydrogen bond interactions with Y37(TM1), Y251(TM6), and E283(TM7), and has favorable hydrophobic interactions with several binding site residues including P84, L104, F109, and I198. The benzene ring of AK530 forms a π-π interaction with W248. (c) The hydrogen bond networks, involving multiple transmembrane domains, define the shape of CCR5 cavity for AK530. There is one network of hydrogen bonds involving Y37 (TM1), M287 (TM7), and Y108 (TM3). Another network involves S180 (ECL2), G163 (TM4), K191 (TM5), and T195 (TM5). The hydrogen bond networks are different for the unbound and inhibitor bound CCR5.
Structural analysis of AK530 and AK317 interactions with CCR5
The structural interactions of AK530 with CCR5 are shown in Fig. 5a–c. The binding cavity of AK530 involves residues from each of the transmembrane domains and the ECL2. The binding cavity is stabilized by several intra-molecular hydrogen bonds involving residues in different trans-membrane regions. Y37, M287, and Y108, which are in transmembrane helices 1, 7, and 3 respectively, form a hydrogen bond network that defines one end of the cavity. Another network involves S180(ECL2), G163(TM4), K191(TM5), and T195(TM5). AK530 binds in a somewhat diagonal fashion, reaching deeper into the cavity of CCR5, and has several polar and non-polar interactions that give rise to its very high affinity (KD = 1.4 nM). The hydroxymethylene of AK530 is predicted to have a hydrogen bond interaction with the side-chain of Y37; and the diketopiperazine ring of AK530 with the side-chains of Y251 and E283. The hydrophobic interactions that stabilize binding involve P84, L104, and I198. There are π-π interactions involving F109 and W248. Indeed, binding affinity of CCR5 species containing Y37A, Y108A, or W248A was drastically reduced in our saturation binding assays and the residues implicated in the binding of AK530 as described above appear to have important structural significance. F109 is present in a cluster of aromatic residues in TM3. W248 is part of a highly conserved set of residues present in TM6 of class A GPCRs. E283 is the fifth residue in TM7. This position has been shown to be involved in the binding of small molecule ligands to several chemokine receptors.33 It is noteworthy that we have demonstrated that the E283 residue is important not only in the binding of other CCR5 ligands such as SCH-C and TAK-779, but is also important in preserving the CC-chemokine/CCR5 interactions and HIV-1-gp120/CD4 complex binding to CCR5.22
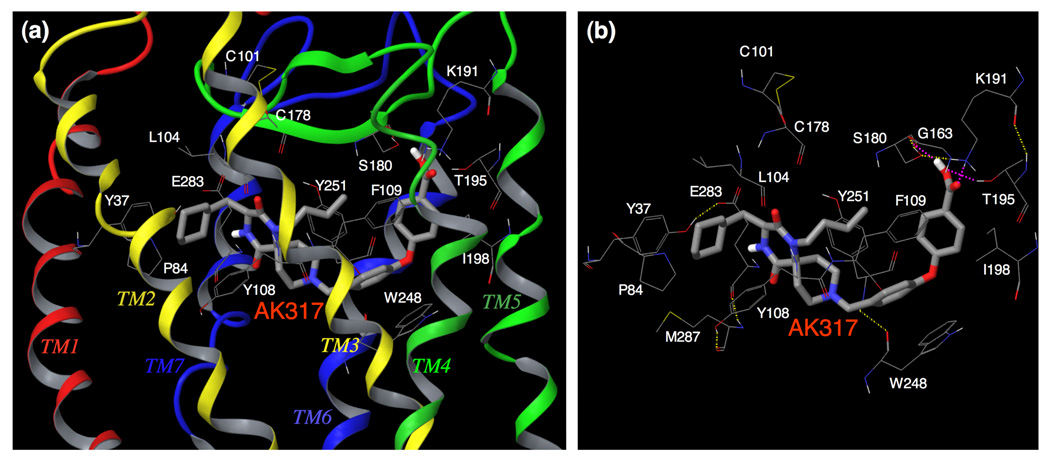
The binding site residues and the binding mode of AK317 are shown in Fig. 6a–b. AK530 and AK317 are analogues and are expected to share certain common features of CCR5 binding. However, important structural differences were identified between the two analogues. AK530 has a cyclohexyl-hydroxymethyl group, whereas AK317 only has a cyclohexylmethyl group. AK530 has a phenyl-pyrazol moiety while AK317 has a phenoxy-benzoic acid group as a substituent. The carboxylate group of the benzoic acid in AK317 forms hydrogen bond interactions with S180, K191, and T195. These three residues, along with G163, are involved in an intra-CCR5 hydrogen bond network. Thus, it appears that K191 and T195 are not only important in defining the shape of the cavity, but are also involved in directly binding with AK317. AK317 has van der Waals interactions with P84, Y108, I198, and Y251, and π-π interactions with F109 and W248. Another intra-CCR5 hydrogen bond network involving Y37(TM1), E283(TM7), M287(TM7), and Y108(TM3) is responsible for defining the binding cavity. As shown in Figures 4a and 4b, the size and shape of the binding cavity of AK530 appear to differ from those of AK317, and the superimposition of the binding modes of AK317 and AK530 clearly shows that although these two inhibitors share certain common binding features, the orientations and topography of these two inhibitors are substantially different (Fig. 4d).
Fig. 6. Amino acid residues forming the binding cavity within CCR5 for AK317.
(a) The binding mode of AK317 within CCR5 is shown. (b) The intramolecular hydrogen bond interactions of CCR5 defining the binding cavity, and the binding interactions of AK317 with CCR5 are shown. AK317 has hydrogen bond interactions with S180, K191, and T195 (shown in pink dotted line). Other residues in the binding cavity are predicted to have hydrophobic interactions with AK317. As in the case of AK530, there are several intra-molecular hydrogen bond networks (shown in yellow dotted line) that define the shape of the CCR5 binding cavity for AK317. There is a network involving, S180 (ECL2), G163 (TM4), K191 (TM5), and T195 (TM5), and another involving Y37 (TM1), E283 (TM7), M287 (TM7), and Y108 (TM3). The conformation involving amino acid residues in the latter network differs from that of the case of AK530 binding. It appears that CCR5 undergoes different conformational changes to accommodate different inhibitors.
