Abstract
Ligand release from the low-density lipoprotein receptor (LDLR) has been postulated to involve a “histidine switch” induced intra-molecular rearrangement that discharges bound ligand. A recombinant soluble low-density lipoprotein receptor (sLDLR) was employed in ligand binding experiments with a fluorescent-tagged variant apolipoprotein E-N-terminal domain (apoE-NT). Binding was monitored as a function of fluorescence resonance energy transfer (FRET) from excited Trp residues in sLDLR to an extrinsic fluorophore covalently attached to Trp null apoE3-NT. In binding experiments with wild type (WT) sLDLR, FRET-dependent AEDANS fluorescence decreased as the pH was lowered. To investigate the role of His190, His562 and His586 in sLDLR on pH dependent ligand binding and discharge, site directed mutagenesis studies were performed. Compared to WT sLDLR, triple His→Ala mutant sLDLR displayed attenuated pH-dependent ligand binding and decreased ligand release as a function of low pH. When these His residues were substituted for Lys, whose positively charged side chain does not ionize over this pH range, ligand binding was nearly abolished at all pH values. When sequential His to Lys mutants were examined, evidence obtained suggested that His562 and His586 function cooperatively. Whereas the sedimentation coefficient for WT sLDLR increased upon lowering the pH from 7 to 5, no such change occurred in the case of the triple Lys mutant receptor or a His562Lys / His586Lys double mutant receptor. The data support the existence of a cryptic, histidine side chain ionization-dependent alternative ligand that modulates ligand discharge via conformational reorganization.
An important feature of cholesterol delivery to cells via the low density lipoprotein receptor (LDLR) pathway relates to the ability of the receptor to recycle to the plasma membrane after releasing its cargo. During the internalization process endocytic vesicles are converted to endosomes and these enclosed vesicular structures are acidified through the action of transmembrane proton pumps (1). The resulting decrease in endosomal pH is considered a key step in ligand release from the receptor. This separation event is critical to both receptor recycling and delivery of lipoprotein cargo to lysosomes for liberation of their cholesterol load. Based on the assumption that receptor-ligand interactions are electrostatic in nature, it is presumed that lowering the pH within the endosome will lead to an alteration in ionizable amino acid side chains that decrease ligand-receptor affinity. Indeed, the X-ray crystal structure of the soluble portion of the LDLR (2) provided a plausible molecular explanation for this process. This structure was determined at pH 5.3, a condition where ligand-binding interactions do not occur. Remarkably, the structure revealed that the β-propeller segment forms contacts with cysteine-rich LDLR type-A (LA) ligand binding repeats 4 and 5 under these conditions, thereby blocking access to potential ligands. The model suggests that, at low pH, an intra-molecular rearrangement occurs wherein the β-propeller segment displaces ligand from ligand binding modules, thereby serving as an alternative ligand. While this cryptic, pH-dependent process emerged from X-Ray structure analysis, it is entirely consistent with previous mutagenesis studies. Russell et al. (3) showed that deletion of the 5th of the seven LA repeats in LDLR abolishes binding activity toward apoE containing lipoprotein particles while deletion of the β-propeller segment produces a mutant receptor that is capable of binding LDL, but fails to release ligand at low pH (4).
In the present study we focused on His residues located in LA repeat 5 or the β-propeller segment (2,5). Two of these His residues (at positions 190 and 562) are mutated to Tyr in familial hypercholesterolemia (6). In other proteins, key His residues have been implicated in regulating ligand release upon endocytosis (7-9) as well as in chaperone interactions with an LDLR family member in the ER and Golgi (10). In a cell-based assay employing flow cytometry to investigate LDL binding to LDLR, Beglova et al. (11) reported evidence that His residues at positions 190, 562 and 586 function in ligand release from the receptor. To investigate this further we sequentially replaced these His with either Ala or Lys. Results from in vitro binding experiments are consistent with a model wherein cooperative electrostatic interactions between the LA repeats and the β-propeller domain contribute to pH dependent structural alterations that not only prevent ligand binding but lead to discharge of bound ligand.
Materials and Methods
Recombinant apoE3-NT and soluble LDLR
Recombinant Trp-null apoE3-NT was produced and isolated from E. coli as described by Fisher et al. (12,13). WT and mutant soluble LDLR (sLDLR; N-terminal residues 1 – 699) were isolated from conditioned media of stably transfected HEK 293 cells as described (14). Site-directed mutagenesis was performed using the QuikChange XL kit from Stratagene according to the manufacturer's instructions. All mutations were verified by dideoxy automated DNA sequencing. All variant sLDLR generated were analyzed by SDS-PAGE under reducing and non-reducing conditions as a measure of native protein folding and disulfide bond formation (15).
ApoE3-NT•phospholipid complexes
1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was purchased from Avanti Polar Lipids (Pelham, AL, USA). The phospholipid was dissolved in chloroform:methanol (3:1; v/v) and dried into a thin film in a glass tube. Following dispersion of the lipid in 50 mM sodium phosphate, pH 7.0, 150 mM NaCl, apoE3-NT was added. This mixture was subject to bath sonication at 24 °C until clear (16,17).
