Abstract
Recent findings suggest bidirectional antagonisms between the K65R mutation and thymidine analogue mutations in human immunodeficiency virus type 1 (HIV-1)-infected, treatment-experienced patients, yet little is known about HIV-2 in this regard. This study addressed the effects of innate polymorphisms in HIV-2 on emergent resistance to nucleoside/nucleotide analogues. Emergent drug resistance profiles in HIV-2 subtypes A (n = 3) and B (n = 1) were compared to those of HIV-1 subtypes B and C. Drug resistance was evaluated with cord blood mononuclear cells (CBMCs) and MT2 cells, using selective pressure with tenofovir (TFV), zidovudine (ZDV), stavudine (d4T), didanosine (ddI), abacavir (ABC), lamivudine (3TC), emtricitabine (FTC), or various dual-drug combinations. Resistance was evaluated using conventional and ultrasensitive sequencing approaches. In agreement with our previous findings, dual-drug combinations of TFV, ddI, ABC, d4T, ZDV, and 3TC preferentially selected for K65R in HIV-1 subtype C isolates. In HIV-1 subtype B, TFV-3TC and ZDV-3TC selected for M184I and D67N, respectively. In contrast, selections with all four HIV-2 cultures favored the development of M184I in dual-drug combinations that included either 3TC or FTC. Since HIV-2 cultures did not develop K65R, an ultrasensitive allele-specific real-time PCR assay was developed to distinguish the presence of 65R from wild-type K65 after 16 cycles with a discriminatory ability of 0.1% against a population of wild-type virus. These results underscore potential differences in emergent drug resistance pathways in HIV-1 and HIV-2 and show that polymorphisms may influence the development of the resistance pathways that are likely to emerge.
Persons infected with human immunodeficiency virus type 2 (HIV-2) usually exhibit slower disease progression, lower rates of vertical transmission, and lower viral loads than patients infected with HIV-1 and are often asymptomatic (10, 33, 47). This has resulted in a more-restricted spread of HIV-2 than HIV-1, mainly to parts of west Africa (25). The reverse transcriptases (RT) of HIV-1 and HIV-2 possess approximately 40% divergence in amino acid sequences (19).
Patients infected with either HIV-1 or HIV-2 are routinely treated with nucleoside and nucleotide RT inhibitors (NRTIs) (1), even though all those drugs were developed for use against HIV-1, and little information on their activity against HIV-2 is available. Although multiple studies have reported the selection of mutations associated with resistance to NRTIs in HIV-1, few studies have been performed on patients infected with HIV-2 (8, 9, 11). Furthermore, no database or algorithm exists for the interpretation of mutations selected during the treatment of HIV-2 disease. Although it is known that HIV-2 shows natural resistance to nonnucleoside RT inhibitors (NNRTIs) and the fusion inhibitor T-20 (42, 51, 53), information on HIV-2 resistance to NRTIs is lacking. Despite the overall sensitivity of HIV-2 to NRTIs, some articles have suggested that HIV-2 may be naturally resistant to certain NRTIs such as zidovudine (ZDV) (41). However, a recent study claimed that HIV-1 and HIV-2 display comparable sensitivities to ZDV and other NRTIs in vitro (48).
Recent findings suggest bidirectional antagonisms between K65R and thymidine analogue mutations (TAMs) in treatment-experienced patients with HIV-1 (37-39). Our own studies have shown a facilitated development of K65R in HIV-1 subtype C, due to a signature KKK motif at codons 64, 65, and 66 (5). This present study addressed the effects of innate polymorphisms (T69N, V75I, V118I, L210N, T215S, and K219E) on emergent resistance to NRTIs in HIV-1 and HIV-2 subtypes using cord blood mononuclear cells (CBMCs) and the MT2 cell line. We also developed an ultrasensitive allele-specific real-time PCR (AS-PCR) assay to detect the possible presence of the K65R mutation in HIV-2 cultures that did not contain this mutation on the basis of regular genotyping.
(This work was performed by M. L. Ntemgwa in partial fulfillment of the requirements for a Ph.D. degree from the Faculty of Graduate Studies and Research, McGill University, Montreal, Quebec, Canada.)
MATERIALS AND METHODS
Virus isolates, cells, and plasmids.
Three HIV-2 subtype A isolates (CBL-20, CBL-23, and MVP-15132) and one HIV-2 subtype B isolate (CDC310319) were obtained from the AIDS Research and Reference Reagent Program of the National Institutes of Health (NIH), Bethesda, MD. CBL-20 and CBL-23 were isolated from symptomatic patients from The Gambia and have been previously described (46), while MVP-15132 was from a German woman who had been infected by a Senegalese man (14). CDC310319 was from a blood donor from the Ivory Coast (35). The HIV-1 subtype B clinical isolate 5512 was obtained with informed consent from a drug-naïve individual at our clinics in Montreal, Canada, while BG-05, an HIV-1 subtype C clinical isolate, was obtained from Botswana; both have previously been described (12, 30). MT2 cell lines were also obtained from the NIH AIDS Research and Reference Reagent Program. CBMCs were obtained through the Department of Obstetrics, Jewish General Hospital, Montreal, Canada. We obtained the HIV-2ROD plasmid courtesy of Andrew Lever, Cambridge University, United Kingdom, with the permission of Tom Schulz, University of Hannover, Germany.
Drugs.
We received lamivudine (3TC) and abacavir (ABC) as gifts from GlaxoSmithKline (Research Triangle Park, NC); ZDV, didanosine (ddI), and stavudine (d4T) were obtained from Sigma-Aldrich, Inc. (Oakville, Ontario, Canada); and tenofovir (TFV) and emtricitabine (FTC) were obtained from Gilead Pharmaceuticals (Foster City, CA).
Virus production.
Viral strains were amplified as described by the coculture of peripheral BMCs from infected patients with uninfected CBMCs (26, 44). The HIV-2 isolates were amplified in CBMCs according to specifications provided by the NIH (2). The pol regions of the isolates were sequenced as described below.
Selections for resistance mutations in CBMCs and MT2 cells.
CBMCs or MT2 cells were infected with viruses (multiplicity of infection of 0.01) for 2 h at 37°C and subsequently washed with RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum to eliminate any unbound virus. Infected CBMCs or MT2 cells were seeded in 24-well plates at a density of 2.5 × 105 per well (16). Selection for resistance in CBMCs or MT2 cells was performed by a standard procedure using increasing concentrations of drugs (TFV, ZDV, d4T, ddI, ABC and 3TC), which were used either as single agents or in combination at starting concentrations below the 50% inhibitory concentration levels of the drugs. Based on RT values in culture fluids at the previous round of replication as a measure of viral replication (31, 40), drug concentrations were increased to levels ranging between 0.25 and 10 μM. At each passage, the culture fluids were harvested and kept at −80°C for a subsequent genetic analysis by sequencing.
