Abstract
In transmembrane (TM) domains, tetraspanin KAI1/CD82 contains an Asn, a Gln, and a Glu polar residue. A mutation of all three polar residues largely disrupts the migration-, invasion-, and metastasis-suppressive activities of KAI1/CD82. Notably, KAI1/CD82 inhibits the formation of microprotrusions and the release of microvesicles, while the mutation disrupts these inhibitions, revealing the connections of microprotrusion and microvesicle to KAI1/CD82 function. The TM polar residues are needed for proper interactions between KAI1/CD82 and tetraspanins CD9 and CD151, which also regulate cell movement, but not for the association between KAI1/CD82 and α3β1 integrin. However, KAI1/CD82 still efficiently inhibits cell migration when either CD9 or CD151 is absent. Hence, KAI1/CD82 interacts with tetraspanin and integrin by different mechanisms and is unlikely to inhibit cell migration through its associated proteins. Moreover, without significantly affecting the glycosylation, homodimerization, and global folding of KAI1/CD82, the TM interactions maintain the conformational stability of KAI1/CD82, evidenced by the facts that the mutant is more sensitive to denaturation and less associable with tetraspanins and supported by the modeling analysis. Thus, the TM interactions mediated by these polar residues determine a conformation either in or near the tightly packed TM region and this conformation and/or its change are needed for the intrinsic activity of KAI1/CD82. In contrast to immense efforts to block the signaling of cancer progression, the perturbation of TM interactions may open a new avenue to prevent cancer invasion and metastasis.
Functionally, KAI1/CD82 suppresses tumor metastasis and cell migration.1,2,3 The role of KAI1/CD82 in metastasis suppression was originally discovered in metastatic prostate cancer,4 and then it was found that KAI1/CD82 expression also suppresses the invasion and/or metastasis of other epithelial malignancies.1,2,3 Although the mechanism remains unclear, recent studies have shown that KAI1/CD82 may inhibit cell motility by regulating the biological activities of its associated proteins and/or reorganizing plasma membrane microdomains.1,2,3 In other words, KAI1/CD82 may suppress cancer metastasis by directly inhibiting cancer cell movement. In addition, KAI1/CD82 overexpression was reported to induce apoptosis of cancer cells5,6 by releasing intracellular glutathione and by accumulating intracellular reactive oxygen intermediates.6 Recently, KAI1/CD82 was found to bind Duffy antigen receptor for chemokine proteins expressing on endothelium, which results in the senescence of KAI1/CD82-positive cancer cells in primary lesions and the metastasis of KAI1/CD82-negative cancer cells to distant organs.7
Structurally, KAI1/CD82 protein is a member of the tetraspanin superfamily, members of which can be found in all eukaryotic organisms and engage in a wide spectrum of biological functions.8,9 Consistent with the role that a fungus tetraspanin plays in cellular invasion, many tetraspanins in humans regulate cell movement.8 A prominent feature of tetraspanins is that they associate with each other and with other transmembrane (TM) and intracellular signaling molecules to form a transmembrane multimolecular complex called tetraspanin web or tetraspanin-enriched microdomain (TEM).8,9 KAI1/CD82 associates with other tetraspanins such as CD9 and CD81 in the plasma membrane.3 In addition, KAI1/CD82 associates with a list of other TM proteins such as integrins, Ig superfamily proteins, and growth factor receptors, which are also the components of the tetraspanin web or TEM.3
Approximately one-third of amino acid residues of KAI1/CD82 are embedded in the lipid bilayer and form four TM domains.10,11,12 Interestingly, the TM domains of most, if not all, tetraspanins contain several conserved polar residues. For example, the first, third, and fourth TM domains of KAI1/CD82 contain respectively, a highly hydrophilic amino acid residue that is fully buried in the membrane lipid bilayer. Although the precise role of these polar residues in tetraspanin function remains unknown, recent studies suggest that the strong polar residues in CD9, CD81, and UPIb are involved in molecular packing, ie, the interactions between TM segments.13,14,15 This notion agrees with earlier observations made from other TM proteins that TM polar residues can mediate peptide-peptide interactions within the lipid bilayer.16,17,18,19,20 Furthermore, studies from T cell receptor demonstrated that the TM polar residues contribute to the assembly, cell surface expression, and signaling of T cell receptor,21,22,23 underscoring the biological significance of TM polar residues. Because the associations between KAI1/CD82 and other TM proteins in tetraspanin web or TEM may not result from the direct protein-protein interaction of either extracellular domains or intracellular domains, we hypothesized that these polar residues in KAI1/CD82’s TM domains play important roles in the interactions between KAI1/CD82 and some of its associated proteins or the formation of KAI1/CD82-containing TEM. Because the associations of KAI/CD82 with the cell adhesion proteins and growth factor receptors in TEM are possibly needed for KAI1/CD82 motility-inhibitory activity, we also hypothesized that the TM polar residues of KAI1/CD82 are also functionally important.
Materials and Methods
Antibodies and Extracellular Matrix
The monoclonal antibodies (mAbs) used in this study were CD82 mAbs M104,24 TS82b25 (Diaclone SAS, Besancon, France), 4F9,12 6D7,12 and 8E412; CD9 mAb MAB726 and C9BB27; CD81 mAb M3824; CD151 mAbs 5C1127 and 8C328; integrin β1 mAb TS2/16 mAb29; and β-tubulin mAb (Sigma, St. Louis, MO). The tetraspanin mAbs were kindly provided directly or indirectly by Drs. O. Yoshie, E. Rubinstein, C. Morimoto, L. Jennings, M. Hemler, and K. Sekiguchi. A mouse IgG2b (clone MOPC 141; Sigma) was used as a negative control antibody in flow cytometry. The polyclonal antibody used in this study was integrin α3 antibody.30 The secondary antibodies were horseradish peroxidase-conjugated goat anti-mouse IgG (Sigma) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibody (Biosource International, Camarillo, CA). The extracellular matrix (ECM) proteins used in this study were human plasma fibronectin (FN; Invitrogen, San Diego, CA), mouse laminin 1 (LN; Invitrogen), and Matrigel (BD Bioscience, San Jose, CA).
Cell Culture and Transfectants
The prostate cancer cell line Du145 and the fibroblastoma cell line HT1080 were obtained from American Type Culture Collection (Manassas, VA) and cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml streptomycin. As previously described,31 the asparagine (N), glutamine (Q), and glutamic acid (E) mutant (NQE) of KAI1/CD82, in which the N17, Q99, and E242 residues are simultaneously replaced by A residues, was generated by PCR-based site-directed mutagenesis using KAI1/CD82 wild-type cDNA as the template and constructed into pcDNA3.1(+) plasmid expression vector (Invitrogen). The introduced mutations were confirmed by DNA sequencing. The Du145 transfectant of Mock, KAI1/CD82 wild-type, and KAI1/CD82 NQE mutant were established as described previously.31 Briefly, plasmid DNAs were transfected into Du145 cells via Lipofectamine 2000 (Invitrogen) and selected under G418 (Invitrogen) at a concentration of 1 mg/ml. Hundreds of G418-resistant clones were pooled, and the KAI1/CD82-positive cells were collected using flow cytometric cell sorting. The pooled Mock or KAI1/CD82-positive clones were the stable transfectants used in all subsequent experiments. HT1080-Mock and KAI1/CD82 stable transfectants were established in the same way as described above.
Flow Cytometry
Cells were harvested with 2 mmol/L EDTA/PBS, washed once with PBS, treated with DMEM supplemented with 5% goat serum at 4°C for 15 minutes, and then incubated with a primary mAb at 4°C for 1 hour. After removing unbound primary mAbs with two washes, cells were additionally incubated with FITC-conjugated goat anti-mouse IgG at 4°C for 30 minutes. The cells were analyzed on a FACSCalibur flow cytometer (BD Bioscience).
Fluorescent Microscopy
As described in earlier studies,30,31 circular glass coverslips (Fisher, Pittsburgh, PA) were coated with the ECM proteins fibronectin (10 μg/ml) or laminin 1 (10 μg/ml) at 4°C overnight and then blocked with 0.1% heat-inactivated bovine serum albumin at 37°C for 45 minutes. Cells were harvested in 2 mmol/L EDTA/PBS, washed once in PBS, and plated on the ECM-coated coverslips in serum-free DMEM at 37°C overnight. Cells were then fixed with 3% paraformaldehyde at room temperature for 15 minutes. If necessary, cells were permeabilized with 0.1% Brij 98 in PBS at room temperature for 2 minutes. Nonspecific binding sites were blocked with 20% goat serum in PBS at room temperature for 1 hour. Primary mAbs were diluted at a final concentration of ∼1 μg/ml in 20% goat serum/PBS and incubated with cells at room temperature for 1 hour followed by three washes with PBS. Cells were then labeled with FITC-conjugated goat anti-mouse IgG at room temperature for 1 hour, followed by four washes with PBS. Finally, the coverslips were mounted on glass slides in FluroSave reagent (Calbiochem, Carlsbad, CA). Digital images were captured using a Zeiss Axiophot fluorescent microscope at a magnification of ×63.
Cell Labeling, Immunoprecipitation, Immunoblot, and Western Blot
For metabolic labeling with S35, Du145 transfectants were incubated with 40 μCi/ml S35 l-methionine (Perkin-Elmer, Boston, MA) in methionine- and cysteine-free DMEM containing 10% dialyzed FCS at 37°C overnight. Cells were lysed in lysis buffer containing 1% Brij 97 (Sigma), 25 mmol/L HEPES, 150 mmol/L NaCl, 5 mmol/L MgCl2, 1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 2 mmol/L sodium orthovanadate, and 2 mmol/L sodium fluoride. Insoluble material was removed by centrifugation at 14, 000 × g for 15 minutes, and the lysates were precleared by incubating with protein A- and G-Sepharose beads (Amersham, Uppsala, Sweden) at 4°C for 6 hours. Then the mAb-preabsorbed protein A- and G-Sepharose beads were incubated with cell lysate at 4°C overnight. Immune complexes were collected by centrifugation followed by four washes in lysis buffer. Immune complexes were eluted from the beads with Laemmli sample buffer and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) under a nonreducing condition. The gels were treated with autoradiography enhancement reagent (Perkin-Elmer), dried, and exposed to film (Kodak, Rochester, NY) at −80°C for 5 to 7 days.
