Abstract
A global collection of plasmids of the IncHI1 incompatibility group from Salmonella enterica serovar Typhi were analyzed by using a combination of DNA sequencing, DNA sequence analysis, PCR, and microarrays. The IncHI1 resistance plasmids of serovar Typhi display a backbone of conserved gene content and arrangement, within which are embedded preferred acquisition sites for horizontal DNA transfer events. The variable regions appear to be preferred acquisition sites for DNA, most likely through composite transposition, which is presumably driven by the acquisition of resistance genes. Plasmid multilocus sequence typing, a molecular typing method for IncHI1 plasmids, was developed using variation in six conserved loci to trace the spread of these plasmids and to elucidate their evolutionary relationships. The application of this method to a collection of 36 IncHI1 plasmids revealed a chronological clustering of plasmids despite their difference in geographical origins. Our findings suggest that the predominant plasmid types present after 1993 have not evolved directly from the earlier predominant plasmid type but have displaced them. We propose that antibiotic selection acts to maintain resistance genes on the plasmid, but there is also competition between plasmids encoding the same resistance phenotype.
The emergence of multiply drug-resistant (MDR) bacteria presents an increasing global threat to the clinical management of infectious diseases (43). Even though they predate the use of antibiotics for treatment of bacterial infection, plasmids are a major vehicle by which bacteria acquire resistance genes (32). Whole resistance plasmids can move between bacteria through conjugation; however, some genes are also independently mobile via the exchangeable gene cassettes of integrons (11) or as simple or composite transposons (28). Indeed, the serial capture of DNA sequences by plasmids using either legitimate or illegitimate recombination systems has played a major role in the evolution of plasmid-encoded MDR phenotypes across many bacterial species (36). Sometimes, as seen with the genomic island of Salmonella enterica serovar Typhimurium DT104, resistance genes associated with plasmids can integrate into the chromosome (3, 37).
Since the early 1970s antibiotic resistance has become established in S. enterica serovar Typhi, the cause of human typhoid (40). Two years after chloramphenicol was first used for treating typhoid fever in 1948 (42), the first resistant serovar Typhi isolate was described (4). However, it took another 22 years before chloramphenicol resistance became a major clinical problem, with the first major outbreak of resistant typhoid fever in 1972 (21). Thereafter, the global spread of chloramphenicol-resistant serovar Typhi occurred very rapidly, and over the next two decades further antibiotic resistance genes were acquired to generate MDR serovar Typhi (40). Although occasionally IncF, IncP, and IncB/O plasmids are found (20), it is striking that the majority of resistance plasmids from serovar Typhi belong to the HI1 incompatibility group (IncHI1). Thus, IncHI1 plasmids appear to have evolved a stable relationship with the serovar Typhi bacterial host so that even in the absence of obvious antibiotic selection, a proportion of the isolates maintain the plasmid (7).
There are three fully sequenced IncHI1 plasmids publicly available: R27 (31), pHCM1 (24), and pAKU1 (13). Holt et al. (13) have reported a detailed comparison of these plasmids showing a 164.4-kb shared IncHI1-associated backbone and multiple antibiotic resistance genes carried within Tn10 and a Tn9/Tn21 composite transposon.
A study of IncHI1 plasmids isolated from outbreaks and sporadic typhoid cases in Vietnam between 1993 and 1996 revealed seven different HindIII-digested restriction fragment length polymorphism (RFLP) patterns (patterns 1 to 7), with pattern 1 being the most common (38). The trend was reversed after 1996, at which point pattern 7 became predominant, while plasmids of other patterns were no longer detected. Based on this study, it was hypothesized that a milestone of IncHI1 plasmid evolution had been captured, during which a newly acquired increase in fitness drove one plasmid type to outcompete all others circulating in the serovar Typhi population in Vietnam.
We extend our analyses here to a global collection of IncH1 plasmids isolated from serovar Typhi over the past several decades using a combination of DNA microarrays, PCR, and sequence analysis. We have devised a multilocus sequence typing (MLST) scheme (17) for the IncHI1 plasmids, which is called plasmid MLST (PMLST).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial isolates and plasmids used in the present study are listed in Table 1 and Table S3 in the supplemental material.
TABLE 1.
