Abstract
Hallmarks of the pathogenesis of autoimmune encephalomyelitis include perivascular infiltration of inflammatory cells into the central nervous system, multifocal demyelination in the brain and spinal cord, and focal neuronal degeneration. Optimal treatment of this complex disease will ultimately call for agents that target the spectrum of underlying pathogenic processes. In the present study, Fn14-TRAIL is introduced as a unique immunotherapeutic fusion protein that is designed to exchange and redirect intercellular signals within inflammatory cell networks, and, in so doing, to impact multiple pathogenic events and yield a net anti-inflammatory effect. In this soluble protein product, a Fn14 receptor component (capable of blocking the pro-inflammatory TWEAK ligand) is fused to a TRAIL ligand (capable of inhibiting activated, pathogenic T cells). Sustained Fn14-TRAIL expression was obtained in vivo using a transposon-based eukaryotic expression vector. Fn14-TRAIL expression effectively prevented chronic, nonremitting, paralytic disease in myelin oligodendrocyte glycoprotein-challenged C57BL/6 mice. Disease suppression in this model was reflected by decreases in the clinical score, disease incidence, nervous tissue inflammation, and Th1, Th2, and Th17 cytokine responses. Significantly, the therapeutic efficacy of Fn14-TRAIL could not be recapitulated simply by administering its component parts (Fn14 and TRAIL) as soluble agents, either alone or in combination. Its functional pleiotropism was manifest in its additional ability to attenuate the enhanced permeability of the blood-brain barrier that typically accompanies autoimmune encephalomyelitis.
Despite a steadily expanding set of treatment options for multiple sclerosis (MS), there remains a pressing need for more effective therapeutic agents to address this debilitating autoimmune disorder of the central nervous system (CNS). Although the precise etiology of MS is unknown, key features of its pathogenesis and clinical evolution are emerging.1,2,3 Among various immune cellular effectors that have been implicated, pathogenic T cells loom large as pivotal drivers of the disease. As a consequence, various therapeutic paths are converging on T effectors as targets, with complementary goals of blocking their activation and re-activation, eliminating them from the larger T-cell reservoir, and interfering with their transit to sites of pathogenesis within the CNS.
A complex interplay of positive and negative intercellular signals regulates activation and maintenance of T-cell effector functions. Proteins of the tumor necrosis factor (TNF) superfamily figure prominently in this matrix of signals, bridging various cells of the immune system, as well as to cells of other organ systems. In so doing, TNF superfamily members contribute to both tissue homeostasis and pathogenesis via effects on cell survival and death, cellular differentiation, and inflammation.4,5 From the standpoint of autoimmune pathogenesis, two especially interesting members of the TNF superfamily are the cell surface ligands TWEAK (TNF-related weak inducer of apoptosis)6,7 and TRAIL (TNF-related apoptosis-inducing ligand).8,9,10,11 TWEAK, a TNF superfamily ligand, and its counter-receptor Fn14 (fibroblast growth factor-inducible 14-kDa protein) are expressed in a range of immune and nonimmune cell types. TWEAK, which is expressed on cells such as macrophages, dendritic cells, NK cells, endothelial cells, microglial cells, and astrocytes,6,12,13 stimulates proliferation of astrocytes and endothelial cells, as well as production of various inflammatory cytokines, chemokines, and adhesion molecules.14,15,16,17,18,19 Moreover, the TWEAK:Fn14 signaling axis has pro-inflammatory effects that go beyond promoting cell proliferation and cytokine production, some of which tie into autoimmune pathogenesis. TWEAK, whose endogenous expression is elevated in the CNS during experimental autoimmune encephalitis (EAE), a murine model for MS, increases the permeability of the neurovascular unit,20 contributing in this way to perivascular inflammatory cell infiltration. Moreover, TWEAK has pro-angiogenic activity,21 which is of interest given the association between angiogenesis and autoimmune pathogenesis.22 TWEAK increases EAE severity and associated neurodegeneration,14,23,24 and circulating TWEAK levels are significantly increased in patients with MS and other chronic inflammatory diseases.6 The induction of inhibitory anti-TWEAK or Fn14 antibody (Ab) in vivo, via vaccination with the extracellular domains of either TWEAK or Fn14, ameliorates EAE manifestations in rat and mouse models.25
TRAIL, a TNF superfamily ligand, binds to several different cognate TNF receptor superfamily receptors, some activating and others decoy. The activating receptors in humans are TRAIL-R1(DR4), TRAIL-R2 (DR5), and osteoprotegrin, whereas in mice, the sole activating receptor is DR5. Virtually all cells of the immune system (including T cells, B cells, natural killer cells, dendritic cells, monocytes, and granulocytes) up-regulate surface TRAIL ligand and/or release soluble TRAIL in response to interferon and other activation signals. TRAIL receptors are primarily expressed on immune cells, such as activated T cells.8,26 Native TRAIL expression attenuates EAE, as is evident from experiments invoking genetic deletion of TRAIL (in TRAIL−/− knockout mice) or administration of TRAIL blocking agents [soluble TRAIL receptor (sDR5) or neutralizing anti-TRAIL mAb].27,28,29 Moreover, embryonic stem cell-derived dendritic cells with enforced co-expression of TRAIL and pathogenic myelin oligodendrocyte glycoprotein (MOG) protein suppress EAE induction.30 Interestingly, soluble TRAIL has emerged as a response marker in MS patients undergoing interferon (IFN)-β therapy,31 with those most likely to respond to treatment showing early and sustained soluble TRAIL induction after therapeutic cytokine administration.
Based on this intriguing constellation of activities associated with the TWEAK:Fn14 and TRAIL:TRAIL-R signaling axes, which impact immunological responses and inflammatory processes, we have now designed an Fn14-TRAIL fusion protein that bridges them. The Fn14 component of this fusion protein has the capacity to bind and block endogenous TWEAK, whereas the TRAIL ligand component, once anchored to TWEAK-bearing cells via the Fn14 bridge, can direct intercellular inhibitory signals to its cognate receptors on TRAIL receptor-bearing cells, such as activated T cells.
Fn14-TRAIL is in essence exchanging TWEAK pro-inflammatory signals into immunoinhibitory TRAIL-driven ones. However, in contrast to another fusion protein that alters intercellular signals, the trans signal converter protein CTLA-4-FasL,32,33,34,35 Fn14-TRAIL is designed to redirect an exchanged negative signal to third-party (TRAIL receptor-bearing) cells. Furthermore, by virtue of the highly pleiotropic functionality of the TRAIL:TRAIL-R and TWEAK:Fn14 signaling axes, Fn14-TRAIL has inherently greater potential for higher order functionality with a net anti-inflammatory output. The present study begins to explore this fusion protein’s functional repertoire by demonstrating Fn14-TRAIL’s capacity to ameliorate MOG-induced EAE.
Materials and Methods
Mice
Four- to six-week-old C57BL/6 female mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and were maintained under pathogen-free conditions at the University of Pennsylvania Animal Facility. All animal experiments were approved by the University of Pennsylvania Animal Care and Use Committee.
Reagents
Plasmids pT2/BH and pNEB193 UbC-SB11 were provided by Dr. Perry Hacket (University of Minnesota, Minneapolis, MN), and murine TRAIL and TWEAK cDNAs were obtained from Dr. Hideo Yagita (Juntendo University School of Medicine, Tokyo, Japan). The plasmid pMFneo was obtained from Dr. Herman Waldmann (University of Oxford, Oxford, UK). Mouse MOG38-50 peptide (GWYRSPFSRVVHL) was synthesized using F-moc solid phase methods and purified by high performance liquid chromatography at Invitrogen Life Technologies (Carlsbad, CA). Pertussis toxin was purchased from EMD Biosciences (San Diego, CA). The following reagents were purchased from BD Pharmingen (San Diego, CA): enzyme-linked immunosorbent assay (ELISA) Ab pairs for mouse interleukin (IL)-2, IL-4, IL-6, IFN-γ, and recombinant mouse IL-2, IL-4, IL-6, IFN-γ. An IL-17 ELISA Ab pair was obtained from Southern Biotechnology (Birmingham, AL), and recombinant mouse IL-17 was purchased from Biosource (Camarillo, CA). PE-anti-mouse TRAIL and PE-anti-mouse TWEAK were purchased from eBioscience (San Diego, CA). Recombinant TRAIL (Super Killer TRAIL) was purchased from Axxora Platform (San Diego, CA).
Plasmid Construction
Chimeric Fn14-TRAIL and Fn14-IgG1(mut) coding cassettes were constructed by polymerase chain reaction (PCR), using partially overlapping synthetic oligonucleotides. cDNA encoding amino acids 1 to 79 of murine Fn14 (Swiss-prot accession number Q9CR75) was joined to cDNA encoding either amino acids 118 to 291 of murine TRAIL (Swiss-prot accession number P50592) or a mutated human IgG1 Fc (Fcγ1) segment, respectively. For the latter, a cDNA encoding human Fcγ136 was modified by PCR-based site-directed mutagenesis, using oligonucleotides configured to mutate C220→S, C226→S, C229→S, N297→A, E233→P, L234→V, and L235→A. To express soluble TRAIL, cDNA encoding amino acids 118 to 291 of murine TRAIL was used. All of these cDNA segments were subcloned into a pMFneo eukaryotic expression vector downstream of an EF1-α promoter region. Coding sequence for luciferase was mobilized with HindIII and BamH1 from pTAL-Luc (BD Biosciences, San Jose, CA), and subcloned into the respective sites of pMFneo. cDNA encoding full-length murine TWEAK was generated by PCR and subcloned into the pcDNA3 eukaryotic expression vector (Invitrogen Life Technologies).
To generate a derivative Sleeping Beauty expression vector that incorporates within the same plasmid both transposon and transposase cassettes, a transposase coding sequence flanked upstream by a ubiquitin C promoter was generated by PCR from pNEB193 UbC-SB11 (base pairs 432 to 2958) and then ligated between the ApaI and XhoI sites of pT2/BH vector, which contains a transposon cassette. This new expression vector, incorporating both transposase and transposon expression cassettes, was designated pSBC21. Next, cDNAs corresponding to Fn14-TRAIL, soluble Fn14, Fn14-IgG1(mut), soluble TRAIL, or luciferase, each linked to the EF1-α promoter, were subcloned from their respective pMFneo expression constructs into the transposon cassette of pSBC21, downstream of the transposase expression module. All subcloned cDNAs were oriented in the same direction as the transposase.
Cell Culture and Transfection
Human 293 kidney cells and CHO cells were cultured in Dulbecco’s modified Eagle’s medium and Ham’s F-12, respectively, supplemented with 100 μg/ml of penicillin, 100 U/ml of streptomycin, and 2% and 10% heat-inactivated fetal bovine serum. 293 cells were transiently transfected with the Fn14-TRAIL, soluble Fn14, Fn14-IgG1(mut), and soluble TRAIL pMFneo expression plasmids, using LipofectAMINE reagent (Invitrogen Life Technologies). Proteins in conditioned media were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Western blot analysis. Anti-mouse Ab used for detecting Fn14 and TRAIL were purchased from eBioscience and R&D Systems (Minneapolis, MN), respectively. CHO cells were transiently transfected with a pcDNA3-based murine TWEAK expression construct. TWEAK expression on transfectants was verified by immunofluorescence and flow cytometry.
Induction and Disease Evaluation of EAE
EAE was induced according to a standard induction protocol.37 Briefly, female C57BL/6 mice were challenged with a total of 300 μg of MOG38-50 peptide (divided into two subcutaneous injections, one on each dorsal flank) in 0.1 ml of phosphate-buffered saline (PBS), emulsified in an equal volume of Complete Freund’s Adjuvant (CFA) containing 4 mg/ml of Mycobacterium tuberculosis H37RA (Difco, Detroit, MI). These mice were simultaneously injected intravenously with 100 ng of pertussis toxin in 0.2 ml of PBS. A second intravenous injection of pertussis toxin (100 ng/mouse) was given 48 hours later. Mice were examined daily for signs of EAE and scored as follows: 0, no disease; 1, tail paralysis; 2, hind limb weakness; 3, hind limb paralysis; 4, hind limb plus forelimb paralysis; 5, moribund or dead.