For cell surface biotinylation, Du145 transfectants were labeled with 100 μg/ml EZlink sulfo-NHS-LC biotin (Pierce, Rockford, IL) in PBS at room temperature for 1 hour followed by three washes with PBS. The biotinylated cells were lysed in 1% Brij 97 lysis buffer, and cell lysates were immunoprecipitated as described above. Immunoprecipitated proteins resolved by SDS-PAGE were transferred to nitrocellulose membrane (Schleicher & Schuell, Keene, NH) and incubated with horseradish peroxidase-conjugated extravidin (Sigma). Blots were visualized by chemiluminescence (Perkin-Elmer).
Immunoprecipitation and immunoblotting were performed as described.32 Briefly, cells were lysed with 1% NP40 or 1% Brij97 lysis buffer. Lysates were immunoprecipitated as described above, and the precipitates were then separated by SDS-PAGE. Proteins were transferred to nitrocellulose membranes and sequentially blotted with a primary antibody and a horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Sigma) secondary antibody followed by chemiluminescence detection. For Western blotting, lysates were directly separated by SDS-PAGE followed by the blotting procedures as indicated above.
Chemical Cross-Linking
CD82 dimerization was assessed as described.33 Briefly, the amino-specific cross-linker dithio-bis-succinimidylproprionate (DSP; Pierce, Rockford, IL) was dissolved in DMSO at 10 mg/ml immediately before use. DSP was added to the cell lysate prepared from 1% Brij97 lysis buffer (25 mmol/L Hepes, pH 7.4, 150 mmol/L NaCl, 2 mmol/L MgCl2, and aforementioned protease inhibitors) to a final concentration of 0.25 mg/ml. DSP was incubated with the cell lysate at 4°C for 1 hour, and 1% NP-40 in 20 mmol/L Tris-HCl was added to cell lysate for further incubation for 15 minutes. CD82 was immunoprecipitated and then immunoblotted with its mAb.
Cell Migration Assay
Migration assays were performed in Transwell filter inserts in 24-well tissue culture plates (BD Bioscience) as described.31 The polycarbonate membrane filters of Transwell contain pores 8 μm in diameter. Filters were spotted with FN (10 μg/ml) or LN-1 (10 μg/ml) on the lower surface of the Transwell inserts at 4°C overnight and then blocked with 0.1% heat-inactivated bovine serum albumin at 37°C for 30 minutes. Cells were detached at 90% confluence, washed once in PBS, and resuspended in serum-free DMEM containing 0.1% heat-inactivated bovine serum albumin. A 300-μl of cell suspension was added to inserts at a density of 3 × 104 cells/insert. DMEM containing 1% FCS was added to the lower wells. Migration was allowed to proceed at 37°C for 3 hours. Cells that did not migrate through the filters were removed using cotton swabs, and cells that migrated through the inserts were fixed and stained with Diff-Quick (Baxter, McGraw Park, IL). The number of migrated cells per microscopic field (at original magnification ×40) was counted visually under a light microscope. Data from five independent experiments were pooled and analyzed using a two-tailed, Student’s t-test.
Cell Invasion Assay
Invasion was measured using Biocoat Matrigel invasion chambers (BD Biosciences) by following the manufacturer’s protocol. Briefly, cells at the 80% to 90% confluent stage were detached. DMEM containing 10% FCS and 10 nmol/L stromal cell-derived factor 1 (R&D, Minneapolis, MN) was placed in the lower well, and 2.5 × 103 cells in 500 μl of serum-free medium were loaded to the upper chamber of the Matrigel-coated insert and incubated at 37°C for 22 hours. Cells that invaded to the lower surface of the filter were stained with Diff-Quick (American Scientific Products, McGraw Park, IL) and quantified with light microscope at original magnification ×100. The data were expressed as the average number of cells from five randomly selected fields and analyzed statistically using a two-tailed, Student’s t-test. The experiments were repeated three times.
Experimental Metastasis Assay
The metastatic potentials of HT1080 transfectants were evaluated in mice using experimental lung metastasis assay as described.34,35 A total of 30 BALB/c nu/nu female mice (8-week old, Harlan, Indianapolis, IN) were included in the assay with 10 mice for each group/transfectant. HT1080 transfectant cells (1 × 106 cells in 0.1 ml PBS) were injected into each nude mouse through tail vein. The mice were sacrificed 3 weeks after the injection. Lungs were dissected, fixed in formalin solution, and photographed. The macrometastatic foci (>0.3 mm) on the lung surface were counted under the dissect microscope. Statistical analysis (Student’s t-test) was performed with StatView software (SAS Institute Inc., Cary, NC), and the results were expressed as mean ± SD.
RNA Interference
The pSIREN-RetroQ retroviral construct containing short hairpin RNAs (shRNAs) targeting the human CD151 mRNA were generated as described.36 The shRNAs of sh2 and sh3 constructs target 5′-GCGAGACCATGCCTCCAACAT-3′ and 5′-AGTACCTGCTGTTTACCTACA-3′ sequences of human CD151, respectively. The sh3 construct effectively silences CD151 expression, but the sh2 cannot silence CD151 and therefore was used as control.36 These constructs were cotransfected with the pVSV-G retroviral coat protein expression vector into GP2-293 packaging cells by using lipofectamine 2000 (Invitrogen). At 48 hours after transfection, retrovirus-containing culture medium was collected, filtered through 0.45-μm filters, mixed with 4 μg/ml polybrene (Sigma), and then used to transduce Du145 cells that were pre-incubated at 4°C for 30 minutes. The stable Du145-sh2 and -sh3 transductants were selected with 2 μg/ml puromycin (Sigma) and maintained in 1 μg/ml puromycin.
Transmission Electron Microscopy
Cells grown in the tissue culture flasks at 70% to 80% confluence were detached with 2 mmol/L EDTA in PBS and spun down at 1000 × g. The cells were washed once with PBS buffer, resuspended in 2.5% glutaraldehyde for fixation at room temperature for 4 hours, and then postfixed in 1% osmium tetraoxide in PBS at room temperature for 4 hours. After postfixation, the cells were rinsed briefly in deionized water, and en bloc stained with 2% uranyl acetate in 0.85% sodium chloride at 4°C overnight. The cell pellets were dehydrated in graded solutions of ethanol, from 30% through 100%, at 1 hour each, infiltrated first with 50% Spurr in 100% ethanol overnight at room temperature, then with 100% Spurr over an 8-hour period involving at least three changes of Spurr. The cells were cured at 60°C for 2 days. One-micron sections were cut on a Reichert Ultracut E microtome and stained with toluidine blue. Areas of interest were selected, sectioned at approximately 75 nm, and poststained with uranyl acetate and lead citrate immediately. The cells were observed and photographed on a JEOL 2000EX transmission electron microscope at 60 kV.
Modeling and Analysis of KAI1/CD82 Transmembrane Regions
An atomic model of KAI1/CD82 was built with the homology modeling method by using the strategy similar to the previously published model of CD81 (PDB code: 2AVZ).14 In the modeling procedure, the four TM helices were constructed based on a fully antisymmetric, left-handed, coiled coil template, which was found in cytochrome oxidase subunit III (PDB code: 2OCC, chain C), and the large extracellular loop (EC2) was constructed based on the crystallographic structure of the tetraspanin CD81 EC2 domain (PDB code: 1G8Q). The homology modeling was performed in SWISS-MODEL, a fully automated protein structure homology-modeling server.37 The homology model was then minimized with the program AMBER838 in implicit solvent. In detail, 1000 steepest descent minimizations were conducted first, followed by 5000 conjugate gradient minimizations to refine the model system. A modified Generalized Born implicit water model39 was then used to mimic the solvated environment. Amber99 force field was used during minimization.38 All of the calculations were performed on an IBM 420-cpu P4 cluster system located at the Hartwell Center for Bioinformatics and Biotechnology of St Jude Children’s Research Hospital.
Results
Generating and Characterizing the TM Hydrophilic Residue Mutant of KAI1/CD82: the NQE Mutant
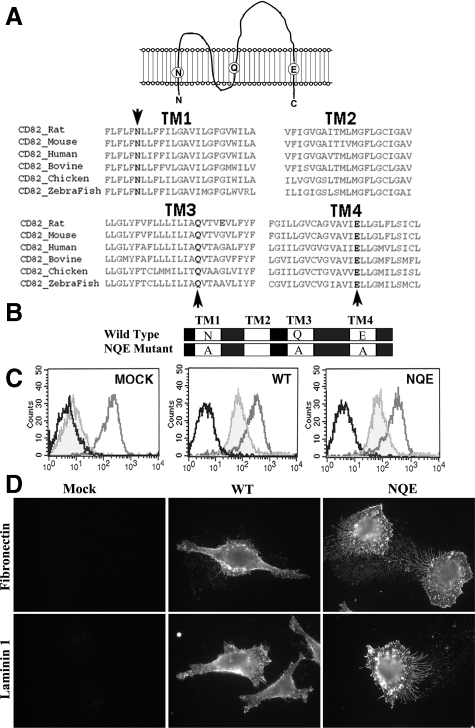
Human KAI1/CD82 TM domains TM1, TM3, and TM4 contain hydrophilic amino acid residue N, Q, and E, respectively (Figure 1A). Sequence alignment shows that the three polar residues are fully conserved in all KAI1/CD82 proteins of vertebrates (Figure 1A, bottom panel), implying the functional importance of this characteristic feature for tetraspanin TM domains. To determine the structure-function relationship, we replaced the polar residues N17, Q99, and E242 with alanines by using recombinant PCR, and the mutant was named after the amino acid abbreviation NQE (Figure 1B). The selection of alanine residue for the replacement was based on its neutrality and small size, which together minimize the adverse effect of the mutation and allow TM helices to still be packed tightly in the presence of the mutation. The stable transfectants of NQE mutant, wild-type, and Mock were established in metastatic prostate cancer cell line Du145, which barely expresses endogenous KAI1/CD82.28 The KAI1/CD82 wild-type and NQE transfectant cells with the equivalent levels of KAI1/CD82 expression at the cell surface were collected by flow cytometric sorting and used as the stable transfectants for the following studies (Figure 1C).
