Abstract
mRNA decapping is a critical step in eukaryotic cytoplasmic mRNA turnover. Cytoplasmic mRNA decapping is catalyzed by Dcp2 in conjunction with its co-activator Dcp1, and is stimulated by decapping enhancer proteins. mRNAs associated with the decapping machinery can assemble into cytoplasmic mRNP granules called processing bodies (PBs). Evidence suggests that PB-associated mRNPs are translationally repressed and can be degraded or stored for subsequent translation. However, whether mRNP assembly into a PB is important for translational repression, decapping or decay has remained controversial. Here we discuss the regulation of decapping machinery recruitment to specific mRNPs and how their assembly into PBs is governed by the relative rates of translational repression, mRNP multimerization and mRNA decay.
Keywords: mRNA turnover, decapping, Dcp2, Dcp1, processing bodies
Introduction
Eukaryotic gene expression is regulated at multiple levels including through the control of mRNA translation and degradation in the cytoplasm. Both processes are modulated by the mRNA 5′ N7-methyl guanosine (m7G) cap, which is critical for translation of most cellular mRNAs and at the same time protects them from 5′ to 3′ exonucleolytic degradation (Cougot et al., 2004b). The removal of the m7G cap by the process of decapping represses gene expression by simultaneously shutting down mRNA translation and activating mRNA degradation (Eulalio et al., 2007a; Parker and Sheth, 2007). The m7G cap is protected from decapping and activates translation through its association with the cytoplasmic cap binding protein, eukaryotic initiation factor 4E (eIF4E), a component of the eIF4F complex (Cougot et al., 2004b). eIF4F forms a tight complex with the mRNA cap, which is further stabilized by an interaction between the eIF4G subunit of eIF4F and cytoplasmic poly(A)-binding protein at the mRNA poly(A)-tail (Amrani et al., 2008; Wells et al., 1998). For an mRNA to be decapped, the complex between eIF4F and the mRNA cap must be antagonized by pro-decapping factors. Once this occurs, translation is inhibited and mRNA decapping can ensue (Cougot et al., 2004b; Eulalio et al., 2007a; Parker and Sheth, 2007). Thus, mRNA decapping and mRNA translation are thought to be competing pathways. Studies over the last few years have revealed that messenger ribonucleoproteins (mRNPs) that are translationally repressed and associated with the cytoplasmic decapping machinery can concentrate in mRNP granules called processing bodies (PBs) (Eulalio et al., 2007a; Parker and Sheth, 2007). However, whether PB formation plays an active role in translational repression and mRNA decay is unclear. In this review, we discuss the mechanisms by which the cytoplasmic mRNA decapping machinery is activated on specific mRNAs and present a kinetic model that predicts the conditions under which the resulting mRNPs assemble into PBs. This kinetic model for PB formation can possibly be extended to understand the functions of other PB-like mRNP granules found in specialized cells such as neurons and germ-line cells.
The cytoplasmic mRNA decapping machinery
The catalytic engine of the yeast cytoplasmic mRNA decapping machinery is composed of the catalytic subunit Dcp2 and its co-activator Dcp1. Dcp1 from the yeast Saccharomyces cerevisiae was the first identified decapping factor and was initially thought to be responsible for catalysis (LaGrandeur and Parker, 1998). However, later studies identified Dcp2 as a high-copy suppressor of temperature-sensitive yeast strains lacking functional Dcp1 (Dunckley and Parker, 1999), and bacterially expressed yeast, human, Caenorhabditis elegans, Arabidopsis thaliana and Drosophila melanogaster Dcp2 was subsequently demonstrated to possess decapping activity in the absence of Dcp1 (Cohen et al., 2005; Iwasaki et al., 2007; Lin et al., 2008; Lykke-Andersen, 2002; Steiger et al., 2003; van Dijk et al., 2002; Wang et al., 2002; Xu et al., 2006). Other factors that catalyze decapping exist, such as DcpS, which hydrolyzes the cap product of 3′ to 5′ exonucleolytically degraded capped RNAs, the nuclear X29 decapping enzyme and virally encoded decapping enzymes (Blanc et al., 1992; Ghosh et al., 2004; Parrish et al., 2007; Tang et al., 2005; Wang and Kiledjian, 2001). However, these will not be discussed further here.
The decapping reaction catalyzed by Dcp2 releases m7GDP and a 5′ monophosphorylated mRNA body. This is thought to be an irreversible process, which targets the mRNA for degradation by the 5′ to 3′ exonuclease Xrn1. Dcp2 contains an N-terminal Nudix/MutT motif, commonly found in pyrophosphatases, which is critical for decapping (Figure 1A) (Dunckley and Parker, 1999; Lykke-Andersen, 2002; van Dijk et al., 2002; Wang et al., 2002). Like other Nudix-domain pyrophosphatases (McLennan, 2006), the catalytic center of Dcp2 contains three conserved glutamate residues, which coordinate a divalent cation responsible for activation of a water molecule for cap hydrolysis (She et al., 2006). Biochemical studies indicate that yeast, human and C. elegans Dcp2 can decap both m7G-capped and m2,2,7G- capped RNAs, but show poor activity on unmethylated G-caps (Cohen et al., 2005; Piccirillo et al., 2003; Steiger et al., 2003; van Dijk et al., 2002). Moreover, short RNAs, or RNAs hybridized to a DNA oligo at their 5′ end, are not efficiently decapped by Dcp2 in vitro (Cohen et al., 2005; Piccirillo et al., 2003; Steiger et al., 2003; van Dijk et al., 2002). These findings suggest that Dcp2 must contact the mRNA in two ways to promote efficient mRNA decapping: i) through the m7G-cap and ii) through contacts with the 5′ end of the mRNA (Figure 1A). This idea has been confirmed by recent crystallographic studies, which revealed that a conserved channel in the Dcp2 Nudix domain interacts with both the cap and the mRNA body through several important residues (Deshmukh et al., 2008; She et al., 2008). Consequently, altering these residues results in a reduction or loss of mRNA decapping in vitro (Deshmukh et al., 2008; She et al., 2008). Thus, it is predicted that anything associated with the cap or the immediate 5′ end of a cellular mRNA, such as the eIF4F complex, will protect the mRNA from decapping.
