Abstract
A novel Klebsiella pneumoniae carbapenemase (KPC) variant, designated blaKPC-5, was discovered in a carbapenem-resistant Pseudomonas aeruginosa clinical isolate from Puerto Rico. Characterization of the upstream region of blaKPC-5 showed significant differences from the flanking regions of other blaKPC variants. Comparison of amino acid sequences with those of other KPC enzymes revealed that KPC-5 was an intermediate between KPC-2 and KPC-4, differing from KPC-2 by a single amino acid substitution (Pro103→Arg), while KPC-4 contained Pro103→Arg plus an additional amino acid change (Val239→Gly). Transformation studies with an Escherichia coli recipient strain showed differences in the properties of the KPC variants. KPC-4 and KPC-5 both had pIs of 7.65, in contrast with the pI of 6.7 for KPC-2. KPC-2 transformants were less susceptible to the carbapenems than KPC-4 and KPC-5 transformants. These data correlated with higher rates of imipenem hydrolysis for KPC-2 than for KPC-4 and KPC-5. However, KPC-4 and KPC-5 transformants had higher ceftazidime MICs, and the enzymes from these transformants had slightly better hydrolysis of this drug than KPC-2. KPC-4 and KPC-5 were more sensitive than KPC-2 to inhibition by clavulanic acid in both susceptibility testing and hydrolysis assays. Thus, KPC enzymes may be evolving through stepwise mutations to alter their spectra of activity.
Klebsiella pneumoniae carbapenemase (KPC) is a molecular class A serine β-lactamase belonging to functional group 2f. KPC was first reported in 2001 after its discovery in a K. pneumoniae clinical isolate collected from North Carolina during the ICARE (Intensive Care Antimicrobial Resistance Epidemiology) surveillance study (35). An amino acid variant of KPC-1, termed KPC-2, was discovered shortly thereafter in Klebsiella spp. (27, 36) and Salmonella enterica (16). However, a recent correction of the blaKPC-1 sequence has indicated that the blaKPC-1 and blaKPC-2 sequences are identical, and thus, KPC-1 and KPC-2 are the same enzyme (35). Mutations in the blaKPC-2 structural gene have resulted in two variants, blaKPC-3 (33) and blaKPC-4 (20).
blaKPC has been detected in several members of the Enterobacteriaceae, not only on the East Coast of the United States (23) but also in several other U.S. locations (5, 13, 15, 24, 26). Most notably, Klebsiella spp. harboring the blaKPC gene have become endemic to New York City (3), and as a result, the New York Health Department has issued an advisory to control the spread of these organisms (8). KPC has since become a global concern, with the identification of this β-lactamase gene in organisms from various locations worldwide (18-20, 28-30). In Colombia alone, Villegas et al. (29) have reported the presence of blaKPC-2 in K. pneumoniae and Citrobacter freundii and the first detection of blaKPC in Pseudomonas aeruginosa (28). The acquisition of blaKPC by different bacterial species may be attributed to its location on transferable plasmids or to dissemination by mobile genetic elements. A study by Naas et al. (17) has described the association of blaKPC-2 with transposon Tn4401 on plasmids of various sizes from different organisms. Similarly, blaKPC-3 has been shown to reside on plasmids that can be moved through conjugal transfer (10, 33).
KPC-2 and KPC-3 hydrolyze several different classes of β-lactams, including the carbapenems (1, 36). Both enzymes were highly efficient at hydrolyzing nitrocefin, cephalothin (cefalotin), and cephaloridine, modestly efficient against the carbapenems and cefotaxime, and inefficient against cefoxitin and ceftazidime. In comparison, KPC-3 had slightly better hydrolytic activity against ceftazidime than KPC-2 (1). Despite the ability to hydrolyze the carbapenems in in vitro assays, KPC-producing organisms do not always display a resistant phenotype; KPC-2- and KPC-3-positive Klebsiella and Enterobacter spp. often have carbapenem MICs below the susceptible breakpoint (3, 4). KPC-positive strains are typically resistant to the penicillins, extended-spectrum cephalosporins, and aztreonam, but MICs of these drugs and the carbapenems are reduced in the presence of clavulanic acid due to enzyme inhibition (33, 36).
In a recent surveillance study of the mechanisms associated with carbapenem resistance in P. aeruginosa strains from Puerto Rico (32), blaKPC was detected in two genetically unrelated strains with high imipenem MICs (>64 μg/ml). The first P. aeruginosa strain, PR100, possessed blaKPC-2, while a novel KPC variant, blaKPC-5, was discovered in the second strain, PR280. The current study describes the characterization of blaKPC-5 and the properties of the encoded enzyme, KPC-5, in comparison with other blaKPC variants.