Selection at a particular drug concentration was defined as complete when a repeated passage revealed that the levels of RT activity in culture fluids had peaked at the same time as that of a control well that did not contain drugs.
Nucleic acid extraction and PCR amplification.
Viral RNA was extracted from culture supernatants using the Qiagen QIAamp viral extraction kit (Mississauga, Ontario, Canada). For the amplification of HIV-2 genetic information, the RNA in culture supernatants was reverse transcribed and the protease (PR) and RT genes amplified by PCR using specific primer pairs, i.e., RT2-reverse and PR1-forward and nested PR3-forward and RT4-reverse (13, 43). The PR and RT genomic regions of the HIV-2 pol gene were amplified from these same PCR products, which were analyzed in 1% agarose gels with ethidium bromide. The resulting PCR-amplified DNA fragments were purified using the QIAquick PCR purification kit. The PCR products were used as templates for the nucleotide sequencing analysis of the RT gene. The amplification of HIV-1 was performed using a previously published protocol (Virco BVBA, Mechelen, Belgium). Oligonucleotides were chemically synthesized and purchased from Invitrogen (Burlington, Ontario, Canada).
DNA sequencing analysis.
The RT genes of the HIV-2 samples were directly sequenced to analyze for NRTI-associated mutations using primer pairs RT3, RT4, RT5, and RT6 (43). The cycle sequencing of both strands was performed on the GeneAmp PCR 9700 instrument with the BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA). Excess dye-labeled terminators were removed from the extension products, and the purified products were sequenced on the ABI Prism 3130 XL genetic analyzer (Applied Biosystems). The nucleotide sequences of the RT genes were then assembled using GeneTool 2.0 autoassembler software. The nucleotides were aligned and assessed in terms of amino acid differences using BioEdit version 7.0 software (20) (reference to HIV-2ROD [Genbank accession number M15390]). Differences in amino acids at sites conferring resistance to NRTIs were then analyzed. The HIV-1 samples were sequenced in both directions using a previously published protocol (Virco) according to the manufacturer's instructions (18, 28). The sequence for each sample was compared to databases of known drug resistance mutations.
Plasmid and primers for site-directed mutagenesis.
We introduced the K65R mutation into the HIV-2ROD plasmid by site-directed mutagenesis by the use of a QuikChange II XL site-directed mutagenesis kit (Stratagene, LaJolla, CA). The introduction of the K65R mutation into the plasmid was confirmed by sequencing and ultimately transformed into DH5α (Invitrogen) for high-yield plasmid production. The following primer pairs were used, in accordance with the manufacturer's instructions: HIV-2ROD (65R) (forward), 5′-CCACATTTGCAATCAAGAGAAAGGACAAAAACAAATGGAG-3′, and HIV-2ROD (65R) (reverse), 5′-CTCCATTTGTTTTTGTCCTTTCTCTTGATTGCAAATGTGG-3′.
Allele-specific PCR assay to detect the K65R mutation.
Since we did not select for the K65R mutation in HIV-2 isolates using NRTIs such as ABC, TFV, and ddI which are associated with this mutation, we designed a novel ultrasensitive AS-PCR assay to determine if the K65R mutation may have been present as a minority species.
The supplemental material includes details of the AS-PCR assay for the detection of the K65R mutation in HIV-2 as well as information related to the discriminatory ability of the assay.
Our novel K65R AS-PCR assay utilized SYTO9 (Molecular Probes, OR), an intercalating dye, and 3′ lock nucleic acid (LNA)-specific primers. The real-time PCRs were performed using 2 μl of the PCR products and three primer pairs that were specific for the K65R mutation, wild-type K65, and the total virus population, i.e., K65R plus the wild type, respectively. The primer sets used were 65R-fw (5′-CCC TAC ATT TGC AAT CAA GA+G-3′) and 65R-rev (5′-TGG AAT TCC TAG CTG AAT TTC TGT-3′) for the mutation-specific reaction; WT-fw (5′-CCC TAC ATT TGC AAT CAA GA+A-3′) and WT-rev (5′-TGG AAT TCC TAG CTG AAT TTC TGT-3′) for the wild-type-specific reaction; and TOT-fw (5′-CCC TAC ATT TGC AAT CAA GA-3′) and TOT-rev (5′-TGG AAT TCC TAG CTG AAT TTC TGT-3′) for the total copy reaction, where “+” represents a LNA. Of note, all reverse primers were the same and all changes were mainly implemented on the forward primer.
Real-time PCRs were enacted for 45 cycles of melting at 94°C for 15 s, annealing at 58°C for 10 s, and extension at 72°C for 30 s. This was followed by two holds, the first at 72°C for 1 min and another at 50°C for 30 s.
For the quantification of various viral populations, DNA standards were prepared and run in parallel with each reaction. The standards were tested in duplicate for each experiment and were prepared in a serial dilution from 109 to 103 copies per reaction. The viral PCR products were tested at concentrations of 107 copies/μl.
The specificity of the detection of the amplified product was checked using the analysis of the melting curve for each reaction. In this study, we used high resolution melt (HRM) analysis, an enhancement to traditional melting analysis that significantly increases the detail and information that can be captured. HRM is a simpler and more cost-effective way to characterize DNA samples according to sequence, GC content, length, or strand complementarity than probes, since it allows for single nucleotide polymorphism genotyping and the genotyping of all single nucleotide polymorphism conversions. The Rotor-Gene 6000 real-time rotary analyzer (Corbett Research, Sydney, Australia) utilized in this study is one of only a few instruments that can carry out this analysis.
The discriminatory ability or limit of detection of our assay was tested by adding 106 copies of wild-type plasmid onto equal volumes of serial dilutions of samples of K65R-containing plasmid (106 to 102).
Replicative ability of HIV-2 and HIV-1.
The replicative abilities of HIV-1 and HIV-2 wild-type isolates were determined as previously described (34). Briefly, wild-type viruses were used to infect CBMCs for 2 h. Infected CBMCs were plated onto 96-well plates containing medium supplemented with interleukin-2. After 3 days of incubation, the cells were refed with fresh medium containing interleukin-2. At day 7, RT assays were performed to determine growth rates (6). In addition, p24 antigen levels were measured in these cultures using the Vironostika HIV-1 Antigen MicroELISA kit (Biomerieux bv, Boxtel, The Netherlands).