Plasmids used for microarray analysis in this study
| Hosta | Plasmid RFLPb | Plasmid | Resistancec | Time (yr) of isolation, location |
|---|---|---|---|---|
| Control* | Not done | R27 | T | 1961, United Kingdom |
| 40R181* | Not done | 40R181 | CSSuT | 1972, Mexico |
| 40R344* | Not done | 40R344 | CSSuT | 1972, India |
| 42R917* | Not done | 42R917 | CSSuT | 1970s, Vietnam |
| 44R311* | Not done | 44R311 | CSSuT | 1970s, Thailand |
| 44R315* | Not done | 44R315 | ACSSuT | 1970s, Thailand |
| PB36† | Not done | pPB36 | CSxtT | 1981, Peru |
| Ty3 | RFLP2 | pSTY2 | ACSSxtT | 1992, Vietnam |
| CT18 | RFLP1 | pHCM1 | ACSSxtT | 1993, Vietnam |
| Ty10 | RFLP1 | pSTY1 | ACSSxtT | 1993, Vietnam |
| Ty7 | RFLP1 | pTy7 | ACSSxtT | 1993, Vietnam |
| Ty5 | RFLP2 | pTy5 | ACSSxtT | 1993, Vietnam |
| Ty49 | RFLP3 | pSTY3 | ACSSxtT | 1993, Vietnam |
| Ty24 | RFLP4 | pSTY4 | ACSSxtT | 1993, Vietnam |
| Ty39 | RFLP5 | pSTY5 | ACSSxtT | 1993, Vietnam |
| Ty55 | RFLP6 | pSTY6 | ACSSxtT | 1993, Vietnam |
| CT53 | RFLP7 | pCB | ACSSxtT | 1994, Vietnam |
| 9541 | RFLP7 | pSTY7 | ACSSxtT | 1996, Vietnam |
| 14200 | RFLP7 | pD6 | ACSSxtT | 1997, Vietnam |
| 61-57-85† | Not done | p61-57-85 | ACSSxtT | 2000, Pakistan |
*, Supplied as E. coli K-12 transconjugants by Henry Smith, HPA, Colindale, United Kingdom. †, DNA received only.
RFLP patterns were obtained from previous study (38).
A, ampicillin; C, chloramphenicol; S, streptomycin; Sxt, trimethoprim-sulfamethoxazole; Su, sulfathiazole; T, tetracycline.
Genomic DNA extraction.
Whole genomic DNA was extracted from 30 ml overnight bacterial cultures, grown in Luria-Bertani broth (Difco, Detroit, MI), by the standard CTAB (cetyltrimethylammonium bromide) method of DNA extraction (1). RNA contamination was removed with 10 μg of RNase (Sigma, Poole, United Kingdom)/ml for 10 min at 37°C, and DNA was quantified as A260/A280.
Microarray experiments.
Pairs of specific oligonucleotide primers were designed for each predicted coding sequence (CDS) based on the EMBL gene sequence files of pHCM1 (AL513383) and pHCM2 (AL513384) using Microarray Design Software (K. Vass, Beatson Institute for Cancer Research, Glasgow, United Kingdom) and the primer design program Primer3 (27). Whole genomic DNA extracted from serovar Typhi CT18 was used as a template in the PCRs. PCR amplification was carried out using standard conditions in a RoboAmp4200 (MWG-Biotech, Germany). The microarray was printed by using MicroGrid II Biorobotics (MWG-Biotech) onto poly-l-lysine-coated slides, with four copies of each CDS per array. Specific PCR products corresponding to six genes located on the serovar Typhi chromosome and Cy3-dCTP- and Cy5-dCTP-labeled nucleotides were printed as controls.
Whole genomic serovar Typhi DNA was used as a template for direct incorporation of fluorescent labeled nucleotides. In each experiment the control DNA (serovar Typhi CT18 whole genomic DNA) was labeled with Cy5-dCTP (Amersham, United Kingdom) and the test DNA with Cy3-dCTP (Amersham). More than 5 μg of whole genomic DNA of control and test isolates and 3 μg of random primers (Roche, Welwyn Garden City, United Kingdom) in 41.5 μl of water were heated at 95°C for 5 min, snap cooled on ice, and centrifuged briefly. Each labeling reaction mix contained 100 μM dA/T/GTP, 40 μM dCTP, 300 μM Cy3-dCTP, or Cy5-dCTP, and 5 U of Klenow reagent in a final volume of 50 μl. Reaction mixes were incubated at 37°C in the dark for 90 min. Microarray slides were incubated in a prehybridization solution (3.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS], and 10 mg of bovine serum albumin/ml in a final volume of 50 ml of H2O) at 65°C for 20 min. After prehybridization slides were rinsed thoroughly in H2O for 1 min and propan-2-ol for 1 min and then dried.
