Abstract
Staphylococcal cassette chromosome mec (SCCmec) is a mobile genetic element characterized by flanking terminal direct and, in most cases, inverted repeat sequences, the mec and ccr gene complexes, and their surrounding DNA regions. Unique combinations of the mec and ccr gene complexes generate various SCCmec types. Six SCCmec types have been reported to date. We describe here a novel SCCmec type identified in a Canadian methicillin-resistant Staphylococcus aureus (MRSA) epidemic strain. MRSA clinical isolates were screened for known SCCmec types by multiplex and conventional PCR methods. Three phenotypically and genotypically identical MRSA clinical isolates with a pulsotype identical to CMRSA9 were identified locally and found to be nontypeable by available SCCmec typing schemes. Complete sequencing of the SCCmec element revealed a nucleotide fragment of 32,168 bp integrated at an identical chromosomal integration site (attBscc) at the 3′ end of the orfX gene. The nucleotide sequences at the chromosome-SCCmec junction regions were typical of other SCCmec types, but the element harbored a unique combination of class A mec and type 4 ccr gene complexes. Sequence recombination analysis suggested that this unique SCCmec type may be derived from homologous recombination between the previously described SCCRP62A of S. epidermidis strain RP62A and SCC composite island of S. epidermidis ATCC 12228, respectively, or via recombination of other staphylococcal strains that carry the same or similar mobile cassettes. We identified a previously undescribed type of SCCmec from isolate C10682, tentatively designated type VIII, and we provide compelling evidence supporting the ability of SCC elements to transfer horizontally or undergo recombination to generate new SCCmec types.
Methicillin resistance in Staphylococcus spp. results from acquisition of mecA, which encodes an additional altered low-affinity penicillin-binding protein (PBP2a), conferring broad resistance to almost all penicillin-related compounds (9). Methicillin-resistant Staphylococcus aureus (MRSA) strains have acquired and integrated into their genome a 21- to 67-kb mobile genetic element, termed the staphylococcal cassette chromosome mec (SCCmec), which harbors the methicillin resistance gene (mecA) and other antibiotic resistance determinants (13, 15, 20).
The SCCmec element integrates into the Staphylococcus aureus chromosome at a unique site (attBscc) located downstream of an open reading frame (ORF) of unknown function known as orfX that is near the origin of replication (14). These elements are characterized by the presence of flanking terminal direct and, in most cases, inverted repeats, two essential genetic components (the mec gene complex and the ccr gene complex), and three joining (J; previously known as junkyard) regions (13, 15, 18, 20). The mec gene complex is composed of IS431 and/or IS1272, mecA, and intact or truncated sets of regulatory genes mecR1 (encoding the signal transducer protein MecR1) and mecI (encoding the repressor protein MecI) (13, 15, 18, 20). The ccr gene complex (ccrA, ccrB, and ccrC) encodes recombinases of the invertase/resolvase family, which mediate the integration of SCCmec into and its excision from the recipient chromosome and are responsible for the mobility of this element. The rest of the SCCmec element is comprised of J regions (J1, J2, and J3) that are located between and around the mec and ccr gene complexes and contain various genes or pseudogenes, including plasmid- or transposon-mediated resistance genes for non-β-lactam antibiotics or heavy metals (16). To date, there are five classes (A, B, C1, C2, and D) of mec gene complexes and five allotypes (types 1, 2, 3, 4, and 5) of ccr gene complexes which have been described (11, 15). Different combinations of these complex classes and allotypes generate various SCCmec types. SCCmec elements are currently classified into types I (combination of the type 1 ccr and the class B mec gene complex; 1B), II (2A), III (3A), IV (2B), V (5C2), and VI (4B), based on the nature of the mec and ccr gene complexes, and are further classified into subtypes according to differences in their J region DNA (13, 15, 20, 26).
A new type (5C1; tentatively designed type VII) has recently been identified in a community-associated MRSA (CA-MRSA) isolate from Sweden (2). We describe here a novel SCCmec type harboring a previously uncharacterized and unique combination of class A mec and type 4 ccr gene complexes (4A), tentatively designated type VIII, identified in a Canadian MRSA (CMRSA) epidemic strain, CMRSA9, which is a typical hospital-associated MRSA (HA-MRSA) epidemic strain commonly found in Quebec and other regions in Canada (4).
(This work was presented in part at the 13th International Symposium on Staphylococci and Staphylococcal Infections, Cairns, Queensland, Australia, 7 to 10 September 2008 [34a]).
MATERIALS AND METHODS
Bacterial strains and isolates.
The MRSA clinical isolates under study were recovered from patients in local hospitals in Calgary, Alberta, Canada. Strain C10682 was isolated in 2003 from a percutaneous endoscopic gastrostomy site subcutaneous abscess in a 58-year-old male patient, C4631was isolated in 2004 from an abdominal wound of a 60-year-old male patient, and C5073 was recovered in 2005 from a screening nasal swab from an 80-year-old female patient.
The SCCmec type prototypic MRSA reference strains, including type I (NCTC10442), type II (N315), type III (85/2082), type IVc (MR108), type IVd (JCSC4469), and type V (WIS [WBG8318]-JCSC3624) strains, were kindly provided by K. Hiramatsu and T. Ito from Juntendo University, Tokyo, Japan (13, 15, 20, 24). The SCCmec type IVa and type IVb strains CA05 and 8/6-3P, respectively, were obtained from R. Daum, University of Chicago, Chicago, IL (20). The SCCmec type VI control strain HDE288 was obtained from H. de Lencastre, Rockefeller University, New York, NY (26). The Canadian epidemic MRSA reference strains CMRSA1 to CMRSA10 were provided by National Microbiology Laboratory, Health Canada, Winnipeg, Manitoba, Canada. The U.S. epidemic MRSA reference strains USA100 to USA800 (NRS382, NRS383, NRS384, NRS123, NRS385, NRS22, NRS386, and NRS387, respectively) were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus program supported under NIAID/NIH contract no. N01-AI-95359.
Identification, SCCmec typing, and molecular characterization of isolates.