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in GenBank under the following accession numbers: 5512 (EU927298), BG-05 (EU927299), CBL-20 (EU927300), CBL-23 (EU927301), MVP-15132 (EU927302), and CDC310319 (EU927303).
RESULTS
Phylogenetic analysis.
After sequencing, the subtype status of HIV-1 and HIV-2 isolates was confirmed by aligning and comparing with the RT regions of various reference strains representing HIV-1 and HIV-2 subtypes, obtained from the Los Alamos database (http://www.hiv.lanl.gov), using BioEdit version 7.0 (20); phylogenetic trees were constructed with MEGA version 3.0 software (27) using the neighbor-joining method. Bootstrap values of >70% were considered significant. The results showed that all three HIV-2 subtype A isolates clustered with other subtype A viruses from the database, while CDC310319 clustered with HIV-2 viruses of subtype B (data not shown).
Natural polymorphisms in HIV-1/HIV-2 RT.
At baseline, the HIV-1 subtype B clinical isolate 5512 and subtype C isolate BG05 had no major mutations associated with resistance to NRTIs. However, isolate 5512 harbored polymorphisms at positions K122E, I135V, S162C, and D177N, while BG05 harbored V35T, T39E, S48T, K122E, D123G, K173V, D177E, T200A, Q207E, R211K, V245K, and E248D. All of the above are minor polymorphisms not associated with HIV-1 resistance (Fig. 1).
FIG. 1.
Baseline polymorphisms in HIV-2 RT compared with HIV-1 subtype B and amino acid alignment of the RT sequences of HIV-2 and HIV-1 that were used in this study. The positions shaded are those involved in HIV-1 drug resistance (T69N, V75I, V118I, L210N, T215S, and K219E for NRTIs and V90I, K101A, K103R, V106I, V179I, Y181I, Y188L, and G190A for NNRTIs). The dots indicate identity with HIV-1 subtype B HXB2. The HIV-1 subtype B sample 5512 possessed the baseline polymorphisms K122E, I135V, S162C, and D177N, while BG-05 harbored V35T, T39E, S48T, K122E, D123G, K173V, D177E, T200A, Q207E, R211K, V245K, and E248D, which are all minor polymorphisms not associated with HIV-1 resistance. The HIV-2 viruses studied included CBL-20, CBL-23, MVP-15132, and CDC310319. The HIV-1 viruses studied were isolates 5512 and BG-05.
The HIV-2 isolates used in this study showed natural polymorphisms at sites known to be associated with HIV-1 resistance to NRTIs (Fig. 1). These polymorphisms included T69N, V75I, V118I, L210N, T215S, and K219E, which are known to be associated with HIV-1 resistance to NRTIs, and also V90I (3/4), K101A, K103R (1/4), V106I, V179I, Y181I, Y188L, and G190A, which are known to be associated with HIV-1 resistance to NNRTIs. Of particular interest are polymorphisms at positions 210, 215, and 219 known to be associated with HIV-1 TAMs that are associated with cross-resistance to all currently approved NRTIs (24, 29).
Viral replication.
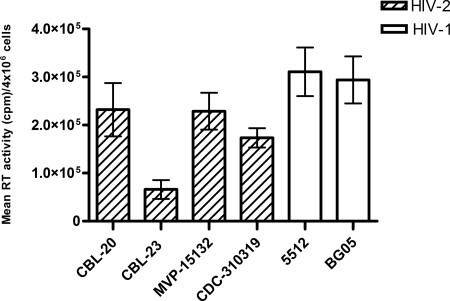
It is known that HIV-2 replicates less well than HIV-1. The results of Fig. 2 show differences in the RT activities per 4 × 106 cells per round of infection for HIV-1 and HIV-2 isolates. The RT activities of the HIV-2 isolates were lower than those of HIV-1, in conformity with published data.
FIG. 2.
RT activities of wild-type HIV-1 and HIV-2 isolates. The bars represent the means ± standard errors of the means of the maximal RT activity of wild-type viruses per round of infection measured each week for 30 weeks. The HIV-2 viruses studied included CBL-20, CBL-23, MVP-15132, and CDC310319. The HIV-1 viruses studied were isolates 5512 and BG-05.
Time to the development of resistance mutations in HIV-1/HIV-2 in CBMCs.
Using single-drug selections, K65R was selected using TFV and ddI in HIV-1 subtype C, while M184V was selected with 3TC as shown in Table 1. M184V appeared within 6 weeks in most HIV-1 and HIV-2 isolates. Some HIV-2 isolates developed novel mutational motifs, such as S134A, V167I, and A174V, under TFV selective pressure. The HIV-2 isolate MVP-15132 that developed the S134A and V167I mutations under TFV pressure showed cross-resistance to TFV, 3TC, ddI, and ABC and high-level resistance to ZDV (data not shown).
TABLE 1.
Time to development of mutations associated with resistance in HIV-1 and HIV-2 using single NRTIs in CBMCsa
| Drug | Wk | Sequence inb:
|
|||||
|---|---|---|---|---|---|---|---|
| HIV-1
|
HIV-2
|
||||||
| Subtype C BG-05 | Subtype B 5512 | Subtype A
|
Subtype B CDC310319 | ||||
| CBL-20 | CBL-23 | MVP-15132 | |||||
| ABC | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | wt | wt | wt | wt | wt | wt | |
| 22-30 | L74V, M184V, or K65R | wt | wt | wt | wt | wt | |
| d4T | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | wt | wt | wt | wt | wt | wt | |
| 22-30 | wt | wt | wt | wt | wt | wt | |
| ddI | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | wt | wt | wt | wt | wt | wt | |
| 22-30 | K65R | wt | wt | wt | wt | wt | |
| TFV | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | K65R | wt | wt | wt | wt | wt | |
| 22-30 | K65R | wt | wt | A174V | S134A, V167I | wt | |
| ZDV | 1-5 | wt | wt | wt | wt | wt | ND |
| 22-30 | wt | wt | wt | wt | wt | ||
| 3TC | 1-5 | wt | wt | wt | wt | wt | wt |
| 6 | M184I | M184V | M184V | M184I | M184V | M184V | |
| 22-30 | M184V | M184V | M184V | M184V | M184V | M184V | |
Viral isolates were serially passaged using increasing doses of ABC, d4T, ddI, TFV, ZDV, or 3TC. Genotypic analysis ascertained the time to the first appearance of resistance-associated mutations. At baseline, all wild-type HIV-2 isolates harbored the following polymorphisms known to be associated with emergent resistance to RT inhibitors in HIV-1: for NRTIs, T69N, V75I, V118I, L210N, T215S, and K219E, and for NNRTIs, V90I, K101A, K103R, V106I, V179I, Y181I, Y188L, and G190A.
wt, wild type; ND, not determined.