Labeled DNA was purified and eluted in a final volume of 21 μl, with a final concentration of 4× SSC and 0.3% SDS. The hybridization mixture was denatured at 95°C for 2 min, allowed to cool, applied to the microarray, and covered with a coverslip. Hybridization was carried out at 65°C for 16 to 20 h in a waterproof hybridization chamber. After hybridization, slides were washed at 65°C in 1× SSC and 0.06% SDS for 2 min, followed by two washes in 0.06× SSC each for 2 min. To ensure reproducibility of data, three separate hybridizations were carried out for each plasmid isolate.
Data input and analysis.
Scanning of microarrays was carried out by using an Affymetrix 428 dual laser array scanner. Scanned data were analyzed and quantified in ImaGene v4.2 (Biodiscovery, Inc.). The quantified data file was extracted and normalized using GeneSpring version 5.1 (Silicon Genetics). Genes were classed as present or absent/highly divergent as follows. Briefly, a list of “core” genes was derived. To allow for divergence in primary DNA sequence or poor hybridization signal, this was defined as genes present in at least 19 of 20 serovar Typhi plasmids hybridized to the microarray. Insertion sequences, transposons, and antibiotic resistance genes were excluded from this analysis since the plasmids were selected because of the presence of resistance and so these genes were necessarily present even though they are not part of the plasmid's survival functions. For each isolate the median of the normalized ratio (baseline) and standard deviation were calculated for the 141 “core” genes (26). Three standard deviations (P ≤ 0.01) above and below the baseline for each isolate were used to set the threshold for those genes defined as “present.” Genes with normalized ratios higher than three standard deviations above the baseline were considered duplicated, whereas ratios that were less than three standard deviations below the baseline were considered absent or highly divergent.
PCR for genetic elements.
Oligonucleotide primers were taken from the literature or designed (Primer3 v.0.4.0 [http://frodo.wi.mit.edu/]) to specifically amplify the elements listed in Table 2. For confirmation of microarray data, long range PCRs were designed (see Table S2 in the supplemental material). Amplification of serovar Typhi whole genomic DNA was carried out using standard PCR conditions, the Tm for each primer is given in Table 2 or in Table S2 in the supplemental material. Specific amplicons were identified by separation on 1.0% agarose gel, followed by staining with ethidium bromide and visualization under UV light.
TABLE 2.
Oligonucleotide primers
| Primer | Sequence (5′-3′) | Tm (°C) | Gene (size [bp]) | Source or reference |
|---|---|---|---|---|
| 5′CS | GGCATCCAAGCAGCAAG | 55.2 | Variable | 25 |
| 3′CS | AAGCAGACTTGACCTGA | 51.4 | ||
| IntI1-F | GCCTTGCTGTTCTTCTACGG | 59.4 | int1 (550) | 16 |
| IntI1-R | GATGCCTGCTTGTTCTACGG | 59.4 | ||
| IntI2-F | CACGGATATGCGACAAAAAGGT | 58.4 | int2 | 18 |
| IntI2-R | GTAGCAAACGAGTGACGAAATG | 58.4 | ||
| IntI3-F | GCCTCCGGCAGCGACTTTCAG | 65.7 | int3 | 18 |
| IntI3-R | ACGGATCTGCCAAACCTGACT | 59.8 | ||
| SulI1-F | CTTCGATGAGAGCCGGCGGC | 65.5 | sul1 (430) | 29 |
| SulI1-R | GCAAGGCGGAAACCCGCGCC | 67.6 | ||
| qacEΔ1-F | ATCGCAATAGTTGGCGAAGT | 55.3 | qac (220) | 30 |
| qacEΔ1-R | CAAGCTTTTGCCCATGAAGC | 57.3 | ||
| P43F | CTGGATTCCCCAGAAAAACA | 59.9 | HCM1.043 (570) | This study |
| P43R | TGAATCACTGCCCGTATCAA | 60.0 | ||
| P54F | CTCCGCCTAGGTGTGTTTGT | 60.0 | HCM1.054 (733) | This study |
| P54R | CGTAATCGCCGTTTTCTTTG | 56.0 | ||
| P64F | ATGTGACCAACACGGAGACA | 58.0 | HCM1.064 (728) | This study |
| P64R | CATCGCCTTCCTGATGATCT | 58.0 | ||