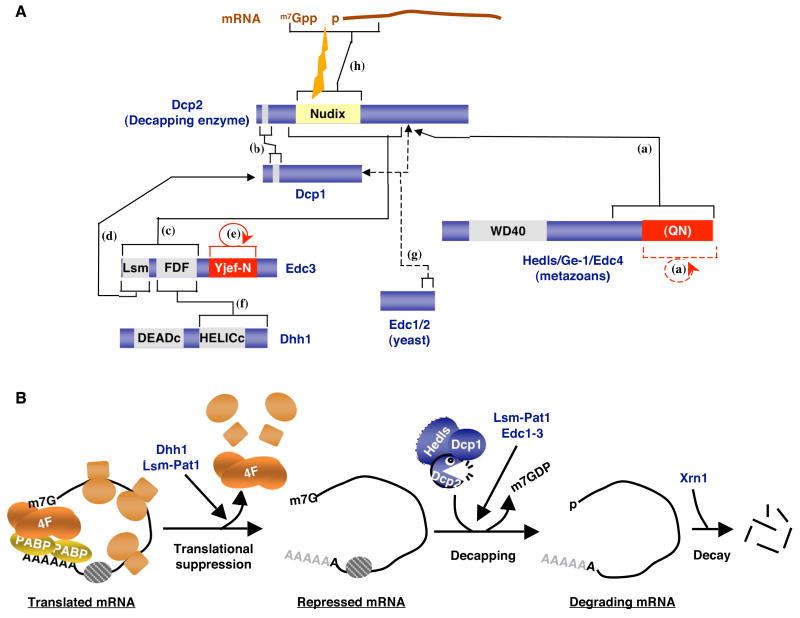
Figure 1. Multiple protein factors promote mRNA decapping.
(A) Interactions between the decapping enzyme Dcp2 and factors that stimulate decapping, Dcp1, Edc3, Dhh1 (Rck/P54, Me31B, CGH-1, Xp54), metazoan-specific Hedls and yeast-specific Edc1/2. Direct interactions between the decapping factors and between Dcp2 and mRNA are indicated by solid lines and brackets. The interaction between Edc1/2 and Dcp1/2 has been shown indirectly, through activation of decapping in vitro and is represented by dashed lines. Putative PB assembly domains are shown in red. Specific protein domains are indicated. (Q/N): Putative glutamine/asparagine-rich region of Hedls. The catalytic Nudix domain of Dcp2 is indicated in yellow. The indicated interactions are based on observations described in: (a): (Fenger-Gron et al., 2005; Xu et al., 2006; Yu et al., 2005), (b): (Deshmukh et al., 2008; She et al., 2006; She et al., 2008), (c): (Decker et al., 2007; Tritschler et al., 2007), (d): (Tritschler et al., 2007; Fenger-Gron et al., 2005), (e): (Decker et al., 2007; Tritschler et al., 2007), (f): (Decker et al., 2007; Tritschler et al., 2007), (g): (Dunckley et al., 2001; Schwartz et al. 2000), and (h): (Deshmukh et al., 2008; She et al., 2006; She et al., 2008). (B) mRNA decapping activation is thought to occur in two distinct steps. First, the cap-binding complex (eIF4F) must be removed from the mRNA, a process that renders the mRNA translationally inactive. This process is stimulated, at least in yeast, by Dhh1 and the Lsm-Pat1 complex. The core decapping complex, including Dcp2, Dcp1 and possibly in metazoans Hedls, can then access and remove the mRNA cap through a process that can be stimulated by Edc1–3. The decapped mRNA is then degraded by Xrn1.
Whereas Dcp2 exhibits decapping activity in vitro, Dcp1 is a critical Dcp2 cofactor in yeast cells (Beelman et al., 1996; Sakuno et al., 2004; Steiger et al., 2003). Moreover, recombinant yeast Dcp2 is stimulated by Dcp1 and alterations in the Dcp2 N-terminal region that impair the interaction with Dcp1 (Figure 1A) prevent efficient decapping in vitro and in vivo (Sakuno et al., 2004; She et al., 2006; She et al., 2004; She et al., 2008; Tharun and Parker, 1999). Recent kinetic studies have provided insights into the mechanism by which Dcp1 promotes Dcp2 activity. Dcp1 strongly stimulates Dcp2 catalytic activity while having little effect on the interaction of Dcp2 with the m7G cap (Deshmukh et al., 2008). Structural evidence suggests that this process involves the transformation of Dcp2 from an inactive open conformation to an active closed conformation, which orients the Dcp2 N-terminus towards the catalytic site and renders Dcp2 catalytically active (Deshmukh et al., 2008; She et al., 2008). This could conceivably be an important mechanism by which Dcp2 activity is regulated in the cell.
Interestingly, yeast Dcp1 interacts with Dcp2 through amino acids that are not highly conserved in metazoans (Deshmukh et al., 2008; She et al., 2006; She et al., 2008). Moreover, human Dcp1 does not stably associate with Dcp2 in vitro or when overexpressed in human embryonic kidney (HEK) 293 cells, and human, C. elegans and D. melanogaster Dcp1 does not stimulate Dcp2 activity in vitro (Cohen et al., 2005; Fenger-Gron et al., 2005; Iwasaki et al., 2007; Lin et al., 2008; Lykke-Andersen, 2002; van Dijk et al., 2002). Instead, Dcp2 activity in these organisms might be stimulated through a metazoan-specific protein called Hedls/Ge-1/Edc4 (hereafter called Hedls) (Fenger-Gron et al., 2005; Xu et al., 2006; Yu et al., 2005), which in human and A. thaliana stimulates both the activity of Dcp2 and the association between Dcp2 and Dcp1 (Figure 1A) (Fenger-Gron et al., 2005; Xu et al., 2006). Future studies should reveal whether Hedls is a ‘Dcp’, i.e. a bona fide core component of the metazoan decapping complex critical for catalysis, or an ‘Edc’, a more peripheral enhancer of decapping.
The competition between translation and decapping
Several lines of evidence suggest that the decapping complex competes with the cytoplasmic translation initiation eIF4F complex for the mRNA cap (Beelman and Parker, 1994; Schwartz and Parker, 1999; Schwartz and Parker, 2000). Thus, the decapping of a cellular mRNP involves at least two distinct steps outlined in Figure 1B: First, the cap-binding translation factors must be displaced, and second, Dcp2 must bind the cap and catalyze decapping. Factors that stimulate decapping could potentially promote either of these steps. As the eIF4F complex is critical for translation initiation of the majority of cellular mRNAs, the first step of decapping involves the repression of mRNA translation. However, since mRNAs can be kept stably in a translationally repressed state, not all mRNAs that are translationally repressed are decapping substrates (Brengues et al., 2005; Teixeira et al., 2005). The simplest hypothesis is that only the translational repression events that cause general destabilization of the eIF4F-cap complex will stimulate decapping. This destabilization could conceivably be achieved by the action of pro-decapping factors that either directly interfere with the eIF4F-cap complex, or that repress translation in a manner that, in a more indirect way, enhances the off-rate of the eIF4F complex. Alternatively, decapping may occur only on those translationally repressed mRNAs that contain cis-elements that attract the decapping machinery.