The HIV-1 subtype C virus BG-05 followed two different mutational pathways under ABC selective pressure, i.e., either the K65R or the L74V/M184V pathway. The K65R mutation was not selected in either the HIV-2 or HIV-1 subtype B isolates under selective pressure with either ABC, TFV, ddI, or d4T or any combination thereof, all of which are known to select K65R in HIV-1 subtype C (Table 1 and 2).
TABLE 2.
Time to the development of mutations associated with resistance in HIV-1 and HIV-2 using dual NRTI combinations in CBMCsa
| Drug combination | Wk | Sequence inb:
|
|||||
|---|---|---|---|---|---|---|---|
| HIV-1
|
HIV-2
|
||||||
| Subtype C BG-05 | Subtype B 5512 | Subtype A
|
Subtype B CDC310319 | ||||
| CBL-20 | CBL-23 | MVP-15132 | |||||
| ZDV-3TC | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | wt | D67N | M184I | ND | M184I | M184I | |
| 22-30 | K65R | D67N | M184I | M184I | M184I | M184I | |
| TFV-ddI | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | wt | wt | wt | wt | wt | wt | |
| 22-30 | K65R | wt | wt | wt | wt | wt | |
| TFV-ABC | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | wt | wt | wt | wt | wt | wt | |
| 22-30 | K65R | wt | wt | wt | wt | wt | |
| TFV-3TC | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | K65R | M184I | M184I | wt | wt | wt | |
| 22-30 | K65R | M184I | M184I | M184I | M184I | M184I | |
| d4T-ddI | 1-5 | wt | wt | wt | wt | wt | wt |
| 12 | K65R | wt | wt | wt | wt | wt | |
| 22-30 | K65R | wt | wt | wt | wt | wt | |
Viral isolates were serially passaged using increasing doses of drug combinations ZDV-3TC, TFV-ddI, TFV-ABC, TFV-3TC, or d4T-ddI. Genotypic analysis ascertained the time to the first appearance of resistance-associated mutations.
wt, wild type; ND, not determined.
In agreement with our previous findings, dual-drug combinations of TFV, ddI, ABC, d4T, ZDV, and 3TC preferentially selected for the K65R mutation in HIV-1 subtype C isolates within 22 weeks, including the drug combination ZDV-3TC (Table 2). In HIV-1 subtype B, TFV-3TC favored the selection of the M184I pathway, while ZDV-3TC selected for TAMs, mostly D67N (Table 2). In contrast, selections with all dual-drug combinations that included 3TC favored the selection of M184I in all four HIV-2 cultures (Table 2).
Time to the development of resistance mutations in HIV-2 in MT2 cell lines.
Previous studies (C. Invernizzi, D. Coutsinos, D. Moisi, M. Oliveira, B. Spira, B. Brenner, and M. Wainberg, presented at the 15th Conference on Retroviruses and Opportunistic Infections, 2008) showed that the facilitated development of K65R in HIV-1 subtype C is due to a signature KKK motif at codons 64, 65, and 66. In comparison with HIV-1 subtype C, this involves differences at codons 64 (AAG→AAA), 65 (AAA→AAG), and 66 (AAA→AAG) that are absent in other subtypes. The K65R resistance mutation arises in HIV-1 by a single A to G transition, AAA→AGA or AAG→AGG in subtypes B and C, respectively.
In this study, we examined this same motif in HIV-2 and observed that all our HIV-2 subtype A isolates had the same amino acids, KKK, at these three key positions, as is the case in HIV-1. At the nucleotide level, all the HIV-2 subtype A isolates were identical to HIV-1 subtype B at the first two codons of the KKK motif, i.e., 64 (AAG) and 65 (AAA), but had a different sequence at codon 66 (AAG). In contrast, the HIV-2 subtype B isolate CDC310319 was similar to HIV-1 subtype C at the first two codons of KKK, i.e., 64 (AAA) and 65 (AAG), but had a different sequence at codon 66 (AGA), which codes for arginine, i.e., KKR. By analogy, the selection of the K65R mutation in HIV-2 should also occur by a single A-to-G transition.
Based on this, we chose two HIV-2 isolates, one subtype A (CBL-20) and one subtype B (CDC310319), and performed drug selections using MT2 cells. In these experiments, we used drug combinations that are currently or are likely to be used in patients infected with HIV-2, i.e., TFV-FTC (Truvada), ZDV-3TC (Combivir), or TFV-3TC.
Table 3 shows the results of the selection experiments for HIV-2 subtypes A and B in MT2 cells with single-drug regimens (ABC, TFV, ddI, 3TC, and FTC) as well as dual-drug combinations (TFV-3TC, TFV-FTC, and ZDV-3TC). The results show that ABC and FTC each selected for the M184I mutation in both HIV-2 subtypes A and B, while M184V was preferentially selected by 3TC. Neither TFV nor ddI selected for any mutations associated with resistance apart from a novel A138M mutation in virus CBL-20; the role of this mutation needs to be confirmed. When dual-drug combinations (TFV-3TC, TFV-FTC, and ZDV-3TC) were used, all HIV-2 viruses followed the M184I pathway, irrespective of subtype. Of note, the K65R mutation was never selected in HIV-2.
TABLE 3.