| P94F | GGAACTAGCGGGATCATGTG | 60.4 | HCM1.094 (562) | This study |
| P94R | GTAAGACCCCGCCACTGTTA | 59.9 | ||
| P99F | AGAAAAACGGGGACCTCAGT | 59.9 | HCM1.099 (590) | This study |
| P99R | GGATTGCTCACGGGAGATTA | 60.0 | ||
| P107F | CAGCATTGAGCATGAAAGGA | 59.9 | HCM1.107 (583) | This study |
| P107R | TTCGCAGCCTCTTTCAATTT | 59.9 | ||
| P116F | TCTTCACCACGCCATATTCA | 56.0 | HCM1.116 (772) | This study |
| P116R | GTATCGTCATGCGGGTCTTT | 58.0 | ||
| P177F | CCTGATGGAGCCTTTGACAT | 60.1 | HCM1.177 (464) | This study |
| P177R | GAATCAGGGTCGATCGAAAA | 60.0 | ||
| P178acF | ACTGAGCTGTTCGCGATTTT | 60.0 | HCM1.178ac (578) | This study |
| P178acR | GCGGGGTGGTTAATGTCTTT | 61.1 | ||
| P259F | GAACGTAATTCCAGCGGAGA | 60.2 | HCM1.259 (599) | This study |
| P259R | CGCATTGTTTATGGCTACGA | 59.7 | ||
| P277F | TGTGCTTTACTGCCTGATGG | 59.8 | HCM1.277 (588) | This study |
| P277R | CGCATGGTTGTTTTGTATCG | 59.9 | ||
| P280F | GGTGTTTGGCGAGTTTAACG | 58.0 | HCM1.280 (576) | This study |
| P280R | CATCAGGTTGTTAGCCACGA | 58.0 | ||
| P286F | GCCCTTGTTCTGCTTTTCAG | 58.0 | HCM1.286 (713) | This study |
| P286R | CTCTCCATCAAACGGATGGT | 58.0 | ||
| HCM1.DF | CGATTTGTGAAGTTGGGTCA | 56.0 | Region D | This study |
| HCM1.DR | AGAAAGGATTCCCGGAAAAA | 54.0 | ||
| HCM1.EF | CCTTTACCATCTCGCCTCAC | 60.0 | Region E | This study |
| HCM1.ER | ACCTTATCGCACCACCAGAC | 60.0 | ||
| Insert1056L | TAGGGTTTGTGCGGCTTC | 56.0 | Ins1056 | This study |
| Insert1056R | CCTTCTTGTCGCCTTTGC | 56.0 | ||
| pSPA-07 | GAAAGGAATCATCCACCTTCA | 57.0 | Region C | This study |
| pSPA-08 | AACTGTCGCTACGCCTGACT | 60.0 | ||
| pSPA-09 | ATCCAGCGTGCAAAGATTTC | 56.0 | Region C | This study |
| pSPA-10 | TGGGGGAGAACACCACTTTA | 58.0 |
PMLST.
Thirteen loci were initially chosen as candidates for the PMLST scheme (Table 2 and Table 3). These were amplified, sequenced, and aligned to assess the variation in 14 plasmids. Six conserved genes were selected for the final PMLST scheme to apply to a set of 36 plasmids. In order to differentiate it from MLST (14, 17), we have termed the method PMLST. Analysis of PMLST data was carried out using eBURST (http://eburst.mlst.net/).
TABLE 3.
Candidate PMLST genes and their annotations
| Coding sequence | Size (bp) | PMLST sequence size (bp) | Gene function |
|---|---|---|---|
| HCM1.043 | 777 | 356 | Hypothetical protein |
| HCM1.054 | 882 | repA, RepHI1B replication initiation protein, an IncHI1 specific replication protein | |
| HCM1.064 | 876 | 527 | repA2, RepHI1A replication initiation protein, an IncHI1 specific replication protein |
| HCM1.094 | 453 | htdA, IncHI1 transfer repressor | |
| HCM1.099 | 1,509 | 417 | trhW, one of nine trh genes (trhALEKBVCPW) essential for H-pilus production |
| HCM1.107 | 1,857 | trhI, S. enterica serovar Typhi putative ATP-dependent helicase | |
| HCM1.116 | 2,013 | 491 | Hypothetical protein |
| HCM1.177 | 711 | Hypothetical protein | |
| HCM1.178ac | 405 | 403 | Probable DNA-binding protein, contains Pfam match to H-NS histone family |
| HCM1.259 | 516 | 393 | Hypothetical protein, contains Pfam match to transglycosylase SLT domain |
| HCM1.277 | 714 | Possible periplasmic protein | |
| HCM1.280c | 546 | Hypothetical protein | |
| HCM1.286 | 1,206 | Possible DNA-binding protein |
eBURST (9) was used to analyze allelic profiles, determined from the PMLST sequences by using the standard MLST approach (17). As an alternative approach to analysis, the individual locus sequences were concatenated to give a single representative sequence for each plasmid. Variant bases were identified by aligning these sequences and recoded into discrete characters for analysis with the MIX algorithm from the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html [March 2008]).