LDLR binding assay
Two μg sLDLR (in 10 mM citric acid, 2 mM CaCl2, 90 mM NaCl; adjusted to specified pH values) was incubated with 1 μg of a Trp-null apoE3-NT previously labeled on Cys112 with the fluorescent probe, N-(iodoacetyl)-N′-(5-sulfo-1-napthyl) ethylenediamine (AEDANS) and complexed with DMPC. Interaction between AEDANS-Trp-null apoE3-NT•DMPC and sLDLR is detected by fluorescence resonance energy transfer (FRET) between excited Trp residues in sLDLR and the AEDANS moiety covalently attached to Trp-null apoE3-NT•DMPC (14). Following incubation, samples were excited at 280 nm and fluorescence emission intensity at 470 nm determined (slit width 5.0 nm) on a Perkin-Elmer Model LS 50B luminescence spectrometer.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted at 22 °C and 35 000 rpm using a Beckman XL-I analytical ultracentrifuge and absorbance optics following the procedures outlined in the instruction manual published by the Spinco Business Center of Beckman Instruments, Inc., Palo Alto, CA (1997). Runs were performed for 5 h during which a maximum of 75 scans were taken. The sedimentation velocity data were analyzed using the Transport Method contained in the Beckman Analysis Program {Optima XL-A/XL-I Data Analysis Software, version 4.0 Copyright © 1997}. The program Svedberg was used to calculate the sedimentation constant, S20,w, for each experiment by analyzing 12 sets of data selected between the 28th and 56th scan. The program Sednterp (Sedimentation Interpretation Program, Version 1.01) was used to calculate the intrinsic sedimentation constant, S020,w and the axial ratio, a/b. Sednterp calculates the partial specific volume and degree of hydration from the amino acid composition of the protein using the methods of Cohn and Edsall (18) and Kuntz (19), and also calculates the solvent density and viscosity using known values from physical tables.
Results
Design and rationale
A model describing a pH-dependent ligand displacement mechanism for discharge of bound ligand from sLDLR has been proposed on the basis of X-ray crystallography and site directed mutagenesis (2,5,20). In the present study a known LDLR ligand, apoE3-NT, was employed in assays of ligand binding and release. When AEDANS-labeled Trp null apoE3-NT binds to the receptor, a stable fluorescence signal is established via intermolecular FRET from excited Trp residues in the receptor to the AEDANS moiety covalently bound to apoE. A corresponding decrease in AEDANS fluorescence intensity upon dissociation of ligand can be monitored as a real-time measure of ligand release (14). This concept provides a general experimental design for investigating structural requirements for ligand binding and may be adaptable to other LDLR family members and their ligand interactions.
pH-dependent binding of apoE3-NT•DMPC disks to sLDLR
To establish that interaction between WT sLDLR and apoE3-NT•DMPC in the present assay system will display pH dependence, FRET between excited Trp in sLDLR and AEDANS bound to Trp-null apoE3-NT was monitored. Initially, ligand stability as a function of solution pH was evaluated by non-denaturing PAGE analysis (Figure 1). The data reveal that, incubation of fluorescent-labeled apoE3-NT•DMPC disk complexes in different pH buffers had no effect on particle migration behavior or band intensity. Thus, we conclude that the ligand employed in these studies is stable to the pH variation examined. In binding experiments with sLDLR at pH 7, the expected enhancement in AEDANS fluorescence intensity was observed (Figure 2). As the solution pH was decreased from 7 to < 5, however, there was a corresponding decrease in AEDANS fluorescence intensity. At pH 5.9 the enhancement in AEDANS fluorescence intensity upon introduction of sLDLR was 21 % of the value observed at pH 7.4. Control experiments showed that AEDANS fluorescence intensity via direct excitation at 349 nm was unaffected by pH over this range. Given the ionization behavior of His side chains over this pH range together with X-Ray crystal structure of sLDLR, these data are consistent with a “Histidine switch” mechanism (21) wherein a conformational change in the receptor facilitated by protonation of key His side chains prevents access of the apoE3-NT•DMPC ligand to LA repeats in LDLR (2).
Figure 1. Native PAGE analysis of apoE3-NT DMPC complexes.
