Abstract
Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), an oncogenic fusion gene protein that is characteristically found in a subset of anaplastic large cell lymphomas, promotes tumorigenesis through its functional and physical interactions with various biologically important proteins. The identification of these interacting proteins has proven to be useful to further our understanding of NPM-ALK-mediated tumorigenesis. For the first time, we performed a proteome-wide identification of NPM-ALK-binding proteins using tandem affinity purification and a highly sensitive mass spectrometric technique. Tandem affinity purification is a recently developed method that carries a lower background and higher sensitivity compared with the conventional immunoprecipitation-based protein purification protocols. The NPM-ALK gene was cloned into an HB-tagged vector and expressed in GP293 cells. Three independent experiments were performed and the reproducibility of the data was 68%. The vast majority of the previously reported NPM-ALK-binding proteins were detected. We also identified proteins that are involved in various cellular processes that were not previously described in association with NPM-ALK, such as MCM6 and MSH2 (DNA repair), Nup98 and importin 8 (subcellular protein transport), Stim1 (calcium signaling), 82Fip (RNA regulation), and BAG2 (proteosome degradation). We believe that these data highlight the functional diversity of NPM-ALK and provide new research directions for the study of the biology of this oncoprotein.
Anaplastic lymphoma kinase-positive anaplastic large-cell lymphoma (ALK+ALCL) is a subtype of T/null-cell non-Hodgkin lymphoma characterized by the consistent expression of CD30 and anaplastic cytologic features.1 The aberrant expression of anaplastic lymphoma kinase (ALK) in approximately 80% of ALK+ALCL tumors is a result of the reciprocal chromosomal translocation that leads to the fusion of the N-terminal portion of the nucleophosmin (NPM) gene at 5q35 to the C-terminal portion of the ALK gene at 2p23.2 NPM-ALK, the resulted fusion gene protein, is a 75-kDa fusion protein containing the first 117 amino acid residues of NPM and the 1058 to 1620 amino acid fragment of ALK.2 This fusion protein is oncogenic.3 More recent studies suggest that NPM-ALK contributes to tumorigenesis by exerting its constitutively active tyrosine kinase embedded in the ALK portion of the fusion gene protein.4,5 Specifically, NPM-ALK is known to inappropriately activate and deregulate several signaling pathways, including those of JAK/STAT3, PI3K/AKT, Ras/MAPK, mTOR, MEK/ERK, and JNK/c-Jun.4,5,6,7,8,9,10,11,12,13,14
The emerging field of proteomics provides a powerful avenue to examine protein-protein interactions. These techniques are highly suitable for studying oncogenic proteins such as NPM-ALK, since the oncogenic potential of NPM-ALK appears to hinge on its functional and physical interactions with its binding partners. The traditional method utilizes immunoprecipitation of the protein of interest, which is subsequently analyzed using mass spectrometry (MS). While this method has been extensively used to study protein-protein interactions, it is limited by a relatively high level of background, attributable to the nonspecific binding of proteins to the agarose beads or immunoglobulins used for immunoprecipitation. This high background may mask specific binding proteins, especially those present in relatively low quantities or those binding to only a small subset of the protein under study. To circumvent this problem, a method called tandem affinity purification (TAP), has been recently developed to improve the specificity and sensitivity of the protein purification process.15,16,17,18 The essence of TAP is to express the protein of interest that is tagged with two specific types of peptides, allowing two rounds of specific capture and purification.19,20
There are a number of affinity tags reported and used in various protein-protein interaction studies involving TAP.21 Kaiser et al recently reported a novel hexahistidine-biotin (HB)-tag vector (Figure 1), consisting of a RGS-hexahistidine tag and a bacterially derived biotinylation signal peptide. The RGS-hexahistidine tag (or RGSH6) is a 9-amino acid peptide including 6 histidine residues that are preceded by three amino acid residues (R,G,S). This design has been shown to result in greatly reduced cross-reactivity with eukaryocytic proteins. The biotinylation signal peptide is a 75-amino acid sequence containing a specific lysine that can be readily biotinylated by endogenous biotin ligase in vitro.15,16 The protein purification process involves the initial capture of the HB-tagged protein by Ni2+-chelate beads that recognize the histidine tag, followed by a second-step purification using immobilized streptavidin beads that recognize the biotin tag. The use of the biotin-streptavidin in the second step also allows ‘on-bead’ tryptic digestion, a technique shown to be superior to either ‘in-gel’ or ‘in solution’ digestion, in both sensitivity and specificity.22,23 In contrast with the traditional immunoprecipitation-based method, the use of the HB-tag approach also decreases the chance of interference with the biological functions of the protein of interest.7
Figure 1.
N-terminal HB tags consisting of a nine amino acid peptide (RGSH6) that can bind to Ni2+ chelate resins and a 75 amino acid sequence (BIO) that can be efficiently biotinylated in mammalian cells through an endogenous biotin ligase. Structure of HB tag; sequence of BIO tag; sequence of TEV protease cleavage site.
In addition to the use of TAP, we also used our recently developed MS method to enhance the detection and identification of the interacting proteins. This highly sensitive technique is based on the use of precursor ion exclusion24 and optimized sample loading25 in a capillary liquid chromatography (LC) quadrupole time-of-flight mass spectrometer. A replicate LC-MS run of the same sample with precursor ion exclusion allows identification of low abundance peptides by excluding the high abundance peptides identified in the first run.24 Optimal sample loading to the capillary LC-MS also has been shown to facilitate the identification of low abundance peptides; thus, we used an automated on-line high performance liquid chromatography-UV (HPLC-UV) to optimize the sample loading, as previously described.25 Improvement in our LC-MS technique has allowed us to identify many proteins with high confidence in cells, tissues, and body fluids. In one of our recent publications, we found a total of 3749 different proteins or protein groups from breast cancer tissues with a false positive identification rate of less than 0.95%, representing the most comprehensive proteome profile for human breast cancer tissues to date.26
With these advances in the proteomics technology, we performed a proteome-wide search for NPM-ALK-interacting proteins. With the use of the HB-tag based TAP and the optimized LC-MS technique, we hypothesize that we will be able to establish a comprehensively profile of NPM-ALK-interacting proteins, including those expressed at low quantities or bound to only a small subset of NPM-ALK. We believe that this new information will allow us to better understand the biology of NPM-ALK and its mechanisms to mediate tumorigenesis.
Materials and Methods
HB-Tagged NPM-ALK Vector and Gene Transfection
The HB-tagged vector was kindly provided by Dr. Peter Kaiser (University of California Irvine, CA). NPM-ALK, amplified from a pcDNA-NPM-ALK plasmid (a kind gift from Dr. S. Morris, St. Jude’s Children Research Hospital, Memphis, TN), was inserted in-frame into the HB-tagged vector using XbaI-containing primers, and the final sequence was confirmed.
Cell Lines, Tissue Culture, and Gene Transfection
GP293, a human embryonic kidney cell line (Clontech, Mountain View, CA), was maintained in Dulbecco’s Modified Eagle’s Medium (Sigma, Ontario, Canada), supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY), antibiotics (10 mg/ml streptomycin and 10,000 U/ml penicillin; Invitrogen, Ontario, Canada). GP293 cells were transfected with either HB-tagged NPM-ALK or HB empty vector using lipofectamine 2000 (Invitrogen, Ontario, Canada) in accordance with the manufacturer’s suggested protocol. The cell culture was supplemented with 4 μmol/L biotin to improve the biotinylation efficiency of HB-tagged NPM-ALK. For co-immunoprecipitation experiments, we used two ALK+ALCL cell lines (Karpas 299 and SUP-M2), both of which have been previously described.27 These two ALK+ALCL cell lines were maintained in RPMI 1640 (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum. All of the cells were cultured under an atmosphere of 95% O2 and 5% CO2 in 98% humidity at 37°C.