As described above, in spite of sharing a common core, the different substituents make the conformation of these two molecules different which gives rise to differences in structural interactions with CCR5. The binding orientations differ the most around transmembranes five and six. The phenoxy-benzoic acid group of AK317 interacts with S180 and K191. S180 is a part of ECL2 whereas K191 in TM5 is likely located above the plane of the extra-cellular region. The pyrazol group of AK530 on the other hand, bends towards the cytoplasmic region, and makes the inhibitor bind deeper into the cavity. There are no direct interactions of AK530 with K191 but it seems to be forming a favorable conformation of the binding pocket by an intra-molecular hydrogen bond network with G163 and T195 (Fig. 5c). K191, located in the upper domain of TM5, can be considered to be an extracellular residue as well since a part of K191 is located above the cellular plane (Fig. 2). We speculate that the direct interactions of AK317 with the ECL2 residues such as S180, and the residues in the extracellular region (such as K191) are responsible for its high antiviral IC50 of 2 nM (Table 1). In spite of being a weaker binder compared to AK530, the interactions of the phenoxy benzoic acid group of AK317 seems to be responsible for its comparable antiviral potency with AK530 which has no direct interaction with ECL2 residues in the vicinity of TM4 and TM5.
The binding cavity of both AK530 and AK317 is mostly lipophilic (Fig. 4a–b). The only strongly hydrophilic regions are towards the extra-cellular loop region. The shape and volume of the cavities are slightly different for these two inhibitors as CCR5 is likely to undergo conformational changes to different extents to accommodate these two inhibitors. The unoccupied regions of the cavity suggest new optimization ideas for these inhibitors. For example, substituents of AK530 that can potentially interact with K191 may increase its anti-viral potency even further.
Determination of interatomic distances between key amino acids when AK317 and AK530 bind to CCR5
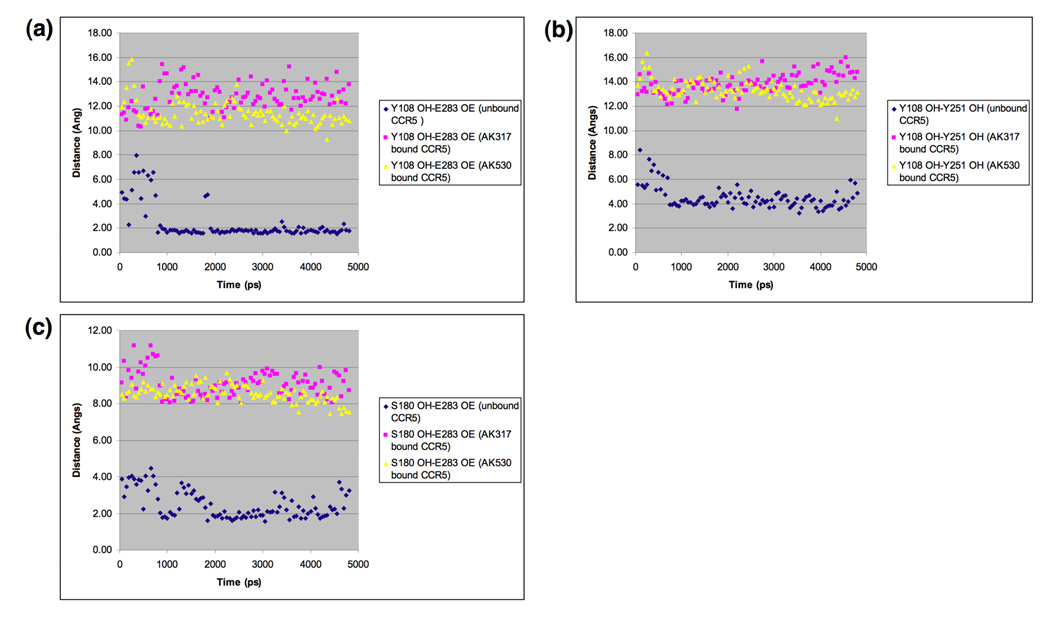
In an attempt to examine whether the binding of AK317 or AK530 to CCR5 causes significant changes in interatomic distances between key amino acids that form the hydrogen bond network seen in the inhibitor-unbound CCR5 as described above (Fig. 3), we carried out molecular dynamics simulation for 4800 ps for AK317- and AK530-bound CCR5, and analyzed critical interatomic distances with the inhibitor-unbound conformation (Fig. 7a–c). In the inhibitor-unbound conformation, Y108 in TM3 was in close proximity of Y251 located in TM6 and had a hydrogen bond interaction with E283 in TM7 (Fig. 3, and Fig 7a–b). In the AK317-bound CCR5, Y108 had rotated away from Y251, and the hydrogen bond interaction between Y108 and E283 was lost. Rotation of Y108 away from Y251, and disruption of hydrogen bond interaction between Y108 and E283 was also observed when AK530 bound to CCR5. TM2 and TM3 can be thought to be in a single plane (above the plane of the paper), and TM6 and TM7 can be thought to be in a different plane (behind the plane of the paper). The rearrangement of Y108 was necessary for the formation of the binding cavity between the transmembrane helices for inhibitor binding. Furthermore, in the unbound CCR5 structure, E283 is hydrogen-bonded to S180, which is part of the ECL2 (Fig. 3). In fact, E283 moved considerably away from S180 in the inhibitor-bound models of CCR5 (Fig. 5c, Fig. 6b, and Fig. 7c). Disruption of hydrogen bonds between transmembrane residues, and a network of hydrogen bonds mediated through water molecules is known to give rise to the change in intracellular loop conformation of bovine rhodopsin.29;30 Change in ionic interactions between transmembrane residues is also thought to be responsible for the activation of β2-adrenergic receptor.34 We speculate that the rotation of Y108 away from the vicinity of Y251, the loss of hydrogen bond interaction between Y108 and E283 and that between E283 and S180 presumably change the conformation of the ECL2 that is crucial for the binding of gp120/CD4 complex. These models of inhibitor-unbound and -bound CCR5 may give further insights to the mechanism of inhibition of CCR5 inhibitors involving transmembrane and extracellular residues that are interacting with each other.
Fig. 7.
Key interatomic distances from molecular dynamics simulation of AK317-bound, AK530-bound, and inhibitor-unbound CCR5. Molecular dynamics simulation for 4800 ps for AK317- and AK530-bound CCR5 was conducted and critical interatomic distances between key amino acids were determined. A hydrogen bond is present if the interatomic distance is less than 3Å. (a) Distance between Y108 (hydroxyl hydrogen, PDB atom type OH) and E283 (carboxylate oxygen, PDB atom type OE). In the inhibitor-unbound conformation, there is a strong hydrogen bond interaction between Y108 in TM3 and E283 in TM7. Y108 and E283 have to move away from each other to form the binding cavity for the inhibitor to bind, and there is no hydrogen bond between these residues after AK317 and AK530 bind. (b) Distance between Y108 and Y251 hydroxyl oxygen): The tyrosines have moved away from each other after inhibitor binding. (c) Distance between E283 (carboxylate oxygen, PDB atom type OE) and S180 (hydroxyl hydrogen, PDB atom type OH). In the unbound conformation, E283 in TM7 has hydrogen bond interactions with S180 in ECL2. This hydrogen bond is disrupted for AK317 and AK530 bound CCR5.