Staphylococcal isolates were identified morphologically and biochemically by standard laboratory procedures as previously described (34). Clinical MRSA isolates were screened for known SCCmec types by multiplex (32) and conventional (13, 18) PCR methods as previously described, as well as with the use of our new type 4 ccr complex primer pair ccr4-F (5′-ATCGCTCATTATGGATACTGC-3′) and ccr4-R (5′-CAAATGACCCTTGTCTATAACG-3′). Isolates were typed using pulsed-field gel electrophoresis following a standardized protocol (23). Isolates were tested for the presence of the Panton-Valentine leukocidin (lukS-PV and lukF-PV) genes with a triplex PCR assay (21). Staphylococcal protein A gene (spa) typing (10, 29), multilocus sequence typing (MLST) (7), and accessory gene regulator (agr) typing (27) were conducted as described elsewhere. Profiles based on 35 common S. aureus virulence genes comprising 20 exotoxin genes, 12 adhesin genes, and 3 exoenzyme genes were determined by PCR amplification using previously described primer pairs (33).
Long-range PCR and random primer sequencing walking strategy.
Frozen bacteria were subcultured twice onto tryptic soy agar plates (Becton Dickinson, Sparks, MD) prior to DNA extraction. Genomic DNA was isolated from 5 ml of bacterial culture by using the QIAamp DNA minikit following protocol D (Qiagen Inc., Mississauga, Ontario, Canada). Two methods were employed for DNA sequencing. Long-range PCR amplification was used to amplify the chromosomal junction-mec complex region as well as the mec-ccr complex region. Briefly, 1× PCR XL-Buffer II, 1.2 mM Mg(OAc)2, 800 μM deoxynucleoside triphosphate (dNTP) blend (200 μM each of dATP, dGTP, dCTP, and dTTP), 1.2 units of rTth DNA polymerase (Applied Biosystems, Foster City, CA), 200 nmol of each primer, and 4 μl of template DNA were combined into a 50-μl reaction mixture. Cycling conditions were as follows: an initial denaturation at 93°C for 2 min was followed by 10 cycles of 93°C for 15 s, 52°C for 30 s, and 68°C for 10 min. A further 20 cycles of 93°C for 15 s, 52°C for 30 s, and 68°C for 10 min (Auto X = +20 s per cycle) were done. Long-range PCR products were gel purified with the MinElute gel extraction kit (Qiagen Inc., Mississauga, Ontario, Canada) and sequenced by primer extension using the Applied Biosystems BigDye terminator sequencing kit and ABI 3730S automated DNA sequencer.
Due to the inability to amplify the J1 (ccr-chromosomal junction) region, a modified random primer sequencing walking strategy was employed using the nested random primers arb1 (5′-GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT-3′) and arb2 (5′-GGCCACGCGTCGACTAGTAC-3′) (3). Round 1 of the random PCR was done with a known primer targeting a region approximately 700 bp from the end of the already sequenced SCCmec element and primer arb1. Two microliters of the extracted genomic DNA was added to 48 μl of PCR mix containing 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 2 mM MgCl2, 200 μM of each dNTP (dATP, dTTP, dGTP, and dCTP) (Invitrogen Inc., Carlsbad, CA), 0.4 μM known primer, 0.2 μM primer arb1, and 2.5 units of Taq DNA polymerase (Invitrogen Inc., Carlsbad, CA). Cycling conditions included a 5-min incubation at 96°C and then five cycles of 94°C for 30 s, 30°C for 30 s, and 72°C for 90 s, followed by 30 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 2 min. A final incubation at 72°C for 5 min was followed by a hold at 4°C. Round two of the random PCR was done with a second known primer designed more proximal to the end of the previously sequenced element. Two microliters of the unpurified round 1 product was added to a 50-μl PCR mix containing 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 3 mM MgCl2, 200 μM of each dNTP (dATP, dTTP, dGTP, and dCTP) (Invitrogen Inc., Carlsbad, CA), 0.4 μM known primer, 0.2 μM primer arb2, and 2.5 units of Taq DNA polymerase (Invitrogen Inc., Carlsbad, CA), with PCR conditions consisting of 29 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s, was followed by an incubation at 72°C for 5 min and a hold at 4°C. All random PCR products larger than 1 kb were gel purified with the Qiagen MinElute gel extraction kit and sequenced with primer extension as described above.
Sequence and recombination analyses.
Sequence homology and identity comparison to other sequences was performed with the NCBI BLAST sequence search (http://www.ncbi.nlm.nih.gov). ORF matching/annotation and sequence translation were accomplished by analysis of the sequence deposited in the GenBank database, using the Vector NTI software program (Invitrogen). The DNASTAR Lasergen version 7.2 software package program (DNAStar Inc.) was used to align the sequences and determine similarity and divergence among the sequences (this analysis also included the ClustalW method). Phylogenetic and bootstrap analyses were conducted using MEGA version 4 (30). For the sequence recombination analyses, the sequence was first subjected to scanning for recombination with the NCBI Genotyping Recombinant Sequence Analysis Program (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi), using all reported types of SCCmec sequences from GenBank. Creation of a distance plot and bootscanning were performed using the SimPlot program to identify informative sites and to select crossover breakpoints (19). These breakpoints were used to divide the alignment into segments for phylogenetic tree construction, and the recombination event was further confirmed by bootstrap testing as described above.
Confirmation tests of other clinical isolates/strains harboring SCCmecC10682.
MRSA isolates harboring the strain C10682 SCCmec element (SCCmecC10682) were further screened from our collection of local MRSA clinical isolates (May 1999 to December 2006) by using the class A mec and type 4 ccr gene complex-specific primers mecI-F/mecA147-R (Table 1) and ccr4-F/ccr4-R, respectively. The PCR products of strains positive for class A mec and type 4 ccr genes were subjected to full-length sequencing with primer sets mecI-F/mecA147-R and ccr-F/ccr-R (Table 1). For further confirmation, PCR amplification targeting various regions spanning the whole SCCmecC10682 element was performed. The detailed sequences and locations of these 12 primers pairs are listed in Table 1, and a schematic is shown in Fig. 1A.
TABLE 1.