TAP
Ni2+-Sepharose beads (Amersham Biosciences, Piscataway, NJ) were pre-equilibrated in buffer #1 (20 mmol/L sodium phosphate, 500 mmol/L sodium chloride, 20 mmol/L imidazole, pH = 7.3). Cell lysates (2 to 3 mg) were incubated with Ni2+-Sepharose beads at 4°C overnight. Ni2+-Sepharose beads were then collected by centrifugation at 500 × g for 5 minutes, followed by washing with buffer #1. Subsequently, proteins were eluted in buffer #2 (20 mmol/L sodium phosphate, 500 mmol/L sodium chloride, 500 mmol/L imidazole, pH = 7.3). The resulting eluate was loaded onto immobilized streptavidin beads that had been pre-equilibrated in buffer #3 (0.1 m/L phosphate, 0.15 m/L sodium chloride, pH = 7.3). After incubation at 4°C overnight, the streptavidin beads were extensively washed with buffer #3. Streptavidin beads were then collected by centrifugation (2500 × g, 5 minutes) and stored in −20°C until tryptic digestion.
On-Bead Tryptic Digestion, Peptide Extraction, and Desalting
On-bead tryptic digestion was performed by using standard digestion techniques described previously with modifications.16,22,23,28 Peptides were desalted and quantified by HPLC (Agilent 1100 HPLC system, 4.6 × 50 mm C18 column, Varian, Ontario, Canada). Tandem mass spectrometric (MS/MS) analysis were performed using a quadrupole time-of-flight premier mass spectrometer (Waters, Manchester, UK) equipped with a nanoACQUITY Ultra Performance LC system (Waters, Milford, MA) as previously described with minor changes.28 The reversed phase LC separation was performed by using a 120-minute gradient. The MS data were recorded as previously described,28 except that peptide precursor ion exclusion strategy was applied to exclude relatively high-abundance peptides identified from the previous run and allow the low-abundant peptides to be identified.24
Mass Spectrometry Analysis and Data Analysis
Database searches by MASCOT were performed as previously described with modifications.28 The database searching was restricted to Homo sapiens in the Swiss-Prot database. The redundant peptides were removed from the proteins list. The single peptide hits with a matching score above the MASCOT threshold score for identity was manually examined and considered as a positive identification if the fragment ions contained more than five continuous g, b, or a ions. Additionally, we applied the target-decoy search strategy to determine false peptide matching.29,30
Immunoprecipitation and Western Blot Analysis
For immunoprecipitation, a standard protocol was used as previously described.31 The complex was subjected to SDS-polyacrylamide gel electrophoresis and Western blotting, and the proteins were visualized using enzyme chemiluminescence (Amersham Biosciences, Piscataway, NJ). The following antibodies were used for immunoprecipitation and immunoblotting: mouse anti-Stat3, anti-Hsp90, polyclonal anti-ALK, Nup98, MCM6, 82Fip, Rac3, and DPM1, (all of which were purchased from Santa Cruz Biotechnology, Santa Cruz, CA); and monoclonal anti-ALK antibody (Zymed, Ontario, Canada); rabbit polyclonal anti-Importin 8 antibody and rabbit polyclonal anti-BAG-2 antibody (IMGENEX, San Diego, CA); mouse monoclonal anti-MSH2 antibody (Calbiochem, Gibbstown, CA); mouse monoclonal anti-Stim1 and anti-Crop antibody (Abnova, Ontario, Canada); and rabbit polyclonal anti-Exportin 5 (Abcam, Cambridge, MA).
Results
Expression of HB-Tagged NPM-ALK in GP293 Cells
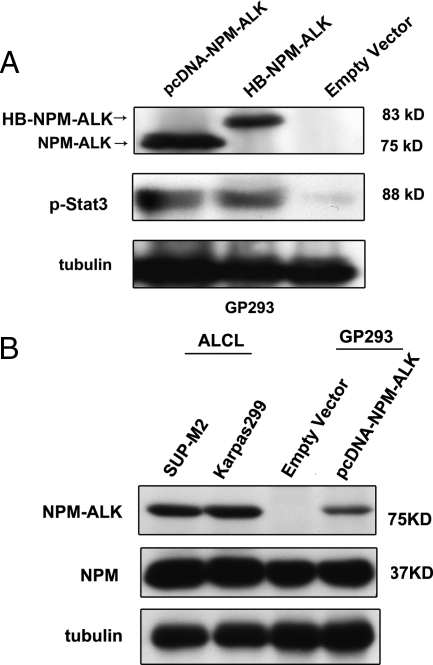
The NPM-ALK construct was inserted into the HB-tagged vector such that NPM-ALK was tagged with RGSH6 and the biotinylation signal sequence at its N-terminus. After the sequence of HB-tagged NPM-ALK construct was verified, the function of the expression vector was validated by Western blot (Figure 2A). As shown in Figure 2A, due to the addition of the HB tag, HB-tagged NPM-ALK migrated slightly slower that the un-tagged NPM-ALK, We also used Stat3, a protein known to be activated and phosphorylated by NPM-ALK,32 as a surrogate marker to assess the functional integrity of HB-tagged NPM-ALK. GP293 cells transfected with HB-tagged NPM-ALK had a dramatic upregulation of phosphorylated Stat3 (pStat3) compared with cells transfected with the HB empty vector, which contained only a relatively low level of endogenous pStat3. The pStat3 band intensity was similar between cells transfected with HB-tagged NPM-ALK and those transfected with pcDNA-NPM-ALK, suggesting that both NPM-ALK constructs are equally efficient in phosphorylating Stat3.
Figure 2.
Confirmation of functional expression of HB-NPM-ALK in GP293. A: Western blot analysis showed expression of NPM-ALK in the pcDNA-NPM-ALK and HB-NPM-ALK transfected GP293 cell line but not in the empty vector transfected GP293 cell line. Expression of functional NPM-ALK proteins was examined by using anti-pStat3 antibody. Western blot analysis probed with anti-pStat3 showed that in the transfectant, Stat3 was phosphorylated. B: Comparison of NPM-ALK transfected GP293 cells with ALK+ALCL cells, demonstrating that similar levels of NPM-ALK were expressed.
In the protein-protein interaction studies, gross overexpression of a protein and subsequent testing of its binding partners may produce different results from those generated under normal expression. However, in the case of NPM-ALK, we found that the relative expression of NPM-ALK in GP293 cells transfected with HB-tagged NPM-ALK and that expressed in ALK+ALCL cell lines at the steady state was similar (Figure 2B). Thus, we believe that our experimental model is representative.