Structural changes in ECL2 caused by CCR5 inhibitors block the binding of CCR5-specific mAbs to CCR5
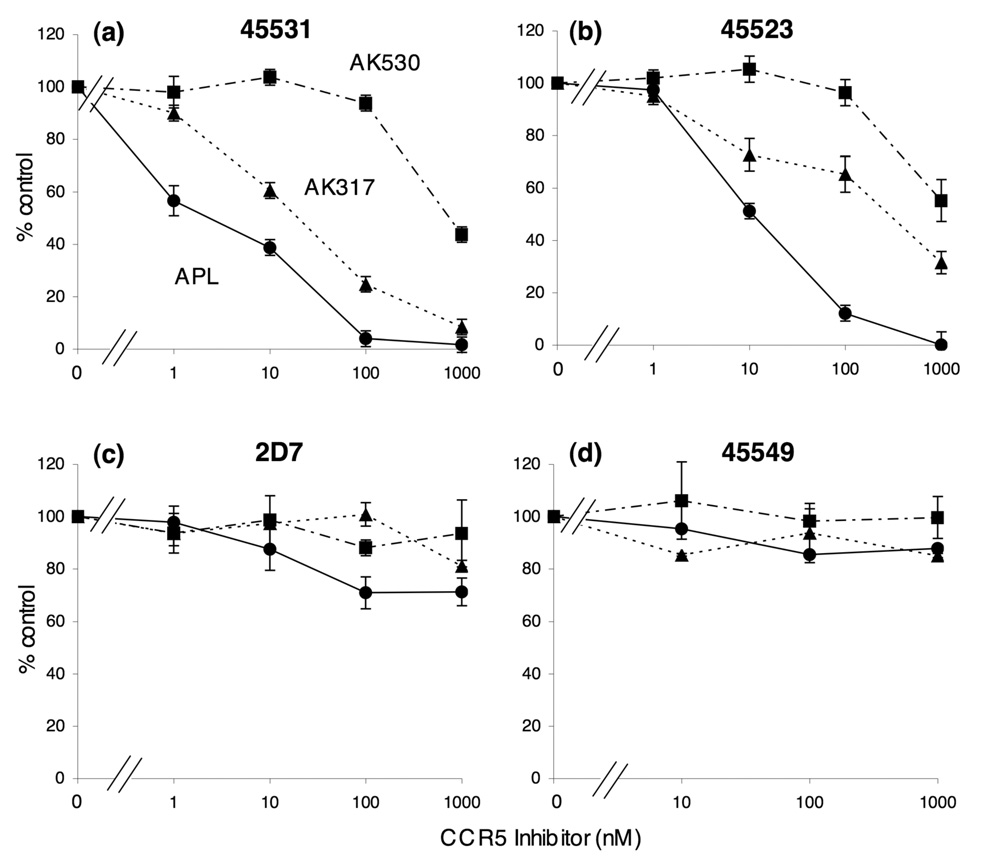
Our structural analyses described above demonstrated that AK530 relatively well maintained its CCR5 binding with the K191A substitution, while AK317 significantly reduced its interactions with CCR5K191A, prompting us to examine whether the different CCR5 binding profiles of these two inhibitors led to different dynamics in their interactions with CCR5-specific monoclonal antibodies. To this end, we examined whether AK530 and AK317 blocked the binding to CCR5 expressed on CHO cells of four monoclonal antibodies (mAbs), 45531 [specific for the C-terminal half of ECL2 (ECL2B)], 45523 (reactive to CCR5 multi-domain), 2D7 [specific for the N-terminus of ECL2 (ECL2A)], and 45549 (reactive to CCR5 multi-domain).25; 35; 36 In this mAb binding blocking assay, the cells were incubated with each inhibitor for 30 min, followed by the addition of a mAb. As shown in Fig. 8, AK317 effectively inhibited the binding of mAb 45531 with an IC50 value of 16.3 nM, while the inhibition by AK530 was much less with an IC50 value of 746 nM. AK317 also blocked the binding of mAb 45523 with an IC50 value of 282 nM, while AK530 did so with an IC50 value of >1 µM. None of the inhibitors blocked the binding of mAbs 2D7 or 45549. When the cells were exposed to a mAb first, followed by the addition of each inhibitor, the resultant inhibition levels were virtually the same (data not shown). Considering that these CCR5 inhibitors are lodged in a hydrophobic pocket at the interface of ECL2 and the upper TM domain (Fig. 4 – Fig. 6), the inhibition of mAb binding by CCR5 inhibitors observed presumably occurred through the allosteric changes secondarily caused by CCR5 binding of the inhibitor but not through their competition over the binding to the antigenic epitope(s) of CCR5.
Fig. 8.
Inhibition of the binding of anti-CCR5 mAbs by AK530, AK317, and APL. Inhibition by three CCR5 inhibitors of the binding of anti-CCR5 mAbs [45531 (a), 45523 (b), 2D7 (c), and 45549 (d)], which recognize the extracellular domain(s) of CCR5, is illustrated. CCR5-overexpressing CHO cells were incubated with each of FITC-conjugated anti-CCR5 mAbs in the presence of various concentrations of a CCR5 inhibitor and the fluorescence intensity on the cells was determined. Each value was compared to that obtained without an inhibitor and is shown as % control.