Primers used for characterization of the SCCmecC10682 element in this study
| Primer | Primer no. | Sequence (5′ to 3′) | Spanning position (bp)a | Product size (bp)a | Reference |
|---|---|---|---|---|---|
| ccr-F | GGMGAACAAGTCARAAATGG | 25060-26828 | 1,769 | This study | |
| ccr-R | TTGCCTTGATAATAGCCTTC | This study | |||
| mecI-F | CCCTTTTTATACAATCTCGTT | 5955-9589 | 3,635 | 32 | |
| mecA147-R | ATGCGCTATAGATTGAAAGGAT | 32 | |||
| J3-F20 | 1 | CTTCCTGTATTTCGTCTATGC | 835-1031 | 197 | This study |
| J3-1R | 2 | TTTAAAAAATGATTATCCATCAGG | This study | ||
| J3-2F | 3 | CAACATCTAACTCCAACCAG | 2451-4360 | 1,910 | This study |
| J3-3R | 4 | TGTCTTTCTTACTTTAATATGACGG | This study | ||
| J3-1F | 5 | CCTTTAAATCTACTTTGTTCTGC | 5090-5234 | 145 | This study |
| J3-R20 | 6 | AATACGATGATTTTATAGTAGGAG | This study | ||
| mecA1 | 7 | GTAGAAATGACTGAACGTCCGATAA | 7080-7389 | 310 | 28 |
| mecA2 | 8 | CCAATTCCACATTGTTTCGGTCTAA | 28 | ||
| mecI-F | 9 | CCCTTTTTATACAATCTCGTT | 9444-9589 | 146 | 32 |
| mecI-R | 10 | ATATCATCTGCAGAATGGG | 32 | ||
| J2-F20 | 11 | AGAAGTCTTACACACTCCAGGC | 12144-12527 | 384 | This study |
| J2-3R | 12 | TGATTGAGTGTTAAAGTTTCG | This study | ||
| J2-3F | 13 | GATGATAAATGCTCCACCTG | 13024-14840 | 1,817 | This study |
| J2-4R | 14 | CTTTATATCTTGATCGATTGC | This study | ||
| tnpB-F | 15 | AACGAGAGTGGAAATTGAAGC | 17462-19803 | 2,342 | This study |
| aad9-R | 16 | ACTTATCATCACACTCTCCCCG | This study | ||
| J2-F9 | 17 | CACACAGAAGCCATATTTAGCG | 21063-22618 | 1,556 | This study |
| tnpA-R | 18 | CTAAGCGATAACACTTCACCG | This study | ||
| J2-F6 | 19 | GATTTGTTAAATATTGTCGAGG | 22420-25261 | 2,842 | This study |
| J2-R9 | 20 | GATTTACAAGATATACAGCACCG | This study | ||
| ccr4-F | 21 | ATCGCTCATTATGGATACTGC | 25079-26828 | 1,750 | This study |
| ccr4B-R | 22 | CAATAGCACGTTGTCTTTGGC | This study | ||
| comp-F5 | 23 | GAATTTTTGAATGATACTGGC | 28303-30163 | 1,861 | This study |
| J1-R2 | 24 | AATGGATTGGAATAATACTAAGCC | This study |
Each primer size and spanning position apply to the forward primer and subsequently listed reverse primer.
FIG. 1.
A unique combination of class A mec and type 4 ccr gene complexes identified in a clinical isolate C10682 of a CMRSA epidemic strain. (A) The 32-kb element with 36 predicted ORFs (ORFs CA001 to copA) could be divided into five distinct regions, including the mec and the ccr gene complexes and their surrounding DNA regions, J1, J2, and J3. Typical direct repeats (DRSCC-R and DRSCC-L), as well as hitherto unreported dyad repeats (DyadR-R and DyadR-L) present at both right and left extremities of the chromosome-SCCmec junctions, are shown, but no discernible inverted repeats were found. Primer locations for the 12 PCR targets chosen to characterize the other identical MRSA strains are indicated with solid arrows and numbers corresponding to primer numbers shown in Table 1. Refer to Table 1 for details of each primer. (B) Schematic diagram of the structure of three cassettes of SCCmecC10682, SCCRP62A, and SCC-CI showing that SCCmecC10682 may be derived from homologous recombination between previously described SCCRP62A and SCC-CI cassettes of S. epidermidis RP62A (CP000029) and ATCC 12228 (AE015929; BK001539) strains, respectively. Sequences derived from SCCRP62A are shaded in black, those from SCC-CI are shaded in light gray, and the sequences of the shared homologous region have a mixed pattern of shading.
Nucleotide sequence accession number.
The complete sequence of the SCCmecC10682 of strain C10682 has been assigned GenBank accession no. FJ390057.
RESULTS
A unique combination of class A mec and type 4 ccr gene complexes identified in a CMRSA epidemic clinical strain.
Upon initial SCCmec type screening of our collection of local MRSA clinical isolates, we discovered a previously uncharacterized combination of mec and ccr gene complexes in a clinical MRSA isolate designated C10682. Using mec and ccr gene complex-specific primers (Table 1), we showed by PCR amplification that this isolate contained a class A mec gene complex and a type 4 ccr gene complex. The DNA fragment encompassing the entire SCCmec element of this strain was amplified by long-range PCR, along with nucleotide sequence determination by primer extension. Complete sequencing of the SCCmec element revealed the presence of a nucleotide fragment insert of 32,168 bp in an identical chromosomal integration site (attBscc) located at the 3′ end of the orfX gene but harboring a unique combination of class A mec and type 4 ccr gene complexes (Fig. 1A).
The complete element, similar to other SCCmec types, could be divided into five distinct regions, including the mec and the ccr gene complexes and their surrounding DNA regions, J1, J2, and J3 (Fig. 1A). The 7,545-bp mec complex fragment was shown to possess a typical class A mec complex arrangement, including the upstream insertion sequence IS431 (harboring complementary 17-bp right and left invert repeat sequences [IR-R and IR-L, respectively] at each end), followed by a region that contains four ORFs (Table 2), mecA (2,007 bp, resulting in 668 amino acids [aa]) and its complete regulatory genes mecR1 (1,758 bp; 585 aa) and mecI (372 bp; 123 aa), i.e., IS431-mecA-mecR1-mecI (Fig. 1A). The entire mec gene complex is 100% identical to the prototypic class A mec gene complex described for the typical SCCmec type II strain of S. aureus N315, as well as S. epidermidis RP62A, but different from the other classes of mec gene complexes (data not shown).
TABLE 2.