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa clinical isolates PR100 and PR280 were collected from the Puerto Rico Medical Center in 2006. Enterobacter cancerogenus strain E624, harboring blaKPC-4, was identified during a bacteremia resistance surveillance program in 2003 (20).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed by the agar dilution methodology according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (7). Results were interpreted (susceptible, intermediate, or resistant) according to breakpoints established by the CLSI. Antimicrobial agents were obtained as follows: cefoxitin and cefotaxime were from Sigma Chemical Co. (St. Louis, MO); piperacillin and tazobactam were from Wyeth Pharmaceuticals (Philadelphia, PA); aztreonam and cefepime were from Bristol-Myers Squibb (Princeton, NJ); ceftazidime was from GlaxoSmithKline (King of Prussia, PA); imipenem and ertapenem were from Merck & Co. (Rahway, NJ); and meropenem was from AstraZeneca (Waltham, MA).
Minimum spanning tree.
A minimum spanning tree was constructed from the aligned blaKPC sequence data using Bionumerics software (version 4.1; Applied Maths, Kortrijk, Belgium) in order to assess the evolutionary relationships between the variants. Linkage priority was assigned, in order of decreasing importance, to types with the highest number of (i) single-locus (base pair) variants, (ii) double-locus variants, and (iii) samples belonging to a single type.
PCR.
The full-length blaKPC genes and upstream regions from strains E624 and PR280 were amplified by PCR as previously described (27) using flanking primers KPC2FF1 (5′-GGTGATTCAGGGGTAAAGTGG-3′) and KPC1R (5′-GACAGTGGTGGTAATCCATGC-3′) or primers KPC1F (5′-GCTACACCTAGCTCCACCTTC-3′) and KPC1R with an annealing temperature of 46°C and 2 mM Mg2+.
Upstream sequence identification of blaKPC-5.
DNA from strain PR280 was obtained from an overnight culture using the Qiagen (Germantown, MD) DNeasy blood and tissue kit according to the manufacturer's instructions. The blaKPC-5 upstream sequence was generated using the GenomeWalker universal kit (Clontech Laboratories Inc., Mountain View, CA). A primary PCR was performed on StuI-digested DNA using the kit adaptor primer (AP1) and the blaKPC-specific primer KPC-2UPSEQ2 (5′-GTTATCACTGTATTGCACGG-3′) by using the parameters recommended in the protocol except that an annealing/extension temperature of 68°C was used in cycle step 1 and 63°C was used in cycle step 2. A nested PCR was performed using the nested adaptor primer (AP2) provided by the manufacturer and the gene-specific primer KPC-2UPSEQ1 (5′-GTCCTGTTCGAGTTTAGCG-3′) with the protocol-recommended parameters except that an annealing/extension temperature of 68°C was used in cycle step 1 and 60°C was used in cycle step 2. All PCRs were performed using Platinum Taq high-fidelity DNA polymerase (Invitrogen, Carlsbad, CA). The nested PCR product was extracted from a 1% agarose gel using the QIAquick gel extraction kit (Qiagen). The extracted product was cloned into vector pCR2.1 (Invitrogen), transformed into Escherichia coli TOP10 cells using the protocol supplied by the manufacturer, and sequenced. The upstream sequence was confirmed by sequencing a PCR product amplified with primers designed based on the GenomeWalker-generated sequence and a DNA template obtained from clinical isolate PR280.
Mapping of the start site of blaKPC-5 transcription.
The start site of blaKPC-5 transcription was determined by a 5′ rapid amplification of cDNA ends (RACE) procedure using a 5′ RACE kit (Invitrogen). Total bacterial RNA was isolated from a logarithmic-phase culture of strain PR280 as previously described (31). Eight micrograms of total RNA was treated with RNase-free DNase (Promega, Madison, WI) (1.25 U/μg of RNA) for 2 h at 37°C. First-strand cDNA synthesis was performed as recommended in the protocol by using 2 μg of DNase-treated RNA and the blaKPC-specific primer KPC5RRTR (5′-CGGCGTTATCACTGTATTGC-3′), which annealed 397 bp downstream of the blaKPC-5 translational start site. The dC-tailed cDNA was amplified by PCR using the Abridged Anchor Primer provided by the manufacturer and a second blaKPC-specific primer, KPC6R (5′-CGTTATCACTGTATTGCACG-3′), which annealed 394 bp downstream of the translational start codon. PCR was conducted for 35 cycles with the following parameters: denaturation at 94°C for 30 s, annealing at 46°C for 30 s, extension at 72°C for 2 min, and a final extension at 72°C for 7 min. The amplified product was purified, cloned, and sequenced as described above.
Cloning of the KPC variants.
blaKPC-2, blaKPC-4, and blaKPC-5 were amplified from their respective templates by PCR using primers KPCFRBS (5′-CAAACAAGGAATATCGTTG-3′) and KPC1R (5′-GACAGTGGTGGTAATCCATGC-3′), which flank the blaKPC structural gene but do not include promoter sequences. The 969-bp amplicons were cloned into the pCR2.1 vector and then subcloned into vector pACYC184 (Fermentas Inc., Glen Burnie, MD) using the restriction enzyme EcoRI (Invitrogen). The EcoRI site of pACYC184 is located within the chloramphenicol resistance gene, cat, and transcription of the cloned blaKPC genes initiates from the cat promoter. This method ensured that expression of the blaKPC genes would be equal between transformants, enabling direct comparisons between the cloned genes. The plasmids containing the blaKPC genes were transformed into competent E. coli TOP10 cells (Invitrogen).