Effects of amino acid substitutions in CCR5 on HIV-1-gp120-elicited cell fusion
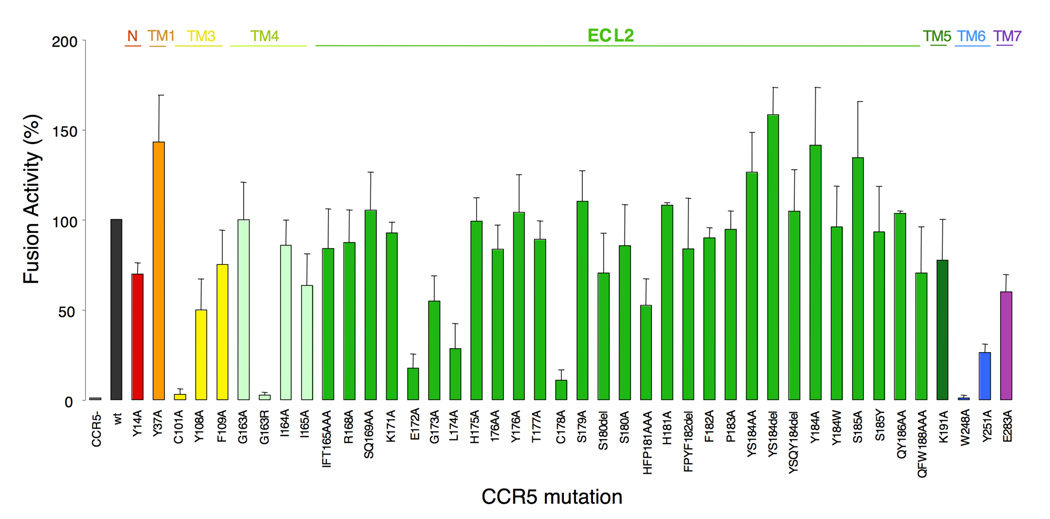
In order to better understand conformational changes arising in ECL2 and to determine amino acid(s) that are critical for the cellular entry of HIV-1, we examined the effects of single and multiple amino acid substitutions in ECL2 on HIV-1-gp120-elicited cell fusion. Fig. 9 shows the magnitude of cell-cell fusion between Tat+,env+ 293T cells and Luc+,CCR5+ MAGI cells in the HIV-1-gp120-elicited cell-cell fusion assay. Seven single amino acid substitutions introduced into CCR5 resulted in a substantial reduction in the HIV-1-envelope protein-elicited fusion level by more than 70% (Fig. 9). Three of these single amino acid substitutions (E172A, L174A, and C178A) are located in ECL2 that has an antiparallel β-hairpin structure, while C101A and G163R are in TM3 and TM4, respectively, and W248A and Y251A are in TM6 (Fig. 2 and Fig. 3). It is of note that E172 is at the NH2-end of the β-sheet structure of ECL2 and C178 also belongs to the β-hairpin structural motif (Fig. 2). This finding strongly suggests that these mutations led to the disruption of ECL2’s β-sheet structure and resulted in a significant reduction in the cell-cell fusion. Thus the β-sheet structure of ECL2 seems to be critical for cellular entry of HIV-1. It is noteworthy that although the two amino acids, E172 and L174, do not interact with CCR5 inhibitors (Fig. 4 – Fig. 6), their mutations significantly reduced cell-cell fusion (Fig. 9).
Fig. 9.
Effects of an amino acid substitution(s) and deletion(s) on HIV-1 gp120-elicited cell-cell fusion. CD4+ MAGI cells which expressed sufficient numbers of wild-type or mutated CCR5 were cultured with HIV-1 env+, tat+ 293T cells for 6 hours, and the fusion efficiency was determined with the luciferase activity (luminescence levels) using the reporter gene activation assay. The magnitude of luminescence levels with mutant CCR5 is expressed as % fusion (% control compared to the luminescence level with wild-type CCR5). Note that three single amino acid substitutions (E172A, L174A, and C178A), which resulted in a substantial reduction in the fusion level (> 70%), are located in ECL2 that has an antiparallel β-hairpin structure (Fig. 2 and Fig. 3), while C101A and G163R are in TM3 and TM4, respectively, and W248A and Y251A are in TM6.
The C101A and C178A substitutions are thought to disrupt the disulfide bond between C101 and C178, which is crucial for the maintenance of the robust ECL2 structure as demonstrated in this study. Thus, both C101A and C178A substitutions probably led to the aborted HIV-1 envelope-elicited fusion. G163, located in TM4, does not interact directly with AK530, AK317 or APL, but is responsible for maintaining the shape of the binding cavity by its hydrogen bond interactions with S180, K191, and T195 (Fig. 5 and Fig. 6). G163R substitution also resulted in virtually complete loss of fusion (Fig. 9). This is consistent with earlier observations that G163R mutation results in the loss of HIV-1 infectivity.22; 31
Amino acids within ECL2 at positions 179 through 190 appeared not to have a crucial role in the HIV-1 envelope-elicited fusion. Indeed, the amino acid substitutions in these positions did not significantly reduce the fusion (Fig. 9). It is noteworthy that these amino acids are located distant from the binding cavity for CCR5 inhibitors (Fig. 2, Fig.5, and Fig. 6). It is also of note that W248 has hydrogen bond interactions with Y251 and is thought to play a role in maintaining the conformation of ECL2 (Fig. 3), which explains the reason why W248A substitution results in loss of fusion.
Interestingly, both C178A and C101A substitutions substantially reduced CCR5 binding of the three CCR5 inhibitors (AK530, AK317, and APL), but not that of SCH-C or TAK-779.22 In particular, APL forms hydrogen bonding with C178 and, additionally, has tight interactions with the intramolecular hydrogen bond network (S180/G163/K191/T195).22 AK317 also has interactions with S180/G163/K191/T195 network (Fig. 6). Thus, SDP-containing CCR5 inhibitors, especially APL and AK317, appear to have greater interactions with ECL2 compared to other non-SDP-containing CCR5 inhibitors such as SCH-C or TAK-779. Our observation that AK530, which has the greatest binding affinity to CCR5 (Table 1), but exerts less potent anti-HIV-1 activity (Table 1) may be related to its reduced interactions with ECL2 (Fig. 5 and Fig. 7). Such tight interactions with ECL2 may represent a feature conferring potent anti-HIV-1 activity on CCR5 inhibitors, different than allosteric changes caused by CCR5 inhibitors.
DISCUSSION
In the present study, we demonstrated that two novel CCR5 inhibitors, AK530, and AK317, are lodged in a hydrophobic cavity located between the upper transmembrane domain and ECL2.CCR5 is a member of GPCRs, the largest superfamily of proteins in the body. The understanding of the structure of human GPCRs would be invaluable in elucidating their roles in a number of biological processes and should also greatly aid designing therapeutics. In particular, the elucidation of detailed structure of human CCR5 and its interactions with HIV-1 envelope glycoprotein should help establish a strategy for HIV-1 intervention. However, no crystal structures of human CCR5 are yet available; bovine rhodopsin and β2-adrenergic receptor are the only GPCR species, for which crystallographic data have been obtained.32; 37 Thus, we have previously explored an alternate approach: combination of site-directed mutagenesis-based saturation binding assay and molecular modeling based on the crystallographic data of bovine rhodopsin.22 The site-directed mutagenesis-based saturation binding assay alone can give certain insights as to which residues of the receptor are implicated in the binding with the inhibitor. However, it does not indicate which specific atoms of the residue are involved. Homology modeling and docking give insights to interactions between atoms, but these methods produce multiple possible solutions, and it is difficult to differentiate between distinct interactions that are within a small energy range. Combining these complementary methods of site-directed mutagenesis and computational protein structure determination have made it possible to conduct robust structural analyses.22 However, certain validation of the model is crucial. Thus, in the present study, we chose five amino acid residues of CCR5 that were judged to be critical for APL binding based on our previous analysis and additional mutagenesis studies were carried out. When an amino acid substitution, P84H, C101A, F109A, T195P, or T195S, was introduced to CCR5WT and saturation binding assay was performed using 3H-APL, all KD values proved to be greater than 100 nM, while the KD value of 3H-APL with wild-type CCR5 was 3.6 nM. This corroborated the notion we previously made 22 that the alternate approach using saturation binding assay combined with our computational protein structure determination can provide reasonably robust structural insights.