ORFs in and around SCCmecC10682
| Genetic region | ORF or key structural componenta | Position (bp) | Size (bp) | Protein size (aa)b | % Identity | Homolog(s) (strain[s]) | Informationc |
|---|---|---|---|---|---|---|---|
| Chromosome | orfX | 1-151 | 151 | NA | 100 | SARCOL0025 (COL) | Conserved hypothetical protein (partial cds) |
| Repeat region | DRSCC-R | 137-148 | 12 | NA | Direct repeat at right extremity of SCCmec | ||
| DyadR-R | 240-254 | 15 | NA | 100; 100 | (COL); (RP62A) | Dyad repeat at right extremity of SCCmec | |
| J3 | CA001 | 329-1624 | 1,296 | 431 | 100 | SERP2528 (RP62A) | Conserved hypothetical protein |
| CA002 | 2039-2278 | 240 | 79 | 100 | SERP2527 (RP62A) | Conserved hypothetical protein | |
| mec gene complex | IR-R* | 2250-2266 | 17 | NA | 100 | (RP62A) | Right inverted complementary repeat of IS431 element |
| tnp* | 2310-2984 | 675 | 224 | 100 | SERP2526 (RP62A) | Transposase for IS431 element | |
| IR-L* | 3025-3041 | 17 | NA | 100 | (RP62A) | Left inverted complementary repeat of IS431 element | |
| CA004 | 3242-3409 | 168 | 55 | 100 | SERP2525 (RP62A) | Truncated probable HMG-CoA synthase | |
| CA005 | 3507-3737 | 231 | 76 | 100 | SERP2524 (RP62A) | Conserved hypothetical protein | |
| CA006 | 4246-4989 | 744 | 247 | 100 | SERP2523 (RP62A) | Glycerophosphoryldiester phosphodiesterase homologue | |
| CA007 | 5086-5514 | 429 | 142 | 100 | SERP2522 (RP62A) | MaoC domain protein dehydratase | |
| mecA | 5560-7566 | 2,007 | 668 | 100 | SERP2521 (RP62A) | Penicillin binding protein 2′ | |
| mecR1 | 7666-9423 | 1,758 | 585 | 100 | SERP2520 (RP62A) | Methicillin resistance protein | |
| mecI | 9423-9794 | 372 | 123 | 100 | SERP2519 (RP62A) | Methicillin resistance regulatory protein | |
| J2 | CA011 | 9895-10107 | 213 | 70 | 100 | SERP2518 (RP62A) | Conserved hypothetical protein |
| xylR | 10302-11411 | 1,110 | 369 | 100 | SERP2517 (RP62A) | Xylose repressor homologue | |
| CA013 | 11525-11785 | 261 | 86 | 100 | SERP2516 (RP62A) | Conserved hypothetical protein | |
| CA014 | 11921-12781 | 861 | 286 | 99 | SERP2516 (RP62A) | Conserved hypothetical protein | |
| CA015 | 12892-13956 | 1,065 | 354 | 100 | SERP2515 (RP62A) | Rhodanese-like domain protein | |
| CA016 | 14092-14352 | 261 | 86 | 100 | SERP2514 (RP62A) | Conserved hypothetical protein | |
| CA017 | 14352-14996 | 645 | 214 | 100 | SERP2513 (RP62A) | Conserved hypothetical protein | |
| CA018** | 15335-15946 | 612 | 203 | 100 | SERP2512 (RP62A) | Methyltransferase type 12 | |
| CA019** | 16356-16556 | 201 | 66 | 100 | SERP2511 (RP62A) | Peptide L | |
| ermA** | 16531-17262 | 732 | 243 | 100 | SERP2510 (RP62A) | rRNA adenine N-6-methyltransferase | |
| aad(9)** | 17388-18170 | 783 | 260 | 100 | SERP2509 (RP62A) | Streptomycin 3′-adenyltransferase | |
| tnpC** | 18321-18698 | 378 | 125 | 100 | SERP2508 (RP62A) | Transposase C | |
| tnpB** | 18705-20597 | 1893 | 630 | 100 | SERP2507 (RP62A) | Transposase B | |
| tnpA** | 20594-21679 | 1,086 | 361 | 100 | SERP2506 (RP62A) | Transposase A | |
| CA025 | 21798-22115 | 318 | 105 | 100 | SERP2505 (RP62A) | Truncated radC DNA repair protein | |
| CA026 | 22136-22642 | 507 | 168 | 96; 87 | SERP2504 (RP62A); SE0052 (ATCC 12228) | Hypothetical protein | |
| CA027 | 22658-22969 | 312 | 103 | 96; 95 | SERP2503 (RP62A); SE0053 (ATCC 12228) | Hypothetical protein | |
| CA028 | 22971-23063 | 93 | 30 | 97; 84 | SERP2502 (RP62A); SE0054 (ATCC 12228) | Conserved hypothetical protein | |
| CA029 | 23056-23406 | 351 | 116 | 90; 98 | SERP2501 (RP62A); SE0055 (ATCC 12228) | Conserved hypothetical protein | |
| ccr gene complex | ccrB4 | 23907-25535 | 1,629 | 542 | 99 | SE0056 (ATCC 12228) | Cassette chromosome recombinase B4 |
| ccrA4 | 25532-26893 | 1,362 | 453 | 100 | SE0057 (ATCC 12228) | Cassette chromosome recombinase A4 | |
| J1 | CA032 | 27080-27661 | 582 | 193 | 100 | SE0058 (ATCC 12228) | Putative membrane protein |
| ccrA | 27937-28056 | 120 | 39 | 100 | SE0059 (ATCC 12228) | Cassette chromosome recombinase A | |
| CA034 | 28168-28527 | 360 | 119 | 100 | SE0060 (ATCC 12228) | Hypothetical protein | |
| CA035 | 29163-29261 | 99 | 32 | 100 | SE0061 (ATCC 12228) | Hypothetical protein | |
| copA | 29381-31315 | 1,935 | 644 | 99 | SE0062 (ATCC 12228) | Copper-transporting ATPase CopA | |
| Repeat region | DyadR-L | 32040-32054 | 15 | NA | Dyad repeat at left extremity of SCCmec | ||
| DRSCC-L | 32310-32321 | 12 | NA | 100; 93 | (HDE288); (COL) | Direct repeat at left extremity of SCCmec | |
| Chromosome | CA037 | 32624-32716 | 93 | 30 | 100 | SACOL0057 (COL) | Conserved hypothetical protein |
| CA038 | 32863-33371 | 509 | NA | 100 | SACOL0058 (COL) | Conserved hypothetical protein (partial cds) |
*, IS431; **, Tn554.
NA, not applicable.
cds, coding region; HMG-CoA, 3-hydroxy-3-methylglutaryl-coenzyme A.