IEF and β-lactamase hydrolysis assays.
Crude β-lactamase extracts were prepared from the blaKPC transformants, and the isoelectric points of the KPC variants were determined by analytical isoelectric focusing (IEF) on an Ampholine polyacrylamide gel (pH range, 3 to 10) according to previously described methods (27). Extracts from the blaKPC transformants were also used to assess the hydrolytic properties of the KPC variants by a spectrophotometric hydrolysis assay (27) using 100 μM concentrations of cephalothin, cefotaxime, and imipenem and a 50 μM concentration of ceftazidime as substrates. For inhibition assays, clavulanic acid (final concentration, 100 μM) was incubated with crude extracts from the blaKPC transformants at 30°C for 5 min in phosphate buffer (pH 7.0) prior to the addition of the substrate.
Nucleotide sequence accession number.
The blaKPC-5 nucleotide sequence was deposited in the GenBank database with accession number EU400222.
RESULTS
Sequence comparisons of the variant KPCs.
During the preparation of this article, the sequence of another novel blaKPC allele, blaKPC-6, was published in GenBank (accession number EU555534) and was included in the sequence analysis for all published blaKPCs below. DNA sequence analysis of the structural gene showed that blaKPC-5 was 99% identical to both blaKPC-2 and blaKPC-4, with only single-nucleotide differences from either β-lactamase gene. blaKPC-5 had the same nucleotide substitution at position 308 (C→G) as blaKPC-4 compared with blaKPC-2, blaKPC-3, and blaKPC-6, resulting in an amino acid change from proline to arginine (Table 1). While blaKPC-4 and blaKPC-6 contained a guanosine at position 716, coding for a glycine amino acid, the blaKPC-2, blaKPC-3, and blaKPC-5 sequences differed at this nucleotide location, with a thymine residue, coding for a valine (Table 1). blaKPC-3 was the only gene to have a thymine substitution at position 814 (coding for tyrosine272), whereas the other blaKPCs possessed a cytosine (coding for histidine272).
TABLE 1.
Nucleotide and amino acid variations between KPC enzymes
| KPC | Nucleotide (amino acid) at positiona:
|
||
|---|---|---|---|
| 308 (103) | 716 (239) | 814 (272) | |
| KPC-2 | C (Pro) | T (Val) | C (His) |
| KPC-3 | C (Pro) | T (Val) | T (Tyr) |
| KPC-4 | G (Arg) | G (Gly) | C (His) |
| KPC-5 | G (Arg) | T (Val) | C (His) |
| KPC-6 | C (Pro) | G (Gly) | C (His) |
Nucleotide (amino acid) positions in blaKPCs.
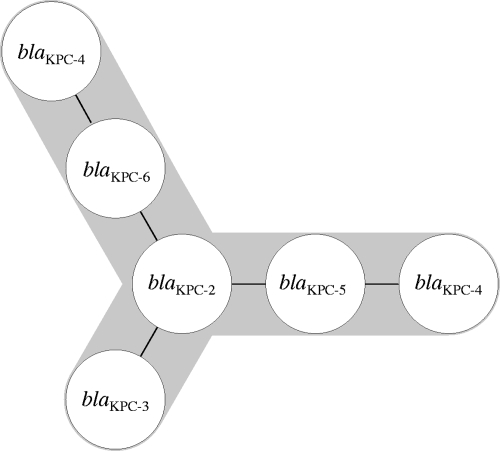
To evaluate the genetic and evolutionary relationships of the blaKPC variants, a minimum spanning tree was constructed on the basis pf sequence divergence. The spanning tree (Fig. 1) identified blaKPC-2 as the founding (i.e., ancestral) sequence, while blaKPC-3, blaKPC-5, and blaKPC-6 formed three separate branches as a result of single-nucleotide polymorphisms (SNPs). blaKPC-5 and blaKPC-6 serve as intermediate steps to the selection of blaKPC-4 (two SNPs), which may emerge through either pathway (Fig. 1). blaKPC-3 and blaKPC-4 have the highest number of nucleotide changes between variants, with three SNPs.
FIG. 1.
Minimum-spanning-tree depiction of interrelationships among blaKPC gene sequences. Each line represents a SNP between variants.
Upstream sequence and transcriptional start site of blaKPC-5.