In the present study, we attempted to obtain a refined structural CCR5 model through molecular dynamics computation. In an inhibitor-unbound CCR5, we identified key interactions between residues located in different transmembranes, and between transmembrane and ECL2 amino acid residues. Amino acids involved in such intramolecular interactions included Y37, H175, Y108, Y251, E283, and S180. The intramolecular hydrogen bond interactions observed among different transmembrane (TM) regions, and between TM and ECL2 loop presumably stabilize the unbound conformation of CCR5 (Fig. 3). In order to determine the structures of AK317, and AK530 complexed to CCR5, we utilized a novel algorithm to incorporate receptor flexibility and induced-fit effects.56 The optimized AK317-CCR5 and AK530-CCR5 complex structures indicated a rotation of Y108 (TM3) away from both Y251(TM6) and E283(TM7) to make room for the inhibitor within the transmembrane helices. As a result of inhibitor binding, the hydrogen bond between Y108 and E283 and that between E283 and S180 seen in the unliganded CCR5 were disrupted, and these residues form hydrogen bond or tight van der Waals interactions with the inhibitors instead (Fig. 5–Fig. 7). The disruption of inter-helix residue interactions may cause movement of the TM helices and changes in the conformation of ECL2. A similar mechanism, involving disruption of inter-helix hydrogen bond interactions, is thought to be responsible for changes in loop conformation in rhodopsin,27; 29 and in the activation of β2–adrenergic receptor.34 It is of note that Y37(TM1) is involved in a hydrogen bond interaction with ECL2 via H175. We have observed that Y37 interacts with SCH-C, and TAK-779.22 These arrangements of hydrogen bond interactions between transmembrane helices and ECL2 should also provide an explanation of the reason these inhibitors exert potent antiviral activity against HIV-1. Thus, the disruption of hydrogen bond interactions between TM helices, and between TM helices and ECL2, should be a mechanism of allosteric inhibition observed by the binding of CCR5 inhibitors to a binding domain residing mostly within the transmembrane residues.
We also examined the interplays of ECL2 and selected amino acid residues that consist of the largest hydrophobic cavity within CCR5, which accommodate small molecule CCR5 inhibitors. Our previously published data 22 and data by others 19; 23 showed that two CCR5 inhibitors (SCH-C, TAK-779) have no direct interactions with amino acid residues in extracellular domain including ECL2. However, all three SDP-based CCR5 inhibitors we examined (AK530, AK317, and APL) had substantial interactions with amino acids in ECL2, in particular with C178 located in the antiparallel β-hairpin structural motif of ECL2 and K191 located at the interface of ECL2 and TM5 (Fig. 4– Fig. 6). The C178A substitution virtually abrogated the binding of the three inhibitors to CCR5. C178 is presumed to form a disulfide bond with C101 of TM3 and seems to be critical for the conformation of ECL2. The disruption of the disulfide bond with C178A mutation may result in decreasing the binding of the three SDP-based inhibitors. Our previous observation,22 and reports from others,19; 23 that the same C178A substitution did not affect the CCR5 binding of two other CCR5 inhibitors, SCH-C and TAK-779, suggest that C178 plays a unique but critical role in the binding of AK530, AK317, and APL. In the present study, K191A substitution also virtually nullified CCR5 binding of AK317 and APL (Table 2), although it did not significantly affect the binding of AK530 probably due to the absence of hydrogen bonding between AK530 and K191 (Fig. 5). Taken together, these data suggest that unlike the cases of SCH-C and TAK-779, at least a part of the hydrophobic cavity, where AK530, AK317, and APL are lodged within CCR5 involves ECL2. The disruption of ECL2’s β-sheet structure by the removal of disulfide bond through C101A and C178A substitutions virtually nullified both the binding of all three CCR5 inhibitors and the HIV-1 gp120-elicited fusion. This strongly suggests that ECL2 plays a crucial role not only in the binding of the three CCR5 inhibitors but also in the interaction of HIV-1 envelope glycoproteins with CCR5. The data also suggest that at least two amino acids in ECL2, C178 and K191, can be potential targets for the design of CCR5 inhibitors.
Several studies have shown that the resistance against a CCR5 inhibitor emerged without the change of coreceptor usage.38–43 Resistant R5-HIV-1 variants were reportedly obtained by passage 22 for AD101,39 passage 43 for TAK-652,40 passage 20 for VVC,42 and passages 12–18 for MVC. 41 However, we have failed in selecting R5-HIV-1 variants resistant to APL even after 60 passages in vitro (over ~1.5 years) (Nakata et al. unpublished), although the possibility of the emergence of HIV-1 variants resistant to APL cannot be ruled out in other settings.44 Pugach et al. demonstrated that one of the mechanisms by which HIV-1 becomes resistant to CCR5 inhibitors such as VVC, is by "noncompetitive resistance", a process in which a resistant virus continues to enter target cells regardless of the concentration of the inhibitor, once HIV-1 acquires an ability to use the inhibitor-bound CCR5 for entry.43 Of note, Westby et al. reported that a resistant virus against maraviroc (MVC) retained susceptibility to APL, suggesting that the virus can use MVC-bound CCR5 for entry but can not use APL-bound CCR5.41 Thus, APL is likely to have such a profile that does not allow or delay HIV-1 acquisition of an ability to utilize the "APL-bound" CCR5 for its cellular entry. This potentially favorable property of APL may be related to the direct interactions of APL with amino acids in ECL2, producing “substantially distorted” ECL2, with which HIV-1 gp120 cannot get engaged for its cellular entry, while certain unique allosteric changes of ECL2 conformation following the binding of APL might also explain the substantial delay or lack of the emergence of APL-resistant HIV-1.
The present data, taken together, demonstrate that structural modeling analysis coupled with CCR5 binding affinity data should help understand structural/molecular mechanism of the inhibition of HIV-1 infection by CCR5 inhibitors. The data should not only help delineate the structural dynamics of CCR5 following ligand binding but also aid in the design of therapeutic inhibitors. The data, in particular, demonstrate that through studying the properties of inhibitor-unbound and -bound CCR5, transmembrane residues such as Y108, Y251, and E283 are important for both gp120 fusion, HIV infectivity,22 and inhibitor binding. The loss of hydrogen bond interactions among these key transmembrane residues and the interactions between E283 and S180, which are essential for the formation and maintenance of the binding pocket for CCR5 inhibitors, might be responsible for changes in ECL2 conformation, providing insights to the mechanism of gp120 inhibition.