The ccr genes contain a total of 2,987 nucleotides which encodes ccrA (1,362 bp; 453 aa) and ccrB (1,629 bp; 542 aa), with 4 nucleotides overlaying each other between the end of ccrB and the beginning of ccrA (Table 2). Our ccrA showed 100% nucleotide sequence identity with ccrA4 in the SCC composite island (SCC-CI) of S. epidermidis ATCC 12228 while sharing only 89.6% homology with ccrA4 in the type VI SCCmec of HDE288.
Our ccrB, on the other hand, was 99.9% homologous to that in the SCC-CI and contained a single nucleotide change at position 926 (A→C), resulting in a glutamic acid-to-alanine substitution at amino acid position 309 (E309A). The gene was 94.5% homologous to the ccrB4 of the type VI SCCmec of HDE288. Phylogenetic and bootstrap analyses of the representative ccrA, ccrB, and ccrC gene complexes confirmed that it belonged to type 4 ccr gene complexes (data not shown).
To our knowledge, the combination of a class A mec gene complex and a type 4 ccr gene complex has never been previously described. A new SCCmec type element (SCCmecC10682) based on identification of this combination of mec and ccr gene complexes in our strain is, therefore, purported.
SCCmecC10682 may be derived from homologous recombination between previously described SCCRP62A and SCC-CI cassettes of S. epidermidis RP62A and ATCC 12228 strains, respectively.
Given the great extent of homology between the present SCCmecC10682 ORFs (sequences) and the previously described SCC cassette of S. epidermidis RP62A (SCCRP62A) and SCC-CI cassette of S. epidermidis ATCC 12228, SCCmec recombination in the present SCCmecC10682 sequence was examined. The complete SCCmecC10682 sequence was first subjected to scanning for recombination with the NCBI Genotyping Recombinant Sequence Analysis Program, using all reported types of SCCmec and SCC sequences as the references, and further confirmed by bootscanning and bootstrap testing. The NCBI program detected a potential recombination event leading to the construction of the SCCmecC10682 element. Such recombination was found to occur between SCCRP62A and the SCC-CI, with the J3, the mec gene complex, and the majority of the J2 region derived from the former and the other sequences (ccr gene complex and J1 region) derived from the latter (Fig. 1B). To estimate the location of the apparent crossover site, bootscanning (19) was performed using the complete sequences of the SCCRP62A and the SCC-CI, and the representative type V SCCmec of strain WIS was used as an outgroup sequence. The most likely breakpoints were located in three sections, i.e., at nucleotide (nt) position 130 to 22375 (22,246 bp) of SCCmecC10682 spanning the 5′ upstream region beginning from the right direct repeat (DRSCC-R) of the right extremity of SCCmecC10682 to the part of ORF CA026 located near the end of J2 region, which was shown to be 100% identical to that of the RP62A strain (nt 564 to 22809 of SCCRP62A) but only 43.8% identical to that of ATCC 12228 (nt 1 to 24570 of the SCC-CI); nt 22376 to 23500 (1,125 bp), representing the last four ORFs located at the end of the J2 region (before the ccr gene complex) with 91.3 to 96.3% identity among these three strains (nt 22810 to 23933 of SCCRP62A; nt 24571 to 25694 of the SCC-CI); and nt 23501 to 32305 (8,805 bp) from the ccr gene complex to the left extremity of SCCmecC10682 (5 bp before the left direct repeat [DRSCC-L]), which was 99.9% identical to that of ATCC 12228 (only 6-bp difference; nt 25695 to 34489 of the SCC-CI) but had only 57.8% homology with that of strain RP62A (nt 23524 to 28612 of SCCRP62A) (Table 2). This recombination event was further confirmed by bootstrap testing by constructing separate phylogenetic trees of the resulting regions with bootstrap support of 100%, 75%, and 100% for the three sections mentioned above, respectively (data not shown).
The boundaries of SCCmecC10682.
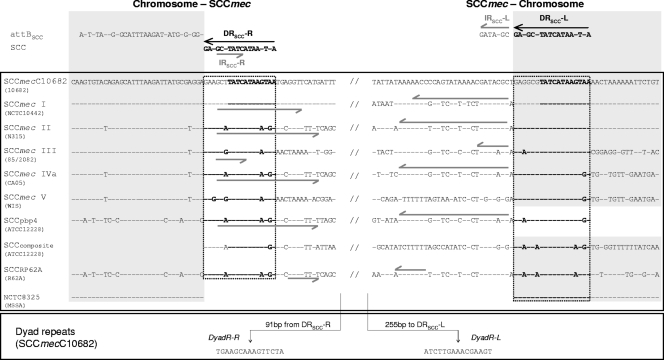
SCCmecC10682 was integrated at exactly the same nucleotide position at the 3′ end of the orfX gene as the type I SCCmec of strain NCTC10442, sharing identical characteristic nucleotide sequences at both left and right extremities of the chromosome-SCCmec junction regions, as well as identical nucleotide sequences of integrated site attBscc, but differing from SCCRP62A and the SCC-CI of ATCC 12228 and other reported SCCmec types (Fig. 2). It contains the same 12-bp (5′-TAT CAT AAG TAA-3′) flanking direct repeat (DRSCC-L and DRSCC-R) at both junctions as strain NCTC10442 but lacks any inverted repeats (IRSCC-L and IRSCC-R) in SCCmecC10682 (Fig. 2). Instead, it harbors a pair of 15-bp dyad repeats, referred to as DyadR-L (5′-ATC TTG AAA CGA AGT-3′) and DyadR-R (5′-TGAAGCAAAGTTCTA-3′), at its junctions, which are 91 bp away from the DRSCC-R and 255 bp from the DRSCC-L, respectively (Fig. 2 and Table 2).
FIG. 2.
The boundaries of SCCmecC10682. SCCmecC10682 was integrated at exactly the same nucleotide position at the 3′ end of orfX gene (with identical characteristic nucleotide sequences at both left and right extremities of the chromosome-SCCmec junction regions as well as an identical nucleotide sequence of integrated site attBscc) as the prototypic type I SCCmec of strain NCTC10442. It differed, though, from SCCRP62A and the SCC-CI of ATCC 12228 and other reported SCCmec types. It contained a pair of hitherto unreported dyad repeats (DyadR-R and DyadR-L) but lacked discernible inverted repeats. Chromosomal sequences are boxed in gray, DRSCC sequences are indicated with bold type and a dotted box, while IRSCC sequences are indicated with a gray arrow. Consensus attBscc, DRSCC-R, IRSCC-R, DRSCC-L, and IRSCC-L sequences are indicated above. Dyad repeats are shown below with nucleotide distance from the direct repeat indicated. MSSA, methicillin-susceptible S. aureus.