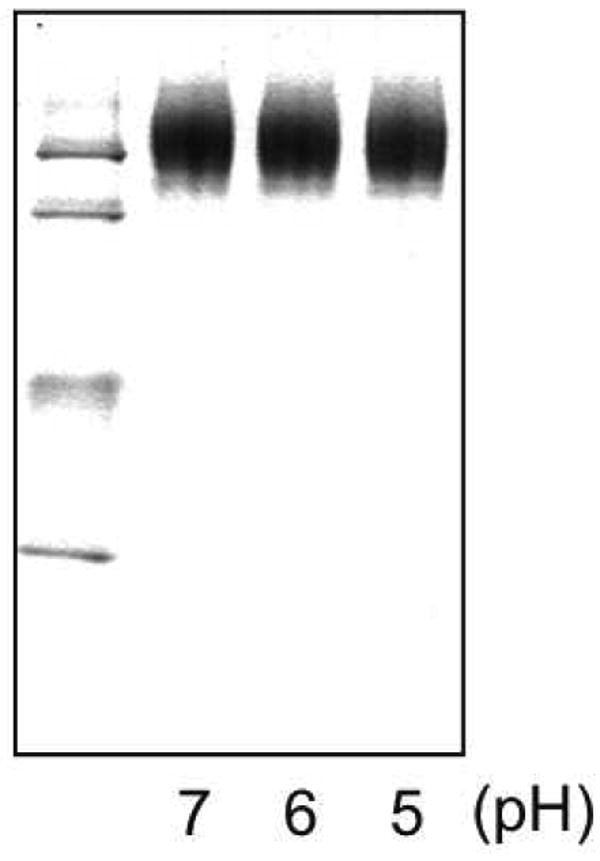
AEDANS-apoE3-NT•DMPC disks were incubated in buffer at the indicated pH values for 1 h at 37 °C. Following this the samples were electrophoresed on a 4 -20 % acrylamide gradient slab gel under non-denaturing conditions and stained with Coomassie Blue. The relative mobility of size standards is shown at the left. From top to bottom: 17.0, 12.2, 9.2, 8.2 and 7.1 nm diameter, respectively.
Figure 2. The effect of solution pH on ligand binding to sLDLR.
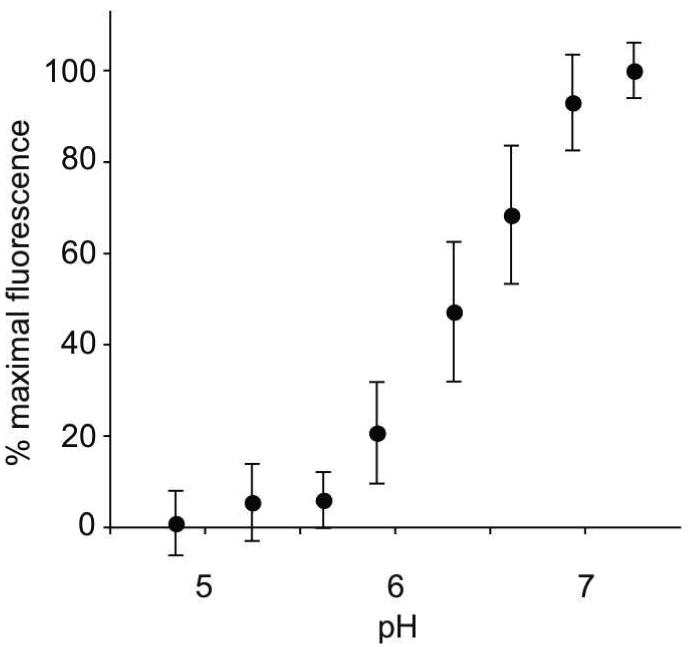
Two μg AEDANS-Trp null apoE3-NT•DMPC and 1 μg sLDLR were incubated in 10 mM citric acid, 2 mM CaCl2, 90 mM NaCl, adjusted to the indicated pH values. Samples (300 μl final volume) were excited at 280 nm and fluorescence emission intensity at 470 nm determined. Values reported are the mean ± S.D. (n = 3).
SDS-PAGE analysis of sLDLR and variants
In order to evaluate the role of His190, His562 and His586 in pH-induced ligand release, recombinant sLDLR were generated in which each of these residues were converted to either Ala or Lys. Prior to performing binding studies, however, isolated variant sLDLR were evaluated by SDS-PAGE. It is known that correctly folded LDLR family members display characteristic migration behavior in the presence and absence of β-mercaptoethanol (15). The pattern observed with WT sLDLR was similar to that reported by others, with slower migration into the gel under reducing conditions (Figure 3). By the same token, both variant sLDLR (triple Ala mutant and triple Lys mutant) behaved in a similar manner, indicating they are correctly folded in solution.
Figure 3. Characterization of sLDLR variants.
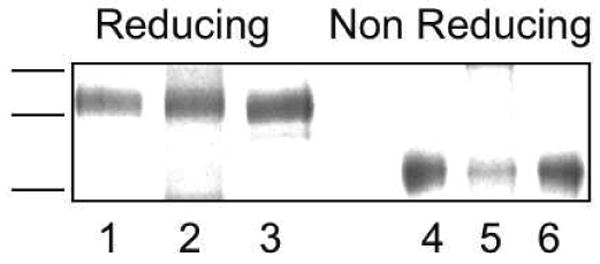
WT and variant sLDLR were subjected to SDS-PAGE under reducing (lanes 1 - 3) and non-reducing (lanes 4 - 6) conditions. Lanes 1 and 4) WT sLDLR; lanes 2 and 5) triple His→Ala mutant and lanes 3 and 6) triple His→Lys mutant. Samples were separated on a 4 – 20 % acrylamide gradient slab gel electrophoresed at 30 mAmp constant current and stained with Coomassie Blue.