Materials and Methods
Reagents
Three spirodiketopiperazine (SDP) derivatives, aplaviroc (APL) [4-[4-[(3R)-1-butyl-3-[(1R)cyclohexylhydroxymethyl]-2,5-dioxo-1,4,9-triazaspiro [5.5] undec-9 ylmethyl] phenoxy] benzoic acid hydrochloride],14; 45 AK530 [(3S)-1-but-2-yn-1-yl-3-[(1S)-cyclohexyl-hydroxymethyl]-9 (3,5-dimethyl-1-phenyl-1H-pyrazol-4-ylmethyl) -1,4,9-triazaspiro [5.5] undecane 2,5-dione dihydrochloride] and AK317 [4-(4-{[(3S)-1-butyl-3-(cyclohexylmethyl)-2,5-dioxo-1,4,9-triazaspiro[5.5] undec-9-yl] methyl} phenoxy) benzoic acid hydrochloride] are discussed in the present report. The methods for the synthesis and physicochemical profiles of AK530 and AK317 will be described elsewhere. Tritiation of these three CCR5 inhibitors was conducted as previously reported 14. The structures of these three CCR5 inhibitors are illustrated in Fig. 1.
Cells, viruses, and anti-HIV-1 assay
The Chinese Hamster Ovary (CHO) cells expressing wild-type CCR5 (CCR5WT-CHO cells) or mutant CCR5 (CCR5MT-CHO cells) 22 were maintained in Ham's F-12 medium (Invitrogen, Carlsbad, CA), supplemented with 10 % fetal calf serum (FCS: HyClone, Logan, UT) in the presence of 100 µg/ml zeomycin (Invitrogen). The MAGI cell line 46 was provided by NIH AIDS Research and Reference Reagent Program and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, 200 µg/ml G418, and 100 µg/ml hygromycin B. MAGI-CCR5 cells 47 were maintained in DMEM supplemented with 10% FCS, 200 µg/ml G418, 100 µg/ml hygromycin B, and 100 µg/ml zeomycin. The 293T cells were cultured in DMEM with 10% FCS. Peripheral blood mononuclear cells (PBM) were isolated from buffy coats of HIV-1 seronegative individuals and activated with 10 µg/ml phytohemagglutinin (PHA) prior to use as previously described.8 Two wild type R5-HIV-1 strains were employed for drug susceptibility assays: HIV-1Ba-L,48 and HIV-1JRFL.49 Antiviral assays using PHA-PBM (p24 assay) and MAGI assay using MAGI-CCR5 cells were also conducted as previously reported.8
FACS Analysis and the mAb displacement assay
CCR5WT-CHO cells (2 × 105) were exposed to differing concentrations (1 nM - 1 µM) of a CCR5 inhibitor for 30 min, followed by the addition of a fluorescein (FITC)-conjugated anti-CCR5 monoclonal antibody, 2D7 (BD PharMingen, San Diego, CA), 45523, 45531, or 45549 (R&D Systems, Minneapolis, MN) and further incubated for 30 min at 4°C. Cells were washed and analyzed on an flow cytometer (FACSCalibur: BD Biosciences, San Jose, CA). Each fluorescent activity in the presence of a drug was compared to that obtained in the absence of inhibitors, and shown as % control.
Saturation binding assay
A panel of mutant CCR5 expressing CHO cells 22 was used for saturation binding assays. The CHO cell lines expressing a mutant CCR5 P84H (CCR5P84H-CHO cells), C101A, L104D, F109A, T195A, T195P, T195S, and W248A were newly generated and used. The saturation binding assay using tritiated CCR5 inhibitors (3H-APL, 3H-AK530, and 3H-AK317) and wild-type or mutant CCR5 expressing CHO cells were conducted as previously described.14 In brief, wild type or mutant CCR5+ CHO cells (1.5 × 105 cells/well) were plated onto 48-well, flat-bottomed culture plates, incubated for 24 hrs, exposed to various concentrations of each 3H-CCR5 inhibitor, washed thoroughly, lysed with 0.5 ml of 1 N NaOH, and the radioactivity in the lysates was measured. The KD (dissociation) values of CCR5 inhibitors and the maximal binding values (Bmax = the number of CCR5/cell) were calculated based on their specific radioactivity using GRAPHPAD PRISM software (Intuitive Software for Science, San Diego, CA).
CCR5 homology model
A homology model of CCR5 was built as follows. The sequence of CCR5 (352 amino acids) was aligned against the sequence of bovine rhodopsin (348 amino acids). The recently determined crystal structure of bovine rhodopsin by Okada et al. was used as the template structure (PDB ID 1U19).27 The alignment was manually adjusted to ensure that the conserved GPCR residues were aligned as follows: in TM1 N55 of bovine rhodopsin to N48 of CCR5; in TM2 D83 to D76; in TM3 E134-R135-Y136 to D125-R126-Y127; in TM4 W161 to W153; in TM5 P215 to P206; in TM6 W265- x266-P267-Y268 to W248- x249-P250-Y251; in TM7 P303 to P294; in H8 F313 to F304. Secondary structure prediction assigned the length of each helix, beta-sheet and loop segments. After building the transmembrane helices, the loops connecting the different transmembrane domains were built by using the ultra-extended sampling protocol in Prime (Prime, version 1.6, Schrödinger, LLC, New York, NY, 2007), which does a more exhaustive sampling of the loop conformations.50 The side chains were predicted using the rotamer library of Xiang and Honig.51 The structure was minimized in implicit water with the OPLS2005 force field 52 as implemented in MacroModel (MacroModel, version 9.1, Schrödinger, LLC, New York, NY, 2005).
All atom molecular dynamics simulations, without using any non-bonded cut-off distances, were carried out on CCR5. A constant temperature of 300K, and SHAKE constraints for bonds to hydrogens were used. The GB/SA continuum solvation model, with water as the solvent was used.53 Using a time step of 1 femto-second, the structures were equilibrated for 100 pico-seconds (ps). The simulation was carried out for 4800 pico-seconds (ps) on a Linux cluster, and structures were monitored at intervals of every 50 ps.
Structural modeling of the interactions of CCR5 inhibitors with CCR5
After building the initial model of CCR5, the CCR5-inhibitor complex structures were further defined with an iterative optimization of CCR5 and ligand structures in the presence of each other, using software tools from Schrödinger (Schrödinger, LLC, New York, NY), as described below. The conformational flexibility of both CCR5 and CCR5 inhibitors were taken into account. The molecular structures of AK530, AK317, and APL were obtained by minimization using the MMFF94 force field as implemented in MacroModel. For each minimized inhibitor configuration, a set of low energy structures was generated by performing a Monte Carlo sampling of their conformations. Thus obtained structures were used as starting structures for docking calculations where their conformations were further refined.