The mechanisms of enhancers of decapping
Several factors that accelerate decapping have been identified. These include enhancers of decapping proteins Edc1, Edc2, and Edc3, of which Edc1 and Edc2 appear to be specific to yeast, whereas Edc3 is conserved throughout eukaryotes (Figure 1A) (Cougot et al., 2004a; Dunckley et al., 2001; Fenger-Gron et al., 2005; Kshirsagar and Parker, 2004; Schwartz et al., 2003). Edc1 and Edc2 show hallmarks of proteins that specifically stimulate the Dcp2 cap-binding/catalysis step (Figure 1B), because, like Dcp1 and Hedls, they stimulate decapping by Dcp2 in vitro on a naked RNA (Schwartz et al., 2003; Steiger et al., 2003). Similarly, although no in vitro stimulation of decapping has been reported, Edc3 most likely stimulates the Dcp2 recruitment/catalysis step (Figure 1B), because cellular depletion of Edc3 impairs decapping of a subset of mRNAs, whereas no effect on translation has been reported (Badis et al., 2004; Coller and Parker, 2005; Kshirsagar and Parker, 2004). Edc3 interacts directly with multiple decapping factors, including Dcp2 and Dcp1 (Figure 1A), and thus might either recruit or activate the decapping complex on target mRNAs (Decker et al., 2007; Tritschler et al., 2007). In contrast to the Edc proteins, two other decapping activators, Pat1 and Dhh1, appear to activate decapping at least in part by promoting translational repression (Figure 1B; the orthologs of yeast Dhh1 are called Rck/p54, Me31D, CGH-1 and Xp54 in humans, D. melanogaster, C. elegans and Xenopus laevis, respectively). Dhh1 has, in addition to its role in decapping, been identified as a general inhibitor of translation (Coller and Parker, 2005; Minshall and Standart, 2004; Nakamura et al., 2001; Navarro et al., 2001). Dhh1 belongs to a family of DexD/H box ATPases (Figure 1A). Thus, a simple hypothesis would be that Dhh1 uses its ATPase activity to release the eIF4F complex from the mRNP, which would simultaneously repress translation and stimulate decapping (Figure 1B). However, Dhh1 also represses translation of reporter mRNAs that do not depend on eIF4F for translation (Coller and Parker, 2005). One possibility is that translational repression by Dhh1 promotes a general destabilization of the eIF4F-mRNA cap complex in a more indirect manner, thus moving the equilibrium towards decapping.
Another factor that both inhibits translation and activates decapping is Pat1. Pat1 together with a complex of Lsm proteins, the Lsm 1–7 complex, promotes decapping of yeast mRNAs that have undergone deadenylation (Bouveret et al., 2000; Tharun et al., 2000; Tharun and Parker, 2001). mRNA deadenylation is a major precursor for decapping in yeast and is often the rate-limiting step in mRNA turnover (Muhlrad et al., 1994). The complex between Lsm 1–7 and Pat1 (hereafter called the Lsm-Pat1 complex) shows inherent affinity for deadenylated mRNA sequences (Bouveret et al., 2000; Chowdhury et al., 2007; Tharun et al., 2000; Tharun et al., 2005; Tharun and Parker, 2001). Pat1 overexpression causes general repression of yeast mRNA translation, and co-depletion of Pat1 and Dhh1 prevents the general translational repression observed upon glucose starvation (Coller and Parker, 2005). Thus, Pat1 might stimulate decapping by promoting translation repression and, directly or indirectly, the release of the mRNA cap-binding complex from the deadenylated mRNA (Figure 1B). Evidence suggests that Pat1 can also stimulate decapping of untranslated mRNA, suggesting that it might also recruit the decapping complex (Coller and Parker, 2005; Pilkington and Parker, 2008) (Figure 1B). However, although the Lsm-Pat1 complex shows RNA-dependent interactions with decapping factors, including Dcp1 and Dcp2 (Tharun et al., 2000; Tharun and Parker, 2001), no direct interactions have been reported.
Whereas deadenylation is a major precursor for decapping of yeast mRNAs (Muhlrad et al., 1994) and deadenylation can trigger decapping in mammalian cells (Yamashita et al., 2005), the link between deadenylation and decapping in organisms other than yeast has not been studied in detail. Clearly, deadenylation does not always lead to decapping as mRNAs can exist stably in a translationally repressed deadenylated form, for example during oocyte maturation in X. laevis and mice and under certain conditions in mammalian neuronal cells (Gray and Wickens, 1998; Huang and Richter, 2004; Paillard and Osborne, 2003). An interesting question for future studies is whether such mRNAs evade decapping owing to a failure to recruit the Lsm-Pat1 complex or by specific repression of one of the downstream steps in activation of decapping outlined in Figure 1B. One possibility is that factors that are involved in translational repression of such mRNAs lock down the eIF4F complex, or some other factor, on the mRNA cap, thus preventing access for the decapping machinery. Alternatively, the identity of the deadenylase(s) that produce the deadenylated mRNA might determine whether the mRNA is handed over to the decapping machinery. Interestingly, the vertebrate deadenylase PARN, which has been implicated in deadenylation of certain stable translationally repressed mRNAs, shows affinity for the mRNA cap (Copeland and Wormington, 2001; Dehlin et al., 2000; Gao et al., 2001; Martinez et al., 2001). Perhaps PARN in this manner prevents decapping of mRNAs with which it stably associates.
Recent evidence suggests that general inhibitors of Dcp2-mediated decapping also exist, as treatment of purified Dcp2 with human cell extracts renders it relatively inactive (Jiao et al., 2006). One such inhibitor called VCX-A has been identified as a testis-specific inhibitor of Dcp2 activity, which appears to prevent decapping by a competition for binding to the cap (Jiao et al., 2006). The biological significance of such general decapping inhibitors is an important topic for future studies.