Differential selection of mutations in HIV-2 subtypes A and B in MT2 cellsa
| Drug | Passage no. | Sequence in HIV-2b
|
|
|---|---|---|---|
| Subtype A CBL-20 | Subtype B CDC310319 | ||
| ABC | 5 | wt | wt |
| 12 | wt | wt | |
| 20 | M184I | M184I | |
| TFV | 5 | wt | wt |
| 12 | A138 M | wt | |
| 20 | A138 M | wt | |
| ddI | 5 | wt | wt |
| 12 | wt | wt | |
| 20 | wt | wt | |
| 3TC | 5 | wt | wt |
| 6 | M184V | M184V | |
| 20 | M184V | M184V | |
| FTC | 5 | wt | wt |
| 12 | M184I | wt | |
| 20 | M184I | M184I | |
| TFV-3TC | 5 | wt | wt |
| 12 | wt | wt | |
| 20 | M184I | M184I | |
| TFV-FTC | 1-5 | wt | wt |
| 12 | wt | wt | |
| 20 | M184I | M184I | |
| ZDV-3TC | 1-5 | wt | wt |
| 12 | wt | wt | |
| 20 | M184I | M184I | |
Viral isolates were serially passaged in increasing doses of ABC, TFV, ddI, 3TC, and FTC or combinations of TFV-3TC, TFV-FTC, or ZDV-3TC in MT2 cells. Genotypic analysis ascertained the time to the first appearance of resistance-associated mutations.
wt, wild type.
All experiments included control wells with no drug. Viral nucleic acid from such wells was also sequenced at different times and did not reveal the presence of any resistance mutations.
Absence of the K65R mutation as a minority species in HIV-2 selections.
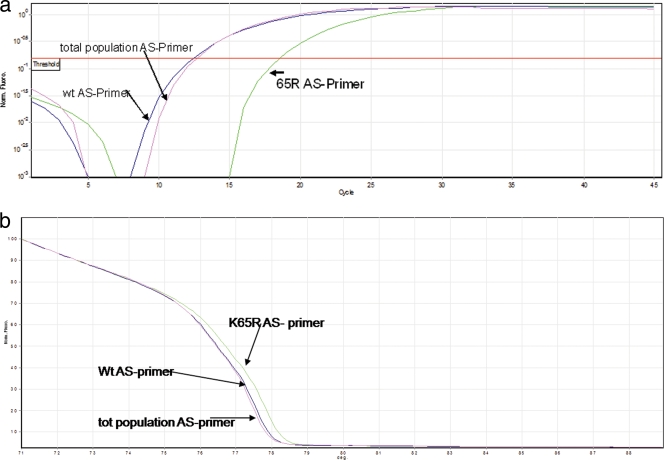
It is known that routine genotyping does not detect populations that are present at frequencies below 20% of the total viral population. In order to determine if the K65R mutation was present as a minority species, our AS-PCR assay was designed with three primer pairs, as described in Materials and Methods. Since our HIV-2 cultures did not develop the K65R mutation, we screened all culture supernatants at the end of the selection using our newly developed ultrasensitive assay. The results obtained using isolate CBL-20 with TFV and ABC are shown in Fig. 3a and show that K65R was absent, even after 30 weeks of culture under TFV pressure. Figure 3a also shows that the total virus population and wild-type curves overlapped, indicating the absence of K65R. The curve based on the K65R-specific primer should have appeared first if even a small percentage of K65R minority virus was present. Similar results were obtained with all other cultures (data not shown), indicating the absence of K65R even as a minority species. These results were further confirmed by HRM analysis as shown in Fig. 3b.
FIG. 3.
Absence of K65R as a minority species in culture supernatants. We tested HIV-2 growth under selection pressure in cells for the presence of the K65R mutation as a minority species using our novel AS-PCR assay as described. The amplification plots (a) and HRM analysis (b) show that K65R was absent even after 30 weeks of culture under TFV drug pressure. Panel a shows that the total virus population and wild-type curves appeared at the threshold level at the same time, indicating the absence of K65R. Similar results were obtained for other samples (data not shown).
DISCUSSION
Recent findings suggest bidirectional antagonisms between K65R and TAMs in HIV-1 treatment-experienced patients (37-39).
HIV-2 expresses several naturally occurring potential drug resistance polymorphisms in RT that might be implicated in emergent drug resistance. Some of these polymorphisms occur at sites that are associated with HIV-1 resistance to NRTIs (T69N, V75I, V118I, L210N, T215S, and K219E), while others are at positions that are implicated in HIV-1 resistance to NNRTIs (V90I, K101A, K103R, V106I, V179I, Y181I, Y188L, and G190A).
In previous studies, treatment-associated changes were shown to occur in the HIV-2 RT gene at sites corresponding to some of those that confer drug resistance in HIV-1 (1, 4, 43). However, these changes could not be directly associated with a particular NRTI, since monotherapy is not an acceptable therapeutic option. In our study, we now show that different mutational pathways may be associated with resistance to different N(t)RTIs or combinations thereof.
Our study has shown that the selection of M184I is the preferred pathway for resistance in HIV-2 with NRTI combinations that include either 3TC or FTC. It is known that the M184V and M184I mutations can be selected both in vitro and in vivo in HIV-1 clinical isolates exposed to 3TC, FTC, or ABC (32, 45, 49, 50, 52). Our study also shows that the time to the development of the M184I/V mutation in HIV-2 under 3TC pressure in vitro with CBMCs is as fast as 6 weeks.
M184V is also the mutation most commonly selected in HIV-2-treated patients receiving 3TC (22, 45, 50).
Some naturally occurring potential drug resistance polymorphisms in HIV-2 compared to those in HIV-1 may confer baseline natural resistance to NRTIs, as has been seen in the case of ZDV (41). This natural resistance has been attributed to sequence differences at positions similar to those implicated in HIV-1 resistance to ZDV and cross-resistance to all approved N(t)RTIs (7, 23), i.e., L210W, T215Y/F, and K219Q/E. In HIV-2, the amino acid changes at these positions are L210N, T215S, and K219E.
In our study, maximal RT values at day 7 in CBMC assays for HIV-2 with ZDV were consistently higher than those with HIV-1. In fact, the maximum ZDV concentration that could be attained for HIV-1 versus HIV-2 was 1.0 μM and 10 μM, respectively, after 30 weeks in culture, while the baseline 50% inhibitory concentrations for HIV-1 and HIV-2 were within the same range. It has been suggested that the mechanisms whereby resistance develops against ZDV are different for HIV-1 and HIV-2 and that diminished incorporation rather than selective excision may play a more important role in HIV-2 (3). They also showed that HIV-1 more readily incorporates ZDV and is more susceptible to ZDV than HIV-2. These findings are consistent with our results that far higher concentrations of ZDV are needed to durably suppress HIV-2 than HIV-1 replication.
Studies are ongoing to revert some of the polymorphisms in HIV-2 at positions 210, 215, and 219 to determine the effect that this will have on HIV-2 resistance and on the development of the K65R mutation in other selection experiments.