Overall structural comparison of SCCmecC10682 with other reported prototypic SCCmec types.
In addition to having unique recombinatory sequences and boundaries, the complete SCCmecC10682 element was revealed by sequence analysis to have a G+C content (of the complete 32,168-bp sequence) of 31.7%, which was lower than that for S. aureus (32.8 to 32.9%). There were a total of 36 ORFs which were either previously described and/or were larger than 100 bp (Fig. 1A and Table 2).
The J1 region (extending from the left chromosome junction-ccr complex) was comprised of five ORFs, including those encoding the copper-transporting ATPase (copA), two hypothetical proteins (CA034 and CA035), a putative membrane protein (CA032), and a truncated ccrA gene (Fig. 1A and Table 2). With the exception of copA, which shows 99% homology, this region is 100% identical to a region between DR-SCCpbp4 and IS431, downstream of ccrAB4 of the SCC-CI element in ATCC 12228. Of note, the truncated ccrA gene (120 bp; 39 aa) has been found only in SCC-CI from the genomic sequence of S. epidermidis ATCC 12228 (35). It contains a single 120-bp coding region, for a hypothetical 39-aa protein, with high degrees of sequence homology to the outer flanking regions of ccrA3 (82.1%), ccrA2 (74.4 to 82.1%), and ccrA1 (76.9%) but lower degrees of homology to ccrA4 (41.0 to 46.2%) and ccrB (7.7 to 19.4%) genes. The putative membrane protein CA032 also shows 91% homology to a protein adjacent to the ccr gene complex in SCCmec type I and 92% homology to a protein adjacent to the ccr gene complex in S. hominis SCC12263.
Region J2 (extending between the ccr and mec gene complexes) was large and contained 19 ORFs, 7 of which (from CA018 to tnpA) belong to the mobile genetic element Tn554, a carrier of genes for erythromycin resistance (ermA) and streptomycin/spectinomycin resistance (aad9) (Fig. 1A and Table 2). ORFs CA026 and CA027 represent two highly conserved sequences found in various SCC elements, including SCCmec types I, II, IV, V, and VI, SCCRP62A, SCCmecZH47, SCC12263, and SCC-CI, with homologies ranging from 85 to 96% for CA9026 and 91 to 96% for CA9027. The remainder of the genes in region J2 show high degrees of homology to genes in the SCCmec type II and III and SCCRP62A elements, with sequence identities ranging from 99 to 100%.
The J3 region (extending from the right chromosome junction to the mec gene complex) is short, consisting of only two ORFs (CA001 and CA002), which code for conserved hypothetical proteins present in SCCRP62A (100% identical) (Fig. 1A and Table 2) as well as in SCCmec types I, II, and IV (99% homology; data not shown).
The size (32 kb) of the SCCmecC10682 element in this HA-MRSA isolate compared with other HA-MRSA strains is similar to that of the type I SCCmec element of NCTC10442 (34 kb) but smaller than the type II (53 kb) and type III (67 kb; now reconsidered a composite of the SCC element containing the mercury resistance operon (SCCmercury) and type III SCCmec [18]) SCCmec elements of strains N315 and 85/2082, respectively (data not shown). As expected, this element is larger than the type IV (21 to 25 kb) and type V (27.6 kb) SCCmec elements typically found in CA-MRSA isolates (although complete sequence information is not available for the type VI element).
Phenotypic, genotypic, and molecular characterization of epidemic strain C10682.
The clinical MRSA isolate C10682 was isolated in 2003 from a percutaneous endoscopic gastrostomy site subcutaneous abscess in a 58-year-old male patient from one of our local hospitals. Further screening of our collection of local MRSA clinical isolates by using the class A mec and type 4 ccr gene complex-specific primers (Table 1) revealed that two additional clinical isolates (C4631 from an abdominal wound in a 60-year-old male patient in 2004 and C5073 from a screening nasal swab in an 80-year-old female patient in 2005), as well as the CMRSA epidemic reference strain CMRSA9, carried the class A mec gene complex along with the type 4 ccr gene complex. Those isolates were found to be nontypeable by available SCCmec typing schemes. The assumption, therefore, was that these strains would also harbor the SCCmecC10682 element. In order to confirm this, PCR amplification targeting various regions spanning the whole SCCmecC10682 element was performed. As seen in Fig. 1A, 12 PCR targets at locations throughout the element, including one spanning the SCCRP62A-SCC-CI recombination junction, were chosen (primers 19 and 20; Table 1). PCR results for all genes tested in each of the three strains were positive (data not shown), indicating that the same SCCmecC10682 element was present in the tested local strains. To further confirm the same unique combination of class A mec and type 4 ccr gene complexes as in isolate C10682, partial ccr and mec gene complex sequencing was conducted, and it indicated that the three tested strains carried the same unique combination as the prototypic isolate C10682.
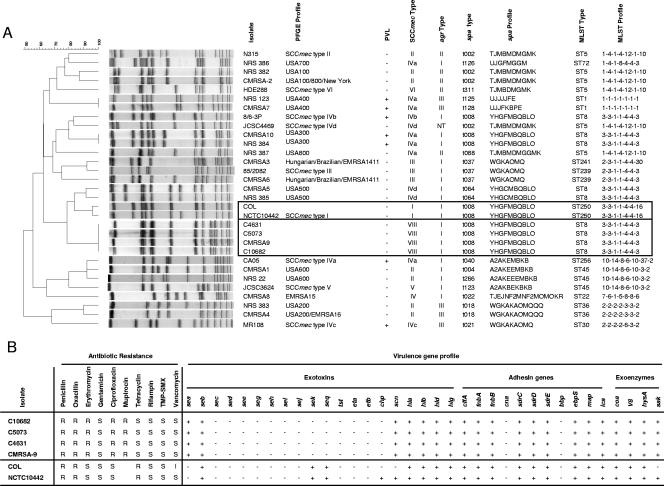
CMRSA9 is a typical HA-MRSA epidemic strain commonly found in Quebec and other regions in Canada (4). These isolates were subjected to further genotypic and phenotypic characterization for comparison with previously reported prototypic SCCmec type strains (types I to VI, including subtypes IVa to IVd), as well as the other major epidemic strains in Canada (strains CMRSA1 to CMRSA10) and the United States (strains USA100 to USA800) (Fig. 3). Molecular typing revealed unique genetic profiles for all four of these isolates (all of which harbored the unique SCCmec type with the class A mec/type 4 ccr gene complex), sharing the same CMRSA9 pulsotype, the same MLST type (ST8), spa type t008, and agr type I, and they lacked Panton-Valentine leukocidin genes (Fig. 3A). This genomic background is similar to those of the prototypic SCCmec type I strains NCTC10442 and COL, but it is different from those of other reference strains (Fig. 3A). We also performed PCR assays to detect 35 of the most common S. aureus virulence genes in these isolates. These four isolates possessed identical virulence gene profiles (Fig. 3B). All isolates also exhibited identical antimicrobial susceptibility and resistance phenotypes, including resistance to β-lactams, erythromycin, ciprofloxacin, and mupirocin but susceptibility to gentamicin, rifampin (rifampicin), trimethoprim-sulfamethoxazole, and vancomycin, which is a typical HA-MRSA antibiotic susceptibility/resistance profile. These data suggest that all four of these isolates belong to the same strain, sharing a similar genomic background and carrying the novel SCCmecC10682 type.