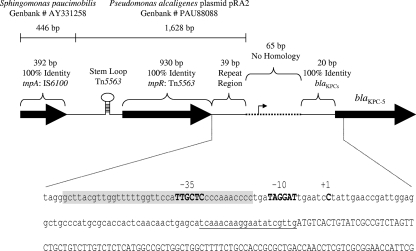
Alignment of the upstream regions from several blaKPC-2 (GenBank accession numbers EU176014, EU176013, EU176012, EU176011, DQ989639, DQ523564, and AF481906) and blaKPC-3 (GenBank accession number AM774409) sequences published in GenBank demonstrated a high degree of sequence identity (data not shown). The sequence flanking the 5′ end of blaKPC-4 has yet to be reported. In order to determine if blaKPC-4 and blaKPC-5 share the same upstream region as the blaKPC genes mentioned above, PCR was performed using flanking primers designed from blaKPC-2. A PCR product was generated for blaKPC-4, and sequence analysis showed that the first 363 bases directly upstream of the translational start codon of blaKPC-4 were identical to the sequence upstream of blaKPC-2 (data not shown). No PCR product could be amplified for blaKPC-5 by using multiple flanking primer sets, suggesting divergence in the flanking sequences. Using a genome-walking technique, 2,159 bases were sequenced upstream of the blaKPC-5 translational start codon (Fig. 2). Although the first 20 bases directly upstream of the blaKPC-5 structural gene were identical to the corresponding region in blaKPC-2, the next 65 bases showed no similarity with the blaKPC-2 flanking sequence or with any sequence in GenBank (Fig. 2). Adjacent to this sequence was a 1,628-bp DNA fragment with 100% identity to a portion of the transposable element Tn5563 in Pseudomonas alcaligenes plasmid pRA2 (GenBank accession number PAU88088). This region contained the 39-bp terminal repeat, resolvase gene (tnpR), and stem-loop found in Tn5563. Although a transposase, tnpA, is also present within Tn5563, the sequence upstream of the 1,628 bp fragment was divergent from this gene. Instead, the remaining 392 bp showed 100% identity to the tnpA2 transposase gene from IS6100, present in Sphingomonas paucimobilis (GenBank accession number AY331258).
FIG. 2.
Genetic organization of the blaKPC-5 upstream region and mapping of the transcriptional start site. The schematic gives the identities of various regions upstream of the blaKPC-5 structural gene to known sequences within GenBank. The nucleotides immediately upstream of the blaKPC-5 translational start codon are shown. The putative −10 and −35 promoter elements are shown in boldface capital letters and labeled. The start site of transcription is indicated by +1 above the cytosine residue (capitalized, in boldface). The bases constituting the 39-bp inverted repeat region of Tn5563 are shaded. The 20-bp fragment that is present in all blaKPCs is identified by a single underline. The blaKPC-5 structural gene sequences are capitalized. The figure is not drawn to scale.
Since the upstream region differed significantly from those of the other blaKPC alleles, 5′ RACE was used to map the transcriptional start site of blaKPC-5 in order to aid in promoter identification. Sequencing of a 424-bp blaKPC-5 cDNA product identified the start site of transcription at a cytosine residue 70 bases upstream of the translational start codon (Fig. 2) and within the 65-bp sequence that showed no homology to any known sequence. A putative −10 promoter element (TAGGAT) was also predicted within this 65-bp region; however, the putative −35 promoter element (TTGCTC) was identified within the 39-bp terminal repeat region (Fig. 2).
The isoelectric points of KPC-4 and KPC-5 differ from that of KPC-2.
IEF of crude extracts from P. aeruginosa strains PR100 and PR280 showed the presence of multiple β-lactamases with pIs of 6.7 (PR100), 7.65 (PR280), and 8.0 (both strains). KPC-2 and KPC-4 have previously been shown to have pIs of 6.7 and 7.65, respectively (25, 36). To determine if the band at the pI of 7.65 in PR280 represented KPC-5, blaKPC-5 was cloned into vector pACYC184 and transformed into E. coli TOP10 cells. Crude extracts from this transformant were analyzed by IEF. For comparison, blaKPC-2 and blaKPC-4 from strains PR100 and E624, respectively, were also cloned into pACYC184, and crude extracts from KPC-2 and KPC-4 transformants were also included. TOP10 transformants harboring blaKPC-4 and blaKPC-5 produced a β-lactamase with a pI of 7.65, whereas a β-lactamase with a pI of 6.7 was present in the blaKPC-2 transformant (data not shown). All three KPCs showed partial inhibition by clavulanic acid (data not shown). Bands other than the one representing KPC enzymes were not visible upon IEF analysis using the extracts obtained from the E. coli transformants.
Antimicrobial susceptibilities of transformants with KPC-2, KPC-4, and KPC-5.