Figure 1.
Schematic presentation of CD82 TM polar residues and establishment of the NQE mutant. A: Sequence alignment of CD82 TM regions. The four TM domains of the vertebrate orthologs of CD82 were aligned by the Clustal W program. Three TM polar residues (in bold and marked with arrow) are fully conserved in all CD82 proteins. B: Schematic presentation of CD82 wild-type and NQE mutant proteins. The NQE mutation denotes that the three conserved TM polar residues, Asn17 (N), Gln99 (Q), and Glu242 (E), are replaced with Ala (A) residues simultaneously. C: Du145 prostate cancer cells were stably transfected with vector (MOCK), CD82 wild-type (WT), and CD82 NQE mutant (NQE). The transfectants were analyzed by flow cytometry for the surface expression of CD82, as shown in light gray. The cells were also stained with a mouse IgG (black) and integrin β1 mAb (medium gray) for negative and positive control, respectively. D: The effect of the NQE mutation on the subcellular distribution of CD82. Du145 transfectant cells cultured on FN (10 μg/ml)- or LN1 (10 μg/ml)-coated glass coverslips were fixed, permeabilized, and incubated with CD82 mAb M104, followed by an FITC-conjugated second Ab incubation. The images were captured under a fluorescent microscope at original magnification ×63.
To assess the effect of NQE mutation on the global structure of KAI1/CD82, we analyzed and compared the binding affinity of different KAI1/CD82 mAbs to wild-type and NQE mutant. Because these mAbs induce different functional effects and can detect KAI1/CD82 proteins under different conditions,10,11,12,24,25,40 they likely recognize different antigen epitopes of KAI1/CD82. Hence, the binding abilities of various mAbs could reflect the global antigen structure. Because most tetraspanin mAbs recognize the larger extracellular loop (LEL) and these KAI1/CD82 mAbs cannot detect KAI1/CD82 antigen under the reduced condition, these antigen epitopes of KAI1/CD82 are most likely present in the LEL, which is framed by disulfide bonds, contains approximately one-half of the KAI1/CD82 amino acid sequence, and is the major structure besides TM regions. As shown in Table 1, KAI1/CD82 NQE mutant exhibited equivalent binding affinity as the wild-type to various KAI1/CD82 mAbs, suggesting that the global structure, especially the structure of LEL, of KAI1/CD82 is not significantly altered by the NQE mutation.
Table 1.
The cell surface expression of integrins and tetraspanins in Du145 transfectants
| Mock | CD82 WT | CD82 NQE | |
|---|---|---|---|
| CD82 | |||
| M104 | 3 | 37.8 | 36.2 |
| 6D7 | 2.8 | 75.5 | 65.8 |
| 4F9 | 2.7 | 168.5 | 160.9 |
| 8E4 | 3.1 | 91.8 | 112.8 |
| TS82b | 2.9 | 84.9 | 89.6 |
| CD151 | 64.8 | 83.5 | 90.8 |
| CD9 | |||
| MAB7 | 102.9 | 104.9 | 118.4 |
| C9BB | 4.2 | 27.6 | 38.2 |
| Integrin β1 | 316.8 | 297.3 | 308.7 |
| Integrin α3 | 324.5 | 354.5 | 372.3 |
The numbers are the mean fluorescence intensity of the cell surface staining of the indicated mAbs in flow cytometry. The analysis was performed multiple times, the same results were obtained, and the results from a representative experiment were presented.
KAI1/CD82 is typically localized at the plasma and intracellular vesicular membranes.3 The subcellular localization of KAI1/CD82 could be functionally important, because the deficiency in function of a KAI1/CD82 palmitoylation mutant was accompanied by the alteration of KAI1/CD82 in subcellular localization.31 To determine whether TM polar residues play any role in KAI1/CD82 subcellular localization, we analyzed the steady-state distributions of KAI1/CD82 WT and NQE proteins by immunofluorescence. We found no apparent difference in the intracellular and plasma membrane distributions of KAI1/CD82 proteins between the wild-type and NQE mutant cells when the transfectant cells were cultured on FN or LN1, although more and longer KAI1/CD82-positive, thin, and long structures or microspikes were found in the NQE mutant (Figure 1D). Also, we found no intracellular aggregates or retention of KAI1/CD82 proteins in NQE mutant, further supporting the above conclusion of the proper folding of KAI1/CD82 NQE mutant proteins.
The TM Polar Residues Are Needed for the Migration-, Invasion-, Metastasis-, and Membrane Protrusion-Inhibitory Activities of KAI1/CD82
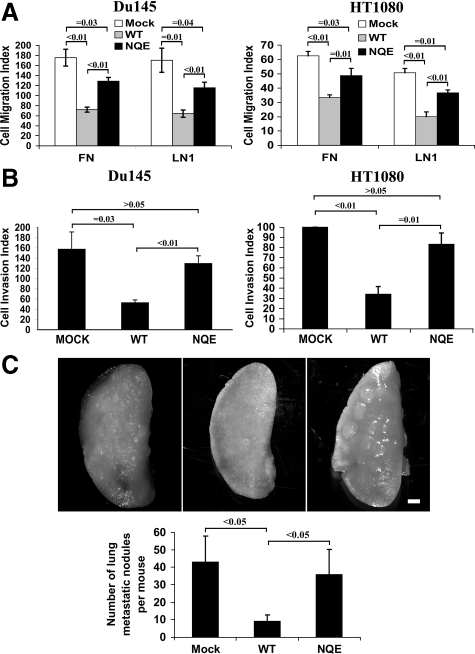
It is well established that the forced expression of KAI1/CD82 in cancer cells inhibits cell migration and invasion.1,2,3,31,32 We investigated the role of the TM polar residues in KAI1/CD82-mediated inhibition of cell movement by analyzing cell migration and invasion using the Mock, KAI1/CD82 wild-type, and KAI1/CD82 NQE transfectants. We analyzed the directional cell migration toward FCS on FN and LN1. As expected, Du145-KAI1/CD82 wild-type cells migrated in markedly lower numbers compared with Du145-Mock cells (Figure 2A, left panel), consistent with previous studies using this or other cancer cell lines.1,2,3 The NQE mutant interrupted the cell migration inhibited by KAI1/CD82 wild-type and migrated approximately onefold better than the wild-type on both ECMs (Figure 2A). We next examined whether the NQE mutation affects the ability of KAI1/CD82 to inhibit cell invasiveness. As shown in the left panel of Figure 2B, the forced expression of KAI1/CD82 wild-type drastically inhibited the cells from invading through Matrigel, the recombinant three-dimensional basement membrane, while the NQE mutant largely interrupted the ability of wild-type to inhibit invasiveness.
Figure 2.
NQE mutation disrupts CD82-mediated suppression of cell migration and invasion. A: Cell migration of Du145 and HT1080 transfectants was analyzed using Transwell inserts coated with FN (10 μg/ml) or LN1 (10 μg/ml). Cells that migrated onto the lower surface were fixed, stained, and then counted. Data are the average cell numbers of five (for Du145 cells) or eight (for HT1080 cells) randomly selected microscopic fields at the magnification ×40, and the results represent the mean ± SE of three (for Du145 cells) or four (for HT1080 cells) independent experiments. For the migration of Du145 transfectants on FN, P value is <0.01 between Mock and wild-type (WT), <0.01 between wild-type and NQE mutant, and = 0.03 between Mock and NQE. For the migration of Du145 transfectants on LN1, P value = 0.01 between Mock and wild-type, <0.01 between wild-type and NQE mutant, and = 0.04 between Mock and NQE. For the migration of HT1080 transfectants on FN, P value is <0.01 between Mock and wild-type, = 0.01 between wild-type and NQE mutant, and = 0.03 between Mock and NQE. For the migration of HT1080 transfectants on LN1, P value <0.01 between Mock and wild-type, <0.01 between wild-type and NQE mutant, and = 0.01 between Mock and NQE. B: Cell invasion of Du145 and HT1080 transfectants was determined using Biocoat Matrigel Invasion chambers (BD). The inserts were primed according to the manufacturer’s directions. Cells (2.5 × 103) were placed in the upper chamber. After incubation for 22 to 24 hours, cells that invaded through Matrigel onto the lower surface of the filter were fixed, stained, and counted. Results obtained from three (for Du145 cells) or four (for HT1080 cells) individual experiments and represent the average invaded cell number of five (for Du145 cells) or eight (for HT1080 cells) microscopic fields. For Du145 transfectants, P value = 0.03 between Mock and wild-type, <0.01 between wild-type and NQE mutant, and >0.05 between Mock and NQE. For HT1080 transfectants, P value <0.01 between Mock and wild-type, = 0.01 between wild-type and NQE mutant, and >0.1 between Mock and NQE. C: Lung metastasis of HT1080 transfectants was analyzed in the murine experimental metastasis model as described in “Materials and Methods.” The lungs of inoculated mice were dissected and photographed. The image of a single lung lobe from each group is shown. The macrometastatic foci (>0.3 mm) on the lung surface were counted under dissecting microscope. The P value <0.05 between Mock and wild-type and between wild-type and NQE mutant and >0.05 between Mock and NQE mutant; for each group, n = 10. Scale bar = 1 mm.
To ensure the effect of the NQE mutation on cell motility, we also analyzed the migratory and invasive abilities of HT1080 transfectants. In contrast to Du145 as a carcinoma line, HT1080 is a sarcoma line. Similar to Du145 cells, HT1080 cells barely express endogenous KAI1/CD82 (see following); and KAI1/CD82 wild-type and NQE mutant proteins were forcedly expressed at equivalent levels at the cell surface in HT1080 transfectants (the mean fluorescence intensity of flow cytometry analysis for wild-type was 152; the mean fluorescence intensity for the NQE mutant was 177). Consistent with the Du145 counterpart, the HT1080-KAI1/CD82 NQE mutant significantly disrupted the inhibitory effects of KAI1/CD82 wild-type on cell migration on FN and LN1 (Figure 2A, right panel). Similarly, the mutant largely disrupted the effect of wild-type on cell invasion through Matrigel (Figure 2B, right panel). The attenuated motility-inhibitory activities of NQE mutant were more evident with the cells harvested from the subconfluent culture than the ones from the confluent culture.