Cytokine and Proliferation Assays
For cytokine assays, splenocytes were cultured at 1.5 × 106 cells per well in 0.2 ml of Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum, in the presence or absence of different concentrations of MOG38-50 peptide, or 1 μg/ml of Con A (Sigma-Aldrich, St. Louis, MO). Conditioned media were collected 40 hours later, and cytokine concentrations were determined by quantitative ELISA using paired mAb specific for the corresponding cytokines per the manufacturer’s recommendations (BD Pharmingen). Proliferation assays were performed using 0.5 × 106 cells per well in 96-well plates. [3H] thymidine was added to the cultures at 48 hours, and cells were harvested 16 hours later. Radioactivity was determined using a flatbed β-counter (Wallac-Perkin Elmer, Waltham, MA).
Hydrodynamic Injection
Mice were injected with pSBC21 vector alone or pSBC21-based expression constructs incorporating Fn14-TRAIL, soluble Fn14, Fn14-IgG1(mut), soluble TRAIL, or luciferase coding sequences. Expression plasmids were dissolved in saline in a volume (in ml) equivalent to 10% of body weight (in g). The entire volume for each animal was injected within 5 seconds via tail veins, according to a published protocol.38 Retro-orbital blood samples were collected using heparinized glass capillaries. After centrifugation, plasma was recovered and kept at −20°C until ELISA assays were performed. For gene transfer experiments, plasmid concentrations were measured spectrophotometrically, and plasmid copy numbers were calculated by using the equation: pmol DNA = (μg DNA) × 106/333g/mol × (no. bp × 2).
Measurement of Recombinant Proteins in Serum
ELISA assays were performed in 96-well microtitration plates. For Fn14-TRAIL, soluble Fn14, and Fn14-IgG1(mut), purified anti-human/mouse Fn14/TWEAK receptor Ab from eBioscience was used as capture Ab; for soluble TRAIL, anti-mouse TRAIL Ab from R&D Systems was used as capture Ab. Detecting Abs were: biotin-anti-mouse TWEAK receptor Ab from eBioscience for Fn14; biotin-anti-mouse TRAIL Ab from eBioscience for Fn14-TRAIL and soluble TRAIL; anti-human IgG, Fcγ fragment-specific Ab from Jackson ImmunoResearch Laboratories (West Grove, PA) for Fn14-IgG1(mut).
Capture Ab diluted in coating buffer (0.1 mol/L carbonate, pH 8.2) was distributed in microtitration plates and incubated at 4°C overnight. After washing twice with 0.05% Tween-20 in PBS, wells were incubated for an additional 2 hours at room temperature with PBS-3% albumin to block nonspecific binding sites. After washing twice again, 100 μl of serum samples were added and incubated at 4°C overnight. After incubation, wells were rinsed four times and incubated for 1 hour with biotinylated detection Ab. For the enzymatic reaction, avidin peroxidase and TMB Microwell peroxidase substrate (KPL, Gaithersburg, MD) were applied sequentially.
In Vivo Bioluminescence Imaging
All of the imaging work was performed at the Small Animal Imaging Facility in the Department of Radiology at the University of Pennsylvania. Images were acquired at 5 hours, 24 hours, 5 days, 22 days, 34 days, 51 days, and 1 year after injection of the luciferase expression plasmid. At the time of imaging, mice were anesthetized with ketamine/xylazine. d-Luciferin (Biotium, Hayward, CA) was dissolved in saline and delivered via intraperitoneal injection before imaging. Mice were then placed in an imaging chamber in which the temperature was maintained at 33°C. Bioluminescent images were acquired using an in vivo imaging system (Xenogen Corp., Alameda, CA). Imaging parameters were field of view of 8 or 10 cm, exposure time of 4 minutes, number of binning 16, and f1/stop of 1. For display, the luminescent image (pseudocolor) was overlaid on a photographic image, which delineated the anatomical landmarks.
Measurement of Blood-Brain Barrier (BBB) Permeability
BBB permeability was assessed essentially as described,39 with some modifications. Briefly, on days 6 and 13 after MOG challenge, 4% Evans blue dye (Sigma-Aldrich) was injected into the tail veins of C57BL/6 mice. After 1 hour, animals were anesthetized and transcardially perfused with saline to remove intravascular dye. After euthanasia, spinal cords, cerebellums/brainstems, and brains were collected. For quantitative measurements, spinal cords were homogenized in 1 ml of PBS. Samples were centrifuged once at 15,800 × g for 30 minutes. Six hundred-μl aliquots of the supernatant were then collected and added to 600 μl of 100% TCA (Sigma-Aldrich). This solution was incubated overnight, and centrifuged at 15,800 × g for 30 minutes. Evans blue extravasation was quantified spectrophotometrically (excitation 630 nm and emission 680 nm) in the supernatants.
Preparation and Analysis of Infiltrating Cells from Spinal Cords
Single cell suspensions of spinal cords were prepared as described previously.40 Briefly, mice were sacrificed and spinal cords were removed, placed in ice-cold RPMI medium containing 27% Percoll, and pressed through a 70-μm Falcon cell strainer. The resulting cell suspension was brought to a volume of 50 ml with additional 27% Percoll, mixed, and centrifuged at 300 × g for 15 minutes. The pellet was kept on ice, while the myelin layer and supernatant were transferred to a new 50-ml tube, homogenized by shaking, and centrifuged again at 300 × g for 15 minutes. The cell pellets were then combined and washed three times in RPMI medium at 4°C. For flow cytometric analysis, single cell suspensions of recovered cells were incubated for 45 minutes with the following Abs: fluorescein isothiocyanate-anti-mouse-IFN-γ, PE-anti-mouse IL-10, APC-anti-mouse IL-17, APC-Alexa flour 750-anti-mouse CD4, Percp-cy5.5-anti-mouse CD8, and PE-cy7-anti-mouse CD69, all purchased from eBioscience.
Molecular Modeling of the Chimeric Fn14-TRAIL Protein
A three-dimensional model of the Fn14-TRAIL protein was generated using the crystal structure of TRAIL (pdb code: 1D0G)41 and a modeled Fn14 molecule. A three-dimensional model of the ligand binding domain of Fn14 was generated using MODELLER.42 Briefly, the starting model of the ligand binding domain of Fn14 was obtained based on the template structure of human TACI (1XU1) and BCMA (1XU2).43 The extended region of Fn14 was treated as a linker between Fn14 and TRAIL. To obtain a stereochemically and energetically favored model, the linker conformation was optimized by short molecular simulation studies using InsightII (Accelrys, Inc., San Diego, CA) as described before.44
Flow Cytometry and MTT Assays
Immunostaining was performed at 4°C with specified Ab suspended in PBS containing 0.5% bovine serum albumin and 0.05% sodium azide (NaN3). All flow cytometric analyses were performed on a FACS Calibur apparatus with Cell Quest software and dual laser (488 and 633 nm) excitation (BD Biosciences). The MTT assay was performed according to the manufacturer’s protocol (American Type Culture Collection, Manassas, VA).
Statistical Analysis
The Student’s t-test or Mann-Whitney U-test was used to determine the statistical significance of differences. A P value of <0.05 was considered to be statistically significant.
Results
Production of Functional Fn14-TRAIL Protein
Recombinant Fn14-TRAIL, along with related control proteins [soluble Fn14, Fn14-IgG1(mut), soluble TRAIL], were produced using a pMFneo eukaryotic expression system. The chimeric Fn14-TRAIL coding sequence linked the full extracellular domains of the Fn14 type I and TRAIL type II membrane proteins, thereby creating a hybrid soluble type I · type II fusion protein. To generate the Fn14-IgG1(mut) coding sequence, several amino acids within the human IgG1 component were mutated (see Material and Methods) to block FcγR binding (and consequent nonspecific depletion of lymphocytes) and to interfere with N-glycosylation (which is important for in vivo effector function of human IgG1).45 The various pMFneo-based expression constructs were transiently transfected into 293 cells, and expression and secretion of the respective proteins were demonstrated by Western blot analysis of conditioned media (Figure 1A).
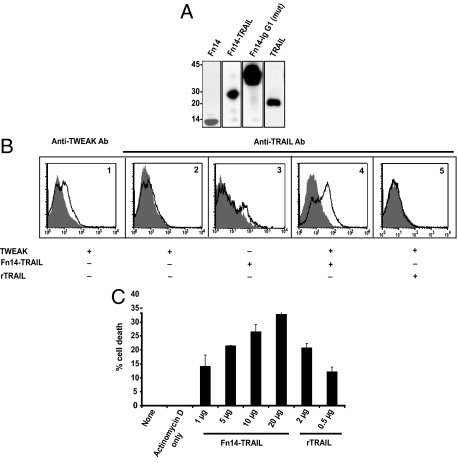
Figure 1.
Expression and functional analysis of Fn14-TRAIL. A: Western blot analysis was performed on conditioned media from 293 cells transfected with expression constructs for Fn14, Fn14-TRAIL, Fn14-IgG1(mut), and TRAIL. Observed bands were consistent with the expected sizes of 8.7 kDa, 27.5 kDa, 34.1 kDa, and 19.0 kDa, respectively. B: CHO cells were transiently transfected with a murine TWEAK cDNA expression construct, and after 48 hours, were incubated at 4°C with purified Fn14-TRAIL or rTRAIL in sodium azide-containing buffer. The presence of membrane-bound TWEAK on transfectants, and the binding of Fn14-TRAIL to them, was verified by flow cytometry, using anti-mouse TWEAK Ab (B1) and anti-mouse TRAIL Ab (B2–B5) as detecting Ab, respectively. B1: TWEAK is expressed on transfected CHO cells, as detected using anti-TWEAK Ab (solid black line) versus isotype control (filled histogram). B2: TRAIL is not detectable on CHO cells, when analyzed using anti-TRAIL Ab (solid black line) versus isotype control (filled histogram). B3 and B4: TRAIL epitopes are enhanced when Fn14-TRAIL is added to TWEAK-expressing, as opposed to TWEAK-negative, CHO cells. Anti-TRAIL Ab and isotype control are represented by solid black line and filled histogram, respectively. B5: TWEAK-transfected cells do not bind to anti-TRAIL Ab (solid black line) in the presence of rTRAIL. Isotype control is shown as filled histogram. C: L929 cells were cultured in flat-bottomed 96-well plates at 2 × 104 cells/well, in 100 μl of AIM-V medium. Sixteen hours later, actinomycin D was added to the cultures at 1 mg/well, and cells were cultured for another 2 hours. Fn14-TRAIL or rTRAIL, as positive control, was then added, and cultures were maintained for an additional 5 hours. The percentage of dead cells was determined by an MTT assay, as described in the Materials and Methods.
To validate the identity of expressed Fn14-TRAIL, we next documented its ability to bind to Fn14’s ligand, TWEAK. To this end, we transiently transfected CHO cells with a murine TWEAK cDNA expression construct (in the pcDNA3 vector), and after 48 hours, transfectants were incubated at 4°C with purified Fn14-TRAIL or soluble TRAIL. Immunofluorescence and flow cytometric analysis of these cells, using anti-mouse TWEAK and anti-mouse TRAIL as detecting Ab, showed significant binding of Fn14-TRAIL, but not soluble TRAIL, to cell surface TWEAK on transfectants (Figure 1B).