Effect of mutations in sLDLR on ligand binding
Whereas Ala substitution introduces a small, uncharged side chain in place of His, the Lys substitution will introduce a positively charged side chain that does not ionize over the pH range 7.0 to 5.0. In control experiments with WT sLDLR the expected decrease in AEDANS fluorescence intensity was observed as the pH was decreased from 7.0 to 6.0 or 5.0 (Figure 4). In the case of the triple His→Ala mutant sLDLR, binding at pH 7.0 was lower than with WT sLDLR and very little change in fluorescence intensity occurred upon decreasing the pH to 6.0. At pH 5.0, AEDANS fluorescence intensity was lower but remained above values observed for WT sLDLR at this pH. On the other hand, in the case of a mutant sLDLR wherein each of the His residues were converted to Lys, little binding was detected at pH 7.0 and essentially no change in AEDANS fluorescence intensity was noted as the pH was decreased to 6.0 or 5.0.
Figure 4. Effect of mutations in sLDLR on pH-dependent ligand binding. (a).
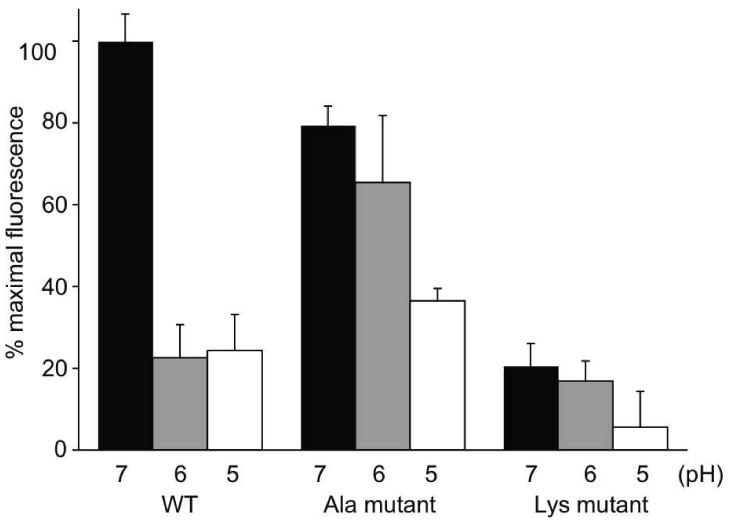
Two μg AEDANS-Trp null apoE3-NT•DMPC and 1 μg of a given sLDLR were incubated in 10 mM citric acid, 2 mM CaCl2, 90mM NaCl, adjusted to pH 7.0, 6.0 or 5.0, respectively. Samples (300 μl final volume) were excited at 280 nm and fluorescence emission intensity at 470 nm determined. Values are mean ± S.D. (n = 6). (b). Two μg AEDANS-Trp null apoE3-NT•DMPC and 2 μg sLDLR were incubated in buffer at pH 7.0. Samples (300 μl final volume) were excited at 280 nm and fluorescence emission intensity at 470 nm determined. Values reported are the mean ± S.D. (n = 6).
To assess the role of individual His residues in ligand binding / release from sLDLR the effect of single Lys mutations on FRET-dependent AEDANS fluorescence were examined. Using WT sLDLR as control, the effect of mutations at His190, His562 and His586, respectively, to Lys were studied at pH 7 (Figure 5). In each case, ligand binding to the single mutant sLDLR was intermediate between the maximal binding observed for WT sLDLR and that for the triple His→Lys mutant. Whereas ligand binding to His190Lys sLDLR was poor, the binding activities of His562Lys or His586Lys sLDLR were only slightly less than control WT sLDLR. However, ligand binding activity at neutral pH was abolished when His562 and His586 were both converted to Lys.
Figure 5. Effect of individual His mutations on ligand binding to sLDLR at neutral pH.
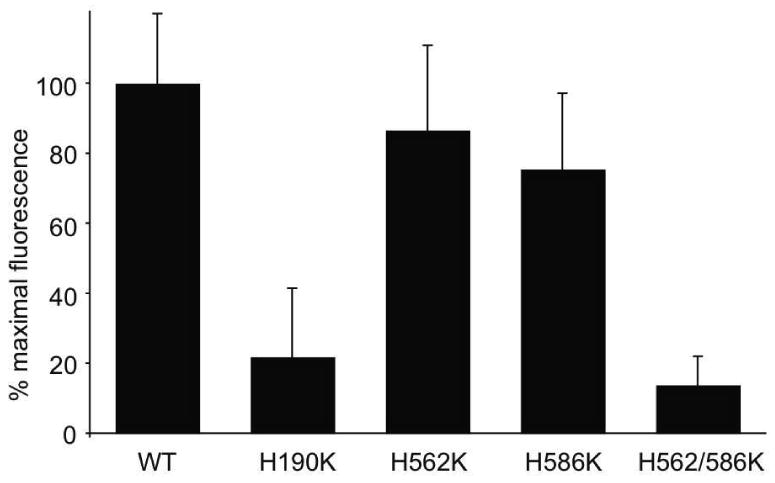
Two μg AEDANS-Trp null apoE3-NT•DMPC and 1 μg of a given sLDLR variant were incubated in 10 mM citric acid, 2 mM CaCl2, 90mM NaCl, adjusted to pH 7.0. Samples (300 μl final volume) were excited at 280 nm and fluorescence emission intensity at 470 nm determined. Values are mean ± S.D. (n = 6).