PMLST allele accession numbers.
We have deposited the DNA sequences of PMLST alleles in the NCBI database under the following accession numbers: HCM1.043 allele 1, FJ183728; HCM1.043 allele 3, FJ183729; HCM1.064 allele 1, FJ183730; HCM1.064 allele 2, FJ183731; HCM1.099 allele 1, FJ183732; HCM1.099 allele 2, FJ183733; HCM1.099 allele 3, FJ183734; HCM1.116 allele 1, FJ183735; HCM1.116 allele 2, FJ183736; HCM1.178ac allele 01, FJ183737; HCM1.178ac allele 02, FJ183738; HCM1.259 allele 01, FJ183739; HCM1.259 allele 02, FJ183740; and HCM1.259 allele 03, FJ183741.
RESULTS
Core genes among IncHI1 plasmids.
Stimulated by the observations of shared IncHI1 backbone by Wain et al. (38-40) and Holt et al. (13), we exploited a custom DNA microarray specifically designed to analyze plasmid variation in S. enterica to determine the shared genome content of IncHI1 plasmids from 20 serovar Typhi isolates selected for their geographic diversity, date of isolation, and patterns of resistance (Table 1). A total of 134 CDSs were detected by microarray in all 20 isolates analyzed (Fig. 1). The regions defined by this analysis were further investigated by PCR, which identified 163 genes present in all isolates (see Table S1 in the supplemental material). Tetracycline resistance encoded by Tn10 was present in all plasmids analyzed but was not always at the same location and so was not considered to be part of the core gene set.
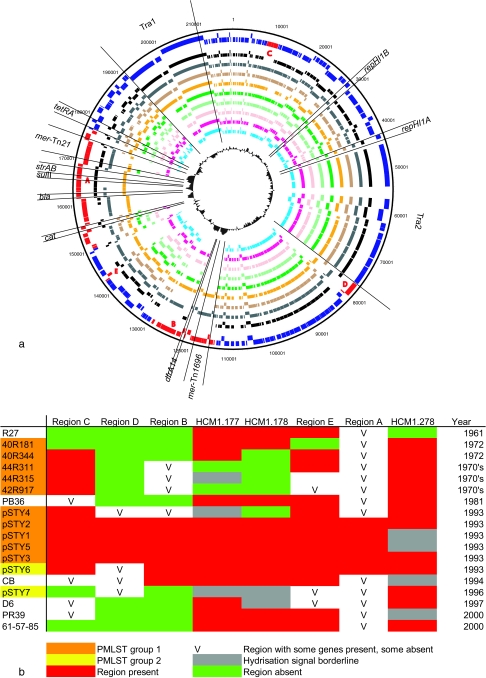
FIG. 1.
Variation in IncHI1 plasmids from serovar Typhi determined by microarray. (a) CDSs are defined in the sequence of pHCM1 in the outer ring (NC_003384). The inner concentric rings each represent the CDSs predicted as present in each different plasmid by microarray. The order, from the inside is as follows: blue, R27 (United Kingdom, 1961); magenta, 40R181 (Mexico, 1972); pink, 40R344 (India, 1972); light green, 40R311 (Thailand, 1970s); dark green, 42R917 (Vietnam, 1970s); yellow, pSTY1 (Vietnam, 1993); sand, pSTY4 (Vietnam, 1993); gray, pSTY6 (Vietnam, 1993); black, pSTY7 (Vietnam, 1996); and red and blue, pHCM1 (Vietnam, 1993) on the outside. The CDSs of pHCM1 are marked blue when present and red when absent from R27 sequence (NC_002305). (b) Details of regions A to E shown in panel a. These are the main regions of variability between plasmids. PMLST clusters are shown in Fig. 3.