The functionality of the TRAIL component of Fn14-TRAIL was determined by evaluating its capacity to induce apoptosis in L929 cells, using an MTT assay. As shown in Figure 1C, Fn14-TRAIL induces apoptosis of L929 cells in a dose-dependent manner in the presence of actinomycin D. Recombinant TRAIL (Super Killer TRAIL) was used as a positive control in this experiment. Of note, no TWEAK was detected by immunofluorescence and flow cytometry on these L929 cells (not shown), arguing against the possibility that the Fn14 component of Fn14-TRAIL drives apoptosis through some kind of back-signaling through surface TWEAK.
Development of a Transposon-Based Expression System for Sustained in Vivo Expression of Fn14-TRAIL
To enable sustained in vivo expression of Fn14-TRAIL (and control proteins), we invoked the transposon-based Sleeping Beauty (SB) expression system. This system combines the advantages of plasmid-mediated gene delivery together with an ability to integrate into the chromosome and provide for sustained transgene expression. To optimize the efficiency of this expression system, we generated a derivative expression vector, designated pSBC21, that combines within a single plasmid both transposon (accommodating the transgene of interest) and transposase expression cassettes (Figure 2A). Because the relative expression level from the two cassettes is important, we screened a number of promoter combinations, and determined that a combination of UBC promoter (driving the transposase) and EF1-α promoter (driving the transposon cassette), arrayed in tandem, affords strong transgene expression (not shown).
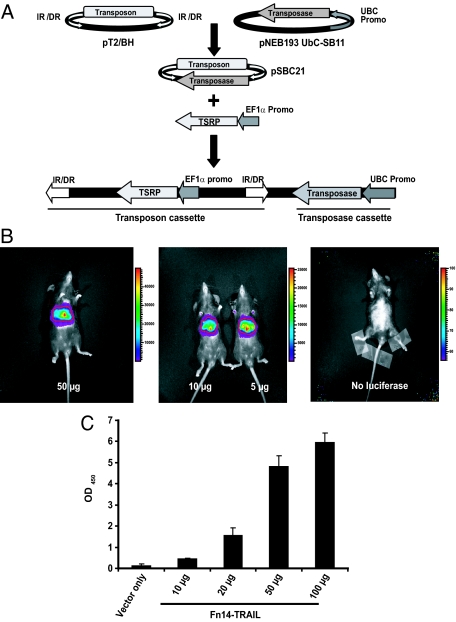
Figure 2.
Functional validation of the dual cassette pSBC21 vector. A: As schematically depicted, the pSBC21 plasmid incorporates in tandem a transposon cassette (with the EF1α promoter driving the trans signal redirecting protein or TSRP) and a transposase expression cassette (driven by the UBC promoter). B: Mice were hydrodynamically injected with either pSBC21 vector only (right) or the indicated concentrations of pLuciferase · SBC21. Bioluminescent images were acquired after intraperitoneal administration of D-Luciferin 22 days after plasmid injection. Color bars represent bioluminescent signal in radiance (p/second/cm2/sr). C: Serum levels of Fn14-TRAIL were determined by ELISA 20 days after the injection of 10, 20, 50, or 100 μg of Fn14-TRAIL · pSBC21 plasmid.
The functionality of this unique dual-cassette transposon/transposase vector derivative (with a UBC/EF1α promoter combination) was validated using a luciferase reporter. A pLuciferase · SBC21 plasmid, at varying concentrations, was administered by hydrodynamic injection to C57BL/6 mice. Hydrodynamic injection of transposon-based expression constructs provides for sustained gene expression in mouse hepatocytes in vivo.38 Bioluminescent images acquired after administration of luciferase’s substrate, d-Luciferin, revealed luciferase expression at 22 days (Figure 2B), with significant levels still detected after 6 months (not shown).
Having documented the functionality of our derivative expression vector using a luciferase reporter, we next invoked this vector for expressing Fn14-TRAIL, specifically asking whether levels of Fn14-TRAIL in serum correlate with the dose of injected pFn14-TRAIL · SBC21 plasmid. C57BL/6 mice (in experimental groups of four) were challenged with MOG38-50 peptide in CFA supplemented with M. tuberculosis, and on day 2 were each treated with a single hydrodynamic injection of empty pSBC21 vector or pFn14-TRAIL · SBC21 plasmid, in escalating doses (10, 20, 50, or 100 μg of plasmid). Serum levels of Fn14-TRAIL were measured by ELISA 20 days after plasmid administration, and a dose-dependent increase in serum Fn14-TRAIL levels was observed, starting with the 10-μg plasmid dose (Figure 2C). Of note, Western blot analysis 20 days after plasmid administration verified that the fusion protein in the serum of Fn14-TRAIL-treated mice was intact (not shown).
Fn14-TRAIL Suppresses MOG-Induced Autoimmune Encephalomyelitis
We next investigated the therapeutic potential of Fn14-TRAIL in a murine EAE disease model. To this end, a single encephalitogenic dose of MOG38-50 peptide was administered to C57BL/6 mice. Two days after peptide injection, we administered by hydrodynamic injection a single dose of pFn14-TRAIL · SBC21 plasmid (50 μg/mouse), or one of four control plasmids [pFn14 · SBC21, pFn14-IgG1(mut) · SBC21, pTRAIL · SBC21, and pSBC21]. Of note, because the expression cassettes represent relatively small components of these various plasmids, plasmid copy numbers for them (within the 50-μg aliquots) fell within a narrow range [Fn14 · pSBC21 = 11.89 pmol, Fn14-TRAIL · pSBC21 = 11.0 pmol, TRAIL · pSBC21 = 11.28 pmol, Fn14-IgG1(mut) · pSBC21 = 10.71 pmol]. By ELISA, we detected comparable serum levels of expressed proteins in animals hydrodynamically injected with each of the respective plasmids (Figure 3A).
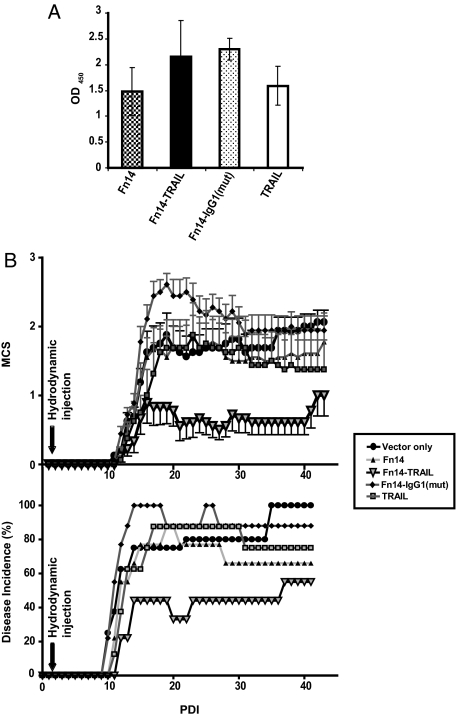
Figure 3.
Fn14-TRAIL suppresses MOG-induced autoimmune encephalomyelitis. A: Serum levels of Fn14, Fn14-TRAIL, Fn14-IgG1(mut), and TRAIL were measured by ELISA 20 days after hydrodynamically injecting 50 μg of the respective pSBC21-based expression plasmids. B: Eight to nine mice per group were challenged with MOG in CFA supplemented with M. tuberculosis as described in the Material and Methods. Forty-eight hours after MOG challenge, mice were hydrodynamically injected with 50 μg of the indicated pSBC21-based expression constructs. Individual mice were scored according to the clinical scale described in the Materials and Methods. Parameters evaluated include mean clinical score (B, top) and disease incidence (B, bottom). The difference between the Fn14-TRAIL-treated versus vector-only control group is statistically significant according to the Mann-Whitney U-test (P < 0.05), where the differences between the other groups shown and the vector-only group are not significant (see Table 1, which is based on the same experiment). The data shown are representative of at least three independent experiments.
Disease progression in the treated mice was monitored by both physical examination (Figure 3 and Table 1) and histological analysis of recovered spinal cords. Fn14-TRAIL expression significantly attenuated EAE manifestations, with decreases in both mean clinical scores (calculated throughout a 43-day period after MOG administration; Figure 3B, top) and cumulative clinical score (calculated throughout a 40-day period after MOG administration; Table 1). Maximum disease score was also significantly lower in the Fn14-TRAIL-treated group, as compared with the various controls (Table 1). Similarly, the disease score at day 35 was also significantly lower for the Fn14-TRAIL group (Table 1).
Table 1.
Clinical Features of EAE
| Treatment groups | Day of onset mean ± SD* | Incidence† | Maximum clinical score mean ± SEM | Maximum clinical score median (range) | Score (d35) mean ± SEM | Score (d35) median (range) | Cumulate score mean ± SEM‡ | Cumulate score median (range) |
|---|---|---|---|---|---|---|---|---|
| Vector (n = 8) | 12.80 ± 4.18 | 8/8 | 2.62 ± 0.42 | 3 (1–4) | 1.933 ± 0.29 | 2 (1–3.5) | 48.00 ± 9.78 | 47.75 (12–89) |
| Fn14 (n = 8) | 13.62 ± 2.72 | 6/9 | 2.44 ± 0.44 | 2.5 (1–4) | 1.750 ± 0.54 | 1 (1–3.5) | 44.00 ± 12.52 | 35.5 (2.5–96.5) |
| Fn14 TRAIL (n = 9) | 14.80 ± 4.7 | 4/9 | 1.22 ± 0.465 | 0.5§ (0–3) | 0.611 ± 0.261 | 0§ (0–2.5) | 18.60 ± 7.45 | 12§ (0–56.5) |
| Fn14-IgG (mut) (n = 9) | 11.66 ± 1.2 | 8/9 | 3.1 ± 0.351 | 3.5 (1–4) | 1.944 ± 0.306 | 2 (0–3.5) | 60.05 ± 7.44 | 66 (10.5–88) |
| TRAIL (n = 8) | 13.42 ± 2.2 | 6/8 | 2.0 ± 0.535 | 2 (0–4) | 1.438 ± 0.448 | 1 (0–3.5) | 42.06 ± 11.08 | 31 (0–82) |
In the calculations of parameters within this table, the scores of all mice are included except for day of onset in which only mice that exhibited signs of EAE were included.
Day of onset is the first day mice showed signs of EAE (after immunization).
Incidence is calculated as number of sick mice/number of mice immunized for each group at day 40.
Cumulative score is the sum of each mouse’s clinical score.
P < 0.05 compared to vector group. The Mann-Whitney U-test was used to determine the statistical significance of differences.
The therapeutic benefit of Fn14-TRAIL was also evident from an analysis of day of disease onset and disease incidence. Fn14-TRAIL extended the mean day of disease onset (14.8 ± 4.7 days), compared with control mice treated with empty vector (12.8 ± 4 days), and typically the latter mice developed EAE starting at ∼10 days after MOG peptide administration (Table 1). The mean day of disease onset for mice receiving pFn14 · SBC21, pFn14-IgG1(mut) · SBC21, pTRAIL · pSBC21 plasmids were 13.63 ± 2.72 days, 11.66 ± 1.2 days, and 13.42 ± 2.2 days, respectively. Although all expressed proteins reduced disease incidence to some extent, remarkably only 50% of Fn14-TRAIL-treated animals showed signs of disease during the course of this experiment (Figure 3B, bottom; Table 1). Of note, because there were no animal deaths in the experiment of Figure 3B (Table 1), the changes in disease incidence that were observed are linked to late onset of disease in some animals and recovery of others.