Transcript-specific decapping activation
Specialized mRNA decay pathways can promote decapping independently of, or in addition to, deadenylation. For example, mRNAs targeted to the nonsense-mediated mRNA decay (NMD) pathway, which degrades mRNAs with premature termination codons, are subjected to rapid decapping (Muhlrad and Parker, 1994) mediated by the central NMD factor Upf1. Upf1 exists in complex with the decapping proteins (Fenger-Gron et al., 2005; He and Jacobson, 1995; Lykke-Andersen, 2002), and therefore likely promotes decapping by recruiting the decapping complex to the target mRNA. NMD also promotes translation repression (Isken et al., 2008; Muhlrad and Parker, 1999), but whether this plays a direct role in decapping is unknown. The importance of decapping relative to other degradation pathways in the decay of NMD substrates appears to vary between organisms and substrate mRNAs (Isken and Maquat, 2007).
Several factors, including TTP, BRF-1 and KSRP, that destabilize mammalian mRNAs containing AU-rich elements (AREs) in their 3′ UTRs have been observed in complex with decapping enzymes, as well as with a number of other mRNA decay enzymes, and are thought to activate decapping by recruiting the decapping complex to target mRNAs (Chou et al., 2006; Fenger-Gron et al., 2005; Stoecklin et al., 2006). This is clearly the case for TTP, which shows ARE-dependent activation of Dcp2 in vitro (Fenger-Gron et al., 2005). ARE-mRNAs might also activate decapping through the Lsm-Pat1 complex, because RNAi-mediated Lsm1 depletion impairs ARE-mRNA decay in human cells (Stoecklin et al., 2006). AREs also can cause translational repression. It remains to be determined whether this repression is mediated directly by ARE-binding proteins or by the recruitment of translational suppressors that promote decapping, such as Dhh1 and Pat1.
mRNA decapping also appears to play an important role in the repression of at least a subset of miRNA target mRNAs, as studies have shown that the RNA induced silencing complex (RISC) can target miRNA substrates for decapping and decay in addition to translational repression (Chu and Rana, 2006; Rehwinkel et al., 2005), and Dhh1 has been implicated in miRNA-mediated silencing (Chu and Rana, 2006; Eulalio et al., 2007c). In addition, PUF proteins, which target associated mRNAs for translational repression and decay, have also been observed in association with decapping factors (Goldstrohm et al., 2006).
Highly transcript-specific mechanisms to recruit the decapping machinery have also been described. For example, in S. cerevisiae the mRNAs for ribosomal protein Rps28b and the nuclear export factor Yra1 are auto-regulated through the recruitment of Edc3, which promotes mRNA decapping (Badis et al., 2004; Dong et al., 2007). The yeast EDC1 mRNA is also regulated through a deadenylation-independent decapping mechanism (Muhlrad and Parker, 2005). Moreover, a recent study identified human mRNAs that directly interact with Dcp2 (Li et al., 2008). One such mRNA, which encodes a component of the 3′ to 5′ exonuclease complex, the exosome, directly recruits Dcp2 in a sequence-specific manner and undergoes Dcp2-dependent mRNA decay (Li et al., 2008). Oligo- and Poly-U tails, which are emerging as signals for mRNA turnover (Furnari et al., 1993; Mullen and Marzluff, 2008; Shen and Goodman, 2004; Song and Kiledjian, 2007), also can activate decapping in vitro (Song and Kiledjian, 2007). Thus, a number of mechanisms exist by which the decapping machinery can be selectively recruited to specific mRNA substrates. An important goal for future studies will be to understand how translational repression is coordinated with activation of decapping in these mRNA decay pathways. Moreover, the importance of decapping relative to other mRNA decay activities in specific mRNA turnover pathways is largely unknown.
mRNAs associated with the decapping machinery can assemble into PBs
In recent years it has become clear that mRNPs that recruit the mRNA decapping machinery acquire the ability to assemble into PBs (Eulalio et al., 2007a; Parker and Sheth, 2007). PBs were first identified following the discovery that Dcp1, Dcp2, the 5′ to 3′ exonuclease Xrn1, and the miRNA pathway component GW182, co-localize in cytoplasmic foci (Bashkirov et al., 1997; Eystathioy et al., 2002; Ingelfinger et al., 2002; Sheth and Parker, 2003; van Dijk et al., 2002). Later studies revealed that a number of other factors that activate decapping also concentrate in PBs (Beckham et al., 2008; Fenger-Gron et al., 2005; Scheller et al., 2007; Yu et al., 2005). In addition, mRNA and protein components of the miRNA, ARE-mRNA decay and NMD pathways can be observed in PBs (Ding et al., 2005; Eystathioy et al., 2003; Franks and Lykke-Andersen, 2007; Kedersha et al., 2005; Liu et al., 2005b; Pillai et al., 2005; Sen and Blau, 2005; Sheth and Parker, 2006; Unterholzner and Izaurralde, 2004). However, the function and importance of the PB in mRNP regulation has been a subject of controversy and many misconceptions. Here we outline the kinetic model for PB formation and disassembly, which serves as a simple framework for predicting the conditions that mediate the assembly of mRNPs into PBs (Figure 2).
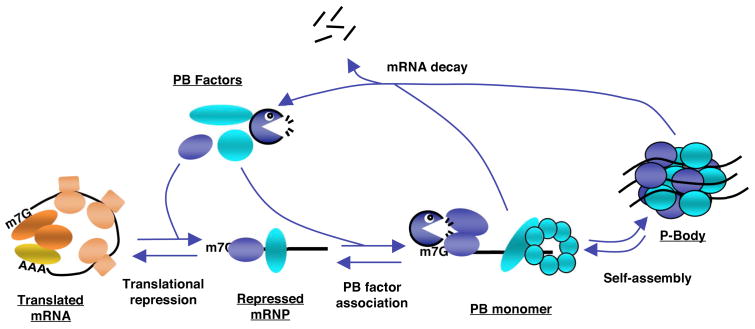
Figure 2. The kinetic model for PB formation.
Model illustrating the hypothesis that the extent of PB formation is directly proportional to the cytoplasmic levels of repressed mRNPs and depends on competing rates of translational repression, PB factor recruitment, mRNP multimerization and mRNA decay. Before mRNAs can assemble into PBs, they must be translationally repressed. Translational silencing factors then promote the formation of a PB “monomer” by recruiting decapping factors and/or other PB assembly components. If mRNA decay enzymes are not limiting and thus mRNA decay is rapid, mRNAs may be degraded prior to PB assembly. However, if decay enzymes are limiting, the repressed mRNAs assemble into PBs where they can be degraded, or released back into the translational pool.