The baseline phenotypes of HIV-2 isolates are generally in the range of those of HIV-1, suggesting that some of the polymorphisms detected in the HIV-2 RT are secondary mutations. These findings underscore that HIV-2 and non-B subtypes of HIV-1 may show differential responses to NRTIs.
Our studies have shown that the facilitated development of K65R in HIV-1 subtype C compared to that in subtype B is due to a signature KKK motif at codons 64, 65, and 66 (5). We have recently shown that changing codons 64 and 65 in HIV-1 subtype B to those present in subtype C led to the rapid selection of the K65R mutation (unpublished data). We have also shown that HIV-2 subtype A and HIV-1 subtype B share the same nucleotides at amino acids 64/65, while HIV-2 subtype B and HIV-1 subtype C share a set of different nucleotides at this same position. However, selection experiments with HIV-2 subtype B did not lead to the selection of K65R with either CBMCs or cell lines.
It is known that the appearance of the K65R mutation can preclude the development of TAMs and vice versa in HIV-1 (37-39). Due to this strong antagonism, the coexistence of K65R and TAMs is rare. The strongest such antagonism seems to exist between K65R and T215Y (37), due to the K65R-mediated reduction of the TAM-related excision process induced by TAMs. Of note, HIV-2 has potential drug resistance polymorphisms at three TAM positions, 210, 215, and 219. However, it is unlikely that HIV-2 RT is similarly affected, since the latter does not apparently enact excision (3).
We were also unable to select for K65R in HIV-2 using CBMCs and MT2 cells, even with drug combinations (i.e., TFV-ABC, TFV-ddI, and d4T-ddI) that would usually select for this mutation with HIV-1. In regard to NRTIs, amino acids at six positions in wild-type HIV-2 are analogous to secondary or accessory drug resistance mutations in HIV-1 (69N, 75I, 118I, 210N, 215S, and 219E). These residues may predispose the virus to distinct evolutionary pathways in response to drug pressure.
We also wondered whether K65R may have been present in HIV-2 but at levels below the 20% that are detectable by routine genotyping (36). Toward this end, we designed an ultrasensitive AS-PCR assay to reveal the presence of K65R and also introduced this mutation into a HIV-2 plasmid by site-directed mutagenesis. Our results showed that the AS-PCR assay was able to distinguish the presence of the K65R mutation from wild-type HIV-2 after 16 cycles of amplification, i.e., a 216 (65,536)-fold decrease in the efficiency of amplification of the incorrect target. Our assay possessed an estimated discriminatory ability of 0.1% for the detection of K65R in a population of wild-type viruses and also showed that the K65R mutation was absent in all culture supernatants at the end of the selection experiments. Thus, both conventional and ultrasensitive approaches have shown that the HIV-2 isolates studied did not select K65R.
Two clinical studies have reported a high frequency of K65R in HIV-2-infected patients receiving NRTIs (9, 11). However, other studies reported that K65R was either rare (only one or two patients) (8, 22) or nonexistent in treated patients (43). In vitro, K65R has rarely been described in HIV-2 and with different isolates than the ones we utilized (41).
Although we did not select for TAMs with HIV-1 subtype B using ZDV, unpublished and published data from many laboratories show that TAMs are preferentially selected when HIV-1 subtype B is put under ZDV selective pressure in cell culture (15-17, 21). In a previous study, HIV-1 was shown to develop the 215Y mutation quickly under ZDV pressure, if the 215S polymorphism was present (41). 215S is naturally present in HIV-2.
It is also important to recognize that the limitations of these investigations include low numbers of virus isolates in in vitro studies compared to what occurs in patients and also the limited time frame of the studies. Although in vitro selection cannot detect all possible mutations, this method has often been predictive of in vivo results. Our observations suggest that TAMs may not often occur in HIV-2 exposed to ZDV. However, pre- and post-ZDV exposure studies in patients infected by HIV-2 should be performed.
Further studies are now being carried out in which the polymorphisms at positions 210, 215, and 219 in HIV-2 are being reverted to wild-type codons found in HIV-1. These viral clones will then be used to evaluate the impact of these reversions on the resistance of HIV-2 to ZDV, and selection experiments will be carried out to determine the impact of these reversions on the selection of K65R in HIV-2.
In this study, we have also observed a novel mutational motif, involving S134A, A138M, and V167I, in some of the HIV-2 isolates selected under TFV drug pressure. Studies on this novel motif will be performed by site-directed mutagenesis to determine the potential clinical relevance.
Supplementary Material
Acknowledgments
This work was sponsored by the Canadian Institutes of Health Research (CIHR) and the Canadian Foundation for AIDS Research (CANFAR). Michel Ntemgwa was the recipient of a Canadian Institutes of Health Research (CIHR) doctoral fellowship award.
We thank Jordana R. Schachter for technical assistance. The HIV-2ROD plasmid was kindly provided by Andrew Lever of Cambridge University with the permission of Tom Schulz of Hannover University, Germany.
Footnotes
Published ahead of print on 8 December 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Adje-Toure, C. A., R. Cheingsong, J. G. Garcia-Lerma, S. Eholie, M. Y. Borget, J. M. Bouchez, R. A. Otten, C. Maurice, M. Sassan-Morokro, R. E. Ekpini, M. Nolan, T. Chorba, W. Heneine, and J. N. Nkengasong. 2003. Antiretroviral therapy in HIV-2-infected patients: changes in plasma viral load, CD4+ cell counts, and drug resistance profiles of patients treated in Abidjan, Cote d'Ivoire. AIDS 17(Suppl. 3):S49-S54. [PubMed] [Google Scholar]
- 2.Beatty, C., M. Bradley, D. Brambilla, F. Breakenridge, J. W. Bremer, J. Dragavon, B. Ladd, D. Livnat, C. Michels, C. Mundy, V. Price, T. Ramacciotti, P. Reichelderfer, B. Staes, C. Starkey, and M. Winters. 1997, posting date. DAIDS virology manual for HIV laboratories. Publication NIH-97-3939. U.S. Department of Health and Human Services, Washington, DC. http://www.niaid.nih.gov/daids/vir_manual/full_vir_manual.pdf.