The protonation states of CCR5 residues were assigned, and residues more than 20 Å from the active site were neutralized. We initially failed to obtain energetically favorable inhibitor-CCR5 complex structures by using a rigid CCR5 structure because of unfavorable steric interaction of the sidechains of the active site residues with the inhibitors. In an attempt to place an inhibitor within CCR5, after analysis of the steric clashes, the active site was artificially enlarged by mutating Y108, C178, E283, and M287 to Ala. The van der Waals radii of inhibitor atoms were scaled by a factor of 0.70 to reduce steric clashes and docked into CCR5. After obtaining an initial “guess” set of CCR5-inhibitor complexes, residues 108, 178, 283, and 287 were mutated back from Ala to their original states. CCR5 atoms within 15 Å of an initially placed inhibitor were subsequently refined. It was achieved by using the rotamer library of Xiang and Honig 51 and optimizing each side chain one at a time holding all other side chains fixed. After convergence, all side chains were simultaneously energy minimized to remove any remaining clashes. The inhibitors were docked again and scored to estimate their relative affinity. The extra-precision mode of Glide,54;55 which penalized unfavorable and unphysical interactions, was used. The docked complexes with higher scores were visually examined along with the mutational data to select the best possible CCR5-inhibitor complex.
Visualization, structural refinement, and docking were performed using Maestro 7.5, MacroModel 9.1, Prime 1.6, Glide 4.5, and IFD script 56 from Schrödinger, LLC (New York, NY 2007). Computations were carried out on a multiprocessor SGI Origin 3400 computer platform and on a Beowulf type Linux cluster.
HIV-1-gp120-elicited cell-cell fusion assay
The entire human CCR5 gene including a stop codon was amplified using pZeoSV-CCR5 47 as a template. The PCR product was ligated into pcDNA6.2/cLumio-DEST vector (Invitrogen), cloned according to the manufacture's recommendation, and termed as pcDNA6.2-CCR5WT (a CCR5 expression vector). A variety of plasmids carrying a mutant CCR5-encoding gene (pcDNA6.2-CCR5MT) were subsequently generated by employing the site-directed mutagenesis technique. An HIV-1 tat expression vector (pcDNA6.2-HIV-tat) was also generated using the same method. For the generation of HIV-1-gp120-overexpressing 293T cells, an HIV-envelope expression vector, pCXN-JRenv,47 was employed. A reporter (luciferase) gene containing plasmid, pLTR-LucE,57 was provided by the NIH AIDS Research and Reference Reagent Program. The envelope expression vector and tat expression vector (0.5 µg each) were co-transfected into 293T cells (2 × 105, 3 ml in 6-well microculture plates) using Lipofectamine 2000 (Invitrogen), while the CCR5WT or mutant CCR5 expression vector and pLTR-LucE (0.5 µg each) were co-transfected into MAGI cells (2 × 105, 3 ml in 6-well microculture plates). On the next day, both the co-transfected cells were harvested and mixed in a well of 96-well plates (2 × 104 cells each). The co-transfected cells were incubated further for 6 hrs and the luciferase activity in each well was detected using Bright-Glo Luciferase Assay System (Promega, Madison, WI) and its luminescence level was measured using Veritas Microplate Luminometer (Turner BioSystmes, Sunnyvale, CA). Non-specific luciferase activity was determined with the luminescence level in the well containing control Tat+, env− 293T cells and Luc+, CCR5+ MAGI cells and the value of the non-specific luminescence level was subtracted from each experimental luminescence level.
Protein Model Data Bank accession code
The coordinates for the models of ligand-free CCR5, CCR5-AK317 and CCR5-AK530 complexes with accession codes PM0075224, PM0075223 and PM0075221 respectively have been deposited in the Protein Model DataBase (http://mi.caspur.it/PMDB).
Acknowledgements
The authors thank Yasuhiro Koh for critical reading of the manuscript. This work was supported in part by the Intramural Research Program of Center for Cancer Research, National Cancer Institute, National Institutes of Health and in part by a Grant for Promotion of AIDS Research from the Ministry of Health, Welfare, and Labor of Japan, and the Grant to the Cooperative Research Project on Clinical and Epidemiological Studies of Emerging and Re emerging Infectious Diseases (Renkei Jigyo: No. 78, Kumamoto University) of Monbu-Kagakusho (HM). We also thank the Center for Information Technology, National Institutes of Health, for providing computational resources on the NIH Biowulf Linux cluster, and Susan Chacko and David Hoover for help with batch job submission configuration on the cluster.
Abbreviations
- APL
aplaviroc
- CCR5
C-C chemokine receptor 5
- ECL2
the second extracellular loop
- HIV-1
human immunodeficiency virus type 1
- MVC
maraviroc
- SDP
spirodiketopiperazine
- TM
transmembrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raport CJ, Gosling J, Schweickart VL, Gray PW, Charo IF. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 4.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co- receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 5.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 6.Kilby JM, Eron JJ. Novel therapies based on mechanisms of HIV-1 cell entry. N Engl J Med. 2003;348:2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- 7.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci U S A. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeda K, Yoshimura K, Shibayama S, Habashita H, Tada H, Sagawa K, Miyakawa T, Aoki M, Fukushima D, Mitsuya H. Novel low molecular weight spirodiketopiperazine derivatives potently inhibit R5 HIV-1 infection through their antagonistic effects on CCR5. J Biol Chem. 2001;276:35194–35200. doi: 10.1074/jbc.M105670200. [DOI] [PubMed] [Google Scholar]
- 9.Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol Pharmacol. 2005;67:1268–1282. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- 10.Tagat JR, McCombie SW, Nazareno D, Labroli MA, Xiao Y, Steensma RW, Strizki JM, Baroudy BM, Cox K, Lachowicz J, Varty G, Watkins R. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. IV. Discovery of 1-[(4,6-dimethyl-5-pyrimidinyl)carbonyl]- 4-[4-[2-methoxy-1(R)-4-(trifluoromethyl)phenyl]ethyl-3(S)-methyl-1-piperaz inyl]- 4-methylpiperidine (Sch-417690/Sch-D), a potent, highly selective, and orally bioavailable CCR5 antagonist. J Med Chem. 2004;47:2405–2408. doi: 10.1021/jm0304515. [DOI] [PubMed] [Google Scholar]
- 11.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seto M, Aikawa K, Miyamoto N, Aramaki Y, Kanzaki N, Takashima K, Kuze Y, Iizawa Y, Baba M, Shiraishi M. Highly potent and orally active CCR5 antagonists as anti-HIV-1 agents: synthesis and biological activities of 1-benzazocine derivatives containing a sulfoxide moiety. J Med Chem. 2006;49:2037–2048. doi: 10.1021/jm0509703. [DOI] [PubMed] [Google Scholar]
- 13.Imamura S, Ichikawa T, Nishikawa Y, Kanzaki N, Takashima K, Niwa S, Iizawa Y, Baba M, Sugihara Y. Discovery of a piperidine-4-carboxamide CCR5 antagonist (TAK-220) with highly potent Anti-HIV-1 activity. J Med Chem. 2006;49:2784–2793. doi: 10.1021/jm051034q. [DOI] [PubMed] [Google Scholar]