Effect of solution pH on binding interaction of single His variant sLDLR
Based on the data presented in Figure 5, wherein the ligand binding ability of the H562K sLDLR and H586K sLDLR variants were generally similar to WT sLDLR, we sought to determine if these variants will display a pH dependence of ligand binding similar to that seen with wild type sLDLR (Figure 6). The observed loss in binding ability as a function of decreasing pH for both single mutant variants is consistent with the concept that the His residues at these positions in WT sLDLR act in concert to induce pH dependent ligand release from the receptor.
Figure 6. Effect of soution pH on ligand binding to single His mutant sLDLR. Effect of mutations in sLDLR on pH-dependent ligand binding.
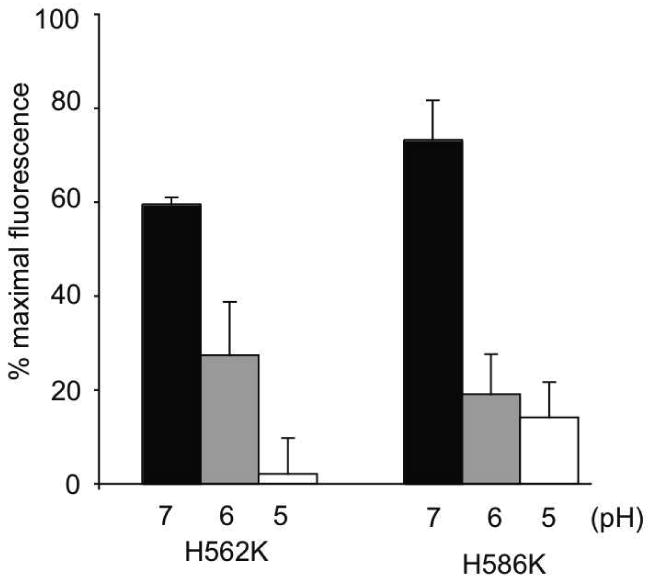
Two μg AEDANS-Trp null apoE3-NT•DMPC and 1 μg of a given sLDLR were incubated in 10 mM citric acid, 2 mM CaCl2, 90 mM NaCl, adjusted to pH 7.0, 6.0 or 5.0, respectively. Samples (300 μl final volume) were excited at 280 nm and fluorescence emission intensity at 470 nm determined. Values are mean ± S.D. (n = 6).
Evaluation of ligand release
Whereas data presented above illustrates the effect of mutations in sLDLR on ligand binding, we sought to evaluate whether these mutations also influence ligand release. pH-dependent ligand release was monitored by FRET as a function of time following a transition from pH 7.0 to 6.0. In the case of WT sLDLR, a rapid decrease in AEDANS fluorescence intensity accompanied the shift in pH. Within seconds of pH lowering, a greater than 80 % decline in AEDANS fluorescence was noted (Figure 7), consistent with discharge of bound ligand from the receptor. Similar to ligand binding data presented in Figure 4, in the case of the triple His→Ala mutant sLDLR, pH-induced changes in AEDANS fluorescence intensity were slower and the end point reached corresponded to approximately 50 % of maximal AEDANS fluorescence intensity (i.e. pH 7). Thus, His→Ala mutations affect both the kinetics and magnitude of pH-induced ligand release from sLDLR.
Figure 7. Effect of solution pH on ligand release from sLDLR.