Proteome-Wide Identification of NPM-ALK Binding Proteins
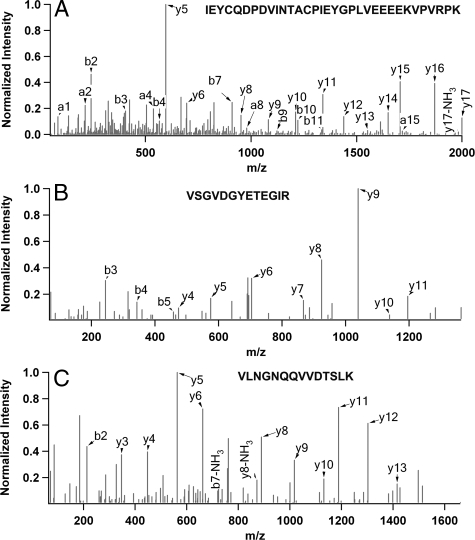
Three independent experiments were performed, each consisting of a negative control sample (cells transfected with the HB empty vector) and an experimental sample (cells transfected with HB-tagged NPM-ALK). Figure 3 illustrates examples of fragment ion spectra from three representative peptides detectable in the purified NPM-ALK protein complex. The vast majority of the sequence-matched spectra were of good quality, allowing us to readily determine their identities with a high degree of confidence. All of the identified peptides were above the MASCOT threshold score for identity with a confidence level of >95%. Moreover, the fragment ion spectra of all of the single peptide hits were manually examined. Using the target-decoy sequence search strategy, the false positive rate is less than 1%.
Figure 3.
ESI MS/MS spectra of representative peptides from three selected proteins. (A) NPM-ALK, (B) MCM6, and (C) 82Fip.
From the three independent experiments, we were able to detect 36 peptides (Figure 4), and the total peptide sequence coverage of NPM-ALK is 72% (averaged 67%). The high peptide sequence coverage of NPM-ALK, the protein bait in this study, supports that high-quality protein purification had been achieved. All three negative control samples showed no evidence of NPM-ALK detectable by MS.
Figure 4.
Sequence coverage of NPM-ALK (72%) from the identified peptides (underlined) from the three independent experiments. The double underlined section represents the overlapped sequences identified from two peptides.
Overall, the data generated from the three independent test samples was reproducible, with 68% of the proteins identified in all three samples. This is comparable to 70% to 75%, a technical reproducibility rate we commonly observed when the same sample was analyzed in triplicate under the same MS analysis conditions.24
To optimize the specificity of our results, proteins detectable in any one of the three independent negative control samples were classified as ‘nonspecific’ and eliminated from our working list. A total of 254 proteins were identified, and 80 (32%) proteins were identified based on the presence of ≥2 peptide fragments in one or more test samples, and/or one peptide present in ≥2 test samples. The remaining 174 proteins were included in the list because manual examination of each of their MS/MS spectra fulfilled the stringent inclusion criteria described in Methods and Materials. Among the ‘single hit’ proteins, there are several previously reported NPM-ALK binding proteins, such as PI3K, mTOR, Jak2, MEKK2, MEKK4, PP2A β, AP1, and Lyn. Additionally, two ‘single hit’ proteins, MSH2 and Importin 8, have been validated by co-immunoprecipitation using ALK+ALCL cells. These ‘high-confidence’ proteins are summarized in Table 1. Additional details of these 254 ‘high-confidence’ proteins, including their protein names, access identification numbers, molecular weights, unique peptide sequences, scores for identity, MASCOT threshold scores for identity, experimental mass of the peptide, calculated mass of the peptide, gene ontology, putative or known biological functions, mass errors, sequence coverage and frequency of these proteins presented in our three independent experiments, were submitted as Supplemental Table S1 (see http://ajp.amjpathol.org). The vast majority of these 254 (234, 92.1%) proteins can be categorized into nine functional groups, as summarized in Table 1. 21 proteins are involved in the apoptotic pathway, and they include PI3K, DEDD, TRAF2, BCLF1, EEA1, Hsp60, VAPA, JIP4, and CROP. 20 proteins are involved in the proteasome degradation pathway, and they include 26S proteasome regulatory subunit, PIAS2, CNOT4, BAG2, Hsp90-α, UBE1, UBP5, CDC37, SENP1, and Hsp70. 11 proteins are involved in regulating protein phosphorylation, and they include PP2Aβ, PTCK3, TNK1, CCNT1, PTN23, PHKG2, and mTOR. Seven proteins are involved in DNA repair, and they include MSH2, PARP1, PCNA, Ku70, Ku86, and MCM6. Forty-three proteins are involved in DNA and RNA processing, and they include IRAK1, eIF-4G1, AP1, LIM protein, MED1, DDX42, and DDX21. Twenty-one proteins are involved in the subcellular protein transport pathway, Nup188, Nup98, Importin 8, RBP2, AAAS, and ZIP10. Twenty-four proteins are involved in the cell cycle regulation pathway, and they include 82Fip, CDC20, SHC1, FGFR2, CDK9, centromere protein J, T53G5, GAK, and LARP7. Forty-seven proteins are involved in various signaling transduction pathways, and they include Ras related proteins, S6 kinase, 14–3-3 protein, MEK kinases, and ATD3A. Forty proteins are involved in biogenesis, and they include DPM1, STT3A, and BCS1. Most of the previously reported NPM-ALK binding proteins were detected, and they include Grb2, Rab, Shc, PI3K, and IRS1.
Table 1.
Summary of NPM-ALK Interacting Proteins Identified by TAP LC-ESI MS/MS and Grouped Based on their Major Biological Functions
| Protein Name (frequency, # of peptides)* |
|---|
| Apoptosis |
| PI3-kinase p85subunit beta (1,3)†; PI3-kinase p85subunit alpha (2,3)†; PI3-kinase p110 subunit beta (1,1)†; Death effector domain-containing protein (1,1); TNF receptor-associated fact3or 2 (1,1); 250 kd substrate of Akt (1,1); C-jun-amino-terminal kinase-interacting protein 4 (1,2); Vesicle-associated membrane protein-associated protein A (1,1); Cisplatin resistance-associated overexpressed protein (CROP) (1,2); Alpha-actinin-1 (1,3); Peroxiredoxin-2 (1,2); Protein kinase C inhibitor protein 1 (1,1); Seladin-1 (1,2); Thioredoxin-related transmembrane protein (1,1); Zinc finger FYVE domain-containing protein 1 (1,2); Cytochrome c oxidasepolypeptide II (2,1); Bcl-2-associated transcription factor 1 (2,7); Heat shock protein 60 (3,25)†; Protein LL5-beta (1,1); Zincfinger FYVE domain-containing protein 2 (1,1); Serine protease11 (2,1) |
| Signal transduction |