Fn14-TRAIL Is More Effective than Its Component Parts, in Combination
Having shown that neither of the components of Fn14-TRAIL, when administered as soluble agents one at a time, are as effective as Fn14-TRAIL in suppressing EAE, we next asked whether the Fn14-TRAIL fusion protein’s therapeutic efficacy can be recapitulated by administering soluble Fn14 and TRAIL proteins simultaneously. This was evaluated in the same EAE model. Two days after administering a single encephalitogenic challenge of MOG38-50 peptide to C57BL/6 mice, single doses of either pFn14-TRAIL · SBC21 plasmid (25 μg/mouse) or a mixture of pFn14 · SBC21 and pTRAIL · SBC21 plasmids (25 μg each/mouse) were hydrodynamically injected into the animals. Although Fn14-TRAIL significantly suppressed EAE as before, with decreases in both mean clinical scores (Figure 4, A and B) and cumulative mean clinical score (Figure 4C), the combination of soluble Fn14 and soluble TRAIL showed no significant therapeutic effect. Serum levels of these various proteins, expressed by hydrodynamic injection of the respective transposon-based expression plasmids, were comparable, as measured by ELISA (Figure 4D). Taken together, these data establish that Fn14-TRAIL has substantial therapeutic benefit in preventing EAE induction, and this effect cannot be replicated by simply administering this fusion protein’s two component elements as soluble agents in combination.
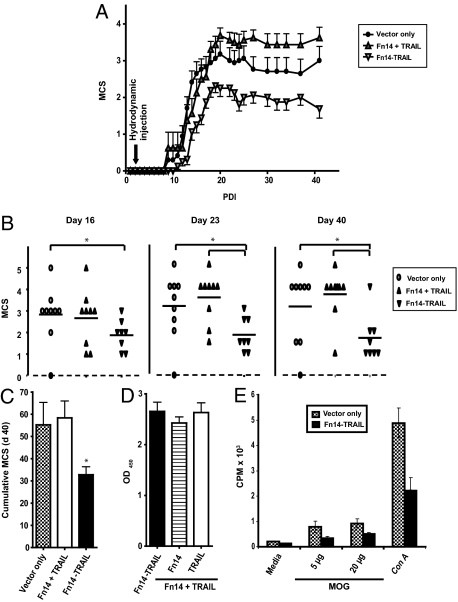
Figure 4.
Fn14 and TRAIL in combination do not achieve Fn14-TRAIL’s therapeutic efficacy. A--C: Forty-eight hours after MOG challenge, eight to nine mice per group were hydrodynamically injected with a single dose of Fn14-TRAIL · pSBC21 plasmid (25 μg/mouse) or a single dose of a mixture of Fn14 · pSBC21 and TRAIL · pSBC21 plasmids (25 μg each/mouse). Mean clinical scores for the indicated groups throughout 40 days (A), and on days 16, 23, and 40 (B) are shown. C: Cumulative MCS of indicated groups at day 40 is also shown. The difference between the Fn14-TRAIL-treated versus vector-only control group is statistically significant according to the Mann-Whitney U-test (*P < 0.05), where the differences between the other groups shown and the vector-only group are not significant. D: Serum levels of Fn14-TRAIL were determined by ELISA 30 days after the injection of combined Fn14 · pSBC21 (25 μg) + TRAIL · pSBC21 (25 μg), or Fn14-TRAIL · pSBC21 (25 μg). E: MOG-challenged mice that had been hydrodynamically injected with pSBC21 vector only or Fn14-TRAIL · pSBC21 were sacrificed 43 days after receiving the therapeutic agent. Splenocytes from each mouse were cultured in the presence or absence of different concentrations of MOG38-50 peptide, and proliferation was evaluated as described in the Materials and Methods. Results are shown as mean ± SD from a total of nine mice per group.
Fn14-TRAIL Blocks Proliferation and Differentiation of Autoreactive T Cells
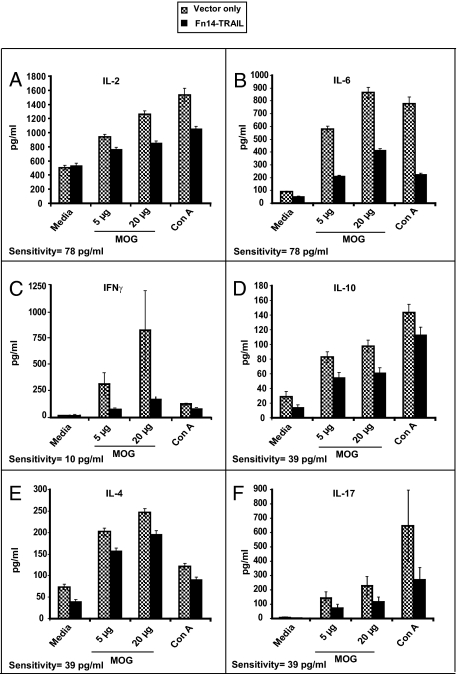
We next assessed the effect of Fn14-TRAIL on the proliferation and differentiation of myelin-specific T cells recovered from treated animals. To this end, splenocytes were recovered 43 days after MOG challenge from both Fn14-TRAIL-treated and control mice receiving vector only. These splenocytes were evaluated in vitro for their proliferation and cytokine production in response to MOG38-50 peptide. Splenocytes from control animals proliferated vigorously in response to MOG peptide (Figure 4E) and produced significant amounts of Th1 (IL-2 and IFN-γ), Th2 (IL-10, IL-4, and IL-6), and Th17 (IL-17) cytokines (Figure 5). In contrast, splenocytes from Fn14-TRAIL-treated animals proliferated to a lesser extent in response to MOG stimulation (Figure 4E) and produced significantly lower levels of these various cytokines (Figure 5). Taken together, these results suggest that both T-cell proliferation and the expression of an array of T-cell cytokines are attenuated by in vivo treatment with Fn14-TRAIL.
Figure 5.
Fn14-TRAIL · pSBC21 inhibits cytokine production in MOG-challenged mice. Mice (eight to nine per group) were challenged with MOG38-50 in CFA supplemented with M. tuberculosis and hydrodynamically injected with pSBC21 vector only or Fn14-TRAIL · pSBC21. Animals were sacrificed 43 days after receiving the therapeutic agent, and splenocytes from each mouse were cultured in the presence or absence of different concentrations of MOG38-50 peptide. Conditioned media were collected 40 hours later, a time point previously shown to be associated with maximal cytokine expression, and concentrations of the indicated cytokines were determined by ELISA (A: IL-2; B: IL-6; C: IFN-γ; D: IL-10; E: IL-4; F: IL-17). Sensitivity of the ELISA for each cytokine is indicated at the bottom of each respective panel.
Fn14-TRAIL Reduces Inflammation in the CNS
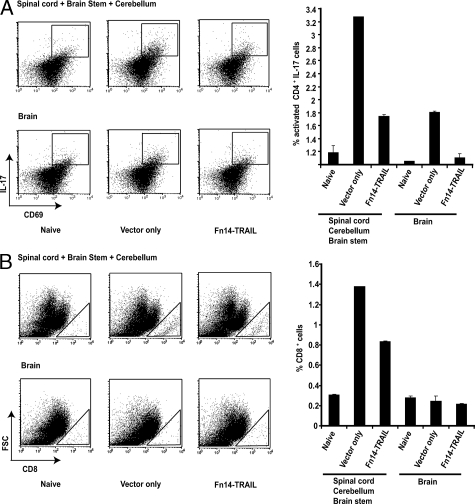
A key pathological feature of EAE is the infiltration of inflammatory cells into the CNS. We next determined Fn14-TRAIL’s effect on this inflammatory process. To this end, we recovered inflammatory cells from CNS tissues, and used immunofluorescence and flow cytometry to characterize these cells in terms of their expression of the early activation marker CD69, the inflammatory cytokine IL-17, and the lymphoid subset markers CD4 and CD8. This combination of markers allowed us to key in on both early-activated CD4+ IL-17-producing cells,46,47 as well as CD8+ T cells (which have been linked to later stages of EAE pathogenesis).48,49 The presence of these cell types was compared 7 days after MOG challenge in Fn14-TRAIL-treated versus vector-only-treated mice, looking separately at brain and spinal cord/cerebellum/brain stem. As shown in Figure 6A, MOG challenge increased the percentage of early-activated CD4+, IL-17-producing lymphocytes in spinal cord/cerebellum/brain stem from 1.2% (in naïve mice, with no MOG challenge) to 3.3%. Fn14-TRAIL treatment of MOG-challenged mice reduced this to 1.8%. By contrast, MOG challenge was associated with fewer of these cells in brain (1.9%), and Fn14-TRAIL treatment brought this down to 1.1%.
Figure 6.
Fn14-TRAIL · pSBC21 treatment reduces early activated CD4+ IL-17-producing cells and CD8+ cells in the CNS of MOG-challenged mice. MOG-challenged mice, treated with either pSBC21 vector only or Fn14-TRAIL · pSBC21 (three mice per group), or naïve mice (no MOG challenge), were sacrificed on day 7 after challenge, and flow cytometry was performed on mononuclear cells isolated from their brains or pooled spinal cords/cerebellums/brain stems. Live lymphocytes were gated based on forward and side scatter. A: Left: Early activated (CD69+) IL-17-producing cells, gated on CD4+ lymphocytes; right: depiction of percentage of CD4+CD69+ IL-17-producing cells. B: Left: CD8+ cells, gating on live lymphocytes; right: depiction of percentage of CD8+ lymphocytes. The data shown are representative of three independent experiments.
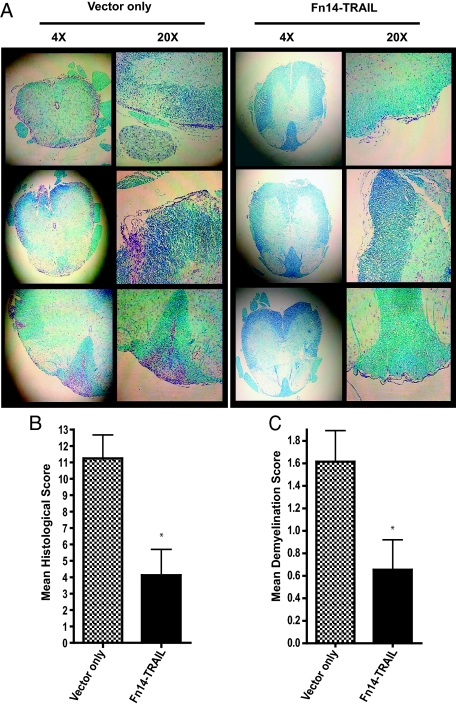
In the case of infiltrating CD8+ T cells, MOG challenge increased the percentage of these inflammatory cells in spinal cord/cerebellum/brain stem from 0.3% (in naïve mice, with no MOG challenge) to 1.3%, and Fn14-TRAIL reduced this to 0.8% (Figure 6B). In contrast, for brain, MOG challenge did not lead to an increase in CD8+ T cells. This lymphoid subset analysis was extended to encompass a broader range of cytokines, encompassing both Th1 and Th2 types. Because the effect of Fn14-TRAIL treatment is more pronounced in spinal cord (as opposed to brain), we focused on inflammatory cells derived from this site. In this experiment, the absolute number of inflammatory cells was measured, along with the percentage of early-activated CD4+ and CD8+ cells and of IFN-γ-, IL-17-, and IL-10-expressing cells, in the spinal cords of Fn14-TRAIL- versus vector-treated EAE mice on days 7 and 14 after MOG challenge. On day 7, all of these cell categories were markedly reduced in the Fn14-TRAIL-treated group (Figure 7, A–D). At day 14 (peak of the disease), the Fn14-TRAIL-associated reduction in absolute numbers of cytokine-expressing cells was still manifest (Figure 7, E and F). These findings with inflammatory cells recovered from spinal cords were consistent with histopathological examination of spinal cord tissues recovered 43 days after MOG challenge. Whereas control vector-only-treated animals uniformly displayed multiple inflammatory foci within their spinal cords, Fn14-TRAIL-treated mice exhibited a dramatic reduction of inflammatory cell infiltration and demyelination in their spinal cords (Figure 8, A–C).
Figure 7.