Through analysis of 20 plasmids by microarray and PCR, we identified a core set of 163 genes common to IncHI1 plasmids. This is >75% of the smallest IncHI1 plasmid thus far described, R27 (31). The annotated functions of the 163 core genes included plasmid DNA replication and modification (HCM1.59 and HCM1.187c) and many genes associated with DNA transfer and mating pair formation, as well as 96 hypothetical coding sequences of entirely unknown function (see Table S1 in the supplemental material). There were also three potential regulatory genes considered core genes. One of these, a histone-like protein (HCM1.178ac), is of particular interest as a potential mechanism for the reduction of biological cost during plasmid acquisition in Salmonella (6). The CDSs HCM1.25, HCM1.22, and the pseudogene HCM1.248 had LysR signatures, suggesting their roles as autoregulatory transcriptional regulators. Other core genes included HCM1.94 (htdA) and HCM1.95 (htdF), encoding transfer repressor proteins. The htdA gene is believed to regulate conjugal transfer of IncHI1 plasmids (41). There was also a putative stable inheritance protein (HCM1.290c) that probably encodes a Hok/Sok postsegregation killing mechanism (34, 35) and may go some way to explaining the stability of IncHI1 plasmids within serovar Typhi. There were also four core CDSs involved in metabolic processes: cobalamin biosynthesis (HCM1.111), a magnesium and cobalt transporter (HCM1.07c), a probable glutamate permease (HCM1.247), and a probable thiol-disulfide interchange protein (HCM1.258c). A probable DNA repair modulator mucA (HCM1.252) and a DNA modification methylase (HCM1.187) were also classified as core genes.
Long-range overlapping PCR confirms microarray data.
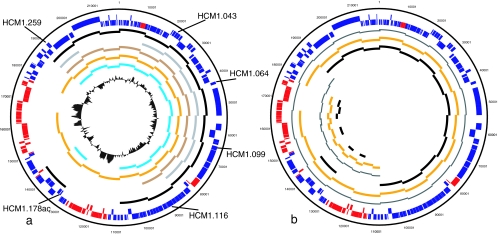
As a complement to the microarray data above, which demonstrate the presence but not the position of core genes in each plasmid, a set of oligonucleotide primers was designed to amplify overlapping fragments around the whole pHCM1 plasmid. Performing the same PCRs with other IncHI1 plasmids gives an indication of those regions similar to pHCM1, where PCR products are obtained, while suggesting which regions show variation, when PCR products are absent or a different size compared to that obtained for pHCM1.
Initially, 20 oligonucleotide pairs were designed to amplify overlapping 10-kb fragments covering the whole of pHCM1 (Fig. 2a). Thereafter, for regions in which variability was indicated, oligonucleotides for overlapping 5-kb fragments were obtained (Fig. 2b). Reactions for oligonucleotide pairs from 2 to 8 and 18 were positive in all plasmids that have been analyzed by 10-kb PCR. These PCRs cover CDSs encoding plasmid maintenance and transfer functions in pHCM1. Three adjacent reactions (1, 19, and 20 in Fig. 2a) were negative in pSTY6; further analysis confirmed a deletion of 10 to 15 kb in this region (data not shown). The regions that did not amplify in the other IncHI1 plasmids are those encoding antibiotic resistance-determining regions in pHCM1. This suggests that the IncHI1 plasmids possess an almost invariable core of genes that code for plasmid maintenance and transfer, the order of which is generally maintained between the plasmids, and variable regions incorporating the antibiotic resistance determinants.
FIG. 2.
PCR confirmation of microarray results. (a) Long-range PCR of ∼10-kb amplicons around IncHI1 plasmids of serovar Typhi. The products for R27 (inner gray ring) are predicted from the DNA sequence, and the primer binding sites are at either end of the gray bars on the inner ring. The order from the inside is R27, pSTY1, pSTY4, pSTY6, and pST7, and pHCM1 is the outside ring. The outmost ring shows positions of six PMLST loci on pHCM1. (b) PCR analysis comparing the two most successful IncHI1 plasmids of serovar Typhi using 5-kb amplicons within regions that do not give products with 10Kb PCR primers. The thin gray lines are predicted products from the pHCM1 sequence. The order from the inside is 5-kb fragments of pSTY7, pSTY1, and pHCM1 and then 10-kb fragments of pSTY7, pSTY1, and pHCM1.
Accessory genes.
Comparison of R27 and pHCM1 previously identified two regions of significant DNA variation between the two plasmids. These regions encoded antibiotic and heavy metal resistance genes, as well as potentially mobile elements, which are likely sites of rapid evolution in these plasmids, driven by antibiotic selection.