| SH3 domain protein 2B (1,3); JAK2 (1,1)†; SLIT-ROBO Rho GTPase-activating protein 1 (2,1); Ras-related protein Rab-10 (1,1); Ras-related protein Rab-2A (1,1); Ras-related protein Rab-14 (1,2); Ras-related protein Rab-7a (1,5); Ran GTPase-activating protein 1 (1,1); Ras-related protein Rab-6A (1,1); Ras-related protein Ral-A precursor (1,1); Ras-related protein Rab-19 (1,1); Ras-related protein Rab-21 (1,1); Ras-related protein Rab-1B (1,3); Ras GTPase-activating-like protein IQGAP1 (1,1); GPI transamidase (1,1); SH2/SH3adapter GRB2 (1,5)†; MAPkinase-activated protein kinase 1b (1,1); SAM68 (1,1); Insulin receptor substrate 1 (1,1)†; 5-hydroxytryptamine receptor 2A (1,1); IL-18 receptoraccessory protein (2,1); AMPK beta-1 chain (1,1); G patch domain-containing protein 8 (1,3); Conserved ERA-likeGTPase (1,1); PDZ-RhoGEF (1,1); Neurotrophic tyrosine kinase receptor type 1 (1,1); Rac3(1,2); ADP-ribosylation factor 6 (1,1); ADP-ribosylation factor-like protein 1 (1,1); ADP-ribosylation factor-like protein 3 (1,1); ADP-ribosylation factor 4 (1,1); Periodic tryptophan protein 2 homolog (1,1); Creatine kinase B-type (1,1); 14–3-3 protein theta (1,1); MARCKS-related protein (1,1); STIM1(1,3); Serine/threonine/tyrosinekinase 1 (1,1); SR-beta (1,1); Stonin-2 (1,1); Transmembrane protein 161A precursor (1,1); Kinesin-like protein KIF13B (1,1); UNR protein (1,1); A-Raf proto-oncogene serine/threonine-protein kinase (1,1); MEK kinase kinase 4 (1,1)†; MEK kinase 2 (1,1)†; ATPase family AAA domain-containing protein 3A (2,9); ATPase family AAA domain-containing protein 3B (1,1) |
| Biogenesis |
| Nucleoplasmin-3 (1,3); Acyl-CoA thioesterase 9 (1,2); Mitochondrial chaperone BCS1 (1,1); Methionine synthase reductase (1,1); Polyadenylate-binding protein 3 (1,1); Retinol dehydrogenase 11 (1,1); Oligosaccharyl transferase subunit STT3B (1,1); STT3-A (2,3); Phosphonoformate immuno-associated protein3 (1,1); NADH-cytochrome b5 reductase 3 (1,1); mPR (1,1); Medium-chain specific acyl-CoA dehydrogenase (1,1); E2K (1,1); Thermostable phenol sulfotransferase (2,2); HDL-bindingprotein (1,2); Glutamate carboxypeptidase 2 (1,1); Alpha-mannosidase 2 (1,1); 17-beta-hydroxysteroid dehydrogenase 4 (1,1); Dihydrolipoamide dehydrogenase (1,1); Peroxin-1 (1,1); Peroxiredoxin-6 (1,5); Adrenodoxin reductase (2,1); PMP70 (1,2); Protoporphyrinogen oxidase (1,1); Ribosome biogenesis protein BOP1 (1,1); Sra-1 (1,1); Dipeptidyl peptidase 9 (2,1); Mitochondrial malonyltransferase (2,2); Palladin (1,1); OSBP-related protein 11 (1,4); OSBP-related protein 10 (1,1); OSBP-related protein 9 (1,3); PRA1 family protein 3 (1,1); PRA1 family protein 2 (1,1); Collagen/fibrinogen domain-containing protein 3 (2,1); Adenine; phosphoribosyltransferase (1,3); Phosphoglucomutase-2 (1,1); NOC3-likeprotein (1,1); Synaptic vesicle membrane protein VAT-1 homolog (1,1); DPM1 synthase (1,2) |
| Proteasome degradation |
| 26S proteasomeregulatory subunit S9 (1,1); E3 SUMO-protein ligase PIAS2 (1,1); E3 ubiquitin-protein ligase CNOT4 (1,1); BAG family molecular chaperone regulator 2 (1,2); Heat shock protein HSP 90-alpha (1,9); Ubiquitin-activating enzyme E1 (1,2); Ubiquitin carboxyl-terminal hydrolase 5 (1,1); Proteasome activator complex subunit 3 (1,1); Hsp90 co-chaperone Cdc37 (1,1); E3 ubiquitin-protein ligase (2,1); Ubiquitin-associated protein 2 (1,1); E3 ubiquitin-protein ligase UBR2 (1,1); E3 ubiquitin-protein ligase RNF216 (1,1); Sentrin/SUMO-specificprotease SENP1 (2,1); Ubiquitin-like product Chap1/Dsk2 (1,1); Proteasome regulatory particle non-ATPase 13 (1,1); SUMO-1-specific protease 3 (1,1); PAB-dependent poly(A)-specific ribonuclease subunit 2 (1,1); F-box only protein 28 (1,1); Heat shock 70 kd protein 1 (3,13) |
| Cell cycle |
| Fe65protein (1,1); WD repeat-containing protein 39 (1,1); ROD1 (1,1); FGFR2 (1,1); SHC-transforming protein 1 (2,4)†; Cyclin-dependent kinase 9 (1,1); Cell division cycle protein 20 homolog (1,1); 82Fip (2,15); Protein TBRG4 (1,1); E2-induced gene 3 protein (2,2); GTP-binding protein NGB (2,4); DHPprotein (1,1); Centromere protein J (1,1); Uncharacterized protein C2orf29 (1,1); TP53-target gene 5 protein (1,1); Interferon-related developmental regulator 1 (1,1); IFIT1 (1,1); Putative helicase MOV-10 (1,1); Cyclin G-associated kinase (2,4)†; Puromycin-sensitive aminopeptidase (1,1); La-related protein 7 (1,3); DLK (1,1); Adipocyte plasma membrane-associated protein (1,1); Tyrosine-protein kinase Lyn (1,1)† |
| DNA Repair |
| KU86 (1,1); KU70 (1,1); PCNA (2,5); FACT complex subunit SPT16 (1,1); DNA mismatch repair protein MSH2 (1,1); PARP-1 (2,2); DNA replication licensing factor MCM6 (2,4); Phosphorylation; Cell division cycle 2-related protein kinase 7 (1,1); PP2A-beta (1,1)†; Serine/threonine-protein kinase PCTAIRE-3 (1,1); Non-receptor tyrosine-protein kinase TNK1 (1,1); PRP4 kinase (1,1); Cyclin-T1 (2,3); mTOR (1,1)†; Phosphorylase kinasesubunit beta (1,1); PSK-C3 (1,1); Hisdomain-containing protein tyrosine phosphatase (1,2); Pyrophosphatase SID6–306 (1,1) |
| DNA and RNA processing |
| NFkB-repressing factor (1,2); Interleukin-1 receptor-associated kinase 1 (1,1); eIF-4G1 (1,4); TAFII-150 (1,1); Hic-2 (1,1); MBP-1 (3,14); CREB/ATF bZIP transcription factor (1,1); LanC-like protein 2 (1,1); Bromodomain adjacent to zinc finger domain protein 1B (1,1); Prohibitin-2 (2,2); Pre-mRNA-splicing factorSRP20 (1,1); Seryl-tRNA(Ser/Sec)synthetase (1,1); Transcription factor AP-1 (1,1)†; EIF-2A (1,1); snRNP-B (2,1); Arginine/serine-rich-splicing factor 14 (1,1); LIM domain-containing protein 1 (1,1); RNase inhibitor (1,1); Glycine–tRNA ligase (1,1); Alanine–tRNAligase (1,2); hRPB8 (1,1); RNase L inhibitor (1,3); PPAR-binding protein (1,1); TAFII-20/TAFII-15 (1,1); Chromosome-associated protein H (1,1); Pre-mRNA 3′-end-processing factor FIP1 (2,1); Proline–tRNA ligase (1,1); ATP-dependent RNA helicase DDX4 (1,9); S1 RNA-binding domain-containing protein 1 (1,1); CPSF 59 kDasubunit (1,2); Putative ribosomal RNA methyltransferase 1 (1,1); Nucleolar RNA helicase 2 (1,2); Splicing factor 45 (1,1); Bic-D 1 (1,1); Zinc finger and SCAN domain-containing protein 29 (1,1); Probable ATP-dependent RNA helicase DHX40 (1,1); Pumilio-2 (1,1); TBR-1(1,1); Zinc finger protein 768 (1,2); Exportin-5 (1,1); Probable helicase with zinc finger domain (1,4); Paraspeckle protein 1 (2,2); Zinc finger protein 503 (1,1) |