- 3.Boyer, P. L., S. G. Sarafianos, P. K. Clark, E. Arnold, and S. H. Hughes. 2006. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS Pathog. 2:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandin, E., L. Lindborg, K. Gyllensten, C. Brostrom, L. Hagberg, M. Gisslen, B. Tuvesson, A. Blaxhult, and J. Albert. 2003. pol gene sequence variation in Swedish HIV-2 patients failing antiretroviral therapy. AIDS Res. Hum. Retrovir. 19:543-550. [DOI] [PubMed] [Google Scholar]
- 5.Brenner, B. G., M. Oliveira, F. Doualla-Bell, D. D. Moisi, M. Ntemgwa, F. Frankel, M. Essex, and M. A. Wainberg. 2006. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS 20:F9-F13. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, B. G., J. P. Routy, M. Petrella, D. Moisi, M. Oliveira, M. Detorio, B. Spira, V. Essabag, B. Conway, R. Lalonde, R. P. Sekaly, and M. A. Wainberg. 2002. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J. Virol. 76:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvez, V., D. Costagliola, D. Descamps, A. Yvon, G. Collin, A. Cecile, C. Delaugerre, F. Damond, A. G. Marcelin, S. Matheron, A. Simon, M. A. Valantin, C. Katlama, and F. Brun-Vezinet. 2002. Impact of stavudine phenotype and thymidine analogues mutations on viral response to stavudine plus lamivudine in ALTIS 2 ANRS trial. Antivir. Ther. 7:211-218. [PubMed] [Google Scholar]
- 8.Colson, P., M. Henry, N. Tivoli, H. Gallais, J. A. Gastaut, J. Moreau, and C. Tamalet. 2005. Polymorphism and drug-selected mutations in the reverse transcriptase gene of HIV-2 from patients living in southeastern France. J. Med. Virol. 75:381-390. [DOI] [PubMed] [Google Scholar]
- 9.Damond, F., S. Matheron, G. Peytavin, P. Campa, A. Taieb, G. Collin, C. Delaunay, G. Chene, F. Brun-Vezinet, and D. Descamps. 2004. Selection of K65R mutation in HIV-2-infected patients receiving tenofovir-containing regimen. Antivir. Ther. 9:635-636. [PubMed] [Google Scholar]
- 10.De Cock, K. M., G. Adjorlolo, E. Ekpini, T. Sibailly, J. Kouadio, M. Maran, K. Brattegaard, K. M. Vetter, R. Doorly, and H. D. Gayle. 1993. Epidemiology and transmission of HIV-2. Why there is no HIV-2 pandemic. JAMA 270:2083-2086. [DOI] [PubMed] [Google Scholar]
- 11.Descamps, D., F. Damond, S. Matheron, G. Collin, P. Campa, S. Delarue, S. Pueyo, G. Chene, and F. Brun-Vezinet. 2004. High frequency of selection of K65R and Q151M mutations in HIV-2 infected patients receiving nucleoside reverse transcriptase inhibitors containing regimen. J. Med. Virol. 74:197-201. [DOI] [PubMed] [Google Scholar]
- 12.Doualla-Bell, F., A. Avalos, B. Brenner, T. Gaolathe, M. Mine, S. Gaseitsiwe, M. Oliveira, D. Moisi, N. Ndwapi, H. Moffat, M. Essex, and M. A. Wainberg. 2006. High prevalence of the K65R mutation in human immunodeficiency virus type 1 subtype C isolates from infected patients in Botswana treated with didanosine-based regimens. Antimicrob. Agents Chemother. 50:4182-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, et al. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, F., L. Yue, P. M. Sharp, and B. H. Hahn. 1993. Genetic typing of HIV-2 from a Senegalese/German heterosexual transmission. AIDS Res. Hum. Retrovir. 9:703-704. [DOI] [PubMed] [Google Scholar]
- 15.Gao, Q., Z. Gu, J. Hiscott, G. Dionne, and M. A. Wainberg. 1993. Generation of drug-resistant variants of human immunodeficiency virus type 1 by in vitro passage in increasing concentrations of 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:130-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, Q., Z. Gu, M. A. Parniak, J. Cameron, N. Cammack, C. Boucher, and M. A. Wainberg. 1993. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:1390-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, Q., Z. X. Gu, M. A. Parniak, X. G. Li, and M. A. Wainberg. 1992. In vitro selection of variants of human immunodeficiency virus type 1 resistant to 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine. J. Virol. 66:12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant, R. M., D. R. Kuritzkes, V. A. Johnson, J. W. Mellors, J. L. Sullivan, R. Swanstrom, R. T. D'Aquila, M. Van Gorder, M. Holodniy, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Accuracy of the TRUGENE HIV-1 genotyping kit. J. Clin. Microbiol. 41:1586-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guyader, M., M. Emerman, P. Sonigo, F. Clavel, L. Montagnier, and M. Alizon. 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326:662-669. [DOI] [PubMed] [Google Scholar]
- 20.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 21.Hu, Z., F. Giguel, H. Hatano, P. Reid, J. Lu, and D. R. Kuritzkes. 2006. Fitness comparison of thymidine analog resistance pathways in human immunodeficiency virus type 1. J. Virol. 80:7020-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jallow, S., S. Kaye, A. Alabi, A. Aveika, R. Sarge-Njie, S. Sabally, T. Corrah, H. Whittle, G. Vanham, S. Rowland-Jones, W. Janssens, and S. J. McConkey. 2006. Virological and immunological response to Combivir and emergence of drug resistance mutations in a cohort of HIV-2 patients in The Gambia. AIDS 20:1455-1458. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2007. Update of the drug resistance mutations in HIV-1: 2007. Top. HIV Med. 15:119-125. [PubMed] [Google Scholar]
- 24.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 25.Kanki, P. J., M. Peeters, and A. Gueye-Ndiaye. 1997. Virology of HIV-1 and HIV-2: implications for Africa. AIDS 11(Suppl. B):S33-S42. [PubMed] [Google Scholar]
- 26.Keulen, W., N. K. Back, A. van Wijk, C. A. Boucher, and B. Berkhout. 1997. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 71:3346-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 28.Kuritzkes, D. R., R. M. Grant, P. Feorino, M. Griswold, M. Hoover, R. Young, S. Day, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Performance characteristics of the TRUGENE HIV-1 genotyping kit and the Opengene DNA sequencing system. J. Clin. Microbiol. 41:1594-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larder, B. A., G. Darby, and D. D. Richman. 1989. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science 243:1731-1734. [DOI] [PubMed] [Google Scholar]