Plasmid pHCM1 contained two different DNA regions encoding mercury resistance, annotated as HCM1.151c to HCM1.159 (similar to mer operon from Tn1696) and HCM1.230 to HCM1.235 (similar to mer operon from Tn21), and neither of these regions was present on R27. Of the five plasmids analyzed from the 1970s, only the Mexican plasmid 40R181 harbored similar mercury resistance determinants. Of the later plasmids, p61-57-85 (isolated in Pakistan in 2000) and pSTY7 (isolated in Vietnam in 1996) gave microarray signatures suggesting the presence of a single mercury resistance determinant, whereas all plasmids from Vietnam isolated from 1993 to 1994 had both operons. Mercury resistance is therefore a phenotype acquired on more than one occasion by the IncHI1 plasmids. The efflux pump encoded by these operons is very specific for mercury and may play a role in protection against mercury in the human digestive tract, from dental filings (33), or in the environment.
Antibiotic resistance is a major phenotype conferred by IncHI1 plasmids in serovar Typhi. The resistance genes identified by microarray analysis consistently match the observed resistance phenotype. It is clear that these plasmids have acquired resistance genes sequentially into the variable regions, probably by the insertion of transposons into transposons (24). In some cases a resistance phenotype is always encoded by the same gene; cat (HCM1.209), tetRA (HCM1.240 and HCM1.241c), blaTEM-1 (HCM1.216), and strAB (HCM1.223 and HCM1.224) are conserved across all plasmids conferring resistance to chloramphenicol, tetracycline, ampicillin, and streptomycin, respectively. The genetic context of these genes in different plasmids, however, is not the same. From genome sequence data we know that tetracycline resistance in R27 is encoded within a complete Tn10 but that the IS10-right, the active transposase from Tn10, and part of the tetracycline resistance operon (tetCD), are not present in pHCM1. The array data are consistent with this and also suggest that plasmids 40R181 (Mexico, 1972) and 44R315 and 44R311 (Thailand, 1970s) possess only the IS10-left transposase (HCM1.194), while plasmids 40R344 (India, 1972) and 42R917 (Vietnam, 1970s) are absent or divergent for the IS10-left transposase (HCM1.194c and HCM1.249c). Sequence data also showed that Tn10 was inserted in different locations in R27 and pHCM1. These sets of evidence suggest that Tn10 has been independently acquired by IncHI1 plasmids on at least three occasions.
Different alleles conferring trimethoprim resistance were present within the IncHI1 plasmids. In pHCM1 the dihydrofolate reductase responsible for trimethoprim resistance is a dfrA14 gene cassette within a truncated version of a class one integron. Truncation has occurred at the 59-base element within a Tn21-like element (8). However, other plasmids isolated in Vietnam from 1992 to 1994 all had an intact class one integron containing the dfrA14 trimethoprim resistance gene cassette (1-kb fragment; Table 4). Later plasmids from Vietnam (pSTY7, pCB, and pD6) had a smaller gene cassette containing dfrA7 gene (0.75-kb fragment; Table 4). It is clear that class one integrons are involved in the acquisition of trimethoprim resistance by capturing different gene cassettes.
TABLE 4.
Presence or absence of common integrase genes as determined by PCR and product size using primers 5′CS and 3′CS to amplify gene cassettes from class 1 integrons
| Plasmid | Date (yr) | Presence (+) or absence (−) of:
|
Size (kb) of cassette | ||||
|---|---|---|---|---|---|---|---|
| intI1 | intI2 | intI3 | qacEΔ1 | sul1 | |||
| pHCM1 | 1994 | + | - | - | - | - | No product |
| pSTY1 | 1993 | + | - | - | + | + | 1 |
| pSTY2 | 1992 | + | - | - | + | + | 1 |
| pSTY3 | 1994 | + | - | - | + | + | 1 |
| pSTY4 | 1994 | + | - | - | + | + | 1 |
| pSTY5 | 1993 | + | - | - | + | + | 1 |
| pSTY6 | 1993 | + | - | - | + | + | 1 |
| pSTY7 | 1996 | + | - | - | + | + | 0.75 |
| pCB | 1994 | + | - | - | + | + | 0.75 |
| pD6 | 1996 | + | - | - | + | + | 0.75 |
It is important to note that plasmids isolated at similar times in similar geographic location, such as 42R311 and 44R315 from Thailand during the 1970s, encode very similar accessory genes (Fig. 1b). Plasmids isolated in Vietnam in between 1993 and 1994 (pSTY1, pSTY2, pSTY3, and pSTY5) encoded the same accessory genes by microarray, even though they displayed differences in restriction profile (38). This suggests that the variation revealed by restriction enzyme analysis is caused not by gene content differences but by rearrangement, single base pair differences in restriction sites, or the acquisition of the same genes in different regions of the plasmid. An overview of the variable regions from selected plasmids containing accessory genes is summarized in Fig. 1b. The two major regions, A and B, present in pHCM1 (Vietnam, 1993) but absent from R27 (United Kingdom, 1961) (38) were also present, in their entirety, in other plasmids isolated in Vietnam from 1993 to 1994. Only one plasmid from the same region and time, pSTY4, showed variation in gene content. However, the more recent plasmid from Vietnam, pSTY7 (from 1996), showed extensive variation in accessory genes compared to the earlier Vietnamese plasmids. To confirm the microarray results, the presence of two variable genes, HCM1.177 and HCM1.178, was investigated by PCR. All results between PCR and microarray were compatible (Fig. 1b).