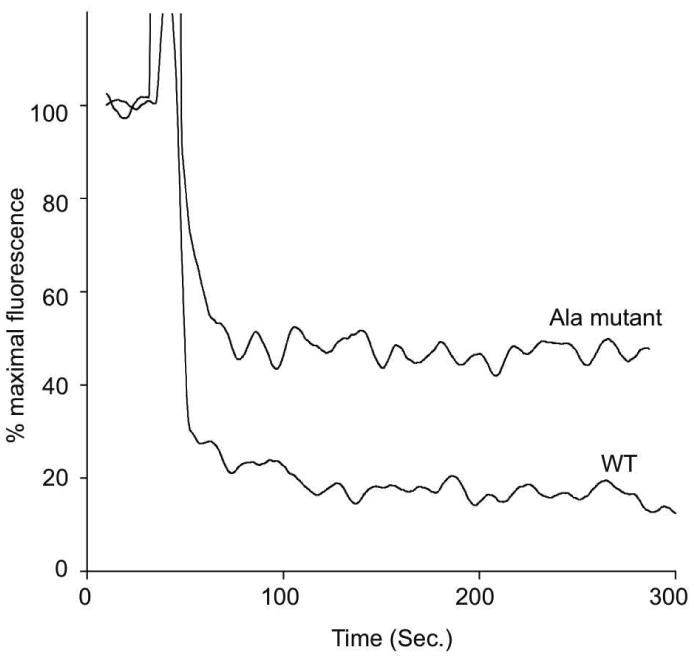
One μg AEDANS-Trp null apoE3-NT•DMPC and 2 μg WT or His→Ala sLDLR were incubated in 10 mM citric acid, 2 mM CaCl2, 90 mM NaCl pH 7.0. After 30 sec the solution pH was changed by adding an aliquot (6 μl) of 2 M citric acid pH = 6.0. Samples (300 μl final volume) were excited at 280 nm and fluorescence emission intensity at 470 nm monitored as a function of time.
Hydrodynamic studies
To evaluate the effect of mutations in sLDLR on the shape of the molecule at different pH values, sedimentation velocity experiments were performed in the analytical ultracentrifuge. As shown in Table 1, the sedimentation coefficient for WT sLDLR increased from 5.3 to 6.1 upon decreasing the solution pH from 7 to 5. The increased sedimentation coefficient and corresponding axial ratio value for WT sLDLR at pH 5 is consistent with adoption of a closed conformation at low pH. In the case of the triple His→Lys mutant receptor, in keeping with an inability to bind ligand at either pH (Figure 4), only minor differences in sedimentation coefficient and axial ratio were observed. A similar result was obtained for an H562K / H586K double mutant sLDLR, revealing that mutations at sites outside the ligand binding domain can not only block ligand binding but also prevent pH dependent changes in molecular shape observed with wild type sLDLR.
Table 1. Hydrodynamic studies of sLDLR variants.
| sLDLR Sample | pH | Concentration (mg/mL) | Sedimentation Coefficient (S20,w) | Axial Ratio (a/b) |
|---|---|---|---|---|
| Wild Type | 7 | 0.41 | 5.3 | 3.9 |
| Wild Type | 5 | 0.36 | 6.1 | 1.3 |
| H562K / H586K | 7 | 0.11 | 5.5 | 3.0 |
| H562K / H586K | 5 | 0.23 | 5.4 | 3.5 |
| H190 K / H562K / H586K | 7 | 0.50 | 5.2 | 4.0 |
| H190 K / H562K / H586K | 5 | 0.25 | 5.1 | 4.3 |
Discussion
X-Ray crystallography studies of sLDLR at endosomal pH provide the framework for a testable model of pH dependent ligand release. The 3-dimensional structure-derived model is consistent with previous mutagenesis studies that demonstrate ligand binding but no release when the β-propeller segment is deleted (4). Likewise, naturally occurring LDLR mutations (22,23) and site directed mutagenesis studies (11) have provided evidence that His residues within the β-propeller and LA repeat 5 are key elements of the pH induced ligand release mechanism. Studies by Beglova et al. (11) using transfected ldlA-7 cells, showed that His190Tyr or His562Tyr LDLR variants bound LDL at neutral pH and released the ligand in response to low pH, albeit with lower efficiency than the native receptor. These authors subsequently changed all three His to Ala or Tyr to assess the cooperativity of the process. Both triple mutants were capable of binding receptor but failed to release ligand at low pH, providing evidence that these His residues participate in concert to promote ligand release. In the present study we have confirmed and extended these findings using a cell-free, solution assay employing sLDLR and apoE•DMPC disk complexes as ligand. The data reveal subtle differences between WT sLDLR and a triple His→Ala mutant sLDLR with the latter displaying an attenuated pH-dependent effect on ligand binding. On the other hand, the triple His→Lys mutant wherein each of the 3 His residues were converted to Lys, failed to bind ligand at any pH examined. Since the pKa of the lysine side chain is ∼10.5, unlike His, these Lys will retain a positive charge at pH 7. Consistent with the postulated role of electrostatics in modulating intra-molecular rearrangement within the receptor, the data suggest a “histidine switch” mechanism is operative in this system. To further refine this system His residues were singly substituted by Lys and the effect on ligand binding at pH 7.0 determined. The data showed that mutation at His190 more effectively blocked ligand binding compared to His562 or His586. Interestingly, however, a double mutant wherein both His562 and His586 were converted to Lys abolished ligand binding. Rudenko et al. (2) showed that, at endosomal pH, His190 interacts with Glu581 while both His562 and His586 interact with Asp149. Thus, it is conceivable that ionization of both these residues is required to stabilize the interaction with Asp149 and thereby, interfere with ligand binding.