FIG. 3.
Phenotypic, genotypic, and molecular characterization of epidemic strain C10682. (A) Comparison of pulsotype and other molecular typing features of clinical isolates harboring SCCmecC10682, with previously reported prototypic SCCmec type strains (types I to VI, including subtypes IVa to IVd), the major epidemic MRSA strains in Canada (strains CMRSA1 to CMRSA10) and the United States (strains USA100 to USA800), as well as the early MRSA strains (isolated from England in the early 1960s) NCTC10442 and COL. PFGE, pulsed-field gel electrophoresis; PVL, Panton-Valentine leukocidin (+, positive; −, negative); agr, accessory gene regulator; NT, nontypeable. (B) Phenotypic and virulence gene profiles of four isolates harboring SCCmecC10682 in comparison to the early MRSA strains NCTC10442 and COL. Exotoxin genes are as follows: sea, seb, sec, sed, see, seg, seh, sei, sej, sek, and seq, staphylococcal enterotoxin A, B, C, D, E, G, H, I, J, K, and Q genes; tst, toxic shock syndrome toxin gene; eta and etb, exfoliative toxin genes A and B; chp, chemotaxis inhibitory protein gene; scn, staphylococcal complement inhibitory protein gene; hla, hlb, hld, and hlg, alpha, beta, delta, and gamma toxin genes. Adhesin genes are as follows: clfA, clumping factor gene; fnbA and fnbB, fibronectin adhesive molecule A and B genes; cna, collagen adhesive molecule gene; sdrC, sdrD, and sdrE, putative adhesin C, D, and E genes; bbp, bone sialoprotein adhesin; ebpS, elastin adhesin; map (eap), major histocompatibility complex class II analogue protein gene; ica, polysaccharide intercellular adhesin gene. Exoenzyme genes are as follows: coa, coagulase gene; V8, serine protease gene; hysA, hyaluronidase gene; sak, staphylokinase gene. S, susceptible; R, resistant; I, intermediate resistant; +, positive; −, negative; TMX-SMX, trimethoprim-sulfamethoxazole.
DISCUSSION
The essential and common traits of the SCCmec elements include (i) carriage of a mec gene complex; (ii) carriage of a ccr gene complex; (iii) integration at a specific nucleotide region (attBscc), which is located at the 3′ end of orfX; and (iv) demarcation by characteristic nucleotides, mostly associated with direct repeats and/or inverted repeats (5, 13, 15, 18, 20). Complete sequencing of SCCmecC10682 reveals that it carries a complete class A mec gene complex and a type 4 ccr gene complex, integrates in an identical chromosomal integration site (attBscc) at the 3′ end of the orfX gene, and is flanked by nucleotide sequences characteristic of direct and dyad repeats at the chromosome-SCCmec junction regions. This new element not only shares all essential and common traits typical of other prototypic SCCmec types but also harbors a previously uncharacterized unique combination of the mec and ccr gene complexes. According to the current criteria for the classification of SCCmec types (5, 13, 15, 18, 20), the SCCmecC10682 element not only qualifies as a SCCmec element but also represents a novel type of SCCmec. After we consulted the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements for the most appropriate name (roman numeral) for this new type of SCCmec element, it was suggested to tentatively assign our SCCmecC10682 (4A) the type SCCmec type VIII, since another new type (5C1; tentatively designed VII) identified in a CA-MRSA isolate from Sweden (2) is currently under review. However, there are only six currently recognized types of SCCmec to date, and they are types I (1B), II (2A), III (3A), IV (2B), V (5C2), and VI (4B) (13, 15, 20, 26).
Our novel SCCmecC10682 was identified from Canadian epidemic MRSA strain CMRSA9. CMRSA9 is a typical HA-MRSA epidemic strain commonly found in Quebec and other regions in Canada (4). Previously, CMRSA9 was reported as being of SCCmec type II (4), presumably because the entire SCCmec element was not sequenced. Instead of sequencing, an older SCCmec multiplex PCR typing assay (25) was used to characterize this SCCmec element (4), in which two (loci C and D) of the four known loci (B, C, D, and G) for SCCmec type II were also found in our SCCmecC10682. Although there is no identical reference strain for CMRSA9, its genomic background is quite similar to those of the strains NCTC10442 (the first MRSA isolate from England in 1961) (17) and COL (an early MRSA strain isolated from England in the early 1960s) (6), with a similar pulsed-field gel electrophoresis pattern and the same spa type, t008 (Fig. 3A). The CMRSA9 strain has an MLST designation of ST8, whereas NCTC10442 and COL have designation ST250 (Fig. 3A). However, they belong to the same clonal complex, CC8, and the only difference between them (ST8 and ST250) is the sequence at one gene, yqi (acetyl coenzyme A acetyltransferase; MLST profile number 3 for CMRSA9 in contrast to 16 for NCTC10442 and COL), while the other six genes are identical (Fig. 3A). They also have similar virulence gene profiles (Fig. 3B). Moreover, they share identical characteristic nucleotide sequences at both left and right extremities of the chromosome-SCCmec junction regions, as well as identical nucleotide sequences for integrated site attBscc, which differ from those for SCCRP62A and SCC-CI of S. epidermidis strains RP62A and ATCC 12228, respectively, as well as for those of the other reported SCCmec types (Fig. 2). Both NCTC10442 (SCCmec type I prototypic strain) and COL carry type I SCCmec; however, CMRSA9 carries a totally different type of SCCmec element. More interestingly, our sequence recombination analyses suggested that this unique SCCmec type might be generated from homologous recombination between the previously described SCCRP62A and SCC-CI of S. epidermidis strains RP62A and ATCC 12228, respectively. However, it is not clear how, where, and from which ancient strain(s) this CMRSA9 strain evolved and how it recently became one of the epidemic strains in Canada.