To evaluate the role of TM polar residues in KAI1/CD82-mediated metastasis suppression, we investigated the lung metastasis capability of HT1080 transfectants by directly inoculating the tumor cell into the tail veil of nude mice. As shown in Figure 2C, KAI1/CD82 expression significantly reduced the numbers of metastatic foci in lung, compared with the group injected with Mock cells, consistent with the earlier studies from elsewhere.41,42 While the lung metastasis nodules detectable on the lung surface in the NQE mutant group was basically equivalent in number to the one in the Mock group (Figure 2C).
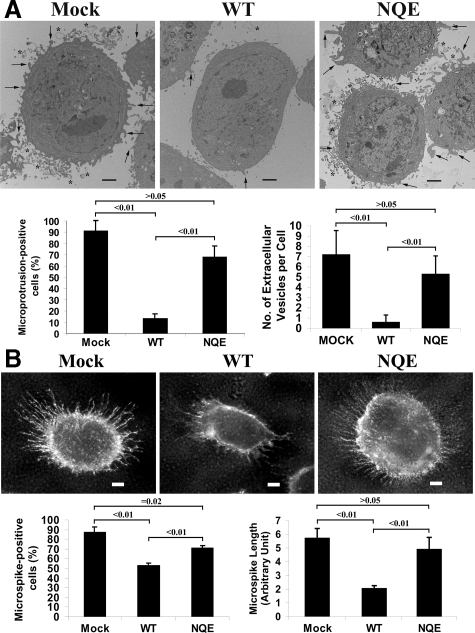
Interestingly, at the ultrastructural level, we found that Du145 cells produced the cellular projections from the plasma membrane (indicated by arrows in Figure 3A) and released the vesicular structures to the cell surroundings (indicated by asterisks in Figure 3A). On KAI/CD82 expression, the number of microprotrusions was dramatically diminished (Figure 3A). In the NQE mutant cells, microprotrusions reappeared, and the number of this ultrastructure was comparable with the one in Mock cells (Figure 3A). Under light microscopy, we found markedly fewer and shorter microspikes in KAI/CD82 wild-type cells compared with those in Mock and NQE cells (Figure 3B). Microspikes are the cellular projections that are present on the dorsal surface of a cell when cultured in complete media, and they are similar in morphology to the microprotrusions observed under electron microscopy. The widths of microprotrusions and microspikes are within the same magnitude but microprotrusions are shorter than microspikes, possibly resulted from the EM sample preparation. Together, KAI1/CD82-mediated inhibition of motility may need and the TM interaction is critical for the formation of these protrusive structures.
Figure 3.
CD82 inhibits the formation of microprotrusions and microspikes and the NQE mutation breaks off the inhibition. A: Du145 transfectant cells were cultured to subconfluence and then detached from dishes. The cells were then fixed, thin-sectioned, stained, and analyzed under a transmission electron microscope, as described in Materials and Methods. The images represent the results from one out of three individual experiments. Arrows indicate the microprotrusions and asterisks indicate microvesicles. Scale bar = 2 μm. The percentages of microprotrusion-positive cells of total cells were quantified from multiple distinct sections in each transfectant and shown in the histogram on the left. A cell with more than five microprotrusions was defined as a microprotrusion-positive cell. The P values: <0.01 between Mock and wild-type and between wild-type and NQE mutant and >0.05 between Mock and NQE mutant. The numbers of microvesicle per cell section were quantified from multiple distinct sections in each transfectant and shown in the histogram on the right. The P values: <0.01 between Mock and wild-type and between wild-type and NQE mutant and >0.05 between Mock and NQE mutant. B: Du145 transfectant cells were fixed and incubated sequentially with integrin α3 mAb X8 and FITC-conjugated second Ab. The images were captured under a fluorescent microscope. Scale bar = 5 μm. The percentages of microspike-positive cells were quantified in each transfectant and shown in the histogram on the left. A cell with more than 20 microspike was defined as a microspike-positive cell. The P value <0.01 between Mock and wild-type, <0.01 between wild-type and NQE mutant, and = 0.02 between Mock and NQE mutant. The average length of microspikes was quantified in 10 cells randomly selected from each transfectant and shown in the histogram on the right. The P value <0.01 between Mock and wild-type, <0.01 between wild-type and NQE mutant, and >0.05 between Mock and NQE mutant.
Roles of TM Hydrophilic Residues in KAI1/CD82-Tetraspanin and -Integrin Associations
Earlier studies showed that the TM polar residues were involved in protein-protein interaction in the membrane lipid bilayer.16,17,18,19,20 Because KAI1/CD82 associates with other tetraspanins, integrins, and IgSF proteins in TEM, we next examined whether the NQE mutation alters the complex formation between KAI1/CD82 and its associated proteins. This analysis also addressed whether KAI1/CD82-TEM interaction correlates the motility-inhibitory activity of KAI1/CD82.
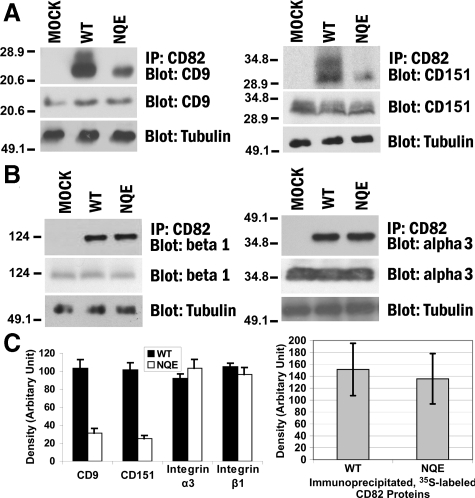
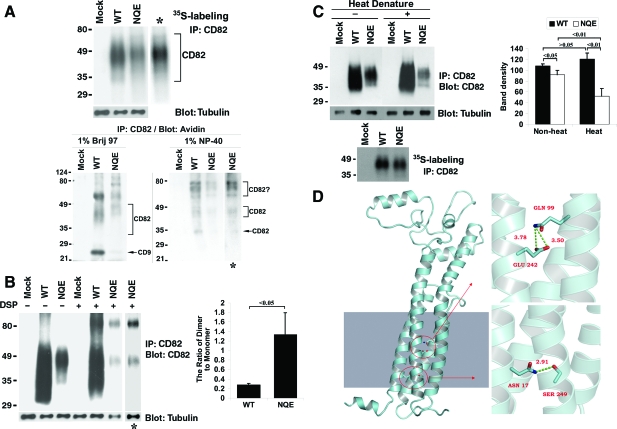
For KAI1/CD82-tetraspanin interactions, we analyzed the associations between KAI1/CD82 and two highly expressed tetraspanins in Du145 cells: CD9 and CD151. After cell lysis, the wild-type and NQE KAI1/CD82 proteins were immunoprecipitated, followed by blotting with CD9 mAb or CD151 mAb (Figure 4A). We found that KAI1/CD82-associated CD9 was markedly reduced in Du145-KAI1/CD82 NQE transfectant cells compared with that in the wild-type cells (Figure 4A, left panel). The mutation also substantially disrupted the KAI1/CD82-CD151 association (Figure 4A, right panel). The total amount of cellular CD9 or CD151 proteins remained equivalent among Mock, KAI1/CD82 wild-type, and KAI1/CD82 NQE transfectants (Figure 4A). CD9 (MAB7 staining) and CD151 were also equivalently expressed at the cell surfaces between Du145-KAI1/CD82 wild-type and NQE transfectants (Table 1). Hence, the fewer associations of NQE mutant with CD9 and CD151 did not result from lower levels of CD9 or CD151 in total or at the cell surface in the NQE mutant. The cell surface levels of KAI1/CD82 protein were almost the same between the wild-type and NQE transfectants (Table 1). For the total cellular KAI1/CD82 proteins, the metabolically labeled KAI1/CD82 proteins acquired by immunoprecipitation from the NQE transfectant were approximately 10% fewer than the ones from the wild-type (Figures 4C), compared with a lot fewer KAI1/CD82-associated CD9 and CD151 in NQE mutant. Thus, decreased association of NQE mutant with CD9 or CD151 was not caused by fewer KAI1/CD82 proteins available in the NQE transfectant.
Figure 4.
The TM polar residues are needed for CD82-TEM interaction. A: The NQE mutation diminished CD82-CD9 and -CD151 associations. Du145-Mock, -CD82 WT, and -NQE transfectant cells were lysed in 1% Brij 97, and the lysates were immunoprecipitated with CD82 mAb M104. The precipitates were resolved in nonreducing SDS-PAGE followed by transfer to nitrocellulose membrane. The membrane was blotted with CD9 mAb MAB7 or CD151 mAb 8C3, and the detected proteins were visualized by chemiluminescence. Tubulin, CD9, or CD151 from the same cell lysates was detected by Western blot and used as internal loading controls, respectively. B: The NQE mutation did not alter CD82-β1 and -α3 integrin associations. Cells were lysed in 1% Brij 97, immunoprecipitated with CD82 mAB TS82b, resolved on SDS-PAGE, and transferred to nitrocellulose membrane. The membrane was blotted with integrin β1 mAb TS2/16 or integrin α3 polyclonal antibody D23. Tubulin, integrin β1, or integrin α3 from the same cell lysates was detected in Western blot and used as internal loading controls, respectively. C: Quantification of CD82-associated proteins (left). The tetraspanins and integrins coprecipitated with CD82, as shown in (A) and (B), were quantified by the densitometry analysis of the bands, and the average band densities ± SD from three experiments were plotted as the histogram. Between wild-type and NQE mutant, P values are <0.01 in CD9 and CD151 and >0.05 in integrin α3 and β1 coprecipitations. The comparison of the CD82 protein loading (right). CD82 proteins were immunoprecipitated from an equal number of S35-labeled Du145-CD82 wild-type and NQE transfectant cells and detected by autoradiography after SDS-PAGE separation. The CD82 protein bands were quantified with densitometry analysis, and the average band densities ± SD from three experiments were plotted as the histogram. P > 0.05.