| Transport |
| AIP (1,1); Importin-8 (1,1); Nucleoprotein TPR (1,3); Rab-11 (1,2); E3 SUMO-protein ligase RanBP2 (1,1); Nup98 (2,3); Sec61 alpha-1 (1,1); mRNA export factor (1,1); hNup188 (1,2); eIF4Etransporter (1,1); Aladin (1,4); Transportin-1 (1,1); ATP-dependent RNA helicase DDX19A (1,1); Cyclin-M1 (1,1)†; SERCA3 (1,1); Probable cation-transporting ATPase 13A1 (1,1); Zinc transporter ZIP10 precursor (1,2); Kinesin-like protein KIF1B (1,1); Zeta-1 COP (1,1); Elongation factor G 1 (1,1); ERGIC-32 (1,1) |
| Others |
| Myomesin-1 (1,1); TP alpha (1,1); WD repeat-containing protein 62 (1,1); WD repeat-containing protein 6 (1,3); WD and tetratricopeptide repeats protein 1 (1,1); EF-hand domain-containing protein C17orf57 (2,1); Plastin-3 (1,2); MHC class I antigen Cw*7 (1,1); YTH domain family protein 3 (1,1); Hornerin (1,1); DSEL (1,1); Dermatan-sulfate epimerase-like protein precursor (2,2); Protein FAM10A5 (1,1); Tumor antigen BJ-HCC-21 (1,2); Synapse-associated protein 1 (1,1); Collagen alpha-1(XII) chain precursor (1,1); Protein EHM2 (1,1); Dynamin-3 (1,1); Neighbor of COX4 (1,1); FAST kinase domain-containing protein 5 (1,1) |
Frequency refers to the number of times a protein was identified in three independent sample replicates (i.e., 2 means the protein was identified two times out of the three samples analyzed); # of peptides refers to the combined number of peptides identified for the protein in all three samples.
Previously reported.
We did not observe a large number of streptavidin tryptic peptides generated by on-bead digestion. Only an average of five streptavidin peptides was detected from the three independent experiments, which did not appear to suppress the sample peptide signals. These results are in keeping with the concept that streptavidin used in the protein purification did not interfere with the MS analysis.
Validation of the NPM-ALK Binding Partner by Immunoprecipitation
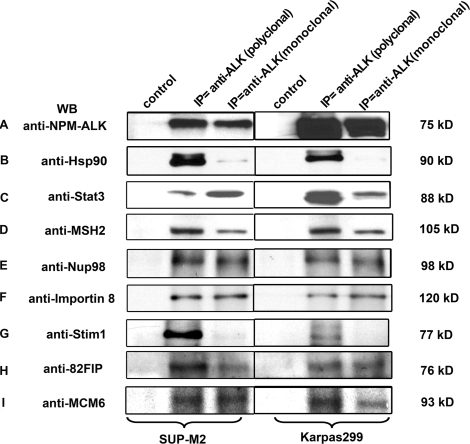
To validate some of the proteins identified by MS, co-immunoprecipitation experiments were performed. Based on how well their biological functions and importance are known, we selected 11 novel, putative NPM-ALK binding proteins for validation and the results are summarized in Table 2. We also included two proteins, Stat3 and Hsp90, which have been previously reported to physically interact with NPM-ALK as a positive control for these co-immunoprecipitation experiments. To document their biological relevance in ALK+ALCL, we used Karpas 299 and SUP-M2, two well-studied ALK+ALCL cell lines. To minimize the bias from the use of any one specific antibody, we used a monoclonal as well as a polyclonal anti-ALK antibody for co-immunoprecipitation. The presence or absence of NPM-ALK on Western blots was revealed by probing with an anti-NPM antibody. As shown in Figure 5A, NPM-ALK at approximately 75 kDa was detectable using immunoprecipitation with either the monoclonal or polyclonal anti-ALK antibody in both cell lines. Both Stat3 and Hsp90 were shown to be co-immunoprecipitated with NPM-ALK. We then probed these blots with antibodies reactive with 7 of the 11 chosen proteins: MSH2, Nup98, Importin 8, Stim1, 82Fip, MCM6, or Exportin 5. Figure 5 illustrates the results for MSH2, Nup98, Importin 8, Stim1, 82Fip, MCM6, all of which co-immunoprecipitated with NPM-ALK. We were unable to detect Exportin 5, which may reflect a failure of antibody-antigen interaction. All of these seven proteins examined were confirmed to be expressed in these two ALK+ALCL cell lines by Western blots (not shown). Of the remaining four proteins, we were unable to perform similar experiments since their molecular weights overlap with that of the immunoglobulin bands.
Table 2.
Summary of Validation Results from Karpas299 and SUP-M2 Cell Lysates by Using Co-Immunoprecipation and Reciprocal Immunoprecipitation
| Candidate name | Interaction validated | No evidence of interaction | Inconclusive |
|---|---|---|---|
| MSH2 | × | ||
| Nup98 | × | ||
| Importin 8 | × | ||
| Stim1 | × | ||
| 82Fip | × | ||
| MCM6 | × | ||
| Exportin 5 | × | ||
| Rac3 | × | ||
| DPM1 | × | ||
| Crop | × | ||
| BAG2 | × |
Figure 5.
Co-immunoprecipitation results from two ALK+ALCL cell lines, SUP-M2 and Karpas299. The monoclonal and polyclonal anti-ALK immunocomplexes were resolved by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane and used for Western blot analysis. Western blot analysis demonstrated the physically association of NPM-ALK (A), Hsp90 (B), Stat3 (C), MSH2 (D), Nup98 (E), Importin 8 (F), Stim1 (G), 82Fip (H), and MCM6 (I).
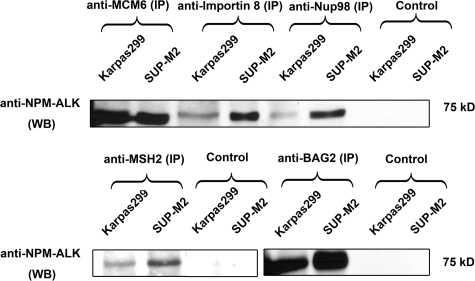
To increase the sensitivity and specificity, we performed ‘reciprocal’ immunoprecipitation experiments using the same two cell lines and various antibodies reactive with nine proteins. First, we confirmed the binding of the four proteins validated by ‘forward’ immunoprecipitation, namely MCM6, MSH2, Nup98, and Importin 8 (Figure 6). Using reciprocal immunoprecipitation, we also confirmed the lack of binding between Exportin 5 and NPM-ALK and found the binding between BAG2 and NPM-ALK. For the remaining three proteins, we were unable to obtain specific antibodies for immunoprecipitation and thus, we cannot confidently exclude them being NPM-ALK binding proteins. Overall, of the eight proteins for which we had sufficient reagents to validate, we successfully confirmed 7 (88%) proteins as NPM-ALK binding partners. We also confidently excluded Exportin 5 as a binding partner since it was found expressed in the whole cell lysates of ALK+ALCL.