A key aspect of the model proposed by Rudenko et al al (2) is that LDLR adopts a closed conformation at endosomal pH that prevents access to the ligand binding site due to interactions with the β-propeller motif that are induced by side chain ionization of key His residues. Consistent with this model, sedimentation velocity experiments on WT sLDLR gave rise to an increase in sedimentation coefficient as well as a decrease in axial ratio as a function of lowering the pH from 7 to 5. Furthermore, consistent with their inability to bind ligand at either pH, the triple His→Lys mutant sLDLR, as well as the H562K / H586K mutant sLDLR did not undergo a corresponding pH dependent change in sedimentation coefficient. Moreover, the calculated axial ratio for both these mutant receptors were largely unchanged at pH 7 and pH 5. The absence of a change in these parameters is consistent with the ionization properties of Lys versus His. Based on the lack of Lys side chain ionization over this pH range it may be predicted that no conformational change will occur if the model is correct. The similar values observed for this mutant receptors at the two pH values is also consistent with their lack of binding activity at either pH. Whether the conformation adopted these mutant sLDLR is the same as that adopted by WT receptor at pH 5, however, remains an open question since the axial ratios observed for both mutant sLDLR studied were more similar to the WT receptor at pH 7. Given this, it appears that the mutant sLDLRs may achieve a unique conformation that results in ligand binding site blockage with retention of more extended conformation. Further studies will be required to elucidate the precise nature of this conformation. Given the successful application of X-ray crystallography to solve the WT receptor structure, such studies should be feasible.
Whereas exploitation of His / Lys substitution mutations has not to our knowledge been used to illustrate effects on receptor – ligand interactions, this general approach has been employed in other systems. For example, Briand et al. (24) showed that replacement of Lys188 of trypsin by His creates a metal chelation site in the substrate binding pocket of this protease. In this case, the His mutation introduced a metal binding ‘switch,’ capable of modulating enzyme activity as a function of pH. Likewise, Nyarko and coworkers (25) reported that the light chain subunit of dynein exists as a dimer at physiological pH but dissociates to a folded monomer at pH < 4.8. By mutating His55 to Lys these authors showed that pH-induced dimer dissociation is reversible and governed by the ionization state of His55. Indeed, mutagenesis of His55 to Lys resulted in a monomer in the pH range of 3-8, while mutation to Ala resulted in a dimer across the same pH range. In other work, Martinez and Bowler (26) investigated the kinetics of an alkaline conformational transition in cytochrome c using pH jump stopped-flow methods to probe the nature of an ionizable “trigger” group for this conformational change. These authors reported that a Lys73His mutation shifted the pKa of the ligand replacing Met80 from about 10.5 to approximately 6.6 in support of the concept that ionization equilibria modulates protein folding in this system. Thus, alteration of the ionization potential of key residues in proteins via mutagenesis represents a useful and discriminating method to examine mechanism.
In the present system, a key factor in experimental design is the availability of a high-resolution structure for the receptor at low pH together with a discriminating assay to probe ligand binding and release. Taken together, the data presented show that the solution-based FRET assay employed recapitulates findings using transfected cells in culture and, further, can be used to dissect the molecular basis of ligand binding and release. The results fit well with the model of pH dependent ligand release proposed by Rudenko et al. (2,5) and reveal the critical role of His side chain ionization in modulation of ligand interactions with the LDLR.
Acknowledgments
The authors thank Jennifer Y. Matsuda for technical assistance.
Abbreviations
- AEDANS
N-(iodoacetyl)-N′-(5-sulfo-1-napthyl) ethylenediamine
- apo
apolipoprotein
- DMPC
dimyristoyl-phosphatidylcholine
- FRET
fluorescence resonance energy transfer
- NT
amino terminal
- sDLR
soluble low density lipoprotein receptor
- WT
wild type
Footnotes
This work was supported by a grant from the National Institutes of Health (HL 64159), the Alberta Cancer Board (CMK) and a Postdoctoral Fellowship Award (15FT-0164) from California Tobacco Related Disease Research Program.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 3.Russell DW, Brown MS, Goldstein JL. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J Biol Chem. 1989;264:21682–21688. [PubMed] [Google Scholar]