The susceptibilities of the E. coli TOP10 transformants containing blaKPC-2, blaKPC-4, and blaKPC-5 to several β-lactams were examined, and results are presented in Table 2. Of note, the structural genes of the blaKPC variants were cloned into a low-copy-number vector, and KPC expression was driven by the chloramphenicol promoter of the vector. This eliminated differences in expression levels, allowing for systematic comparison between variants. The MICs of all drugs tested, except cefoxitin, were at least 16-fold higher for the transformants than for the host strain. Only a modest decrease in susceptibility to cefoxitin was observed, with two- to eightfold increases in the MICs for the transformants. In comparison to each other, KPC transformants had similar MICs of cefotaxime, cefepime, and aztreonam but differed in their susceptibilities to the remaining drugs. MICs of cefoxitin and all three carbapenems were at least four- to eightfold higher for the KPC-2 transformant than for the KPC-4 transformant, while MICs for the KPC-5 transformant were two- to fourfold lower than those for the KPC-2 transformant. The KPC-2 transformant was also fourfold less susceptible to piperacillin-tazobactam than the KPC-4 transformant. The KPC-4 transformant, however, was less susceptible to ceftazidime than the KPC-2 transformant, with MICs differing at least eightfold. As before, the MIC of ceftazidime for the KPC-5 transformant (128 μg/ml) fell between the MICs for the KPC-2 (32 μg/ml) and KPC-4 (>128 μg/ml) transformants. While MICs for the KPC-2 transformant were interpreted as intermediate to meropenem and resistant to imipenem, ertapenem, and cefoxitin, MICs of each carbapenem and cefoxitin for the KPC-4 transformant were in the susceptible range. The KPC-5 transformant was resistant to ertapenem and intermediate to cefoxitin but susceptible to imipenem and meropenem.
TABLE 2.
Antimicrobial susceptibilities of clinical isolates and blaKPC transformants
| Antimicrobial agent | MIC (μg/ml) for the following strain (KPC)a:
|
||||||
|---|---|---|---|---|---|---|---|
| PS10 (KPC-2) | E624 (KPC-4) | PS28 (KPC-5) | E. coli TOP10 | TOP10 (KPC-2) | TOP10 (KPC-4) | TOP10 (KPC-5) | |
| Piperacillin + tazobactam | >256 | >64 | 128 | 1 | >256 | 128 | >256 |
| Ceftazidime | 64 | >64 | 128 | 0.12 | 32 | >128 | 128 |
| Cefotaxime | NDb | >64 | ND | <0.06 | 64 | 64 | 64 |
| Cefoxitin | ND | >64 | ND | 4 | 32 | 8 | 16 |
| Cefepime | ND | 64 | ND | <0.06 | 16 | 8 | 16 |
| Aztreonam | ND | >64 | ND | <0.12 | >128 | >128 | >128 |
| Imipenem | >64 | >32 | >64 | 0.12 | 16 | 2 | 4 |
| Imipenem + clavulanic acid | ND | ND | ND | 0.12 | 4 | <0.06 | 0.12 |
| Meropenem | >64 | >32 | >64 | <0.06 | 8 | 1 | 2 |
| Ertapenem | ND | >16 | ND | <0.06 | 16 | 2 | 8 |
Strains include the E. coli recipient strain TOP10 and transformants harboring the pACYC184 vector with cloned blaKPCs.
ND, not determined.
The addition of clavulanic acid (4 μg/ml) reduced the imipenem MIC for the KPC-2 transformant fourfold (Table 2). Interestingly, MICs of imipenem decreased at least 32-fold for the KPC-4 and KPC-5 transformants in the presence of clavulanic acid, suggesting that KPC-4 and KPC-5 were more sensitive to inhibition by clavulanic acid.
Comparison of hydrolytic activities.
The differences in the susceptibilities of the transformants, described above, suggested that the KPC variants may have different hydrolytic efficiencies. To test this, the hydrolysis rates for several substrates were examined by using a spectrophotometric assay and extracts from the E. coli transformants containing KPC-2, KPC-4, or KPC-5. KPC-2 hydrolyzed cephalothin at levels approximately 7.5- and 3.5-fold higher than KPC-4 and KPC-5, respectively (Table 3). In addition, KPC-2 was more active against imipenem, with hydrolysis rates approximately 6- and 3.5-fold higher than those of KPC-4 and KPC-5, respectively. KPC-4 and KPC-5 were more efficient at hydrolyzing ceftazidime than KPC-2. KPC-5 showed slightly more activity against cephalothin (∼2-fold) and imipenem (∼1.7-fold) than KPC-4. KPC-2, KPC-4, and KPC-5 hydrolyzed cefotaxime with similar efficiencies (Table 3).
TABLE 3.
Hydrolysis rates of KPC variants determined by using extracts from KPC E. coli transformants
| Substrate | Hydrolysis rate (nmol/min/mg)
|
||
|---|---|---|---|
| KPC-2 | KPC-4 | KPC-5 | |
| Cephalothin | 1,310 ± 8 | 172 ± 22 | 368 ± 58 |
| Cephalothin + Clava | 487 ± 54 | 20 ± 1 | 48 ± 4 |
| Cefotaxime | 65 ± 12 | 53 ± 4 | 50 ± 5 |
| Cefotaxime + Clav | 25 ± 1 | 6 ± 0.4 | 7 ± 3 |
| Ceftazidime | 3 ± 1 | 7 ± 1 | 6 ± 0.2 |
| Imipenem | 412 ± 12 | 69 ± 4 | 116 ± 5 |
Clav, clavulanic acid at 100 μM.