Fn14-TRAIL · pSBC21 treatment reduces the number of activated and cytokine-producing cells in spinal cords of MOG-challenged mice on days 7 and 14 of disease. Mice were sacrificed on days 7 or 14 of disease, and flow cytometry was performed on the mononuclear cells isolated from spinal cords of pSBC21 vector-only or Fn14-TRAIL · pSBC21-treated mice (three mice per group), as described in the Material and Methods. Live lymphocytes were gated based on forward and side scatter. CD8 and CD69 markers were measured on gated lymphocytes, and analysis of cells producing each of the cytokines shown was for gated CD4+ lymphocytes. Absolute cell numbers were calculated by multiplying the total number of live mononuclear cells times the percentage of each indicated cell type (determined by flow cytometry). Data are representative of three independent experiments with similar results. A and B: Percentage and absolute number of activated (CD69+) inflammatory cells on day 7 after MOG challenge. C and D: Percentage and absolute number of CD4+ and CD8+ cells, and of IFN-γ-, IL-17-, and IL-10-expressing cells (amid activated CD69+ cells or the total cell pool) on day 7 after MOG challenge. E and F: Percentage and absolute number of CD8+ cells and of IFN-γ-, IL-17-, and IL-10-expressing cells on day 14 after MOG challenge.
Figure 8.
Fn14-TRAIL · pSBC21 treatment reduces spinal cord inflammation in MOG-challenged mice. MOG-challenged mice (eight to nine per group), hydrodynamically injected with SBC21 vector only or Fn14-TRAIL · pSBC21, were perfused transcardially with PBS and 10% formalin phosphate. Spinal cords were removed, cut into six pieces, embedded in paraffin, transversely sectioned at 5 μm, and stained with luxol fast blue and cresyl violet. A: Left: Representative spinal cord sections of vector-only-treated mice at ×4 and ×20 magnifications. Right: Representative spinal cord sections of Fn14-TRAIL-treated mice at ×4 and ×20 magnifications. B: Tissue sections from each of the six spinal cord segments were analyzed for each animal. Scores of inflammation were assigned to individual sections based on the following criteria: 0, no inflammation; 1, <5%; 2, 5 to 20%; 3, 20 to 50%; and 4, >50% of the white matter is infiltrated by leukocytes. For each mouse, the histological score is the sum of scores from the six spinal cord sections. C: Demyelination scores were assigned to individual sections for every mouse, and these represented an average percentage of white matter demyelination in each group. The scoring was as follows: 0, no demyelination; 1, 0 to 10% demyelination; 2, 10 to 25% demyelination; 3, 25 to 50% demyelination; 4, >50% demyelination. The differences between the Fn14-TRAIL-treated group versus the control group are statistically significant according to the Mann-Whitney U-test (*P < 0.05).
Fn14-TRAIL Attenuates BBB Permeability
TWEAK is known to increase the permeability of the neurovascular membrane unit by inducing MMP-9 (matrix metalloproteinase-9) expression.20 We posited that the decreased infiltration of inflammatory cells into the CNS seen in Fn14-TRAIL-treated mice could be a consequence, at least in part, of a reduction in TWEAK-dependent enhancement of blood brain barrier (BBB) permeability. BBB integrity was evaluated by a conventional approach, according to which CNS penetration of intravenously introduced Evans blue dye is monitored.39 We measured Evans blue in the CNS of EAE mice before (6 days after MOG administration) or during (13 days after MOG administration) the peak of the disease. Dye was quantitated in homogenates of various dissected CNS structures. Whereas there was no significant difference in detectable dye between brains of control vector-only and Fn14-TRAIL-treated animals at either time point (Figure 9, A and B), significantly more dye was detected for the vector-only-treated animals in the other CNS structures (spinal cord, cerebellum, and brain stem) at both days 6 (Figure 9A) and 13 (Figure 9B). Of note, this brain versus spinal cord/cerebellum/brain stem difference in BBB permeability mirrored the difference between these tissues in the accumulation of cytokine-expressing inflammatory cells after MOG challenge (Figure 6).
Figure 9.
Fn14-TRAIL treatment reduces BBB permeability. MOG-challenged mice (7 to 10 mice per group), hydrodynamically injected with SBC21 vector only or Fn14-TRAIL · pSBC21, were injected with Evans blue dye on days 6 (A) or 13 (B) after MOG challenge. Evans blue was quantitatively analyzed in extracts of the indicated tissues, as described in the Materials and Methods. The results represent the specific absorbance of Evans blue at 630 nm calculated as ng/g of tissue. In C, concentrations of absorbed dye in spinal cords of vector-only versus Fn14-TRAIL-treated mice with matched mean clinical scores (0 or 1) on day 13 after MOG challenge. *P < 0.05, as determined by Student’s t-test.
We next correlated the concentration of dye penetrated into the spinal cords with the respective EAE mean clinical scores on the day of dye application. Significantly, even for mice with the same EAE mean clinical scores (0 or 1) on day 13, control vector-only-treated mice showed higher concentrations of dye in their spinal cords compared with Fn14-TRAIL-treated mice (Figure 9C). This finding, coupled with the findings of reduced inflammatory cell infiltration in Fn14-TRAIL-treated spinal cords, supports the notion that Fn14-TRAIL attenuates infiltration of inflammatory cells across the endothelial BBB by suppressing the progressive increase in BBB permeability that accompanies encephalomyelitis.
Molecular Model of the Chimeric Fn14-TRAIL Protein
We next asked whether Fn14-TRAIL’s significant therapeutic efficacy might stem, at least in part, from the way in which it engages and bridges TWEAK ligand and TRAIL receptor (DR5) molecules. To this end, we modeled the Fn14-TRAIL protein, both as a monomer (Figure 10A) and as a trimer (Figure 10, B and C). Moreover, we visualized the putative interaction of trimeric Fn14-TRAIL at its opposite ends with TWEAK and DR5, respectively (Figure 10D). Significantly, the modeled TWEAK:Fn14-TRAIL:DR5 complex showed that: i) the TWEAK-binding domains of Fn14, when forced into an artificial trimeric configuration by the chimerized, naturally trimeric TRAIL component, are favorably positioned to interact with their cognate partners within trimeric TWEAK; ii) the DR5-binding domains of trimeric TRAIL are favorably positioned to interact with their cognate partners within trimeric DR5; and iii) the carboxy-terminus of the Fn14 component within the fusion protein serves as a surrogate linker that is sufficiently rigid to keep the Fn14 and TRAIL domains apart, with no propensity for collapsing. Interestingly, the ligand binding domains of Fn14 and TRAIL are separated by ∼60 Å, raising the possibility that this fusion protein could act like a spacer between the interacting cells and limit local cell-to-cell contact. Taken together, the modeling analysis verified that FN14-TRAIL can indeed simultaneously engage both TWEAK and DR5 on opposing cells. Moreover, the enforced Fn14 neo-trimer assumes a configuration that allows for binding to the natural TWEAK trimer, perhaps creating a particularly stable higher order structure.
Figure 10.
Structural model of the TWEAK:Fn14-TRAIL;DR5 complex. Three-dimensional models, generated as described in the Materials and Methods, are shown for Fn14-TRAIL as a monomeric unit (A, Fn14 in blue and TRAIL in white), the Fn14-TRAIL trimer (B, as ribbon model; C, as space-filling model), and the TWEAK:Fn14-TRAIL:DR5 complex (D, with Fn14-TRAIL as space-filling model, TWEAK trimer as ribbon model at top, and DR5 trimer as ribbon model at bottom).
Discussion
Many immunotherapeutic proteins feature a capacity to bind and block molecules essential for immunological responses, for example, co-stimulators, pro-inflammatory cytokines, and adhesins. However, their efficacy is limited by the inherently transient nature of passive blockade, as well as by functional redundancies that often permit bypass of individual blocked signals. This limitation beckons the development of new classes of immunotherapeutic proteins with higher order functionality.50 In this vein, we previously introduced trans signal converter proteins (TSCPs) as a new class of immunomodulatory fusion proteins that combine passive blockade with active inhibition. Such proteins alter the cross talk between two interacting cells, for example, converting an activating APC-to-T cell signal into an inhibitory one. The paradigmatic TSCP for us was CTLA-4-FasL,32 and its efficacy, along with the overall robustness of the intercellular signal conversion concept, have been documented in the contexts of both alloimmunity33,34,35,51 and autoimmunity.52
In the present study, we describe yet another type of fusion protein that can impact intercellular signaling. This one exchanges intercellular signals, and furthermore, redirects converted neo-signals to other cell types for therapeutic benefit. Hence, such proteins might be designated trans signal redirecting proteins (TSRPs). The TSRP described here, Fn14-TRAIL, bridges two prominent intercellular signaling axes: TWEAK-to-Fn14 and TRAIL-to-TRAIL receptor (TRAIL-R; in mice, DR5). Pro-inflammatory TWEAK signals, emanating from a range of TWEAK-bearing immune and nonimmune cell types, are converted by Fn14-TRAIL into inhibitory TRAIL ones. Significantly, the opposing (anti-inflammatory TRAIL) neo-signals are by definition turning the TWEAK-bearing cell’s attention, and in effect redirecting signaling, from Fn14+TRAIL-R− to Fn14−TRAIL-R+ activated T cells driving autoimmune pathogenesis. The functional pleiotropism of the TWEAK:Fn14 and TRAIL:TRAIL-R signaling axes, especially the former, spanning an array of immune and nonimmune cell types,6,7,8,53,54 inheres in Fn14-TRAIL extensive functional possibilities for impacting and expanding cellular networking.
Key findings of the present study include: i) both building blocks of Fn14-TRAIL retain their functionality after chimerization, as documented in vitro by the fusion protein’s ability to bind to enforced TWEAK on transfected CHO cells (through its Fn14 end) and induce apoptosis in actinomycin D-treated, TRAIL-R+ L929 cells (through its TRAIL end); ii) Fn14-TRAIL attenuates MOG-induced EAE in vivo (as documented by a range of classical disease metrics for this model), and significantly, displays therapeutic efficacy that is substantially greater than that of either of its component parts when used as soluble derivatives [soluble Fn14, Fn14-IgG1(mut), soluble TRAIL], either in isolation or in combination; and iii) Fn14-TRAIL, as a TWEAK blocker, can reduce BBB permeability and inflammatory cell transit into the CNS, possibly contributing further to the high therapeutic efficacy of this fusion protein in the autoimmune encephalomyelitis model.