- 14.Maeda K, Nakata H, Koh Y, Miyakawa T, Ogata H, Takaoka Y, Shibayama S, Sagawa K, Fukushima D, Moravek J, Koyanagi Y, Mitsuya H. Spirodiketopiperazine-based CCR5 inhibitor which preserves CC-chemokine/CCR5 interactions and exerts potent activity against R5 human immunodeficiency virus type 1 in vitro. J Virol. 2004;78:8654–8662. doi: 10.1128/JVI.78.16.8654-8662.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fatkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, Saag MS, Goebel FD, Rockstroh JK, Dezube BJ, Jenkins TM, Medhurst C, Sullivan JF, Ridgway C, Abel S, James IT, Youle M, Zavolan M, van der Ryst E. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 16.Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76:8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondru R, Zhang J, Ji C, Mirzadegan T, Rotstein D, Sankuratri S, Dioszegi M. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol Pharmacol. 2008;73:789–800. doi: 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
- 19.Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, Moore JP. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci U S A. 2000;97:5639–5644. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikawa M, Takashima K, Nishi T, Furuta RA, Kanzaki N, Yamamoto Y, Fujisawa J. Analysis of binding sites for the new small-molecule CCR5 antagonist TAK-220 on human CCR5. Antimicrob Agents Chemother. 2005;49:4708–4715. doi: 10.1128/AAC.49.11.4708-4715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seibert C, Ying W, Gavrilov S, Tsamis F, Kuhmann SE, Palani A, Tagat JR, Clader JW, McCombie SW, Baroudy BM, Smith SO, Dragic T, Moore JP, Sakmar TP. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology. 2006;349:41–54. doi: 10.1016/j.virol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 22.Maeda K, Das D, Ogata-Aoki H, Nakata H, Miyakawa T, Tojo Y, Norman R, Takaoka Y, Ding J, Arnold GF, Arnold E, Mitsuya H. Structural and molecular interactions of CCR5 inhibitors with CCR5. J Biol Chem. 2006;281:12688–12698. doi: 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- 23.Tsamis F, Gavrilov S, Kajumo F, Seibert C, Kuhmann S, Ketas T, Trkola A, Palani A, Clader JW, Tagat JR, McCombie S, Baroudy B, Moore JP, Sakmar TP, Dragic T. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J Virol. 2003;77:5201–5208. doi: 10.1128/JVI.77.9.5201-5208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakata H, Maeda K, Miyakawa T, Shibayama S, Matsuo M, Takaoka Y, Ito M, Koyanagi Y, Mitsuya H. Potent anti-R5 human immunodeficiency virus type 1 effects of a CCR5 antagonist, AK602/ONO4128/ GW873140, in a novel human peripheral blood mononuclear cell nonobese diabetic-SCID, interleukin-2 receptor gamma-chain-knocked-out AIDS mouse model. J Virol. 2005;79:2087–2096. doi: 10.1128/JVI.79.4.2087-2096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L, LaRosa G, Kassam N, Gordon CJ, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore JP, Mackay CR. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samson M, LaRosa G, Libert F, Paindavoine P, Detheux M, Vassart G, Parmentier M. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 27.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 28.Gether U, Kobilka BK. G protein-coupled receptors. II. Mechanism of agonist activation. J Biol Chem. 1998;273:17979–17982. doi: 10.1074/jbc.273.29.17979. [DOI] [PubMed] [Google Scholar]
- 29.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 30.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siciliano SJ, Kuhmann SE, Weng Y, Madani N, Springer MS, Lineberger JE, Danzeisen R, Miller MD, Kavanaugh MP, DeMartino JA, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 32.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 33.Rosenkilde MM, Schwartz TW. GluVII:06--a highly conserved and selective anchor point for non-peptide ligands in chemokine receptors. Curr Top Med Chem. 2006;6:1319–1333. doi: 10.2174/15680266106061319. [DOI] [PubMed] [Google Scholar]
- 34.Ballesteros JA, Jensen AD, Liapakis G, Rasmussen SG, Shi L, Gether U, Javitch JA. Activation of the beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J Biol Chem. 2001;276:29171–29177. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- 35.Olson WC, Rabut GE, Nagashima KA, Tran DN, Anselma DJ, Monard SP, Segal JP, Thompson DA, Kajumo F, Guo Y, Moore JP, Maddon PJ, Dragic T. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee B, Sharron M, Blanpain C, Doranz BJ, Vakili J, Setoh P, Berg E, Liu G, Guy HR, Durell SR, Parmentier M, Chang CN, Price K, Tsang M, Doms RW. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 38.Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78:2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trkola A, Kuhmann SE, Strizki JM, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J, Palani A, Shapiro S, Clader JW, McCombie S, Reyes GR, Baroudy BM, Moore JP. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci U S A. 2002;99:395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baba M, Miyake H, Wang X, Okamoto M, Takashima K. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob Agents Chemother. 2007;51:707–715. doi: 10.1128/AAC.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marozsan AJ, Kuhmann SE, Morgan T, Herrera C, Rivera-Troche E, Xu S, Baroudy BM, Strizki J, Moore JP. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338:182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 43.Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361:212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaBranche C, Kitrinos K, Howell R, McDanal C, Harris S, Jeffrey J, Demarest J. Targeting HIV Entry - 1st International Workshop. Bethesda, MD: 2005. Dec 2–3, Abstract 9. [Google Scholar]
- 45.Nishizawa R, Nishiyama T, Hisaichi K, Matsunaga N, Minamoto C, Habashita H, Takaoka Y, Toda M, Shibayama S, Tada H, Sagawa K, Fukushima D, Maeda K, Mitsuya H. Spirodiketopiperazine-based CCR5 antagonists: Lead optimization from biologically active metabolite. Bioorg Med Chem Lett. 2007;17:727–731. doi: 10.1016/j.bmcl.2006.10.084. [DOI] [PubMed] [Google Scholar]
- 46.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maeda Y, Foda M, Matsushita S, Harada S. Involvement of both the V2 and V3 regions of the CCR5-tropic human immunodeficiency virus type 1 envelope in reduced sensitivity to macrophage inflammatory protein 1alpha. J Virol. 2000;74:1787–1793. doi: 10.1128/jvi.74.4.1787-1793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 49.Koyanagi Y, O'Brien WA, Zhao JQ, Golde DW, Gasson JC, Chen IS. Cytokines alter production of HIV-1 from primary mononuclear phagocytes. Science. 1988;241:1673–1675. doi: 10.1126/science.241.4873.1673. [DOI] [PubMed] [Google Scholar]
- 50.Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 51.Xiang Z, Honig B. Extending the accuracy limits of prediction for side-chain conformations. J Mol Biol. 2001;311:421–430. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- 52.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WJ. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 53.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 54.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 55.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 56.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 57.Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]