The kinetic model for PB assembly
Several lines of evidence suggest that PBs are highly dynamic structures formed by the self-assembly and disassembly of translationally repressed mRNPs. This hypothesis is illustrated in the kinetic model for PB assembly shown in Figure 2. A central prediction from this model is that PB formation is directly proportional to the cytoplasmic concentration of translationally repressed mRNPs. This idea is supported by a number of observations. First, the entrapment of mRNAs in polysomes through treatment with drugs such as cycloheximide, which depletes the cellular pool of ribosome-free mRNPs, causes rapid disappearance of visible PBs (Cougot et al., 2004a; Eulalio et al., 2007b; Teixeira et al., 2005). Importantly, PBs can be restored under these conditions when an exogenous mRNA that fails to become trapped in polysomes due to a 5′UTR hairpin is expressed at high levels (Franks and Lykke-Andersen, 2007). Second, several manipulations of human, yeast and Drosophila cells that cause the release of mRNA from polysomes, thus increasing the cellular pool of ribosome-free mRNPs, result in enhanced PB formation (Brengues et al., 2005; Cougot et al., 2004a; Eulalio et al., 2007b; Teixeira et al., 2005). However, it is worth noting that when mRNAs are released from polysomes under conditions of cell stress, such repressed mRNPs assemble instead into mRNP granules related to PBs called stress granules (see below). Third, manipulations that slow down enzymatic steps of mRNA decay that occur after the release of the mRNA from polysomes, thus leading to an increase in the cellular pool of repressed mRNPs committed to decay, cause accumulation of PBs. For example, depletion of the decapping enzyme Dcp2 or the 5′ to 3′ exonuclease Xrn1, both PB factors, causes enhanced PB assembly (Cougot et al., 2004a; Fenger-Gron et al., 2005; Franks and Lykke-Andersen, 2007; Sheth and Parker, 2003). Even the depletion of a component of the exosome, which is not observed in PBs, causes accumulation of ARE-mRNA in PBs (Franks and Lykke-Andersen, 2007), likely due to an increase in the cellular concentration of the translationally repressed ARE-mRNP. Finally, the integrity of PBs is critically dependent on RNA, as treatment with RNase in vitro or in permeabilized cells causes the disassembly of PBs (Eulalio et al., 2007b; Teixeira et al., 2005). These observations taken together demonstrate a direct correlation between the cellular concentration of translationally repressed mRNPs and the extent of PB assembly in the cytoplasm and suggest a model in which PBs form by the self-assembly of repressed mRNPs (Figure 2). A very important aspect of this model is that PBs should not be considered as pre-formed ‘organelles′ to which individual mRNPs become localized, but rather as self-assembling clusters of mRNPs, which form when mRNAs are released from polysomes but do not rapidly degrade and thus accumulate as repressed mRNPs in the cytoplasm. The evidence for the individual steps that lead to PB formation depicted in Figure 2 are discussed below.
How does translational repression lead to PB assembly?
Several lines of evidence suggest mRNPs must be translationally repressed and recruit cytoplasmic PB factors to form a PB (Figure 2) (Coller and Parker, 2005; Decker et al., 2007; Eulalio et al., 2007b; Franks and Lykke-Andersen, 2007; Teixeira et al., 2005). These steps might be highly coupled in yeast through the PB factors Dhh1 and Pat1, which, as discussed above, serve a dual function as general repressors of translation and activators of mRNA decapping (Bonnerot et al., 2000; Bouveret et al., 2000; Coller and Parker, 2005; Coller et al., 2001; Fischer and Weis, 2002; Tharun et al., 2000; Tharun and Parker, 2001). In support of this idea, recent studies indicate that yeast strains lacking Dhh1 and Pat1 display defects in translational repression as well as PB formation in response to glucose starvation (Coller and Parker, 2005). By contrast, over-expression of Dhh1 and Pat1, which results in general release of mRNAs from polysomes, also promotes the formation of large PBs (Coller and Parker, 2005). These observations suggest that a competition between the translation machinery and antagonistic factors such as Pat1 and Dhh1 regulate global translation and PB formation. In addition, Pat1 and Dhh1 are likely among the first PB factors recruited to an mRNA and thus play an important role in promoting the formation of a repressed mRNP that is competent to assemble into a PB (Figure 2).
However, whereas translational repression is clearly required for the assembly of an mRNP into a PB, it might not always be sufficient. mRNAs that are translationally repressed by 5′UTR hairpins or puromycin treatment in human and Drosophila cells, do not assemble into PBs unless they contain cis-elements that recruit cytoplasmic PB factors (Eulalio et al., 2007b; Franks and Lykke-Andersen, 2007). Thus, translational repression and PB factor recruitment could, at least in some cases, constitute independent steps, although whether this is true for endogenous mRNAs remains unknown. Thus, an important question for future studies is whether all cellular translationally repressed mRNPs are competent for PB assembly or whether this is only true for a specific subset.
What factors direct the assembly of mRNPs into PBs?