As in the susceptibility data, clavulanic acid was more inhibitory against KPC-4 and KPC-5 than against KPC-2 in the hydrolysis assays (Table 3). The rates of cephalothin and cefotaxime hydrolysis by KPC-2 decreased approximately 2.6-fold in the presence of clavulanic acid (100 μM). However, more-substantial reductions in hydrolytic activity were observed for KPC-4 against cephalothin (∼8.6-fold decrease) and cefotaxime (∼8.8-fold decrease) with the addition of clavulanic acid. Similarly, rates of cephalothin and cefotaxime hydrolysis by KPC-5 declined 7.7-fold and 7.1-fold, respectively, in the presence of clavulanic acid.
DISCUSSION
Since the discoveries of the synonymous KPC-1 and -2 enzymes, the KPC family has grown to include four additional variants, KPC-3 through KPC-6. While blaKPC-2 and blaKPC-3 have been detected in several members of the Enterobacteriaceae and P. aeruginosa (blaKPC-2 only), blaKPC-4 has been found in only two bacterial species, Enterobacter cancerogenus (20) and K. pneumoniae (25). blaKPC-5 and blaKPC-6 represent newly discovered variants in P. aeruginosa and K. pneumoniae, respectively. Using the aligned sequences of the blaKPCs, a minimum spanning tree indicated that blaKPC-2 was the founding (i.e., ancestral) sequence from which variants have arisen through nucleotide substitutions in this gene. The fact that blaKPC-2-positive organisms were described in numerous reports prior to the discovery of the first variant, blaKPC-3 (23), also supports the hypothesis that blaKPC-2 is the prototype sequence. KPC-3, KPC-5, and KPC-6 each differ from KPC-2 by a single amino acid change. KPC-5 and KPC-6 also differ from KPC-4 by a single amino acid variation. As shown by the spanning tree, KPC-4, a two-amino-acid variant of KPC-2, may have emerged through a sequential process where KPC-5 and/or KPC-6 represents an intermediate step.
The mutations in the variant KPCs affect the characteristics of the enzymes. The shift in the pI from 6.7 (KPC-2 and KPC-3) to 7.65 (KPC-4 and KPC-5) can be explained by the amino acid substitution of arginine (basic, positively charged) for proline (polar, uncharged) at position 103. An increase in pI has also been observed between the β-lactamases TEM-35 (pI 5.2) and TEM-39 (pI 5.4) and between CTX-M-39 (pI 7.0) and CTX-M-26 (pI 8.0), where the enzymes differ by a single arginine substitution (6, 11). However, the substitution of glycine for valine at position 239 in KPC-4 and KPC-6 should not influence the pI of either enzyme, since both amino acids are nonpolar and uncharged. Indeed, KPC-4 and KPC-5 shared the same pI despite the Gly239 change in KPC-4.
The amino acid changes between KPCs also appear to influence the kinetic properties of the enzymes. In comparison to KPC-2, KPC-3 was shown to have a higher rate of hydrolysis for ceftazidime but a lower affinity for cefoxitin (1). Similarly, KPC-4 and KPC-5 have spectra of activity for certain β-lactams different from that of KPC-2. In this study, the blaKPC-2 transformant was less susceptible than other blaKPC transformants to the carbapenems, which can be attributed to more efficient hydrolysis of this drug class. Conversely, extracts from blaKPC-4 and blaKPC-5 transformants had slightly better rates of ceftazidime hydrolysis (twofold), which may explain why their MICs were higher than that for the blaKPC-2 transformant, although increased affinity for ceftazidime may also play a role, as in KPC-3 (1). Interestingly, the susceptibility and hydrolysis results for KPC-5 always fell between the ranges of KPC-2 and KPC-4, with most similarity to the KPC-4 results. These data suggest an evolutionary transformation of KPCs from an efficient carbapenemase (KPC-2) to variants with better ceftazidimase activity (KPC-4) through stepwise mutation. This situation is reminiscent of the extended-spectrum β-lactamases TEM and SHV, which have modified activities to include ceftazidime as a result of mutation (2). A reduction in KPC activity, however, appears to be the cost for this conversion. While the susceptibility and hydrolysis assays suggest variability in the enzymatic properties of the KPC enzymes, we cannot rule out the possibility that the kinetic differences result from altered protein levels due to the mutation(s) within KPC-5 and KPC-4. Other studies have demonstrated that specific mutations in β-lactamases affect protein stability (12, 22) or translation efficiency (12), ultimately reducing the amount of enzyme. Since β-lactamase levels were not measured in the crude lysates from our study, the results of the hydrolysis assays should be confirmed using purified enzymes.