Interestingly, the levels of a CD9 antigen epitope that is detected by CD9 mAb C9BB were drastically increased in both wild-type and NQE mutant in Du145 cells (Table 1). Unlike CD9 mAb MAB7 that probes all CD9 molecules, C9BB is a low-affinity CD9 mAb and recognizes only homoclustered CD9.43 The increased number of C9BB epitopes indicates that KAI1/CD82 expression results in markedly more CD9 homoclusters in Du145 cells and the NQE mutation does not disrupt, but even further enhances, the formation of CD9 homoclusters. In PC3 prostate cancer cells KAI1/CD82 expression however decreased the C9BB epitope level but still inhibited cell motility (data not shown). Hence, KAI1/CD82 expression-induced formation of CD9 homoclusters in Du145 cells is independent of the motility-inhibitory activity of KAI1/CD82.
For KAI1/CD82-integrin associations, earlier studies demonstrated that KAI1/CD82 associates with β1 integrins such as α4β1, α3β1, and α6β1.3 We determined the effect of NQE mutation on the association of KAI1/CD82 with β1 integrins. We found that the NQE mutation did not alter the association between KAI1/CD82 and β1 integrins (Figure 4B, left panel). Because α3β1 integrin is the predominant β1 integrin in Du145 cells,32 we also analyzed the KAI1/CD82-α3β1 integrin association and found no alteration in this association either (Figure 4B, right panel). The total amount of cellular integrin β1 or α3 subunit proteins remained equivalent among Mock, KAI1/CD82 wild-type, and the NQE transfectants (Figure 4B). Also, integrin β1 and α3 subunits were equivalently expressed among Du145 transfectants (Table 1). Hence, the TM polar residues appear not to be critical for KAI1/CD82-integrin interaction.
The levels of KAI1/CD82 proteins precipitated by its mAb are equivalent between the wild-type and NQE transfectants as revealed by the average quantity of 35S-labeled KAI1/CD82 proteins from several immunoprecipitation experiments (Figure 4C).
Tetraspanins CD9 and CD151 Are Not Required for CD82 Function in Motility Inhibition
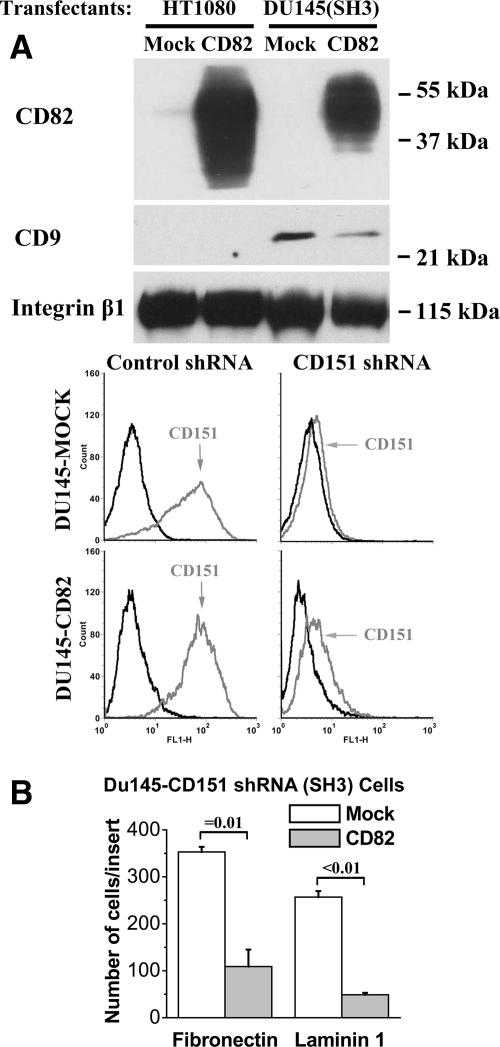
Besides KAI1/CD82, other tetraspanins also regulate cell movement.8,9 Du145 cells express tetraspanins CD9 and CD151. For CD9, data regarding its role in cell movement are mixed: inhibitory in many cases but also promoting in a few cases.8,9,44 For CD151, lines of evidence indicate that it enhances cell movement.8,9 These tetraspanins along with KAI1/CD82 are present in TEM.3,8,9 Hence, based on the diminished KAI1/CD82-tetraspanin association in NQE mutant, it is possible that KAI1/CD82 inhibits cell movement by either up-regulating the motility-inhibitory activity of CD9 or down-regulating the motility-promoting activity of CD151. To test these possibilities, we assessed whether KAI1/CD82 can still inhibit cell migration when CD9 or CD151 is absent. HT1080 fibroblastoma cells do not express CD9 (Figure 5A)45,46 or KAI1/CD82 (Figure 5A). The forced expression of KAI1/CD82 in HT1080 cells still resulted in the significant inhibition of cell migration on both FN and LN1 (Figure 2A right panel), indicating that CD9 is not required for the motility-inhibitory activity of KAI1/CD82. Because Du145 cells express CD151, we specifically silenced CD151 in Du145 transfectant cells (Figure 5A).36 When CD151 was silenced, we found again that KAI1/CD82 could markedly inhibit cell migration on different ECMs (Figure 5B, indicating that CD151 is not essential for KAI1/CD82 function either.
Figure 5.
Tetraspanins CD9 and CD151 are not required for the motility-inhibitory activity of CD82. A: The expression of tetraspanins in HT1080 and Du145 cells. Top: tetraspanin CD9, CD82, and CD151 proteins were analyzed using the cell lysates generated from HT1080-Mock, HT1080-CD82, Du145-Mock/Control shRNA, Du145-CD82/Control shRNA, Du145-Mock/CD151 shRNA, and Du145-CD82/CD151 shRNA transfectants by Western blot. The β1 integrin proteins were used as a protein loading control. Bottom: the flow cytometry analysis of CD151 expression in Du145-Mock and -CD82 cells that stably express control and CD151 shRNA. Black line: negative control mAb mouse IgG; gray line: CD151 mAb 5C11. B: The inhibition of cell migration by CD82 is independent of CD151. The Transwell migration onto FN (10 μg/ml) and LN1 (10 μg/ml) was measured in Du145-Mock/CD151 shRNA and -CD82/CD151 shRNA transfectants as described above. The results represent the mean ± SE number of three independent experiments; P = 0.01 between the Mock/CD151 shRNA and CD82/CD151 shRNA transfectant cells on FN and <0.01 on LN1.
TM Polar Residues Do Not Play Essential Roles in the Glycosylation and Homodimerization of KAI1/CD82
KAI1/CD82 is glycosylated.10,11,12 It has been reported that glycosylation correlates with the surface expression and motility-inhibitory activity of KAI1/CD82.10,24 Because we found in this study that the TM polar residues of KAI1/CD82 are also needed for the motility-inhibitory activity of KAI1/CD82, we next analyzed the role of TM polar residues in the glycosylation of KAI1/CD82. Both KAI1/CD82 wild-type and NQE mutant proteins, which were immunoprecipitated from 35S-labeled Du145 transfectants, exhibited smear bands in SDS-PAGE (Figure 6A, top panel). The intensity of the KAI1/CD82 band in the NQE mutant is slightly lower than the one in wild-type when the same amounts of cell lysates were analyzed, probably because of the less stable antigen epitope of NQE mutant proteins to the 1% NP40 detergent extraction. But both bands displayed a smear-like shape, which typically represents the various degrees of glycosylation. Neither smear showed any significant difference in length and position when the KAI1/CD82 band in the NQE mutant was adjusted to the same intensity (the lane with an asterisk in Figure 6A, top panel) as the one in wild-type. From the immunoprecipitation profile of biotinylated or the cell surface KAI1/CD82 (Figure 6A, bottom panel), we found that, under the 1% Brij 97 lysis condition, the surface KAI1/CD82 in the NQE mutant appears to be highly glycosylated with molecular weights slightly less than 49 kDa. The surface KAI1/CD82 in wild-type exhibits as diffused bands with molecular weights higher than 35 kDa (Figure 6A, bottom panel). These results suggest that the Brij 97-soluble, TEM-associated, cell surface KAI1/CD82 proteins are possibly differentially glycosylated in wild-type and NQE transfectants. Other bands found under the 1% Brij 97 lysis condition likely represent KAI1/CD82-associated proteins such as CD9, because these bands were diminished under the cell lysis condition of 1% NP40, a more stringent detergent that disrupts KAI1/CD82-associated TEM. Under the 1% NP-40 lysis condition, the immunoprecipitation profiles of KAI1/CD82 mAb were basically identical between wild-type and NQE mutant (Figure 6A, bottom panel). Because the KAI1/CD82-associated cell surface proteins typically dissociate from KAI1/CD82 under the 1% NP-40 lysis condition,3 the KAI1/CD82 mAb-precipitated proteins are very likely to be the KAI1/CD82 per se that had reached the cell surface and were glycosylated to various degrees.
Figure 6.