A combination of considerations—evolutionary conservation, breadth of tissue distribution, and functional pleiotropism—argue for the larger biological significance of the TWEAK-to-Fn14 signaling axis and make it an intriguing target for therapeutic modulation. This signaling axis is evolutionarily ancient. Both TWEAK and Fn14 are found in diverse species, as early as zebrafish, and an Fn14-like sequence has been found in the protochordate Branchiostoma floridae, suggesting that the origin of this pathway may have preceded vertebrate evolution.6 Diverse reports, albeit nonsystematic, have pointed to the widespread expression of both TWEAK and Fn14.7 Significantly, both TWEAK and Fn14 tissue expression are modulated by pathogenic processes. For example, TWEAK expression is increased locally in target tissues in the settings of acute injury, inflammatory disease, and cancer, all of which are associated with infiltration of inflammatory cells and/or activation of resident innate immune cell types. Similarly, Fn14 is induced dramatically in injured and diseased tissues, such as in spinal cord after induction of autoimmune encephalomyelitis.55
The functional pleiotropism of the TWEAK-to-Fn14 signaling axis is of particular interest here with respect to inflammation. This signaling axis drives a number of pro-inflammatory events, including direct induction of certain cytokines and chemokines (IL-6, IL-8, RANTES, IP-10, MCP-1), and potentiation of some others (TNF-α, IL-1β).14,16 TWEAK-to-Fn14 signaling also induces matrix metalloproteinases (MMP-1, MMP-9), and promotes prostaglandin E2 secretion.16,17,18,19,56 Furthermore, it has other biological effects that indirectly contribute to inflammation, such as promoting angiogenesis.21 Of note, there is a single published study which has reported that Tweak-null mice have an enhanced innate inflammatory response.13 However, a recent review points out that independently generated Tweak-null mice do not exhibit this in vivo immune phenotype, and the preponderance of published data certainly affirm TWEAK’s broad set of pro-inflammatory activities.7
The impact of the TRAIL:TRAIL-R signaling axis on inflammatory events is primarily via inhibition of a range of immune functions—T-cell cycle progression, autoreactive T-cell proliferation, pro-inflammatory cytokine production, Ab production, and inflammatory reactions.27,28,29,30,57,58,59 This range of immunoinhibitory effects clearly extends beyond CD4+ T cells per se, encompassing CD8+ T cells, B cells, monocytes, and dendritic cells.8,60,61,62,63 The precise nature of TRAIL’s effects on these various cell types in not fully resolved. For instance, although there is evidence that TRAIL can induce apoptotic cell death in macrophages and neutrophils under certain conditions,64,65,66 its pro-apoptotic activity is less clear for T cells.67,68 Interestingly, there is evidence that cell surface-anchored TRAIL, when expressed on dendritic cells primed with MOG, induces regulatory T cells.69 Furthermore, physiological changes in TRAIL receptor expression may translate into special advantages for immunotherapeutic fusion proteins incorporating TRAIL ligand. TRAIL receptor is up-regulated on T cells on activation,26,70 and thus, TRAIL-driven effects of Fn14-TRAIL are expected to be focused on activated, as opposed to naïve, lymphoid effectors.
An important dimension to the present study is the effect of Fn14-TRAIL on BBB permeability in the inflammatory setting. TWEAK contributes actively to regulation of BBB permeability and leukocyte recruitment into the brain in inflammatory conditions, and blockade of TWEAK can interfere with inflammatory recruitment into the CNS,55 and neuronal loss.24 Specifically, there is direct evidence that TWEAK increases the permeability of the neurovascular unit through MMP-9 expression.20 Moreover, endothelial cells secrete CCL2, MCP-1, and IL-8, as well as up-regulate ICAM-1 and E-selectin, in response to TWEAK treatment in vitro.15 Interestingly, the CCL2 chemokine and its receptor CCR2 have been shown to play crucial nonredundant roles in the pathogenesis of EAE.71 CCL2 expression in general correlates with the severity of inflammation and has been shown to increase during the course of chronic relapsing EAE.72 Hence, Fn14-TRAIL, by blocking TWEAK, could potentially impact more than one step involved in inflammatory cell transit into the CNS.
A key observation was a reduction in the absolute number of inflammatory cells, including early activated T cells, in the spinal cords of Fn14-TRAIL-treated animals on day 7 after immunization. Moreover, later on at the peak of disease, the absolute number of inflammatory cytokine-expressing cells remained lower for animals treated with the fusion protein. CNS inflammation correlated with BBB permeability in an intriguing way. Not only did spinal cords, brain stems, and cerebellums of Fn14-TRAIL-treated mice show attenuated BBB permeability, but these mice exhibited less BBB permeability even when compared with vector-only-treated mice with matched disease scores. This dovetails nicely with our observation that Fn14-TRAIL-treated mice, even when compared with vector-only-treated mice with comparable disease scores at the onset of disease, stay healthier in the later stages of disease and exhibit lower cumulative disease scores. Hence, although this EAE model does not permit one to tease apart the contributions of lymphoid cell suppression from reduced lymphoid cell transit into the CNS, the data certainly point to some contribution of the latter to suppression of inflammation that is likely attributable to Fn14-TRAIL antagonism of TWEAK.
A pivotal finding in the present study is that the fusion protein proved to be far more therapeutically active in vivo than its component protein parts (Fn14 and TRAIL), when the latter are administered as soluble entities either alone or in combination. The limited nature of the literature bearing on these particular components when used as soluble agents makes it difficult to compare our findings. Although one group has reported that Ab blockade of TWEAK improves the clinical course of EAE and decreases CNS-specific inflammation,23 the only study invoking Fn14 per se as a blocking agent involved a full-length Fn14, apparently encompassing the transmembrane and cytoplasmic regions of a rat Fn14 protein.25 In the case of TRAIL, there is again only a single report in the literature dealing with human TRAIL and its use as a soluble agent (in this case, produced in bacteria) in an EAE therapeutic context.29 As alternatives, however, surrogates for soluble TRAIL have been invoked, namely, agonistic anti-DR5 Ab27,73 and dendritic cells with enforced TRAIL expression.30 In each of these latter cases, the TRAIL receptor trigger may be achieving a higher effective valency. This, along with dosing factors, could explain why soluble TRAIL in our hands, expressed by in vivo gene transfer, displayed no therapeutic benefit when administered as a control alongside Fn14-TRAIL.
Chimerizing the Fn14 and TRAIL components within a single fusion protein may elicit special and sometimes complementary functional properties that could augment the efficacy of the two components, mask negative effects of one or the other, which might otherwise limit their therapeutic utility, and confer particular advantages in the EAE context. These include the following:
1) By linking Fn14, a protein that is naturally monomeric, to TRAIL (naturally a trimer), one is in effect creating a trimeric variant of Fn14. As it turns out, predictive structural modeling suggests that this Fn14 neo-trimer could neatly dock with Fn14’s naturally-trimeric TWEAK counterreceptor (Figure 10). Moreover, at the other end of the Fn14-TRAIL fusion protein, the trimeric TRAIL is docking with the trimeric DR5 receptor for TRAIL. Hence, these structural complementarities, generated de novo by chimerizing monomeric Fn14 to trimeric TRAIL, may actually be creating a uniquely stable TWEAK-Fn14-TRAIL-DR5 molecular bridge allowing for greater potency in blocking TWEAK and driving DR5 on cells facing each other.
2) TRAIL ligand is in effect being anchored to the cell surface via an Fn14 bridge, which serves to augment its effective valency. In a sense, this is replicating the situation with dendritic cell neo-expressing surface TRAIL, which have been shown to be effective in suppressing EAE.30
3) There is evidence that TRAIL-mediated inhibition may function primarily at the EAE priming phase (during T-cell triggering and expansion),29 whereas TWEAK-blockade’s benefit may be more focused on downstream pro-inflammatory events (including preventing breakdown of the BBB).55 Thus, by combining the two, one has a single agent that could in principle impact both priming and later phases of the disease.
4) One group has reported that TRAIL may contribute to death of neurons after the priming phase,70,73 in keeping with previous studies showing that both primary human neurons74 and oligodendrocytes75 are susceptible to TRAIL-induced cell death. By coupling Fn14 to TRAIL, one is providing an agent that sustains the integrity of the BBB (via TWEAK blockade) and thereby mitigates TRAIL’s access to the CNS. Moreover, because TWEAK has been reported to trigger neuronal cell death,24 Fn14-TRAIL would be expected to protect neurons by blocking this TWEAK activity as well.
5) During EAE, the number of vessels correlates with both disease scores and pathological measures for inflammation, leukocyte infiltration, and demyelination.22 Because TWEAK promotes angiogenesis21 and TRAIL inhibits it,76 there is the intriguing possibility that Fn14-TRAIL may inhibit angiogenesis at inflammatory sites by a dual mechanism, that is, by antagonizing TWEAK’s pro-angiogenic activity and synergistically reinforcing this effect through TRAIL’s anti-angiogenic activity.
6) Yet another synergy could stem from modulation of endothelial cell function and chemokine expression. As stated above, blockade of TWEAK may interfere with chemokine expression by, and leukocyte adhesion to, endothelial cells. TRAIL is also linked to these processes. TRAIL counteracts TNF-α-induced leukocyte adhesion to endothelial cells by down-modulating CCL8 and CXCL10 chemokine expression and release.77 Thus, Fn14-TRAIL could impact leukocyte attraction and adhesion through both its Fn14 and TRAIL ends.
Although the present study centers around the autoimmune encephalomyelitis model, Fn14-TRAIL’s therapeutic benefit may well extend to other autoimmune conditions. TWEAK has emerged as a potent arthritogenic ligand, with anti-TWEAK blocking Ab reducing disease severity in the collagen-induced arthritis model.78,79 TWEAK has also been linked to systemic lupus erythematosus, with TWEAK blocking Ab again showing a beneficial effect with respect to renal damage in mice with induced lupus.80 Adding TRAIL-mediated immunoinhibitory capacity to a TWEAK-blocking agent would be expected to augment therapeutic efficacy in these other autoimmune settings.
Fn14-TRAIL may ultimately prove to have functions that extend beyond those described so far. The observation that Fn14 can stimulate neurite outgrowth in peripheral neurons independent of TWEAK81 suggests the likelihood that there are additional ligands for the Fn14 receptor. If so, Fn14-TRAIL’s effects could go well beyond functions associated with the TWEAK ligand per se, some of which could be serving to reinforce the autoimmune benefit, as well as point to clinical applications beyond autoimmune therapies. Also, because there is now some evidence that TWEAK may bind to at least one additional receptor other than Fn14, namely CD163,82 Fn14-TRAIL would interfere with biological effects stemming from TWEAK interacting with such a non-Fn14 receptor. Furthermore, therapeutic fusion proteins can be functionally more than the mere sum of their parts. This has proven to be the case for the TSCP CTLA-4-FasL, which was shown to be able to uniquely block the up-regulation of the anti-apoptotic c-FLIP protein after T-cell activation.35 Dissecting out just how a multifunctional therapeutic TSRP such as Fn14-TRAIL perturbs immune cellular networking from multiple angles and through this achieves a net anti-inflammatory therapeutic effect should be informative down the road. What may emerge is a sense of how fusion proteins with multifunctional potential can be best deployed to tackle diseases with multidimensional pathogenesis.
Acknowledgments
We thank the University of Pennsylvania Cancer Center Flow Cytometry Core for expert assistance with FACS analysis.
Footnotes
Address reprint requests to Dr. Mark L. Tykocinski, Office of the Dean, Jefferson Medical College, Thomas Jefferson University, 1025 Walnut St., Suite 100, Philadelphia, PA, 19107. E-mail: mark.tykocinski@jefferson.edu.
Supported in part by the National Institutes of Health (RO1 AI031044 and RO1CA 074958 to M.L.T.).
A patent is being filed around Fn14-TRAIL and its uses, and there are active efforts ongoing to license this intellectual property.