While the Pro103→Arg mutation may confer more activity against ceftazidime, this change renders the enzyme more sensitive to inhibition by clavulanic acid. Susceptibility to imipenem increased at least 32-fold in the blaKPC-4 and blaKPC-5 transformants with the addition of clavulanic acid, but only a 4-fold increase was observed for transformants expressing blaKPC-2. These changes in susceptibility were due to the different sensitivities of the enzymes to clavulanic acid inhibition, as demonstrated by substrate hydrolysis. The most dramatic decrease in the hydrolysis rates occurred for KPC-4 in the presence of clavulanic acid (100 μM), followed by KPC-5. Among the three enzymes tested, KPC-2 was the least affected by clavulanic acid. KPC-4 and KPC-5 are unique among β-lactamase variants in that they became more sensitive to inhibition instead of exhibiting a more dramatic switch from inhibitor sensitivity to inhibitor resistance (2) or a transition from inhibitor resistance to inhibitor sensitivity (9).
The flanking regions of the blaKPC genes sequenced thus far, excluding blaKPC-5, were related to Tn4401 (17). Conversely, blaKPC-5 had a unique upstream region, with components from both Tn5563 and IS6100A present. Tn5563 is a class II transposon flanked on either side by 39-bp terminal inverted repeats, encodes a resolvase and a transposase, and carries three open reading frames with similarities to genes encoding mercuric ion transport proteins MerP and MerT and the regulatory protein MerR (34). While the first terminal inverted repeat and the complete resolvase gene, tnpR, were directly upstream of blaKPC-5, the remaining sequence was identical to that of the transposase gene, tnpA2, of IS6100A. IS6100A may have inserted into the backbone of Tn5563 in a manner similar to the insertion of the class I integron In32 into Tn5563 (21).
blaKPC-5 transcription initiates at a cytosine residue located near the 39-bp terminal repeat of Tn5563. A putative −10 promoter element was identified just downstream of the terminal repeat, but the putative −35 promoter element was located within the terminal repeat. Outwardly directed −35 promoter elements have been discovered within the terminal inverted repeats of several insertion sequences (14), causing the overexpression of neighboring genes when resident −10 promoter elements are proportionally spaced.
As demonstrated in this study, the KPC variants have different susceptibilities and hydrolytic properties. This may contribute to the difficulty that clinical laboratories have encountered in identifying KPC-producing organisms. Detecting the presence of these enzymes in clinical isolates is imperative, regardless of their susceptibility patterns, because of the potential for the spread and evolution of these enzymes in response to local selective pressures.
Acknowledgments
This research was conducted with support from the Investigator-Sponsored Study Program of AstraZeneca.
We thank the Puerto Rico Medical Center Bacteriology Laboratory for collecting the isolates, in particular G. J. Vazquez, I. E. Robledo, and M. I. Sante.
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Alba, J., Y. Ishii, K. Thomson, E. S. Moland, and K. Yamaguchi. 2005. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 49:4760-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin. Infect. Dis. 39:55-60. [DOI] [PubMed] [Google Scholar]
- 4.Bratu, S., M. Mooty, S. Nichani, D. Landman, C. Gullans, B. Pettinato, U. Karumudi, P. Tolaney, and J. Quale. 2005. Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob. Agents Chemother. 49:3018-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castanheira, M., H. S. Sader, L. M. Deshpande, T. R. Fritsche, and R. N. Jones. 2008. Antimicrobial activities of tigecycline and other broad-spectrum antimicrobials tested against serine carbapenemase- and metallo-beta-lactamase-producing Enterobacteriaceae: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 52:570-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chmelnitsky, I., Y. Carmeli, A. Leavitt, M. J. Schwaber, and S. Navon-Venezia. 2005. CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob. Agents Chemother. 49:4745-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed., approved standard M7-A6, vol. 28. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Deshpande, L. M., P. R. Rhomberg, H. S. Sader, and R. N. Jones. 2006. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: report from the MYSTIC Program (1999-2005). Diagn. Microbiol. Infect. Dis. 56:367-372. [DOI] [PubMed] [Google Scholar]
- 9.Doi, Y., J. Wachino, M. Ishiguro, H. Kurokawa, K. Yamane, N. Shibata, K. Shibayama, K. Yokoyama, H. Kato, T. Yagi, and Y. Arakawa. 2004. Inhibitor-sensitive AmpC beta-lactamase variant produced by an Escherichia coli clinical isolate resistant to oxyiminocephalosporins and cephamycins. Antimicrob. Agents Chemother. 48:2652-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dortet, L., I. Radu, V. Gautier, F. Blot, E. Chachaty, and G. Arlet. 2008. Intercontinental travels of patients and dissemination of plasmid-mediated carbapenemase KPC-3 associated with OXA-9 and TEM-1. J. Antimicrob. Chemother. 61:455-457. [DOI] [PubMed] [Google Scholar]