The TM polar residues are not essential for CD82 glycosylation and dimerization but are needed for its conformational stability. A: The NQE mutant is glycosylated. Top: Cells were labeled overnight with S35-methionine, lysed in 1% NP-40, and then immunoprecipitated with CD82 mAb TS82b. After SDS-PAGE, the immunoprecipitates were visualized by autoradiography. Bottom: Du145 transfectant (Mock, CD82 WT, and NQE mutant) cells were biotinylated at 4°C for 1 hour and lysed in either 1% Brij 97 or 1% NP-40. The lysates were co-immunoprecipitated with CD82 mAb M104. The precipitates were resolved in nonreducing SDS-PAGE followed by transfer to nitrocellulose membrane. The membrane was blotted with Extravidin, and the detected proteins were visualized by chemiluminescence. B: The NQE mutant forms dimer. Left: Du145 transfectant cells were cross-linked with DSP as described in Materials and Methods, treated with 1% NP-40, immunoprecipitated with CD82 mAb, and then immunoblotted with the same mAb. β1-tubulin in the same cell lysates was used as an internal loading control. The asterisks indicate the longer exposure of the NQE lanes. Right: The ratios of dimer to monomer were quantified based on the densities of dimer and monomer bands and are presented as the average ± SD of the results from 3 experiments. P < 0.05. C: The TM polar residues are needed for the conformational stability of CD82. Left: Du145-Mock, -CD82 wild-type, and -CD82 NQE transfectant cells in equal number were lysed with 1% Brij 97 lysis buffer. CD82 in the lysates was immunoprecipitated by its mAb TS82b and then treated with or without denature at 95°C for 5 minutes. After nonreducing SDS-PAGE separation and transferring to nitrocellulose membrane, CD82 proteins on the membrane were probed with its mAb TS82b, followed by the peroxidase-conjugated second Ab blot and chemiluminescence. β-tubulin from the same cell lysates, detected by Western blot, was used as an internal loading control. CD82 proteins immunoprecipitated from an equal number of S35-labeled Du145-Mock, -wild-type, and -NQE transfectant cells and revealed by autoradiography after SDS-PAGE separation were also included as loading control. Right: The CD82 bands were quantified with densitometry analysis, the band density of CD82 was normalized with the one of tubulin, and the average band densities of CD82 ± SD from three experiments were plotted as the histogram. P values are indicated in the histogram. D: The interactions of polar residues in homology model of CD82. An atomic model of CD82 was constructed with homology model followed by 5000-step energy minimization in AMBER, shown in the left section. The shaded area in the left mimics the membrane. The detailed interactions of polar residues N17, Q99, and E242 were displayed in enlarged images in the right part. The upper right image shows the locations of Q99 in TM3 and E242 in TM4, both of which point to inside TM helix bundle. The bottom right one represents the interaction between N17 in TM1 and S249 in TM4, which forms a hydrogen bond with a distance of 2.91Å.
It has been previously demonstrated that tetraspanins naturally undergo oligomerization and form mainly homodimer33 and that the membrane-proximal portions of large extracellular loop, TM domains, and probably cytoplasmic domains confer the intermolecular interaction to form dimers.13 We analyzed the effect of the NQE mutation on the dimerization of KAI1/CD82. To determine the dimerization, we treated Du145-Mock, -KAI1/CD82 wild-type, and -KAI1/CD82 NQE transfectant cells with a chemical cross-linker DSP under the mild cell lysis condition (1% Brij 97) in which tetraspanin dimerization is sustained.13 Then 1% NP-40, a stringent detergent, was added to the DSP-treated and -untreated cell lysates to disrupt tetraspanin dimers that had not been covalently cross-linked by DSP,13 followed by KAI1/CD82 immunoprecipitation and immunoblot. We found that the NQE mutant could also be dimerized (Figure 6B, left panel). After DSP cross-linking, the KAI1/CD82 antigen epitope in NQE mutant seemed to become even less stable, evidenced by less NQE protein monomer in the DSP-treated group than in the untreated group. In terms of the ratio of KAI1/CD82 dimer and monomer, it was much higher in the NQE mutant probably as a result of more dimerization (Figure 6B, right panel). However, if we consider the structural vulnerability of NQE mutant proteins, it is more likely that the antigen epitopes in dimers are more stable than in monomers. If so, the difference between wild-type KAI1/CD82 and the NQE mutant in the ratio of dimer to monomer may not be significant. At least it is unlikely that this ratio is lower in the NQE mutant than in wild-type.
TM Polar Residues Are Needed for the Conformational Stability of KAI1/CD82 Proteins
The TM polar residues could be involved in protein folding by affecting the interactions between TM helices.47 As elucidated above, by using various CD82 mAbs against different antigen epitopes located in CD82 LEL, we found that the NQE mutation does not significantly alter either the global structure of CD82 or the conformation of these antigen epitopes in LEL under the native condition (Table 1). We then further determined the role of TM polar residues in maintaining the conformation stability of CD82. From lysates of the metabolically labeled transfectant cells, CD82 mAb TS82b precipitated equivalent or sometimes slightly lower amounts of CD82 proteins from the wild-type and NQE transfectants (Figure 6A top panel, Figure 6C left panel, and Figure 4C right histogram), indicating that the TS82b antigen epitope of CD82 NQE remains largely stable during cell lysis with 1% NP-40 detergent and immunoprecipitation. However, after nonreducing SDS-PAGE separation, the higher molecular weight or more glycosylated form (approximately 40 to 50 kDa) of NQE mutant proteins became less detectable in the immunoblot of the same CD82 mAb while the lower molecular weight or less glycosylated species (approximately 30 to 40 kDa) became barely detectable (Figure 6C left panel) or undetectable (data not shown). The heat denaturation (95°C, 5 minutes) made CD82 proteins in the NQE mutant even less detectable in the immunoblot, compared with the one without heat denature (Figure 6C). This result suggests that the NQE mutant, at least the TS82b epitope, becomes unstable or labile and cannot resist denaturation treatments of heat and/or SDS. Although wild-type and NQE were expressed equivalently on the cell surface, the NQE mutant proteins became much less biotinylated compared with the wild-type proteins (Figure 6A, bottom panel), suggesting that either the ε-amino group of lysine residues, the target of biotinylation, in the KAI1/CD82 large extracellular loop becomes inaccessible to biotin after NQE mutation or the antigen epitope of NQE mutant becomes labile after biotinylation. Together, the TM polar residues appear to be needed for conformational stability, although the global structure of the KAI1/CD82 large extracellular loop was not significantly altered on NQE mutation.
The Predicted Roles of TM Polar Residues of KAI1/CD82 in TM Helix Interaction
Collectively, the above observations strongly suggest that TM polar residue-mediated TM interactions sustain the conformational stability of KAI1/CD82. To further assess the roles of the TM polar residues, we analyzed the TM interactions mediated by the N17, Q99, and E242 residues of KAI1/CD82 by using homology modeling. As shown in Figure 6D, homology modeling predicted that the β-amine group of Asn17 (N) residues at the face of TM1 helix forms a hydrogen bond with the β-hydroxyl group of the Ser249 (S) residue at the face of TM4 helix. The molecular modeling also predicts that the δ-amine group of the Gln99 (Q) residue in TM3 and the δ-oxygen group of the Glu242 (E) residue in TM4 project toward the core of four tightly packed helical bundles and form the charged interaction between each other. Together, the modeling predicts that the TM polar residues N, Q, and E of KAI1/CD82 mediate mainly the intramolecular helix-helix interaction.
Discussion
A Critical Role of the TM Polar Residues in CD82 Function
Three strong TM polar residues Asn, Gln, and Asp, one (Asp) with an acidic side chain and two (Asn, Gln) with neutral ones, are highly conserved among all vertebrate orthologs of CD82. Besides Asn, Gln, and Glu residues, there are weak polar residues such as Thr and Ser in CD82 TM regions. Because earlier studies showed no contribution of isolated Thr and Ser polar residues to the TM helix interactions16,17 and the Thr and Ser polar residues in CD82 TM regions are not located in SxxSSxxT and SxxxSSxxT motifs,48 we did not include them in this study. Many other tetraspanins also contain various numbers of Asn, Gln, Asp, and Glu residues in different TM domains, and in many cases these polar residues are also located in TM1, TM3, and TM4 helices. Despite the prevalence of polar residues, the biochemical and biological roles of these conserved TM polar residues in tetraspanins have not been assessed systematically. A mutation of the Asn (N) residue in the TM1 domain of tetraspanin CD9 results in less homodimerization of CD9,13 while the biochemical roles of the other two TM polar residues in CD9 remain to be determined. A Glu → Ala mutation in the TM3 domain of tetraspanin UPIb causes its endoplasmic reticulum (ER) retention,15 probably because of the misfolding of the proteins. However, the roles of TM polar residues in biological functions of CD9 and UPIb remain unknown. In this study, a mutation that replaces all three TM polar residues in CD82 partially or largely attenuates the migration-, invasion-, and metastasis-suppressive activities of CD82. Thus, the TM regions and the interactions within the lipid bilayers mediated by the TM regions are critical for CD82 function.
The mechanism by which CD82 inhibits cell migration is still largely unknown.1,2,3 One possibility is that CD82 per se is a solicitor that initiates or transduces a signal to inhibit cell motility, and another is that CD82 alters the membrane compartmentalization and trafficking of its associated TEM that contains key regulators for cell motility such as other tetraspanins, cell adhesion molecules, and growth factor receptors. We found earlier that, in PC3 metastatic prostate cancer cells, the reversal of cell motility-inhibition of a palmitoylation-deficient mutant of CD82 correlates with the diminished association of this mutant with tetraspanins CD9 and CD81.31 Thus, CD82 may inhibit cell movement by attenuating the biological activities of its associated tetraspanins 3, some of which are well documented in regulating cell movement. The mutation of CD82 TM polar residues also provides us an opportunity to determine which of two possible mechanisms comes into play. Although the diminutions of CD82-CD9 and -CD151 associations correlate well with the attenuated motility-inhibitory activity of the NQE mutant, we have demonstrated in this study that CD82 does not inhibit cell movement through CD9 or by suppressing CD151 function. We did not assess CD81, another major tetraspanin associating with CD82,3 because the expression of CD81 is relatively low, compared with CD9 and CD151, in Du145 cells (our unpublished data). This observation, however, does not exclude the possibility that other TEM components, which are not necessarily tetraspanins, are needed for CD82 function. In fact, the TM domains of CD82 may play a critical role in binding CD82 to membrane lipids such as cholesterol and GM2.48,49 Whether the NQE mutation affects CD82-lipid interaction remains to be determined. It is puzzling that CD82-integrin α3β1 association remained unchanged in the NQE mutant when CD82-CD151 association was markedly diminished, because every integrin α3β1 molecule directly binds to CD151.27 Since some cellular CD151 proteins are free of integrin α3β1 binding,28 we would extrapolate that free CD151 is the major pool associated with the NQE mutant. In other words, the NQE mutation did not alter the association with the integrin α3β1-bound CD151, CD82-integrin α3β1 association is unlikely mediated by CD151, and the NQE mutation may affect the functional cross talk between CD82 and integrins and then integrin-dependent cell movement.
Thus, not only are the TM regions of CD82 actively involved in the physical interactions inside the lipid bilayer, but also the proper TM helix interactions in CD82 protein are needed for maintaining the stability of a function-competent conformation of CD82.