References
- Zamvil SS, Steinman L. The T lymphocyte in experimental allergic encephalomyelitis. Annu Rev Immunol. 1990;8:579–621. doi: 10.1146/annurev.iy.08.040190.003051. [DOI] [PubMed] [Google Scholar]
- Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- Hemmer B, Nessler S, Zhou D, Kieseier B, Hartung HP. Immunopathogenesis and immunotherapy of multiple sclerosis. Nat Clin Pract Neurol. 2006;2:201–211. doi: 10.1038/ncpneuro0154. [DOI] [PubMed] [Google Scholar]
- Mackay F, Kalled SL. TNF ligands and receptors in autoimmunity: an update. Curr Opin Immunol. 2002;14:783–790. doi: 10.1016/s0952-7915(02)00407-7. [DOI] [PubMed] [Google Scholar]
- Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- Burkly LC, Michaelson JS, Hahm K, Jakubowski A, Zheng TS. TWEAKing tissue remodeling by a multifunctional cytokine: role of TWEAK/Fn14 pathway in health and disease. Cytokine. 2007;40:1–16. doi: 10.1016/j.cyto.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov. 2008;7:411–425. doi: 10.1038/nrd2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauli G, Secchiero P. The role of the TRAIL/TRAIL receptors system in hematopoiesis and endothelial cell biology. Cytokine Growth Factor Rev. 2006;17:245–257. doi: 10.1016/j.cytogfr.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Aktas O, Prozorovski T, Zipp F. Death ligands and autoimmune demyelination. Neuroscientist. 2006;12:305–316. doi: 10.1177/1073858405285208. [DOI] [PubMed] [Google Scholar]
- Schaefer U, Voloshanenko O, Willen D, Walczak H. TRAIL: a multifunctional cytokine. Front Biosci. 2007;12:3813–3824. doi: 10.2741/2354. [DOI] [PubMed] [Google Scholar]
- Anel A, Bosque A, Naval J, Pineiro A, Larrad L, Alava MA, Martinez-Lorenzo MJ. Apo2L/TRAIL and immune regulation. Front Biosci. 2007;12:2074–2084. doi: 10.2741/2212. [DOI] [PubMed] [Google Scholar]
- Wiley SR, Winkles JA. TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev. 2003;14:241–249. doi: 10.1016/s1359-6101(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Maecker H, Varfolomeev E, Kischkel F, Lawrence D, LeBlanc H, Lee W, Hurst S, Danilenko D, Li J, Filvaroff E, Yang B, Daniel D, Ashkenazi A. TWEAK attenuates the transition from innate to adaptive immunity. Cell. 2005;123:931–944. doi: 10.1016/j.cell.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Saas P, Boucraut J, Walker PR, Quiquerez AL, Billot M, Desplat-Jego S, Chicheportiche Y, Dietrich PY. TWEAK stimulation of astrocytes and the proinflammatory consequences. Glia. 2000;32:102–107. [PubMed] [Google Scholar]
- Harada N, Nakayama M, Nakano H, Fukuchi Y, Yagita H, Okumura K. Pro-inflammatory effect of TWEAK/Fn14 interaction on human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2002;299:488–493. doi: 10.1016/s0006-291x(02)02670-0. [DOI] [PubMed] [Google Scholar]
- Chicheportiche Y, Chicheportiche R, Sizing I, Thompson J, Benjamin CB, Ambrose C, Dayer JM. Proinflammatory activity of TWEAK on human dermal fibroblasts and synoviocytes: blocking and enhancing effects of anti-TWEAK monoclonal antibodies. Arthritis Res. 2002;4:126–133. doi: 10.1186/ar388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Burkly LC, Gao HX, Berman JW, Su L, Browning B, Zheng T, Schiffer L, Michaelson JS, Putterman C. Proinflammatory effects of TWEAK/Fn14 interactions in glomerular mesangial cells. J Immunol. 2006;176:1889–1898. doi: 10.4049/jimmunol.176.3.1889. [DOI] [PubMed] [Google Scholar]
- Xu H, Okamoto A, Ichikawa J, Ando T, Tasaka K, Masuyama K, Ogawa H, Yagita H, Okumura K, Nakao A. TWEAK/Fn14 interaction stimulates human bronchial epithelial cells to produce IL-8 and GM-CSF. Biochem Biophys Res Commun. 2004;318:422–427. doi: 10.1016/j.bbrc.2004.04.036. [DOI] [PubMed] [Google Scholar]
- Ando T, Ichikawa J, Wako M, Hatsushika K, Watanabe Y, Sakuma M, Tasaka K, Ogawa H, Hamada Y, Yagita H, Nakao A. TWEAK/Fn14 interaction regulates RANTES production. BMP-2-induced differentiation, and RANKL expression in mouse osteoblastic MC3T3–E1 cells. Arthritis Res Ther. 2006;8:R146. doi: 10.1186/ar2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polavarapu R, Gongora MC, Winkles JA, Yepes M. Tumor necrosis factor-like weak inducer of apoptosis increases the permeability of the neurovascular unit through nuclear factor-kappa B pathway activation. J Neurosci. 2005;25:10094–10100. doi: 10.1523/JNEUROSCI.3382-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski A, Browning B, Lukashev M, Sizing I, Thompson JS, Benjamin CD, Hsu YM, Ambrose C, Zheng TS, Burkly LC. Dual role for TWEAK in angiogenic regulation. J Cell Sci. 2002;115:267–274. doi: 10.1242/jcs.115.2.267. [DOI] [PubMed] [Google Scholar]
- Kirk SL, Karlik SJ. VEGF and vascular changes in chronic neuroinflammation. J Autoimmun. 2003;21:353–363. doi: 10.1016/s0896-8411(03)00139-2. [DOI] [PubMed] [Google Scholar]
- Desplat-Jégo S, Varriale S, Creidy R, Terra R, Bernard D, Khrestchatisky M, Izui S, Chicheportiche Y, Boucraut J. TWEAK is expressed by glial cells, induces astrocyte proliferation and increases EAE severity. J Neuroimmunol. 2002;133:116–123. doi: 10.1016/s0165-5728(02)00368-5. [DOI] [PubMed] [Google Scholar]
- Potrovita I, Zhang W, Burkly L, Hahm K, Lincecum J, Wang MZ, Maurer MH, Rossner M, Schneider A, Schwaninger M. Tumor necrosis factor-like weak inducer of apoptosis-induced neurodegeneration. J Neurosci. 2004;24:8237–8244. doi: 10.1523/JNEUROSCI.1089-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AM, Pedre X, Kleiter I, Hornberg M, Steinbrecher A, Giegerich G. Targeting fibroblast growth factor-inducible-14 signaling protects from chronic relapsing experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;159:55–65. doi: 10.1016/j.jneuroim.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Ren X, Ye F, Jiang Z, Chu Y, Xiong S, Wang Y. Involvement of cellular death in TRAIL/DR5-dependent suppression induced by CD4(+)CD25(+) regulatory T cells. Cell Death Differ. 2007;14:2076–2084. doi: 10.1038/sj.cdd.4402220. [DOI] [PubMed] [Google Scholar]
- Hilliard B, Wilmen A, Seidel C, Liu TS, Goke R, Chen Y. Roles of TNF-related apoptosis-inducing ligand in experimental autoimmune encephalomyelitis. J Immunol. 2001;166:1314–1319. doi: 10.4049/jimmunol.166.2.1314. [DOI] [PubMed] [Google Scholar]
- Lamhamedi-Cherradi SE, Zheng S, Tisch RM, Chen YH. Critical roles of tumor necrosis factor-related apoptosis-inducing ligand in type 1 diabetes. Diabetes. 2003;52:2274–2278. doi: 10.2337/diabetes.52.9.2274. [DOI] [PubMed] [Google Scholar]
- Cretney E, McQualter JL, Kayagaki N, Yagita H, Bernard CC, Grewal IS, Ashkenazi A, Smyth MJ. TNF-related apoptosis-inducing ligand (TRAIL)/Apo2L suppresses experimental autoimmune encephalomyelitis in mice. Immunol Cell Biol. 2005;83:511–519. doi: 10.1111/j.1440-1711.2005.01358.x. [DOI] [PubMed] [Google Scholar]
- Hirata S, Senju S, Matsuyoshi H, Fukuma D, Uemura Y, Nishimura Y. Prevention of experimental autoimmune encephalomyelitis by transfer of embryonic stem cell-derived dendritic cells expressing myelin oligodendrocyte glycoprotein peptide along with TRAIL or programmed death-1 ligand. J Immunol. 2005;174:1888–1897. doi: 10.4049/jimmunol.174.4.1888. [DOI] [PubMed] [Google Scholar]
- Wandinger KP, Lunemann JD, Wengert O, Bellmann-Strobl J, Aktas O, Weber A, Grundstrom E, Ehrlich S, Wernecke KD, Volk HD, Zipp F. TNF-related apoptosis inducing ligand (TRAIL) as a potential response marker for interferon-beta treatment in multiple sclerosis. Lancet. 2003;361:2036–2043. doi: 10.1016/S0140-6736(03)13641-0. [DOI] [PubMed] [Google Scholar]
- Huang JH, Tykocinski ML. CTLA-4-Fas ligand functions as a trans signal converter protein in bridging antigen-presenting cells and T cells. Int Immunol. 2001;13:529–539. doi: 10.1093/intimm/13.4.529. [DOI] [PubMed] [Google Scholar]
- Elhalel MD, Huang JH, Schmidt W, Rachmilewitz J, Tykocinski ML. CTLA-4 · FasL induces alloantigen-specific hyporesponsiveness. J Immunol. 2003;170:5842–5850. doi: 10.4049/jimmunol.170.12.5842. [DOI] [PubMed] [Google Scholar]
- Dranitzki-Elhalel M, Huang JH, Rachmilewitz J, Pappo O, Parnas M, Schmidt W, Tykocinski ML. CTLA-4.FasL inhibits allogeneic responses in vivo. Cell Immunol. 2006;239:129–135. doi: 10.1016/j.cellimm.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Orbach A, Rachmilewitz J, Parnas M, Huang JH, Tykocinski ML, Dranitzki-Elhalel M. CTLA-4 · FasL induces early apoptosis of activated T cells by interfering with anti-apoptotic signals. J Immunol. 2007;179:7287–7294. doi: 10.4049/jimmunol.179.11.7287. [DOI] [PubMed] [Google Scholar]
- Brunschwig EB, Levine E, Trefzer U, Tykocinski ML. Glycosylphosphatidylinositol-modified murine B7-1 and B7-2 retain costimulator function. J Immunol. 1995;155:5498–5505. [PubMed] [Google Scholar]
- Stromnes IM, Goverman JM. Active induction of experimental allergic encephalomyelitis. Nat Protoc. 2006;1:1810–1819. doi: 10.1038/nprot.2006.285. [DOI] [PubMed] [Google Scholar]
- Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Prasad R, Giri S, Nath N, Singh I, Singh AK. 5-Aminoimidazole-4-carboxamide-1-beta-4-ribofuranoside attenuates experimental autoimmune encephalomyelitis via modulation of endothelial-monocyte interaction. J Neurosci Res. 2006;84:614–625. doi: 10.1002/jnr.20953. [DOI] [PubMed] [Google Scholar]
- Hilliard B, Samoilova EB, Liu TS, Rostami A, Chen Y. Experimental autoimmune encephalomyelitis in NF-kappa B-deficient mice: roles of NF-kappa B in the activation and differentiation of autoreactive T cells. J Immunol. 1999;163:2937–2943. [PubMed] [Google Scholar]
- Hymowitz SG, Christinger HW, Fuh G, Ultsch M, O'Connell M, Kelley RF, Ashkenazi A, de Vos AM. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell. 1999;4:563–571. doi: 10.1016/s1097-2765(00)80207-5. [DOI] [PubMed] [Google Scholar]
- Martí-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Hymowitz SG, Patel DR, Wallweber HJ, Runyon S, Yan M, Yin J, Shriver SK, Gordon NC, Pan B, Skelton NJ, Kelley RF, Starovasnik MA. Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J Biol Chem. 2005;280:7218–7227. doi: 10.1074/jbc.M411714200. [DOI] [PubMed] [Google Scholar]
- Swaminathan P, Hariharan M, Murali R, Singh CU. Molecular structure, conformational analysis, and structure-activity studies of Dendrotoxin and its homologues using molecular mechanics and molecular dynamics techniques. J Med Chem. 1996;39:2141–2155. doi: 10.1021/jm950579p. [DOI] [PubMed] [Google Scholar]