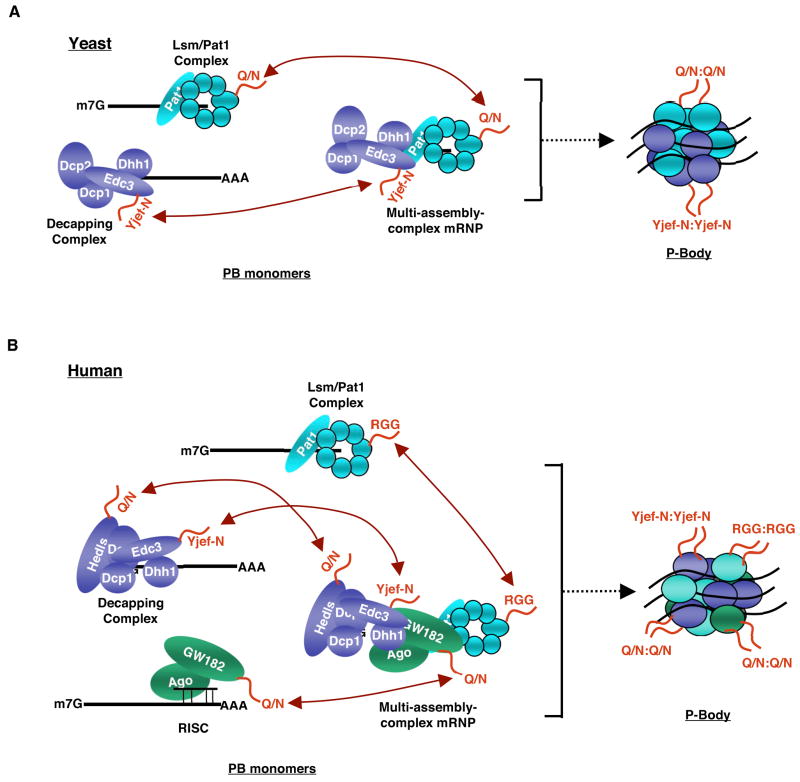
Two protein components of yeast PBs, Edc3 and Lsm4, were recently identified as factors that promote the physical interactions between mRNPs required for PB assembly (Decker et al., 2007; Reijns et al., 2008). The co-deletion of a C-terminal dimerization domain of Edc3 (called Yjef-N; Figure 1A) and a C-terminal Glutamine/Asparagine (Q/N)-rich domain of Lsm4, similar to those found in prions, led to a complete failure in PB assembly, even though translation and mRNA decay appeared to be unaffected (Decker et al., 2007; Reijns et al., 2008). Interestingly, Edc3 and Lsm4 are components of two separate complexes, the decapping complex and the Lsm-Pat1 complex, respectively (Figure 3A) (Bouveret et al., 2000; Decker et al., 2007; Tharun et al., 2000). These findings suggest a mechanism for PB assembly in yeast in which Dhh1 and Pat1 trigger translational silencing and promote the assembly of individual mRNPs into a PB through interactions with Edc3 (as part of the decapping complex) and Lsm4 (as part of the Lsm-Pat1 complex), respectively (Figure 3A) (Decker et al., 2007; Pilkington and Parker, 2008). Edc3 likely has a similar function in eukaryotes other than yeast, as the Yjef-N dimerization domain is highly conserved and Edc3 depletion in Drosophila S2 cells results in the disappearance of visible PBs (Tritschler et al., 2007). By contrast, the prion-like domain of yeast Lsm4 has been replaced by an RGG-domain in most metazoans suggesting that either Lsm4 does not promote PB formation in these organisms, or does so by a different mechanism than in yeast (Decker et al., 2007). Interestingly however, an RGG domain in a different protein, G3BP, has previously been implicated in the assembly of stress mRNP granules (Tourriere et al., 2003).
Figure 3. Model for the assembly step in PB formation.
(A) Yeast Edc3 and Lsm4 contain Yjef-N and Q/N-rich domains, respectively (indicated by red tails), which are protein-protein interaction domains important for the assembly of mRNPs into the PB (Decker et al., 2007; Reijns et al., 2008). Edc3 and Lsm4 are part of separate complexes, the decapping complex and the Lsm-Pat1 complex, respectively. These complexes might promote the assembly of their associated mRNPs into PBs through homomeric interactions as indicated by arrows. (B) In human cells, Hedls and GW182 contain putative Q/N-rich prion-like domains (indicated by red tails), which might promote PB formation in cooperation with, or independently of, Edc3 or Lsm4. It is possible that the assembly of PBs containing a heterogeneous pool of mRNPs requires that a subset of PB mRNPs recruit multiple assembly complexes, which serve as scaffolds for assembly with mRNPs containing single assembly complexes as indicated by arrows. Alternatively, assembly domains may form heterologous interactions (e.g. between Hedls and GW182) to assemble heterogeneous PBs (see text for details).
One interesting question is whether all repressed mRNPs assemble into PBs using the same protein domain, or whether multiple protein domains can direct PB assembly. In support of the latter possibility, several PB components contain predicted Q/N-rich regions similar to that found in yeast Lsm4 (Decker et al., 2007; Reijns et al., 2008). In metazoans, this includes the proteins Hedls and GW182. In human and Drosophila cell lines, depletion of either Hedls or GW182 leads to loss of microscopically visible PBs (Eulalio et al., 2007b; Jakymiw et al., 2005; Liu et al., 2005a; Yu et al., 2005). However, it has not been determined whether this is due to a direct effect on the PB assembly step rather than an indirect effect of modulating translation or mRNA decay. Moreover, overexpression of Hedls, but not a mutant Hedls protein lacking its Q/N-rich region, causes massive aggregation of PB-like structures in human cells (Fenger-Gron et al., 2005; Yu et al., 2005). However, whereas Hedls associates with activators of the NMD and ARE-mRNA decay pathways (Fenger-Gron et al., 2005), GW182 is thought to function specifically in the miRNA silencing pathway (Jakymiw et al., 2005; Liu et al., 2005a; Ding et al., 2005; Rehwinkel et al., 2005). These observations support the idea that different mRNP repression pathways utilize diverse protein complexes to assemble into PBs (Figure 3B). Recruitment of a single one of these assembly complexes might be sufficient for any given mRNP to associate with the PB.
How do mRNPs disassemble from PBs?
Evidence suggests that mRNPs assembled into PBs can undergo one of two fates: they can either be released to re-enter the translational pool or they undergo mRNA decay (Figure 2). Either case would lead to the release of the mRNP proteins from the PB, which is consistent with the rapid cycling of most PB factors observed in Fluorescence Recovery After Photobleaching (FRAP) experiments (Aizer et al., 2008; Andrei et al., 2005; Kedersha et al., 2005; Leung et al., 2006). Evidence that mRNAs can be released from the PB into the translational pool include the observations that yeast mRNAs that assemble into PBs upon glucose starvation engage in translation once glucose is added back (Brengues et al. 2005), and that a human mRNA, which is targeted for PB association by a miRNA, can re-enter translation upon association with the protein HuR (Bhattacharyya et al., 2006). The other possible fate for an mRNA associated with the PB is believed to be its degradation. However, while this is consistent with the observation that many mRNA decay enzymes concentrate in PBs and that the depletion of mRNA decay enzymes leads to accumulation of mRNAs in PBs, direct evidence that mRNAs can degrade while assembled into the PB has not yet been presented. A testable prediction is that the cycling of mRNP proteins out of PBs, as can be measured in photoactivation experiments, is dependent on the catalytic activity of mRNA decay enzymes.