- 11.Henquell, C., C. Chanal, D. Sirot, R. Labia, and J. Sirot. 1995. Molecular characterization of nine different types of mutants among 107 inhibitor-resistant TEM beta-lactamases from clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 39:427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hujer, A. M., K. M. Hujer, M. S. Helfand, V. E. Anderson, and R. A. Bonomo. 2002. Amino acid substitutions at Ambler position Gly238 in the SHV-1 beta-lactamase: exploring sequence requirements for resistance to penicillins and cephalosporins. Antimicrob. Agents Chemother. 46:3971-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, R. N., J. T. Kirby, and P. R. Rhomberg. 2008. Comparative activity of meropenem in US medical centers (2007): initiating the 2nd decade of MYSTIC program surveillance. Diagn. Microbiol. Infect. Dis. 61:203-213. [DOI] [PubMed] [Google Scholar]
- 14.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marschall, J., R. J. Tibbetts, W. M. Dunne, V. J. Fraser, and D. K. Warren. 2007. Presence of the KPC carbapenemase gene in Enterobacteriaceae bacteremia, correlation with carbapenem susceptibility, and impact on clinical outcomes, abstr. C2-1935. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 16.Miriagou, V., L. S. Tzouvelekis, S. Rossiter, E. Tzelepi, F. J. Angulo, and J. M. Whichard. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47:1297-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naas, T., P. Nordmann, G. Vedel, and C. Poyart. 2005. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49:4423-4424. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, M. J. Schwaber, D. Schwartz, and Y. Carmeli. 2006. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob. Agents Chemother. 50:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palepou, M. F., N. Woodford, R. Hope, M. Colman, J. Glover, M. Kaufmann, C. Lafong, R. Reynolds, and D. M. Livermore. 2005. Novel class A carbapenemase, KPC-4, in an Enterobacter isolate from Scotland, abstr. 1134_01_20. Prog. Abstr. 15th Eur. Cong. Clin. Microbiol. Infect. Dis., Copenhagen, Denmark.
- 21.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrosino, J. F., and T. Palzkill. 1996. Systematic mutagenesis of the active site omega loop of TEM-1 beta-lactamase. J. Bacteriol. 178:1821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasheed, J. K., J. W. Biddle, K. F. Anderson, L. Washer, C. Chenoweth, J. Perrin, D. W. Newton, and J. B. Patel. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robledo, I. E., E. S. Moland, E. A. Aquino, G. J. Vazquez, M. I. Sante, J. Bertran, and N. D. Hanson. 2007. First report of a KPC-4 and CTX-M producing K. pneumoniae isolated from Puerto Rico (PR), abstr. C2-1933. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 26.Sellen, T. J., D. C. Halstead, J. Adams, D. A. Dossenback, J. Abid, and D. Paterson. 2007. Emergence of carbapenemase-producing Klebsiella pneumoniae (KPC) in Northeast Florida, abstr. D1553. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 27.Smith Moland, E., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51:711-714. [DOI] [PubMed] [Google Scholar]
- 28.Villegas, M. V., K. Lolans, A. Correa, J. N. Kattan, J. A. Lopez, and J. P. Quinn. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob. Agents Chemother. 51:1553-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villegas, M. V., K. Lolans, A. Correa, C. J. Suarez, J. A. Lopez, M. Vallejo, and J. P. Quinn. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 50:2880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei, Z. Q., X. X. Du, Y. S. Yu, P. Shen, Y. G. Chen, and L. J. Li. 2007. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob. Agents Chemother. 51:763-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2005. AmpC and OprD are not involved in the mechanism of imipenem hypersusceptibility among Pseudomonas aeruginosa isolates overexpressing the mexCD-oprJ efflux pump. Antimicrob. Agents Chemother. 49:4763-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolter, D. J., G. J. Vazquez, I. E. Robledo, M. I. Sante, R. V. Goering, and N. D. Hanson. 2007. Detection of KPC carbapenem-hydrolyzing beta-lactamase in Pseudomonas aeruginosa from the Puerto Rico Medical Center Hospitals (PRMCHs), abstr. C2-1928. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 33.Woodford, N., P. M. Tierno, Jr., K. Young, L. Tysall, M. F. Palepou, E. Ward, R. E. Painter, D. F. Suber, D. Shungu, L. L. Silver, K. Inglima, J. Kornblum, and D. M. Livermore. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 48:4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo, C. C., J. M. Tham, S. M. Kwong, S. Yiin, and C. L. Poh. 1998. Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 165:253-260. [DOI] [PubMed] [Google Scholar]
- 35.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. (Erratum, 52:809, 2008.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yigit, H., A. M. Queenan, J. K. Rasheed, J. W. Biddle, A. Domenech-Sanchez, S. Alberti, K. Bush, and F. C. Tenover. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing beta-lactamase KPC-2. Antimicrob. Agents Chemother. 47:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]