Figure 6.
Reciprocal immunoprecipitation of Karpas299 and SUP-M2 cell lysates performed with anti-MCM6, anti-Importin 8, anti-Nup98, anti-MSH2, and anti-BAG2 antibodies, followed by Western blot analysis.
Discussion
It is widely accepted that NPM-ALK plays a central role in the pathogenesis of ALK+ALCL. In the current model, NPM-ALK transforms cells by physically and functionally interacting with a variety of proteins, many of which are involved in cellular signaling. Equipped with constitutive tyrosine kinase activity, NPM-ALK deregulates a large number of cellular signaling pathways, thereby promoting cell survival and cell proliferation. Studies of NPM-ALK have shed important insights into the significance of aberrant cellular signaling in tumorigenesis, ALK+ALCL has proven to be a highly useful model for cancer research in this regard.
To fully understand the biology of NPM-ALK, comprehensive profiling of proteins physically interacting with this fusion gene oncoprotein has been demonstrated to be a useful approach. Crockett et al used MS to examine NPM-ALK binding proteins.33 To our knowledge, this is the first and only study of NPM-ALK binding proteins using MS. Many of the reported 46 proteins, such as Stat3, PLCγ, Shc, and PI3K, had been previously described as NPM-ALK binding proteins, largely based on data from immunoprecipitation experiments performed by a number of research groups. Importantly, this MS study also revealed a number of novel NPM-ALK binding partners, including EphA1, MEK kinase 1, Elf4B, and Hsp60. These results not only have provided important insights into NPM-ALK, they also have provided the proof of principle that analysis of NPM-ALK binding proteins using MS is a feasible and constructive approach to study the biology of this oncogenic fusion protein.
Since the publication of Crocket et al, the field of proteomics has advanced greatly, which has translated into increased sensitivity and specificity. We believe that the technical differences between our study and that done by Crockett et al33 likely account for the differences in our results. First, we used TAP for protein purification, as opposed to the traditional immunoprecipitation-based method. The advantages of using TAP have been described in detail in the Introduction. The fact that we were able to identify an average of 67% of the peptide sequences of NPM-ALK, as compared with 19% reported in the previous study, is in keeping with the theoretical advantages of TAP. Second, “in-gel” digestion followed by peptide extraction in a gel-based proteomic analysis platform is relatively inefficient, compared with ‘on-bead’ digestion that was used in our study.22,23 The greatly reduced background achieved by ‘on-bead’ digestion effectively unmasks lower abundance proteins, and allows for their identification as specific binding proteins. Third, we used the peptide precursor ion exclusion strategy and an optimal sample loading method, both of which were recently developed LC-MS techniques that facilitate the detection of proteins expressed at relatively low levels.24,25 Fourth, we used a proteome-wide based search, using the Swiss-Prot Homo sapiens database that contains over 19,000 proteins represented in the proteome. This is in contrast with the previous study, which used a selected and targeted protein database built on results from various previous studies of NPM-ALK.33 We believe these four important advances have permitted us to comprehensively survey the human proteome for NPM-ALK binding proteins with an unprecedented sensitivity and specificity.
In addition to the technical differences discussed above, our studies also differ in the choice of the biological system used for the experiments. Whereas we used a system in which the NPM-ALK is enforced to be expressed in GP293 cells, an immortalized embryonic kidney cell line, the previous study used two ALK+ALCL cell lines in the initial search for NPM-ALK binding proteins. As shown in Figure 2, the expression level of NPM-ALK in both systems is not dramatically different and thus, one cannot attribute the differences in our results to a dramatic difference in the expression levels of NPM-ALK in these two systems. We do consider the possibility that the use of immortalized cells may have avoided the possibility for missing important NPM-ALK binding proteins whose expression may have been aberrantly silenced in cancerous cell lines. Lastly, our approach also has allowed us to include a biological negative control, which is not possible when ALK+ALCL cell lines were used. Specifically, we generated a negative control protein list by merging all of the data derived from the three negative control samples. We believe that this approach, in conjunction with the use of TAP, allowed us to minimize the background. This is especially important to detect proteins present in low quantities or proteins bound to a small subset of NPM-ALK.
Our method generated a total of 255 proteins including NPM-ALK. This number is not surprising to us, since other studies using large scale TAP have similar results.34,35 From the biological aspect, we also expect that a relatively large number of proteins can be identified in our purified protein complex. Recently, NPM-ALK was reported to have a relatively long half-life (>48 hours), and this property may result in a relatively stable protein complex with its binding partners.36 In addition, as discussed above, our assay was designed to capture proteins even when they are present in low abundance, and this factor likely contributes to the relatively high number of proteins detected.37 We believe the validity of our results is supported by several observations. First, as discussed above, the high peptide sequence coverage of the core protein NPM-ALK, which was consistently absent in the three negative control samples, points to the high efficiency and specificity of our protein purification method. The second evidence came from the high reproducibility of our data among the three independent test samples, as described above. Third, of the 254 proteins identified, 80 were identified by the presence of multiple peptides in the same test sample and/or the presence of the same peptides in more than one test sample. In our experience, the identification of these proteins is considered to carry high confidence. Fourth, a number of proteins that have been previously reported to interact with NPM-ALK, such as Grb2, Rab, Ras, Shc, PI3K, IRS1, LIM protein, Hsp90, Hsp60, PP2A, cyclin G associated kinase, AP1, Jak2, and MEK kinase, were detectable only in the test samples, but not in the negative control samples. Lastly, of those remaining 174 proteins identified by a single peptide sequence in one test sample, we examined each of their MS/MS spectra, all of which were of high quality. Furthermore, to minimize false positivity, we searched the MS/MS spectra of these proteins against the forward and reverse human proteome database. The overall false positive matching rate was <1%, an acceptable rate commonly reported in the literature.38,39
To validate the biological relevance of these identified proteins in ALK+ALCL cells, we further evaluated 13 proteins using co-immunoprecipitation. Stat3 and Hsp90, two known NPM-ALK binding proteins, were included as positive controls. These 11 potential NPM-ALK binding proteins were chosen from the 7 different biological pathways or processes, including apoptosis protein Crop, nuclear transport proteins Importin 8 and Nup98, DNA repair proteins MSH2 and MCM6, proteasome degradation protein BAG2, RNA processing Exportin 5, biogenesis protein DPM1, and the signal transduction proteins RAC3 and Stim1. Our results further supported the interaction of NPM-ALK and 7 of these 11 proteins studied, including MSH2, Nup98, Importin 8, Stim1, 82Fip, MCM6, and BAG2. For additional three proteins (RAC3, DPM1, Crop), conclusive co-IP data were not obtained. There is paucity of commercial reagents available for these three proteins and the antibodies available have not been tested for IP. These proteins migrate with the abundant heavy and light chain components of the precipitating antibody, masking their detection. Exportin 5 was not successfully validated using these cells and reagents. Therefore, of the 8 protein that were conclusively validated, 7 (88%) were found to associate with NPM-ALK in the ALK+ALCL cell lines using co-IP.