Assuming that mRNAs can in fact degrade while associated with the PB, an important question is how a subset of PB-associated mRNAs evade mRNA decay. Presumably they either fail to recruit or to activate the mRNA decay enzymes that act on unstable PB-associated mRNPs. This hypothesis is supported by observations suggesting that the yeast NMD pathway can target both normal and aberrant mRNAs for PB association (Sheth and Parker, 2006). However, once associated with PBs these mRNAs appear to have different fates as the NMD substrates are degraded whereas the normal mRNAs can be released for translation. An important goal for future studies will be to understand how PB-associated mRNPs are ‘marked’ for degradation or for storage and later re-entry into the translational pool. An interesting hypothesis is that the recently observed structural rearrangement required for Dcp2 activation (Deshmukh et al., 2008; She et al., 2008) constitutes a key step in determining whether or not Dcp-associated mRNPs are subjected to decapping and degradation or remain stable. Alternatively, stable mRNPs could assemble into PBs through protein complexes that do not associate with mRNA decay factors (Figure 3).
Some consequences of the kinetic model for PB formation
An important question that arises from the model of PBs illustrated in Figure 2 is how many mRNPs are needed to form a PB? It simply isn’t known. However, we would like to argue that PBs should not be defined as simply those structures that can be observed in the microscope, as this definition would cause a PB observed in one microscope to cease being a PB when observed in a less powerful microscope. Rather we favor a definition based on function. However, it is not yet clear how to define PB function. So, how should experiments that manipulate PBs be interpreted? Since a key prediction from the model presented in Figure 2 is that any manipulation of a cell (such as an external stimulus or the depletion or over-expression of a protein) that affects the levels of translationally repressed mRNPs will cause an overall alteration of cellular PB morphology, such manipulations could have interfered with any one of the steps illustrated in Figure 2. Moreover, as PBs consist of a heterogeneous pool of repressed mRNPs, it can be predicted that any manipulation that causes a sub-fraction of the mRNP pool to shift in or out of the PB will affect the association of all other mRNPs with PBs simply by affecting the overall concentration of repressed mRNPs available for PB formation. For example, the observations that microscopically visible PBs disappear when the miRNA pathway is repressed in human and Drosophila cells (Eulalio et al., 2007b; Jakymiw et al., 2005; Liu et al., 2005a; Pauley et al., 2006), or when deadenylation is repressed in yeast and mammalian cells (Sheth and Parker, 2003; Zheng et al., 2008), does not necessarily reflect a requirement for these processes in PB assembly. Instead, according to the model in Figure 2, these effects are due to the generally decreased abundance of repressed mRNPs available for cytoplasmic PB formation, which would slow down the PB assembly step and thereby fail to create PBs observable by microscopy. If this interpretation is correct, it is predicted that the overexpression of an mRNA that is translationally repressed in a process independent of deadenylation or miRNAs will restore PBs under these conditions, similarly to how a non-translatable ARE-mRNA can restore PBs when other mRNPs are trapped in polysomes by cycloheximide (Franks and Lykke-Andersen, 2007).
Another important consequence of the model in Figure 2 is that it is not possible using current light microscopy techniques to quantify the fraction of a given mRNP or protein that localizes in PBs versus the cytoplasm. As PBs might be anything from an assembly of two or more mRNPs, what can be classified as a PB via light microscopy might be only the tip of the iceberg. Moreover, as mRNPs can cycle rapidly in and out of PBs with half-lives of less than a minute (Aizer et al., 2008; Andrei et al., 2005; Kedersha et al., 2005; Leung et al., 2006), techniques for separating mRNPs in PBs from those in polysomes following the preparation of cell extracts should be interpreted with care as the act of preparing a cell extract most likely alters the mRNP equilibrium. Instead, techniques that use rapid fixation of complexes prior to extract preparation should be employed, although such techniques may exaggerate the levels of mRNPs in PBs by preventing the release of mRNPs from the PB during incubation.
The relation between PBs and other mRNP granules
A number of mRNP granules that appear to be closely related to PBs have been identified in eukaryotes, including stress granules (Anderson and Kedersha, 2008), various germ cell granules (Anderson and Kedersha, 2006), specific forms of neuronal granules (Barbee et al., 2006) and mRNP granules important in the secretory pathway (Decker and Parker, 2006). These mRNP granules are generally thought to promote temporally and/or spatially restricted mRNA translational repression. However, similarly to PBs, many of these mRNP granules also contain mRNA decay factors, including those promoting decapping. An important goal for future studies is to understand whether mRNAs associated with these specialized granules are subject to mRNA decay in addition to translational repression. Perhaps the turnover of mRNAs associated with some of these granules, together with constant replenishment of newly transcribed mRNA, maintains a steady state of localized mRNA, which can be released for local translation upon specific cues. Alternatively, a subset of the granule-associated mRNPs are stored with mRNA decay enzymes to ensure rapid degradation following their translational activation as a mechanism to prevent sustained signaling, as has been proposed for the Arc mRNA that localizes in neuronal granules (Giorgi et al., 2007).
Another interesting question about the various mRNP granules is how some can co-exist in the same cytoplasm. For example, several mRNP granules exist separately in the C. elegans gonad (Audhya et al., 2005; Boag et al., 2005; Lall et al., 2005; Navarro and Blackwell, 2005), and PBs and stress granules, which form under cell stress, can exist separately in the mammalian cell cytoplasm (Kedersha et al., 2005). The simplest hypothesis is that the protein domains responsible for the assembly of mRNPs into the various granules form mutually exclusive interactions. For example, the putative Lsm4, Edc3, Hedls and GW182 assembly domains of PBs might have the ability to assemble with each other (Figure 3B), whereas the TIA-1/TIAR Q/N-rich prion-like domain and the G3BP RGG domain, which are responsible for assembling translationally repressed mRNPs into stress granules (Gilks et al., 2004; Tourriere et al., 2003), might not interact with the PB factors. An alternative possibility is that all assembly domains are restricted to homomeric associations, but a subset of mRNPs in PBs associate with multiple PB assembly complexes, thereby ensuring co-assembly of the various mRNPs through a network of homomeric interactions (Figure 3B). This possibility is consistent with the observations of separate GW182 and decapping granules observed under certain conditions (Schneider et al., 2006; Vasudevan and Steitz, 2007). Moreover, PBs and stress granules can associate with each other, and these interactions are strongly enhanced upon TTP or BRF1 overexpression (Kedersha et al., 2005). A simple explanation would be that under these conditions, a small subset of repressed mRNPs recruit both a PB and a stress granule assembly complex, which in effect tethers the stress granule to the PB through a subset of the associated mRNPs. Alternatively, the observed association between PBs and stress granules could reflect the maturation of a subset of mRNPs from the monomeric form of one granule to the other. The mechanisms that ensure recruitment of specific assembly complexes to individual mRNPs, thereby targeting them to specific mRNP granules, and the functional consequence of this, should be important topics of future investigation.