S. epidermidis RP62A (ATCC 35984) is a methicillin-resistant biofilm-producing isolate (12). Its complete genome (∼2.6 Mb) has been sequenced and carries a type II SCCmec which is 98% identical at the nucleotide level and identical in gene organization (with the exception of the region from pUB110 flanked by IS431-mec) to that of the S. aureus type II SCCmec of prototypic strain N315 (8). Another S. epidermidis strain, ATCC 12228, is a non-biofilm-forming, non-infection-associated strain (1). Although it is a methicillin-susceptible strain, analysis of its genomic sequence (∼2.5 Mb) reveals that it harbors an SCC element named SCCpbp4 which lacks the methicillin resistance determinant gene (mecA) but contains two pairs of ccrA and ccrB genes along with multiple insertion sequence elements, a restriction-modification system (hsdS and hsdM), and genes encoding penicillin binding protein 4 (pbp4) and conferring resistance to mercury and cadmium (Fig. 1B) (8, 22, 35). The two strains, although different in pathogenicities and structures of SCC islands, share 85% identity in predicted protein-coding sequences (31). Our sequence recombination analyses revealed that the sequences of the 5′ upstream region (carrying the mec gene complex) and the 3′ downstream region (carrying the ccr gene complex) of the new SCCmecC10682 cassette were identical to those of the SCCmec of RP62A (SCCRP62A) and the SCC element of ATCC 12228 (SCC-CI), whereas the middle segment shared 91.3 to 96.3% identity among these strains. These findings suggest that this unique SCCmec type might be derived from homologous recombination between S. epidermidis strains RP62A and ATCC 12228. Therefore, we hypothesize that the CMRSA9 strain may be derived from an early ancestral MRSA strain(s) such as NCTC10442 or COL and replaced its original type I SCCmec with the new SCCmecC10682 type through a recombination event with similar S. epidermidis or other staphylococcal strains (other coagulase-negative staphylococci, MRSA, or methicillin-susceptible S. aureus) that carry the mobile cassette SCCRP62A or SCC-CI, as in the RP62A and ATCC 12228 strains, while the ancestral MRSA strain was evolving under the selective pressures.
Although further detailed studies are needed to support this hypothesis, the present study readily provides compelling evidence supporting the ability of SCC elements to transfer horizontally or undergo recombination to generate new SCCmec types.
Acknowledgments
We thank T. Ito and K. Hiramatsu for the kind gift of the SCCmec control strains NCTC10442, N315, 85/2082, MR108, JCSC4469, and WIS; R. Daum for strains CA05 and 8/6-3P; and H. de Lencastre for the strain HDE288.
This work was partially supported by the Centre for Antimicrobial Resistance (CAR), Calgary Health Region/Calgary Laboratory Services/University of Calgary.
Footnotes
Published ahead of print on 8 December 2008.
REFERENCES
- 1.Arciola, C. R., L. Baldassarri, and L. Montanaro. 2001. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39:2151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglund, C., T. Ito, M. Ikeda, X. X. Ma, B. Soderquist, and K. Hiramatsu. 2008. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob. Agents Chemother. 52:3512-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caetano-Anollés, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-94. [DOI] [PubMed] [Google Scholar]
- 4.Christianson, S., G. R. Golding, J. Campbell, and M. R. Mulvey. 2007. Comparative genomics of Canadian epidemic lineages of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 45:1904-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lencastre, H., D. Oliveira, and A. Tomasz. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 10:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyke, K. G., M. P. Jevons, and M. T. Parker. 1966. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aureus. Lancet i:835-838. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackbarth, C. J., and H. F. Chambers. 1989. Methicillin-resistant staphylococci: detection methods and treatment of infections. Antimicrob. Agents Chemother. 33:995-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmsen, D., H. Claus, W. Witte, J. Rothganger, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 12.Hussain, M., M. Herrmann, C. von Eiff, F. Perdreau-Remington, and G. Peters. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updat. 6:41-52. [DOI] [PubMed] [Google Scholar]
- 17.Jevons, M. P. 1961. “Celbenin”-resistant staphylococci. Br. Med. J. 1:124-125. [Google Scholar]
- 18.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClure, J. A., J. M. Conly, V. Lau, S. Elsayed, T. Louie, W. Hutchins, and K. Zhang. 2006. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococci. J. Clin. Microbiol. 44:1141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mongkolrattanothai, K., S. Boyle, T. V. Murphy, and R. S. Daum. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulvey, M. R., L. Chui, J. Ismail, L. Louie, C. Murphy, N. Chang, and M. Alfa. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:3481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira, D. C., C. Milheirico, and H. de Lencastre. 2006. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 50:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70:4987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryffel, C., W. Tesch, I. Birch-Machin, P. E. Reynolds, L. Barberis-Maino, F. H. Kayser, and B. Berger-Bachi. 1990. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene 94:137-138. [DOI] [PubMed] [Google Scholar]
- 29.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 31.Wei, W., Z. Cao, Y. L. Zhu, X. Wang, G. Ding, H. Xu, P. Jia, D. Qu, A. Danchin, and Y. Li. 2006. Conserved genes in a path from commensalism to pathogenicity: comparative phylogenetic profiles of Staphylococcus epidermidis RP62A and ATCC12228. BMC Genomics 7:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, K., J. A. McClure, S. Elsayed, J. Tan, and J. M. Conly. 2008. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J. Infect. Dis. 197:195-204. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, K., J. Sparling, B. L. Chow, S. Elsayed, Z. Hussain, D. L. Church, D. B. Gregson, T. Louie, and J. M. Conly. 2004. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42:4947-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Zhang, K., J.-A. McClure, S. Elsayed, and J. M. Conly. 2008. 13th Int. Symp. Staphylococci Staphylococcal Infect., abstr. 660.
- 35.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]