- 4.Davis CG, Goldstein JL, Sudhof TC, Anderson RG, Russell DW, Brown MS. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 1987;326:760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- 5.Rudenko G, Deisenhofer J. The low-density lipoprotein receptor: ligands, debates and lore. Curr Opin Struct Biol. 2003;13:683–689. doi: 10.1016/j.sbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Villeger L, Abifadel M, Allard D, Rabes JP, Thiart R, Kotze MJ, Beroud C, Junien C, Boileau C, Varret M. The UMD-LDLR database: additions to the software and 490 new entries to the database. Hum Mutat. 2002;20:81–87. doi: 10.1002/humu.10102. [DOI] [PubMed] [Google Scholar]
- 7.Doi T, Kurasawa M, Higashino K, Imanishi T, Mori T, Naito M, Takahashi K, Kawabe Y, Wada Y, Matsumoto A. The histidine interruption of an alpha-helical coiled coil allosterically mediates a pH-dependent ligand dissociation from macrophage scavenger receptors. J Biol Chem. 1994;269:25598–25604. [PubMed] [Google Scholar]
- 8.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 9.Wragg S, Drickamer K. Identification of amino acid residues that determine pH dependence of ligand binding to the asialoglycoprotein receptor during endocytosis. J Biol Chem. 1999;274:35400–35406. doi: 10.1074/jbc.274.50.35400. [DOI] [PubMed] [Google Scholar]
- 10.Lee D, Walsh JD, Mikhailenko I, Yu P, Migliorini M, Wu Y, Krueger S, Curtis JE, Harris B, Lockett S, Blacklow SC, Strickland DK, Wang YX. RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi. Mol Cell. 2006;22:423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Beglova N, Jeon H, Fisher C, Blacklow SC. Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor. Mol Cell. 2004;16:281–292. doi: 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Fisher CA, Wang J, Francis GA, Sykes BD, Kay CM, Ryan RO. Bacterial overexpression, isotope enrichment, and NMR analysis of the N-terminal domain of human apolipoprotein E. Biochem Cell Biol. 1997;75:45–53. [PubMed] [Google Scholar]
- 13.Yamamoto T, Ryan RO. Role of leucine zipper motif in apoE3 N-terminal domain lipid binding activity. Biochim Biophys Acta. 2006;1761:1100–1106. doi: 10.1016/j.bbalip.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto T, Lamoureux J, Ryan RO. Characterization of low density lipoprotein receptor ligand interactions by fluorescence resonance energy transfer. J Lipid Res. 2006;47:1091–1096. doi: 10.1194/jlr.D600001-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Bajari TM, Strasser V, Nimpf J, Schneider WJ. LDL receptor family: isolation, production, and ligand binding analysis. Methods. 2005;36:109–116. doi: 10.1016/j.ymeth.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto T, Choi HW, Ryan RO. Apolipoprotein E isoform-specific binding to the low-density lipoprotein receptor. Anal Biochem. 2008;372:222–226. doi: 10.1016/j.ab.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn EJ, Edsall JT. Proteins, Amino Acids and Peptides as Ions and Dipolar Ions. Rheinhold; New York: 1943. [Google Scholar]
- 18.Kuntz ID. Hydration of macromolecules. IV. Polypeptide conformation in frozen solutions. J Am Chem Soc. 1971;93:514–516. doi: 10.1021/ja00731a037. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, Ryan RO. Anionic phospholipids inhibit apolipoprotein E--low-density lipoprotein receptor interactions. Biochem Biophys Res Commun. 2007;354:820–824. doi: 10.1016/j.bbrc.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 20.Beglova N, Blacklow SC. The LDL receptor: how acid pulls the trigger. Trends Biochem Sci. 2005;30:309–317. doi: 10.1016/j.tibs.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Blacklow SC. Versatility in ligand recognition by LDL receptor family proteins: advances and frontiers. Curr Opin Struct Biol. 2007;17:419–426. doi: 10.1016/j.sbi.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopkins PN, Wu LL, Stephenson SH, Xin Y, Katsumata H, Nobe Y, Nakajima T, Hirayama T, Emi M, Williams RR. A novel LDLR mutation, H190Y, in a Utah kindred with familial hypercholesterolemia. J Hum Genet. 1999;44:364–367. doi: 10.1007/s100380050179. [DOI] [PubMed] [Google Scholar]
- 23.Sun XM, Patel DD, Webb JC, Knight BL, Fan LM, Cai HJ, Soutar AK. Familial hypercholesterolemia in China Identification of mutations in the LDL-receptor gene that result in a receptor-negative phenotype. Arterioscler Thromb. 1994;14:85–94. doi: 10.1161/01.atv.14.1.85. [DOI] [PubMed] [Google Scholar]
- 24.Briand L, Chobert JM, Tauzin J, Declerck N, Leonil J, Molle D, Tran V, Haertle T. Regulation of trypsin activity by Cu2+ chelation of the substrate binding site. Protein Eng. 1997;10:551–560. doi: 10.1093/protein/10.5.551. [DOI] [PubMed] [Google Scholar]
- 25.Nyarko A, Cochrun L, Norwood S, Pursifull N, Voth A, Barbar E. Ionization of His 55 at the dimer interface of dynein light-chain LC8 is coupled to dimer dissociation. Biochemistry. 2005;44:14248–14255. doi: 10.1021/bi0512694. [DOI] [PubMed] [Google Scholar]
- 26.Martinez RE, Bowler BE. Proton-mediated dynamics of the alkaline conformational transition of yeast iso-1-cytochrome c. J Am Chem Soc. 2004;126:6751–6758. doi: 10.1021/ja0494454. [DOI] [PubMed] [Google Scholar]