- Isaacs JD, Greenwood J, Waldmann H. Therapy with monoclonal antibodies. II. The contribution of Fc gamma receptor binding and the influence of C(H)1 and C(H)3 domains on in vivo effector function. J Immunol. 1998;161:3862–3869. [PubMed] [Google Scholar]
- Komiyama Y, Nakae S, Matsuki T, Nambu A, Ishigame H, Kakuta S, Sudo K, Iwakura Y. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J Immunol. 2006;177:566–573. doi: 10.4049/jimmunol.177.1.566. [DOI] [PubMed] [Google Scholar]
- Hofstetter HH, Ibrahim SM, Koczan D, Kruse N, Weishaupt A, Toyka KV, Gold R. Therapeutic efficacy of IL-17 neutralization in murine experimental autoimmune encephalomyelitis. Cell Immunol. 2005;237:123–130. doi: 10.1016/j.cellimm.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Weiss HA, Millward JM, Owens T. CD8+ T cells in inflammatory demyelinating disease. J Neuroimmunol. 2007;191:79–85. doi: 10.1016/j.jneuroim.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Gold R, Linington C, Lassmann H. Understanding pathogenesis and therapy of multiple sclerosis via animal models: 70 years of merits and culprits in experimental autoimmune encephalomyelitis research. Brain. 2006;129:1953–1971. doi: 10.1093/brain/awl075. [DOI] [PubMed] [Google Scholar]
- Tykocinski ML, Chen A, Huang JH, Weber MC, Zheng G. New designs for cancer vaccine and artificial veto cells: an emerging palette of protein paints. Immunol Res. 2003;27:565–574. doi: 10.1385/IR:27:2-3:565. [DOI] [PubMed] [Google Scholar]
- Jin YZ, Wang GM, Li AL, Xie SS. [Adenovirus-mediated CTLA4-FasL gene transfer induces long-term survival of cardiac allograft in rats]. Zhonghua Yi Xue Za Zhi. 2003;83:1968–1974. [PubMed] [Google Scholar]
- Jin Y, Qu A, Wang GM, Hao J, Gao X, Xie S. Simultaneous stimulation of Fas-mediated apoptosis and blockade of costimulation prevent autoimmune diabetes in mice induced by multiple low-dose streptozotocin. Gene Ther. 2004;11:982–991. doi: 10.1038/sj.gt.3302260. [DOI] [PubMed] [Google Scholar]
- Vince JE, Silke J. TWEAK shall inherit the earth. Cell Death Differ. 2006;13:1842–1844. doi: 10.1038/sj.cdd.4402027. [DOI] [PubMed] [Google Scholar]
- Cretney E, Shanker A, Yagita H, Smyth MJ, Sayers TJ. TNF-related apoptosis-inducing ligand as a therapeutic agent in autoimmunity and cancer. Immunol Cell Biol. 2006;84:87–98. doi: 10.1111/j.1440-1711.2005.01413.x. [DOI] [PubMed] [Google Scholar]
- Desplat-Jégo S, Creidy R, Varriale S, Allaire N, Luo Y, Bernard D, Hahm K, Burkly L, Boucraut J. Anti-TWEAK monoclonal antibodies reduce immune cell infiltration in the central nervous system and severity of experimental autoimmune encephalomyelitis. Clin Immunol. 2005;117:15–23. doi: 10.1016/j.clim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kang YJ, Kim WJ, Woo DK, Lee Y, Kim DI, Park YB, Kwon BS, Park JE, Lee WH. TWEAK can induce pro-inflammatory cytokines and matrix metalloproteinase-9 in macrophages. Circ J. 2004;68:396–399. doi: 10.1253/circj.68.396. [DOI] [PubMed] [Google Scholar]
- Lamhamedi-Cherradi SE, Zheng SJ, Maguschak KA, Peschon J, Chen YH. Defective thymocyte apoptosis and accelerated autoimmune diseases in TRAIL−/− mice. Nat Immunol. 2003;4:255–260. doi: 10.1038/ni894. [DOI] [PubMed] [Google Scholar]
- Song K, Chen Y, Goke R, Wilmen A, Seidel C, Goke A, Hilliard B, Chen Y. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is an inhibitor of autoimmune inflammation and cell cycle progression. J Exp Med. 2000;191:1095–1104. doi: 10.1084/jem.191.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi QS, Ly D, Lamhamedi-Cherradi SE, Salojin KV, Zhou L, Grattan M, Meagher C, Zucker P, Chen YH, Nagle J, Taub D, Delovitch TL. Blockade of tumor necrosis factor-related apoptosis-inducing ligand exacerbates type 1 diabetes in NOD mice. Diabetes. 2003;52:1967–1975. doi: 10.2337/diabetes.52.8.1967. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Yamaguchi N, Abe M, Hirose S, Shirai T, Okumura K, Yagita H. Suppression of antibody production by TNF-related apoptosis-inducing ligand (TRAIL). Cell Immunol. 2002;219:82–91. doi: 10.1016/s0008-8749(02)00602-0. [DOI] [PubMed] [Google Scholar]
- Hayakawa Y, Screpanti V, Yagita H, Grandien A, Ljunggren HG, Smyth MJ, Chambers BJ. NK cell TRAIL eliminates immature dendritic cells in vivo and limits dendritic cell vaccination efficacy. J Immunol. 2004;172:123–129. doi: 10.4049/jimmunol.172.1.123. [DOI] [PubMed] [Google Scholar]
- Janssen EM, Droin NM, Lemmens EE, Pinkoski MJ, Bensinger SJ, Ehst BD, Griffith TS, Green DR, Schoenberger SP. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- You RI, Chang YC, Chen PM, Wang WS, Hsu TL, Yang CY, Lee CT, Hsieh SL. Apoptosis of dendritic cells induced by decoy receptor 3 (DcR3). Blood. 2008;111:1480–1488. doi: 10.1182/blood-2007-09-114850. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Takeda K, Hayakawa Y, Peschon JJ, van den Brink MR, Yagita H. Nature’s TRAIL—on a path to cancer immunotherapy. Immunity. 2003;18:1–6. doi: 10.1016/s1074-7613(02)00502-2. [DOI] [PubMed] [Google Scholar]
- Kaplan MJ, Ray D, Mo RR, Yung RL, Richardson BC. TRAIL (Apo2 ligand) and TWEAK (Apo3 ligand) mediate CD4+ T cell killing of antigen-presenting macrophages. J Immunol. 2000;164:2897–2904. doi: 10.4049/jimmunol.164.6.2897. [DOI] [PubMed] [Google Scholar]
- Renshaw SA, Parmar JS, Singleton V, Rowe SJ, Dockrell DH, Dower SK, Bingle CD, Chilvers ER, Whyte MK. Acceleration of human neutrophil apoptosis by TRAIL. J Immunol. 2003;170:1027–1033. doi: 10.4049/jimmunol.170.2.1027. [DOI] [PubMed] [Google Scholar]
- Lünemann JD, Waiczies S, Ehrlich S, Wendling U, Seeger B, Kamradt T, Zipp F. Death ligand TRAIL induces no apoptosis but inhibits activation of human (auto)antigen-specific T cells. J Immunol. 2002;168:4881–4888. doi: 10.4049/jimmunol.168.10.4881. [DOI] [PubMed] [Google Scholar]
- Zhang XR, Zhang LY, Devadas S, Li L, Keegan AD, Shi YF. Reciprocal expression of TRAIL and CD95L in Th1 and Th2 cells: role of apoptosis in T helper subset differentiation. Cell Death Differ. 2003;10:203–210. doi: 10.1038/sj.cdd.4401138. [DOI] [PubMed] [Google Scholar]
- Hirata S, Matsuyoshi H, Fukuma D, Kurisaki A, Uemura Y, Nishimura Y, Senju S. Involvement of regulatory T cells in the experimental autoimmune encephalomyelitis-preventive effect of dendritic cells expressing myelin oligodendrocyte glycoprotein plus TRAIL. J Immunol. 2007;178:918–925. doi: 10.4049/jimmunol.178.2.918. [DOI] [PubMed] [Google Scholar]
- Nitsch R, Pohl EE, Smorodchenko A, Infante-Duarte C, Aktas O, Zipp F. Direct impact of T cells on neurons revealed by two-photon microscopy in living brain tissue. J Neurosci. 2004;24:2458–2464. doi: 10.1523/JNEUROSCI.4703-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahad DJ, Ransohoff RM. The role of MCP-1 (CCL2) and CCR2 in multiple sclerosis and experimental autoimmune encephalomyelitis (EAE). Semin Immunol. 2003;15:23–32. doi: 10.1016/s1044-5323(02)00125-2. [DOI] [PubMed] [Google Scholar]
- Kennedy KJ, Strieter RM, Kunkel SL, Lukacs NW, Karpus WJ. Acute and relapsing experimental autoimmune encephalomyelitis are regulated by differential expression of the CC chemokines macrophage inflammatory protein-1alpha and monocyte chemotactic protein-1. J Neuroimmunol. 1998;92:98–108. doi: 10.1016/s0165-5728(98)00187-8. [DOI] [PubMed] [Google Scholar]
- Aktas O, Smorodchenko A, Brocke S, Infante-Duarte C, Topphoff US, Vogt J, Prozorovski T, Meier S, Osmanova V, Pohl E, Bechmann I, Nitsch R, Zipp F. Neuronal damage in autoimmune neuroinflammation mediated by the death ligand TRAIL. Neuron. 2005;46:421–432. doi: 10.1016/j.neuron.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Nitsch R, Bechmann I, Deisz RA, Haas D, Lehmann TN, Wendling U, Zipp F. Human brain-cell death induced by tumour-necrosis-factor-related apoptosis-inducing ligand (TRAIL). Lancet. 2000;356:827–828. doi: 10.1016/S0140-6736(00)02659-3. [DOI] [PubMed] [Google Scholar]
- Matysiak M, Jurewicz A, Jaskolski D, Selmaj K. TRAIL induces death of human oligodendrocytes isolated from adult brain. Brain. 2002;125:2469–2480. doi: 10.1093/brain/awf254. [DOI] [PubMed] [Google Scholar]
- Cantarella G, Risuglia N, Dell'eva R, Lempereur L, Albini A, Pennisi G, Scoto GM, Noonan DN, Bernardini R. TRAIL inhibits angiogenesis stimulated by VEGF expression in human glioblastoma cells. Br J Cancer. 2006;94:1428–1435. doi: 10.1038/sj.bjc.6603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchiero P, Corallini F, di Iasio MG, Gonelli A, Barbarotto E, Zauli G. TRAIL counteracts the proadhesive activity of inflammatory cytokines in endothelial cells by down-modulating CCL8 and CXCL10 chemokine expression and release. Blood. 2005;105:3413–3419. doi: 10.1182/blood-2004-10-4111. [DOI] [PubMed] [Google Scholar]
- Perper SJ, Browning B, Burkly LC, Weng S, Gao C, Giza K, Su L, Tarilonte L, Crowell T, Rajman L, Runkel L, Scott M, Atkins GJ, Findlay DM, Zheng TS, Hess H. TWEAK is a novel arthritogenic mediator. J Immunol. 2006;177:2610–2620. doi: 10.4049/jimmunol.177.4.2610. [DOI] [PubMed] [Google Scholar]
- Kamata K, Kamijo S, Nakajima A, Koyanagi A, Kurosawa H, Yagita H, Okumura K. Involvement of TNF-like weak inducer of apoptosis in the pathogenesis of collagen-induced arthritis. J Immunol. 2006;177:6433–6439. doi: 10.4049/jimmunol.177.9.6433. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Burkly LC, Campbell S, Schwartz N, Molano A, Choudhury A, Eisenberg RA, Michaelson JS, Putterman C. TWEAK/Fn14 interactions are instrumental in the pathogenesis of nephritis in the chronic graft-versus-host model of systemic lupus erythematosus. J Immunol. 2007;179:7949–7958. doi: 10.4049/jimmunol.179.11.7949. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Bonilla I, Winkles JA, Strittmatter SM. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci. 2003;23:9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bover LC, Cardo-Vila M, Kuniyasu A, Sun J, Rangel R, Takeya M, Aggarwal BB, Arap W, Pasqualini R. A previously unrecognized protein-protein interaction between TWEAK and CD163: potential biological implications. J Immunol. 2007;178:8183–8194. doi: 10.4049/jimmunol.178.12.8183. [DOI] [PubMed] [Google Scholar]