- 30.Loemba, H., B. Brenner, M. A. Parniak, S. Ma'ayan, B. Spira, D. Moisi, M. Oliveira, M. Detorio, M. Essex, and M. A. Wainberg. 2002. Polymorphisms of cytotoxic T-lymphocyte (CTL) and T-helper epitopes within reverse transcriptase (RT) of HIV-1 subtype C from Ethiopia and Botswana following selection of antiretroviral drug resistance. Antivir. Res. 56:129-142. [DOI] [PubMed] [Google Scholar]
- 31.Loemba, H., B. Brenner, M. A. Parniak, S. Ma'ayan, B. Spira, D. Moisi, M. Oliveira, M. Detorio, and M. A. Wainberg. 2002. Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against nonnucleoside inhibitors of RT. Antimicrob. Agents Chemother. 46:2087-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margot, N. A., J. M. Waters, and M. D. Miller. 2006. In vitro human immunodeficiency virus type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrob. Agents Chemother. 50:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marlink, R., P. Kanki, I. Thior, K. Travers, G. Eisen, T. Siby, I. Traore, C. C. Hsieh, M. C. Dia, E. H. Gueye, et al. 1994. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 265:1587-1590. [DOI] [PubMed] [Google Scholar]
- 34.Ntemgwa, M., B. G. Brenner, M. Oliveira, D. Moisi, and M. A. Wainberg. 2007. Natural polymorphisms in the human immunodeficiency virus type 2 protease can accelerate time to development of resistance to protease inhibitors. Antimicrob. Agents Chemother. 51:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owen, S. M., D. Ellenberger, M. Rayfield, S. Wiktor, P. Michel, M. H. Grieco, F. Gao, B. H. Hahn, and R. B. Lal. 1998. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J. Virol. 72:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer, S., M. Kearney, F. Maldarelli, E. K. Halvas, C. J. Bixby, H. Bazmi, D. Rock, J. Falloon, R. T. Davey, Jr., R. L. Dewar, J. A. Metcalf, S. Hammer, J. W. Mellors, and J. M. Coffin. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parikh, U. M., L. Bacheler, D. Koontz, and J. W. Mellors. 2006. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. J. Virol. 80:4971-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parikh, U. M., D. C. Barnas, H. Faruki, and J. W. Mellors. 2006. Antagonism between the HIV-1 reverse-transcriptase mutation K65R and thymidine-analogue mutations at the genomic level. J. Infect. Dis. 194:651-660. [DOI] [PubMed] [Google Scholar]
- 39.Parikh, U. M., S. Zelina, N. Sluis-Cremer, and J. W. Mellors. 2007. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS 21:1405-1414. [DOI] [PubMed] [Google Scholar]
- 40.Petrella, M., M. Oliveira, D. Moisi, M. Detorio, B. G. Brenner, and M. A. Wainberg. 2004. Differential maintenance of the M184V substitution in the reverse transcriptase of human immunodeficiency virus type 1 by various nucleoside antiretroviral agents in tissue culture. Antimicrob. Agents Chemother. 48:4189-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid, P., H. MacInnes, M. E. Cong, W. Heneine, and J. G. Garcia-Lerma. 2005. Natural resistance of human immunodeficiency virus type 2 to zidovudine. Virology 336:251-264. [DOI] [PubMed] [Google Scholar]
- 42.Ren, J., L. E. Bird, P. P. Chamberlain, G. B. Stewart-Jones, D. I. Stuart, and D. K. Stammers. 2002. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc. Natl. Acad. Sci. USA 99:14410-14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodes, B., A. Holguin, V. Soriano, M. Dourana, K. Mansinho, F. Antunes, and J. Gonzalez-Lahoz. 2000. Emergence of drug resistance mutations in human immunodeficiency virus type 2-infected subjects undergoing antiretroviral therapy. J. Clin. Microbiol. 38:1370-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salomon, H., A. Belmonte, K. Nguyen, Z. Gu, M. Gelfand, and M. A. Wainberg. 1994. Comparison of cord blood and peripheral blood mononuclear cells as targets for viral isolation and drug sensitivity studies involving human immunodeficiency virus type 1. J. Clin. Microbiol. 32:2000-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saracino, A., L. Monno, L. Scudeller, D. C. Cibelli, A. Tartaglia, G. Punzi, C. Torti, S. Lo Caputo, F. Mazzotta, G. Scotto, G. Carosi, and G. Angarano. 2006. Impact of unreported HIV-1 reverse transcriptase mutations on phenotypic resistance to nucleoside and non-nucleoside inhibitors. J. Med. Virol. 78:9-17. [DOI] [PubMed] [Google Scholar]
- 46.Schulz, T. F., D. Whitby, J. G. Hoad, T. Corrah, H. Whittle, and R. A. Weiss. 1990. Biological and molecular variability of human immunodeficiency virus type 2 isolates from The Gambia. J. Virol. 64:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon, F., S. Matheron, C. Tamalet, I. Loussert-Ajaka, S. Bartczak, J. M. Pepin, C. Dhiver, E. Gamba, C. Elbim, J. A. Gastaut, et al. 1993. Cellular and plasma viral load in patients infected with HIV-2. AIDS 7:1411-1417. [DOI] [PubMed] [Google Scholar]
- 48.Smith, R. A., G. S. Gottlieb, D. J. Anderson, C. L. Pyrak, and B. D. Preston. 2008. Human immunodeficiency virus types 1 and 2 exhibit comparable sensitivities to zidovudine and other nucleoside analog inhibitors in vitro. Antimicrob. Agents Chemother. 52:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sosa, N., C. Hill-Zabala, E. Dejesus, G. Herrera, A. Florance, M. Watson, C. Vavro, and M. Shaefer. 2005. Abacavir and lamivudine fixed-dose combination tablet once daily compared with abacavir and lamivudine twice daily in HIV-infected patients over 48 weeks (ESS30008, SEAL). J. Acquir. Immune Defic. Syndr. 40:422-427. [DOI] [PubMed] [Google Scholar]
- 50.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuaillon, E., M. Gueudin, V. Lemee, I. Gueit, P. Roques, G. E. Corrigan, J. C. Plantier, F. Simon, and J. Braun. 2004. Phenotypic susceptibility to nonnucleoside inhibitors of virion-associated reverse transcriptase from different HIV types and groups. J. Acquir. Immune Defic. Syndr. 37:1543-1549. [DOI] [PubMed] [Google Scholar]
- 52.Wainberg, M. A. 2004. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev. Anti-Infect. Ther. 2:147-151. [DOI] [PubMed] [Google Scholar]
- 53.Witvrouw, M., C. Pannecouque, W. M. Switzer, T. M. Folks, E. De Clercq, and W. Heneine. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir. Ther. 9:57-65. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.