The Critical Biochemical and Structural Features for CD82 Function
Because the NQE mutant is functionally compromised, it becomes a useful tool to identify the structural element(s) critical for CD82 function by comparing biochemical properties of this mutant with CD82 wild-type. Like other tetraspanins, CD82 possesses the biochemical features such as TEM association, disulfide bond formation and glycosylation in the LEL, palmitoylation in the interface of TM and cytoplasmic domains, and, possibly, dimerization.
CD82 is N-glycosylated, and three glycosylation sites are located in the LEL.5,24 Ono et al found that, in a CHO cell line deficient in UDP-Glc 4-epimerase, CD82 inhibited cell migration only when galactose was added to the media. This observation suggests that CD82 functions only when it is glycosylated in the cells that synthesize GM3 ganglioside.5 Because glycosylation of many proteins and GM3 synthesis were affected in that cell line, it is hard to discern that the phenotype resulted from the simple lack of glycosylation of CD82 or the combinatory effect of deficiencies in GM3 synthesis and universal glycosylations including CD82 glycosylation. Thus, whether the glycosylation of CD82 per se is required for CD82’s inhibitory activity still remains elusive. The NQE mutant, with less motility-inhibitory activity, does not display a significant alteration in total glycosylation of CD82 (Figure 6A, top panel), suggesting that glycosylation en bloc is not directly involved in CD82 function. A glycosylated CD82 species that may directly associate with TEM at the plasma membrane could not be detected from the cell surface of NQE mutants under Brij 97 lysis condition (Figure 6A, bottom panel). Notably, this low molecular weight or less glycosylated form of CD82 is the structurally unstable one (Figure 6C, left pamel) but likely to be the functionally important form because it is present only in wild-type. Thus, glycosylation stabilizes CD82 conformation, and a specifically glycosylated species of CD82 is its functional form.
Earlier studies demonstrated that tetraspanins RDS, ROM1, CD9, UPIa, and UPIb could form either a homodimer33,50 or a heterodimer.51 Based on the study of CD9, it has been proposed that the dimer is the functional unit of tetraspanin.33 We found that CD82 is dimerized and the NQE mutation results in relatively more dimerization. Relatively more dimerization of the NQE mutant could result from either a higher tendency of the NQE mutant to form a dimer or the higher structural instability of the NQE monomer than the one of dimer in SDS-PAGE and immunoblot. Hence, CD82 dimerization may not be required for the motility-inhibitory activity of CD82 and for the associations of CD82 with CD9 and CD151. Although the TM polar residue with a carboxyl or carboxamide side chain (Glu, Asp, Gln, or Asn) can stabilize TM helix interactions and form SDS-resistant homodimers or homotrimers,16,17 they may not be critical for CD82 homodimerization but rather important for the TEM association. This phenotype is compatible with the roles of TM polar residues of T cell receptor α and β chains, ie, not needed for the dimerization of α and β chains but critical for CD3 association.18,19,20,21,22
Why Are the TM Polar Residues Important for the Function of CD82?
The strong polar residues in TM peptide helices play a crucial role in the physical association between TM peptide helices.16,17,47 The interactions between TM peptides could be intramolecular or intermolecular.18,19,20,21,22 Hence, besides affecting TM protein conformational stability, TM polar residues contribute to the assembly of multiple TM protein complexes such as T cell receptors.18,19,20,21 The conserved TM polar residues are typically located on the outer faces of the TM helices and likely form interhelical hydrogen bonds.16,17,47 Besides polar residues, the tight packing of bulky residues against Gly residues along two TM helices also contributes to TM domain interaction.47 For example, the specific packing of bulky residues against small residues helps define the TM1-TM2 interaction in CD9.13
The NQE mutation appears not to affect the global structure of CD82 proteins, namely the proper folding of the LEL. Because CD82 mAbs recognize various antigen epitopes and the recognitions are sensitive to reduction, we predict that these epitopes are localized in the LEL that is framed by three pairs of disulfide bonds. In flow cytometry and immunoflorescence, different CD82 mAbs appear to bind to the native wild-type and NQE mutant CD82 proteins equally well. Also, from the cell lysate extracted with 1% NP-40 detergent from the metabolically labeled cells, CD82 mAb immunoprecipitated similar amount of wild-type and mutated CD82 proteins, indicating that the antigen epitope in the LEL of the NQE mutant is largely retained in 1% NP-40 mediated cell lysis. Moreover, once the interactions between the TM domains are disrupted, tetraspanins form protein aggregates intracellularly likely as a result of destabilized or misfolded conformation.15,33 The NQE mutant did not form aggregates inside cells and expressed equally at the cell surface as the wild-type, further suggesting that the global structure of CD82 remains intact. However, the conformation of CD82 becomes less stable. Therefore, we conclude that CD82 contains function-competent conformation(s), the TM polar residues are required for sustaining this conformation, and this conformation depends on TM interactions.
If we classify TM interactions into polar residue-dependent and -independent interactions, the former are needed for the motility-inhibitory activity of CD82. The TM polar residue-dependent interactions of CD82 TM helices include both intramolecular and intermolecular ones. The labile antigen epitopes in the LEL of the NQE mutant likely result from aberrant intramolecular TM interactions. Although the diminished associations of CD82 NQE mutant with CD9 and CD151 suggest that TM polar residues possibly engage in intermolecular TM interactions, they are more likely to be the secondary effect of aberrant intramolecular TM interactions. Nevertheless, our observation underscores that the TM interaction-dependent tetraspanin conformation plays a critical role in the tetraspanin-tetraspanin interactions within TEM or in maintaining the integrity of TEM.
The hydrogen bond could form between the side chains of two TM polar residues.47 It could also form between the side chain of one TM polar residue and the side chain or backbone of a residue at the corresponding position of an adjacent TM helix.47 Based on molecular modeling, we predict that the N, Q, and E polar residues are mainly involved in intramolecular TM helix-helix interactions of CD82. The helical bundle interaction between TM3 and TM4 is likely needed for the proper folding of LEL.52,53 On the NQE mutation, the TM3-TM4 interaction likely becomes weakened, which subsequently causes the less stable conformation of LEL. The first TM domain of CD82 has been shown to be required for the ER exit.54 The contribution of TM polar residues to the ER exit was also found in tetraspanin UPIb, in which a mutation of the Glu residue in the third TM domain causes the ER retention of UPIb.15 We found in this study that the Asn17 residue in the first TM region of CD82 unlikely plays a role of CD82’s first TM region in the ER exit. In addition, the Asn17 residue appears not to be needed for the dimerization of CD82, although the corresponding polar residue in the CD9 TM1 helix is involved in the homodimerization of CD9.33
Together, the polar residue-mediated TM interactions are essential for sustaining CD82 in a functionally competent conformation and therefore critical for its function as a motility suppressor. The novelty and significance of this study are multiple. First, CD82 suppresses cell motility directly rather than through its associated TM proteins. Also, due to the tight connection of tetraspanins to cancer,8,9 TM interactions may be needed for the proper function of other tetrasapnins in cancer progression. Moreover, since many proteins that promote cancer progression are the TM proteins and the TM regions can regulate the functions of TM proteins, the under-appreciated TM regions can be used as target sites for cancer therapeutics. For example, tetraspanins CD151 and CO-029 promote cancer invasion and metastasis, so perturbation of their TM polar resides may halt cancer progression. Last, CD82 inhibits the microprotrusion formation, and the TM polar residues are critical for this inhibition. Microprotrusions are distinct from filapodia in morphology and number,45 but they may share a similar morphogenetic mechanism because of their protrusive nature. A plausible prediction would be that tetraspanins control motility by modulating the formation of microprotrusions. Because the CD82 conformation determined by TM interactions regulates these membrane-bending structures, the role of tetraspanins in curving membranes becomes an intriguing issue deserving further exploration.
Acknowledgments
We thank Drs. Lisa Jennings and David Armbruster for constructive suggestions and critical review of the manuscript. We thank Drs. Kiyotoshi Sekiguchi, Masashi Yamada, Eric. Rubinstein, Chikao Morimoto, Osamu Yoshi, Lisa Jennings, Martin Hemler, and all other colleagues who provided the antibodies and constructs used in this study.
Footnotes
Address reprint requests to Xin A. Zhang, Vascular Biology Center, Coleman Building Room H300, 956 Court Avenue, Memphis, TN, 38163. E-mail: xazhang@utmem.edu.
Supported by National Institutes of Health Research Grant CA-96991 and Department of Defense Research Grant W81XWH-04-1-0156 (to X.A.Z).