Does mRNP granule formation regulate translational repression, decapping or mRNA decay?
A major unresolved question is what role, if any, does the assembly of individual mRNPs into mRNP granules, including the PB, play in promoting translational repression, decapping and/or mRNA decay. Why is the ability of repressed mRNPs to assemble into mRNP granules an evolutionarily conserved property of eukaryotic cells? Recent studies have failed to identify mRNA decay and translational repression pathways that are inhibited by the loss of visible PBs (Decker et al., 2007; Eulalio et al., 2007b; Stoecklin et al., 2006). Thus, at least for the tested mRNAs, their ability to assemble into PBs does not appear to be important for their repression and degradation, although it is important to keep in mind, as discussed above, that the loss of visible PBs does not necessarily mean a loss of functional PBs. However, PBs might serve some function that cannot be easily measured using mRNA decay and translation assays. For example, PBs might prevent promiscuous mRNA decapping and decay in the cytoplasm by sequestering mRNA decay enzymes away from translating mRNAs. Thus, general mRNA decay might be enhanced when macroscopic PBs disappear, rather than being inhibited as many would predict. Moreover, it is possible that PBs, by concentrating mRNA decay enzymes, enhance only mRNA decay pathways for which mRNA decay enzymes are limiting. For example, concentrating mRNPs in the PB could provide a mechanism to produce a high local Dcp2 concentration that can in effect jump from one mRNP to the other within the PB. This idea is consistent with the poor cycling of Dcp2 from PBs observed by FRAP (Aizer et al. 2008). A simple prediction from this hypothesis is that only mRNA decay pathways that can be observed to be enhanced upon Dcp2 overexpression will be impaired under conditions where the mRNPs fail to assemble into PBs. Thus, a number of important questions regarding the importance in cytoplasmic mRNA metabolism of decapping and the assembly of repressed mRNPs into mRNP granules remain unresolved. The combination of biochemical, biophysical, molecular and cell biological approaches that are currently ongoing should provide important insights into these processes in the near future.
Acknowledgments
We thank Dr. Tom Blumenthal for comments on the manuscript. Work on decapping and P-bodies in our laboratory is supported by funding from grant R01 GM077243 from the National Institutes of Health to JL. TF has been supported by National Institutes of Health NRSA Institutional Training Grant no. GM-07135 from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The Dynamics of Mammalian P Body Transport, Assembly and Disassembly In Vivo. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C, Hilliker A, Cziko AM, Noueiry A, Ramaswami M, Parker R. The DEAD-Box RNA Helicase Ded1p Affects and Accumulates in Saccharomyces cerevisiae P-Bodies. Mol Biol Cell. 2008;19:984–993. doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Blanc A, Goyer C, Sonenberg N. The coat protein of the yeast double-stranded RNA virus L-A attaches covalently to the cap structure of eukaryotic mRNA. Mol Cell Biol. 1992;12:3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CF, Mulky A, Maitra S, Lin WJ, Gherzi R, Kappes J, Chen CY. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol Cell Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LS, Mikhli C, Jiao X, Kiledjian M, Kunkel G, Davis RE. Dcp2 Decaps m2,2,7GpppN-capped RNAs, and its activity is sequence and context dependent. Mol Cell Biol. 2005;25:8779–8791. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland PR, Wormington M. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA. 2001;7:875–886. doi: 10.1017/s1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004a;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, van Dijk E, Babajko S, Seraphin B. ‘Cap-tabolism’. Trends Biochem Sci. 2004b;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehlin E, Wormington M, Korner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh MV, Jones BN, Quang-Dang DU, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol Cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Tucker M, Parker R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007c;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Adams MD, Pagano JS. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc Natl Acad Sci U S A. 1993;90:378–382. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wilusz CJ, Peltz SW, Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T, Peterson B, Tomasevic N, Peculis BA. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol Cell. 2004;13:817–828. doi: 10.1016/s1097-2765(04)00127-3. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- Huang YS, Richter JD. Regulation of local mRNA translation. Curr Opin Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Takeda A, Motose H, Watanabe Y. Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett. 2007;581:2455–2459. doi: 10.1016/j.febslet.2007.04.051. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jiao X, Wang Z, Kiledjian M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol Cell. 2006;24:713–722. doi: 10.1016/j.molcel.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur TE, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Song MG, Kiledjian M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol. 2008;28:939–948. doi: 10.1128/MCB.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MD, Jiao X, Grima D, Newbury SF, Kiledjian M, Chou TB. Drosophila processing bodies in oogenesis. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.07.033. [DOI] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Ren YG, Nilsson P, Ehrenberg M, Virtanen A. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J Biol Chem. 2001;276:27923–27929. doi: 10.1074/jbc.M102270200. [DOI] [PubMed] [Google Scholar]
- McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-->3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development. 2001;128:3221–3232. doi: 10.1242/dev.128.17.3221. [DOI] [PubMed] [Google Scholar]
- Paillard L, Osborne HB. East of EDEN was a poly(A) tail. Biol Cell. 2003;95:211–219. doi: 10.1016/s0248-4900(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci U S A. 2007;104:2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol Cell Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Araki Y, Ohya Y, Kofuji S, Takahashi S, Hoshino S, Katada T. Decapping reaction of mRNA requires Dcp1 in fission yeast: its characterization in different species from yeast to human. J Biochem. 2004;136:805–812. doi: 10.1093/jb/mvh190. [DOI] [PubMed] [Google Scholar]
- Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, Diez J. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim Biophys Acta. 2007;1773:1786–1792. doi: 10.1016/j.bbamcr.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Schneider MD, Najand N, Chaker S, Pare JM, Haskins J, Hughes SC, Hobman TC, Locke J, Simmonds AJ. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Decker CJ, Parker R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–251. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- She M, Decker CJ, Chen N, Tumati S, Parker R, Song H. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat Struct Mol Biol. 2006;13:63–70. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Sundramurthy K, Liu Y, Chen N, Parker R, Song H. Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat Struct Mol Biol. 2004;11:249–256. doi: 10.1038/nsmb730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Naitow H, Gardner NA, Kolesar A, Tang L, Wickner RB, Johnson JE. The structural basis of recognition and removal of cellular mRNA 7-methyl G ‘caps’ by a viral capsid protein: a unique viral response to host defense. J Mol Recognit. 2005;18:158–168. doi: 10.1002/jmr.724. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S, Muhlrad D, Chowdhury A, Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol. 2007;27:8600–8611. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]