To our knowledge, NPM-ALK has never been reported to be associated with the DNA repair pathway. Dysregulation of DNA repair is strongly associated with tumorigenesis due to the inability of cells to respond appropriately to acquire coding-errors in the genome. The DNA repair proteins associated with NPM-ALK (Ku86, Ku70, PCNA, MSH2, PARP1, and MCM6) are a logically assembly and interaction have been previously documented, such as Ku70/86/PARP1,40 MSH2/PCNA,41 PARP1/PCNA,42 and PCNA/p16(lnk4a)/MCM6.43 PCNA appears to be a constant binding-mediator, and phosphorylation of tyrosine 211 on PCNA is associated with increased cellular proliferation.44 Moreover, interaction of a different diffusion tyrosine kinase, BCR-ABL, has recently been found to suppress the activity of DNA mismatch repair.45 MSH2 is a critical protein in their repair system and its inactivation is highly tumorigenic.46 It is highly possible that it may play roles in the pathogenesis of ALK+ALCL via its interaction with NPM-ALK. Similarly, little is known regarding NPM-ALK and nuclear transport. NPM-ALK is known to be localized to the nucleus as well as cytoplasm, and its nuclear localization is believed to be attributed to its heterodimerization with NPM1, which carries a nuclear localization signal.47 Nevertheless, it remains possible that the nuclear transport proteins identified in this study also contribute to the nuclear localization of NPM-ALK, and perhaps other biologically important proteins.48 Lastly, while recent data implicate that NPM-ALK is degradable via the proteosome degradation pathway,49,50 the biology is not well characterized. In this study, we identified several E3 ubiquitination ligases, such as E3 ubiquitin-protein ligases CNOT4, E3 SUMO-protein ligase PIAS2, E3 ubiquitin-protein ligase RNF216. BAG2, which has been reported to inhibit chaperone-assisted proteosome degradation pathway and one of the main components of CHIP,51,52 were also detectable. Thus, our data support that model that the protein level of NPM-ALK can be regulated by various proteins involved in the proteosome degradation pathway.
In conclusion, we used the recently developed HB-tag TAP protein purification, coupled with a sensitive LC-MS technique, to perform proteome-wide identification of NPM-ALK binding proteins. In addition to those previously described, we have identified a large number of novel proteins that are involved in various biological processes, many of which have not been previously described in association with NPM-ALK. Our results have revealed the functional diversity of NPM-ALK. We also believe that these new data have provided a number of new research directions in the study of NPM-ALK biology.
Supplementary Material
Acknowledgments
We thank Peter Kaiser (University of California Irvine) for providing the HB-tagged vector and Andy Lo of Dr. Li’s lab for valuable comments on the manuscript.
Footnotes
Address reprint requests to Dr. Liang Li, Department of Chemistry, University of Alberta, Edmonton, Alberta, Canada, T6G 2G2, E-mail: Liang.Li@ualberta.ca or Dr. Raymond Lai, Department of Laboratory Medicine and Pathology, Cross Cancer Institute and University of Alberta, Edmonton, Alberta, Canada, T6G 1Z2, E-mail: raymondl@cancerboard.ab.ca.
Supported by operating research grants from the Alberta Cancer Foundation and Canadian Institute for Health Research awarded to R.L. and the Natural Sciences and Engineering Research Council of Canada and Canada Research Chairs Program to L.L. L.L. is a Canada Research Chair in Analytical Chemistry. P.W. is a recipient of Alberta Cancer Board legacy graduate studentship.
F.W. and P.W. contributed equally to the work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
R.L. designed the experiments and participated in manuscript writing. L.L. combined the TAP with a sensitive LC-MS method developed in his lab for this mass spectrometry study. F.W. and P.W. were responsible for collection and analysis of data and manuscript writing. L.C.Y. was responsible for plasmid preparation and the transfection experiments.
References
- Delsol G, Ralfkiaer E, Stein H, Wright D, Jaffe ES. Jaffe ES, Harris NL, Stem H, Vardiman J, editors. lyon, France: IARC press; Pathology and Genetics Tumors of Haematopoietic and Lymphoid tissuesWorld Health Organization Classification of Tumors. 2001:pp. 230–235. [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene. ALK, to a nucleolar protein gene NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Drexler HG, Gignac SM, von Wasielewski R, Werner M, Dirks WG. Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia. 2000;14:1533–1559. doi: 10.1038/sj.leu.2401878. [DOI] [PubMed] [Google Scholar]
- Amin HM, Lai R. Pathobiology of ALK+ anaplastic large-cell lymphoma. Blood. 2007;110:2259–2267. doi: 10.1182/blood-2007-04-060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B, Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, Mandelli F, Mecucci C, Martelli MF. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007;92:519–532. doi: 10.3324/haematol.11007. [DOI] [PubMed] [Google Scholar]
- Piva R, Chiarle R, Manazza AD, Taulli R, Simmons W, Ambrogio C, D'Escamard V, Pellegrino E, Ponzetto C, Palestro G, Inghirami G. Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas. Blood. 2006;107:689–697. doi: 10.1182/blood-2005-05-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Albom MS, Lu L, Quail MR, Becknell NC, Weinberg LR, Reddy DR, Holskin BP, Angeles TS, Underiner TL, Meyer SL, Hudkins RL, Dorsey BD, Ator MA, Ruggeri BA, Cheng M. Anaplastic lymphoma kinase activity is essential for the proliferation and survival of anaplastic large-cell lymphoma cells. Blood. 2006;107:1617–1623. doi: 10.1182/blood-2005-08-3254. [DOI] [PubMed] [Google Scholar]
- Han Y, Amin HM, Franko B, Frantz C, Shi X, Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood. 2006;108:2796–2803. doi: 10.1182/blood-2006-04-017434. [DOI] [PubMed] [Google Scholar]
- Slupianek A, Nieborowska-Skorska M, Hoser G, Morrione A, Majewski M, Xue L, Morris SW, Wasik MA, Skorski T. Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res. 2001;61:2194–2199. [PubMed] [Google Scholar]
- Slupianek A, Skorski T. NPM/ALK downregulates p27Kip1 in a PI-3K-dependent manner. Exp Hematol. 2004;32:1265–1271. doi: 10.1016/j.exphem.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Turner SD, Yeung D, Hadfield K, Cook SJ, Alexander DR. The NPM-ALK tyrosine kinase mimics TCR signalling pathways, inducing NFAT and AP-1 by RAS-dependent mechanisms. Cell Signal. 2007;19:740–747. doi: 10.1016/j.cellsig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Marzec M, Kasprzycka M, Liu X, El-Salem M, Halasa K, Raghunath PN, Bucki R, Wlodarski P, Wasik MA. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–5614. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- Marzec M, Kasprzycka M, Liu X, Raghunath PN, Wlodarski P, Wasik MA. Oncogenic tyrosine kinase NPM/ALK induces activation of the MEK/ERK signaling pathway independently of c-Raf. Oncogene. 2007;26:813–821. doi: 10.1038/sj.onc.1209843. [DOI] [PubMed] [Google Scholar]