References
- Jackson P, Marreiros A, Russell PJ. KAI1 tetraspanin and metastasis suppressor. Int J Biochem Cell Biol. 2005;37:530–534. doi: 10.1016/j.biocel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Tonoli H, Barrett JC. CD82 metastasis suppressor gene: a potential target for new therapeutics? Trends Mol Med. 2005;11:563–570. doi: 10.1016/j.molmed.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Liu WM, Zhang XA. KAI1/CD82, a tumor metastasis suppressor. Cancer Letter. 2006;240:183–194. doi: 10.1016/j.canlet.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Dong J-T, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Issacs JT, Barrett JC. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268:884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- Ono M, Handa K, Withers DA, Hakomori S. Motility inhibition and apoptosis are induced by metastasis-suppressing gene product CD82 and its analogue CD9, with concurrent glycosylation. Cancer Res. 1999;59:2335–2339. [PubMed] [Google Scholar]
- Schoenfeld N, Bauer MKA, Grimm S. The metastasis suppressor gene C33/CD82/KAI1 induces apoptosis through reactive oxygen intermediates. FASEB J. 2003;18:158–160. doi: 10.1096/fj.03-0420fje. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Zhan R, Chaudhuri A, Watabe M, Pai SK, Hirota S, Hosobe S, Tsukada T, Miura K, Takano Y, Saito K, Pauza ME, Hayashi S, Wang Y, Mohinta S, Mashimo T, Iiizumi M, Furuta E, Watabe K. Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nat Med. 2006;12:933–938. doi: 10.1038/nm1444. [DOI] [PubMed] [Google Scholar]
- Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Imai T, Fukudome K, Takagi S, Nagira M, Furuse M, Fukuhara N, Nishimura M, Hinuma Y, Yoshie O. C33 antigen recognized by monoclonal antibodies inhibitory to human T cell leukemia virus type 1-induced syncytium formation is a member of a new family of transmembrane proteins including CD9, CD37, CD53, and CD63. J Immunol. 1992;149:2879–2886. [PubMed] [Google Scholar]
- Gil ML, Vita N, Lebel-Binay S, Miloux B, Chalon P, Kaghad M, Marchiol-Fournigault C, Conjeaud H, Caput D, Ferrara P. A member of the tetra spans transmembrane protein superfamily is recognized by a monoclonal antibody raised against an HLA class I-deficient, lymphokine-activated killer-susceptible, B lymphocyte line. cloning and preliminary functional studies. J Immunol. 1992;148:2826–2833. [PubMed] [Google Scholar]
- Nojima Y, Hirose T, Tachibana K, Tanaka T, Shi L, Doshen J, Freeman GJ, Schlossman SF, Morimoto C. The 4F9 antigen is a member of the tetra spans transmembrane protein family and functions as an accessory molecule in T cell activation and adhesion. Cell Immunol. 1993;152:249–260. doi: 10.1006/cimm.1993.1285. [DOI] [PubMed] [Google Scholar]
- Kovalenko OV, Metcalf DG, DeGrado WF, Hemler ME. Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct Biol. 2005;5:11. doi: 10.1186/1472-6807-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane proteins and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin family. Biophysical J. 2006;90:212–227. doi: 10.1529/biophysj.105.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Kong XP, Sun TT, Kreibich G. Integrity of all four transmembrane domains of the tetraspanin uroplakin Ib is required for its exit from the ER. J Cell Sci. 2006;119:5077–5086. doi: 10.1242/jcs.03285. [DOI] [PubMed] [Google Scholar]
- Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- Blumberg RS, Alarcon J, Sancho J, McDermott FV, Lopez P, Breitmeyer J, Terhorst C. Assembly and function of the T cell antigen receptor. Requirement of either the lysine or arginine residues in the transmembrane region of the alpha chain. J Biol Chem. 1990;265:14036–14043. [PubMed] [Google Scholar]
- Alocover A, Mariuzza RA, Ermonval M, Acuto O. Lysine 271 in the transmembrane domain of the T-cell antigen receptor beta chain is necessary for its assembly with the CD3 complex but not for alpha/beta dimerization. J Biol Chem. 1990;265:4131–4135. [PubMed] [Google Scholar]
- Fuller-Espie S, Hoffman Towler P, Wiest DL, Tietjen I, Spain LM. Transmembrane polar residues of TCR beta chain are required for signal transduction. Int Immunol. 1998;10:923–933. doi: 10.1093/intimm/10.7.923. [DOI] [PubMed] [Google Scholar]
- Fukudome K, Furuse M, Imai T, Nishimura M, Takagi S, Hinuma Y, Yoshie O. Identification of membrane antigen C33 recognized monoclonal antibodies inhibitory to HTLV-1 induced syncytium formation: altered glycosylation of C33 antigen in HTLV-1–positive cells. J Virol. 1992;66:1394–1401. doi: 10.1128/jvi.66.3.1394-1401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, Le Naour F, Oualid M, Billard M, Faure G, Hanash SM, Boucheix C, Rubinstein E. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J Biol Chem. 2001;276:14329–14337. doi: 10.1074/jbc.M011297200. [DOI] [PubMed] [Google Scholar]
- White MM, Foust JT, Mauer AM, Robertson JT, Jennings LK. Assessment of lumiaggregometry for research and clinical laboratories. Thromb Haemost. 1992;67:572–577. [PubMed] [Google Scholar]
- Yauch RL, Berditchevski F, Harler MB, Reichner J, Hemler ME. Highly stoichiometric, stable and specific association of integrin α3β1 with CD151 provides a major link to PI 4-kinase and may regulate cell migration. Mol Biol Cell. 1998;9:2751–2765. doi: 10.1091/mbc.9.10.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi R, Sanzen N, Nada S, Sumida Y, Wada Y, Okada M, Takagi J, Hasegawa H, Sekiguchi K. Potentiation of the ligand-binding activity of integrin α3β1 via association with CD151. Proc Natl Acad Sci USA. 2005;102:1939–1944. doi: 10.1073/pnas.0409493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler ME, Sanchez-Madrid F, Flotte TJ, Krensky AM, Burakoff SJ, Bhan AK, Springer TA, Strominger JL. J Immunol. 1984;132:3011–3018. [PubMed] [Google Scholar]
- Zhang XA, Bontrager AL, Stipp CS, Kraeft SK, Bazzoni G, Chen LB, Hemler ME. Phosphorylation of a conserved integrin alpha 3 QPSXXE motif regulates signaling, motility, and cytoskeletal engagement. Mol Biol Cell. 2001;12:351–365. doi: 10.1091/mbc.12.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility-inhibitory activity. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- Zhang XA, He B, Zhou B, Liu L. Requirement of p130CAS-Crk coupling in KAI1/CD82-mediated suppression of cell migration. J Biol Chem. 2003;278:27319–27328. doi: 10.1074/jbc.M303039200. [DOI] [PubMed] [Google Scholar]
- Kovalenko OV, Yang X, Kolesnikova TV, Hemler ME. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem J. 2004;377:407–417. doi: 10.1042/BJ20031037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse C, Ishida Y, Hikita T, Asai T, Oku N. Junctional adhesion molecule-C promotes metastatic potential of HT1080 human fibrosarcoma. J Biol Chem. 2007;282:8276–8283. doi: 10.1074/jbc.M608836200. [DOI] [PubMed] [Google Scholar]
- Schweinitz A, Steinmetzer T, Banke IJ, Arlt MJ, Stürzebecher A, Schuster O, Geissler A, Giersiefen H, Zeslawska E, Jacob U, Krüger A, Stürzebecher J. Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents. J Biol Chem. 2004;279:33613–33622. doi: 10.1074/jbc.M314151200. [DOI] [PubMed] [Google Scholar]
- Winterwood NE, Varzavand A, Meland MN, Ashman LK, Stipp CS. A critical role for tetraspanin CD151 in {alpha}3{beta}1 and {alpha}6{beta}4 integrin-dependent tumor cell functions on laminin-5. Mol Biol Cell. 2006:2707–2721. doi: 10.1091/mbc.E05-11-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Research. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- Onufriev A, Bashford D, Case DA. Modification of the generalized born model suitable for macromolecules. J Phys Chem B. 2000;104:3712–3720. [Google Scholar]
- Iwata S, Kobayashi H, Miyake-Nishijima R, Sasaki T, Souta-Kuribara A, Nori M, Hosono O, Kawasaki H, Tanaka H, Morimoto C. Distinctive signaling pathways through CD82 and beta1 integrins in human T cells. Eur J Immunol. 2002;32:1328–1337. doi: 10.1002/1521-4141(200205)32:5<1328::AID-IMMU1328>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Hinoda Y, Sato S, Itoh F, Adachi M, Hareyama M, Imai K. Reduced invasive and metastatic potentials of KAI1-transfected melanoma cells. Jpn J Cancer Res. 1998;89:397–404. doi: 10.1111/j.1349-7006.1998.tb00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wei LL, Tang C, Slack R, Mueller S, Lippman ME. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 2001;61:5284–5288. [PubMed] [Google Scholar]
- Yang XH, Kovalenko OV, Kolesnikova TV, Andzelm MM, Rubinstein E, Strominger JL, Hemler ME. Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J Biol Chem. 2006;281:12976–12985. doi: 10.1074/jbc.M510617200. [DOI] [PubMed] [Google Scholar]
- Longhurst CM, Jacobs JD, White MM, Crossno JT, Jr, Fitzgerald DA, Bao J, Fitzgerald TJ, Raghow R, Jennings LK. Chinese hamster ovary cell motility to fibronectin is modulated by the second extracellular loop of CD9. Identification of a putative fibronectin binding site. J Biol Chem. 2002;277:32445–32452. doi: 10.1074/jbc.M204420200. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Berditchevski F. Function of alpha3 beta1-tetraspanin protein complexes in tumor cell invasion. Evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2). J Cell Biol. 1999;146:1375–1389. doi: 10.1083/jcb.146.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L. EWI2/PGRL Associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 2003;63:2665–2674. [PubMed] [Google Scholar]
- Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struc Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- Charrin S, Rubinstein E. A physical and functional link between cholesterol and tetraspanins. Eur J Immunol. 2003;33:2479–2489. doi: 10.1002/eji.200323884. [DOI] [PubMed] [Google Scholar]
- Todeschini AR, Dos Santos JN, Handa K, Hakomori SI. Ganglioside GM2/tetraspanin CD82 complex inhibits Met activation, and its cross-talk with integrins: basis for control of cell motility through glycosynapse. J Biol Chem. 2007;282:8123–8133. doi: 10.1074/jbc.M611407200. [DOI] [PubMed] [Google Scholar]
- Goldberg AF, Moritz OL, Molday RS. Heterologous expression of photoreceptor peripherin/rds and Rom-1 in COS-1 cells: assembly, interactions, and localization of multisubunit complexes. Biochemistry. 1995;34:14213–14219. doi: 10.1021/bi00043a028. [DOI] [PubMed] [Google Scholar]
- Wu XR, Medina JJ, Sun TT. Selective interactions of UPIa and UPIb, two members of transmembrane 4 superfamily, with distinct single transmembrane-domained proteins in differentiated urothelial cells. J Biol Chem. 1995;270:29752–29759. doi: 10.1074/jbc.270.50.29752. [DOI] [PubMed] [Google Scholar]
- Min G, Wang H, Sun TT, Kong XP. Structural basis for tetraspanin functions as revealed by the cryo-EM structure of uroplakin complexes at 6-A resolution. J Cell Biol. 2006;173:975–983. doi: 10.1083/jcb.200602086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienstock RJ, Barrett JC. KAI1, a prostate metastasis suppressor: prediction of solvated structure and interactions with binding partners; integrins, cadherins, and cell-surface receptor proteins. Mol Carcinog. 2001;32:139–153. doi: 10.1002/mc.1073. [DOI] [PubMed] [Google Scholar]
- Cannon KS, Cresswell P. Quality control of transmembrane domain assembly in the tetraspanin CD82. EMBO J. 2001;20:2443–2453. doi: 10.1093/emboj/20.10.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]