- Leventaki V, Drakos E, Medeiros LJ, Lim MS, Elenitoba-Johnson KS, Claret FX, Rassidakis GZ. NPM-ALK oncogenic kinase promotes cell-cycle progression through activation of JNK/cJun signaling in anaplastic large-cell lymphoma. Blood. 2007;110:1621–1630. doi: 10.1182/blood-2006-11-059451. [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Zhang H, Wang X, Larsen LS, Lathrop RH, Hatfield GW, Auer B, Huang L, Kaiser P. HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast. 2006;23:623–632. doi: 10.1002/yea.1380. [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Gregan J, Riedel CG, Petronczki M, Cipak L, Rumpf C, Poser I, Buchholz F, Mechtler K, Nasmyth K. Tandem affinity purification of functional TAP-tagged proteins from human cells. Nat Protoc. 2007;2:1145–1151. doi: 10.1038/nprot.2007.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang IF. Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics. 2006;6:6158–6166. doi: 10.1002/pmic.200600225. [DOI] [PubMed] [Google Scholar]
- Doucette A, Craft D, Li L. Protein concentration and enzyme digestion on microbeads for MALDI-TOF peptide mass mapping of proteins from dilute solutions. Analytical Chemistry. 2000;72:3355–3362. doi: 10.1021/ac000176j. [DOI] [PubMed] [Google Scholar]
- Doucette A, Craft D, Li L. Mass spectrometric study of the effects of hydrophobic surface chemistry and morphology on the digestion of surface-bound proteins. Journal of the American Society for Mass Spectrometry. 2003;14:203–214. doi: 10.1016/S1044-0305(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Wang N, Li L. Exploring the precursor ion exclusion feature of liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry for improving protein identification in shotgun proteome analysis. Anal Chem. 2008;80:4696–4710. doi: 10.1021/ac800260w. [DOI] [PubMed] [Google Scholar]
- Wang N, Xie CH, Young JB, Li L: Off-line two-dimensional liquid chromatography with maximized sample loading to reversed-phase LC-ESI tandem mass spectrometry for shotgun proteome analysis. Anal Chem (in press) [DOI] [PubMed] [Google Scholar]
- Gong Y, Wang N, Wu F, Cass CE, Damaraju S, Mackey JR, Li L. Proteome profile of human breast cancer tissue generated by LC-ESI-MS/MS combined with sequential protein precipitation and solubilization. J Proteome Res. 2008;7:3583–3590. doi: 10.1021/pr800229j. [DOI] [PubMed] [Google Scholar]
- Morgan R, Smith SD, Hecht BK, Christy V, Mellentin JD, Warnke R, Cleary ML. Lack of involvement of the c-fms and N-myc genes by chromosomal translocation t(2;5)(p23;q35) common to malignancies with features of so-called malignant histiocytosis. Blood. 1989;73:2155–2164. [PubMed] [Google Scholar]
- Wang N, Mackenzie L, De Souza AG, Zhong H, Goss G, Li L. Proteome profile of cytosolic component of zebrafish liver generated by LC-ESI MS/MS combined with trypsin digestion and microwave-assisted acid hydrolysis. J Proteome Res. 2007;6:263–272. doi: 10.1021/pr060367o. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Prediction of error associated with false-positive rate determination for peptide identification in large-scale proteomics experiments using a combined reverse and forward peptide sequence database strategy. J Proteome Res. 2007;6:392–398. doi: 10.1021/pr0603194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Amin HM, Medeiros LJ, Ma Y, Feretzaki M, Das P, Leventaki V, Rassidakis GZ, O'Connor SL, McDonnell TJ, Lai R. Inhibition of JAK3 induces apoptosis and decreases anaplastic lymphoma kinase activity in anaplastic large cell lymphoma. Oncogene. 2003;22:5399–5407. doi: 10.1038/sj.onc.1206849. [DOI] [PubMed] [Google Scholar]
- Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M, Levy DE, Inghirami G. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. 2002;21:1038–1047. doi: 10.1038/sj.onc.1205152. [DOI] [PubMed] [Google Scholar]
- Crockett DK, Lin Z, Elenitoba-Johnson KS, Lim MS. Identification of NPM-ALK interacting proteins by tandem mass spectrometry. Oncogene. 2004;23:2617–2629. doi: 10.1038/sj.onc.1207398. [DOI] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14–3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Benzinger A, Muster N, Koch HB, Yates JR, 3rd, Hermeking H. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol Cell Proteomics. 2005;4:785–795. doi: 10.1074/mcp.M500021-MCP200. [DOI] [PubMed] [Google Scholar]
- Hubinger G, Wehnes E, Xue L, Morris SW, Maurer U. Hammerhead ribozyme-mediated cleavage of the fusion transcript NPM-ALK associated with anaplastic large-cell lymphoma. Exp Hematol. 2003;31:226–233. doi: 10.1016/s0301-472x(02)01084-6. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Meierhofer D, Wang X, Huang L. Tandem affinity purification combined with mass spectrometry to identify components of protein complexes. Methods Mol Biol. 2008;439:309–326. doi: 10.1007/978-1-59745-188-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalarski CE, Haas W, Dephoure NE, Gygi SP. The effects of mass accuracy, data acquisition speed, and search algorithm choice on peptide identification rates in phosphoproteomics. Anal Bioanal Chem. 2007;389:1409–1419. doi: 10.1007/s00216-007-1563-x. [DOI] [PubMed] [Google Scholar]
- Ballif BA, Carey GR, Sunyaev SR, Gygi SP. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J Proteome Res. 2008;7:311–318. doi: 10.1021/pr0701254. [DOI] [PubMed] [Google Scholar]
- O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- Lau PJ, Kolodner RD. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. J Biol Chem. 2003;278:14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- Frouin I, Maga G, Denegri M, Riva F, Savio M, Spadari S, Prosperi E, Scovassi AI. Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1, and p21waf1/cip1. A dynamic exchange of partners. J Biol Chem. 2003;278:39265–39268. doi: 10.1074/jbc.C300098200. [DOI] [PubMed] [Google Scholar]
- Souza-Rodrigues E, Estanyol JM, Friedrich-Heineken E, Olmedo E, Vera J, Canela N, Brun S, Agell N, Hubscher U, Bachs O, Jaumot M. Proteomic analysis of p16ink4a-binding proteins. Proteomics. 2007;7:4102–4111. doi: 10.1002/pmic.200700133. [DOI] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, Hung MC. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- Stoklosa T, Poplawski T, Koptyra M, Nieborowska-Skorska M, Basak G, Slupianek A, Rayevskaya M, Seferynska I, Herrera L, Blasiak J, Skorski T. BCR/ABL inhibits mismatch repair to protect from apoptosis and induce point mutations. Cancer Res. 2008;68:2576–2580. doi: 10.1158/0008-5472.CAN-07-6858. [DOI] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin’s lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol Cell Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Bonvini P, Gastaldi T, Falini B, Rosolen A. Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), a novel Hsp90-client tyrosine kinase: down-regulation of NPM-ALK expression and tyrosine phosphorylation in ALK(+) CD30(+) lymphoma cells by the Hsp90 antagonist 17-allylamino,17-demethoxygeldanamycin. Cancer Res. 2002;62:1559–1566. [PubMed] [Google Scholar]
- Bonvini P, Dalla Rosa H, Vignes N, Rosolen A. Ubiquitination and proteasomal degradation of nucleophosmin-anaplastic lymphoma kinase induced by 17-allylamino-demethoxygeldanamycin: role of the co-chaperone carboxyl heat shock protein 70-interacting protein. Cancer Res. 2004;64:3256–3264. doi: 10.1158/0008-5472.can-03-3531. [DOI] [PubMed] [Google Scholar]
- Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell. 2005;16:5891–5900. doi: 10.1091/mbc.E05-07-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, Takayama S, Younger JM, Ren HY, Cyr DM, Patterson C. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280:38673–38681. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.