PMLST, a typing scheme with phylogenetic implications.
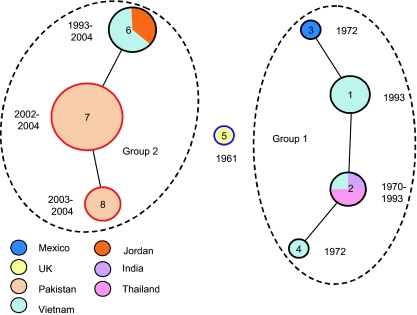
The presence of conserved regions among these plasmids provides material for a molecular typing scheme. Here we apply the principle of MLST (17) to this group of plasmids (PMLST). Thirteen loci not associated with mobile elements were sequenced and analyzed for fourteen of the plasmids to evaluate their usefulness in the PMLST scheme (Table 3). Four loci (HCM1.054, HCM1.094, HCM1.107, and HCM1.277) were excluded from the PMLST as they showed no variation. Three more loci (HCM1.177, HCM1.280c, and HCM1.286) showed inconsistent PCR amplification among the plasmids analyzed and were therefore excluded. The final PMLST set includes 6 loci: HCM1.043, HCM1.064, HCM1.099, HCM1.116, HCM1.178ac, and HCM1.259. Upon sequencing in 36 plasmids, two to three alleles (variants) of each locus were detected, the combinations of which define eight unique plasmid sequence types (STs) (Fig. 3 and see Table S3 in the supplemental material). Figure 3 shows the relatedness of 36 IncHI1 plasmids based on their STs, determined by using eBURST. The eBURST program grouped STs sharing five out of six identical loci, resulting in two groups and a singleton ST. Group 1 contains ST1, ST2, ST3, and ST4, while group 2 includes ST6, ST7, and ST8. Plasmid R27 was assigned ST5 and stood as a singlet, sharing no more than four identical loci with any ST in either group. ST1 includes pHCM1 and four other plasmids, all isolated from sporadic typhoid cases in Vietnam in 1993. Plasmids from ST2 were isolated earlier in the 1970s from Thailand (40R311 and 40R315) and India (40R344) except for pSTY4, which was isolated in Vietnam in 1993. ST3 and ST4 have only one member each, isolated in the 1970s from Mexico (40R181) and Vietnam (42R917), respectively. In group 2, ST6 contains plasmids from Vietnam and Jordan, all isolated after 1993. ST7 consists of 13 plasmids isolated from S. enterica serovar Paratyphi A from Karachi, Pakistan (2002 to 2004). The serovar Paratyphi A plasmid AKU1, which has recently been sequenced and analyzed (13), also belongs to this ST. ST8 contains four other S. Paratyphi A plasmids from Karachi (2003 to 2004) and differs from ST7 by the deletion of locus HCM1.116.
FIG. 3.
Modified eBURST diagram for PMLST. The numbers inside the circles represent the STs. Lines connecting circles represent single-locus variants. The dotted circles represent groups of related plasmids. Plasmids in group 1 are all from before 1993, whereas plasmids from group 2 are from 1993 to 2004. The singlet ST is the first described IncHI1 plasmid R27.
Although these plasmids were grouped purely by sequence data, it is worth noting that group 1 contains plasmids isolated from 1993 backward, while group 2 has plasmids from 1993 onward. This chronological division is interesting since it suggests a competition between two groups that subsequently led to a replacement of group 1 by group 2, presumably due to some selective advantage acquired by the ancestor of the group 2 plasmids.
Differences within the conserved regions.
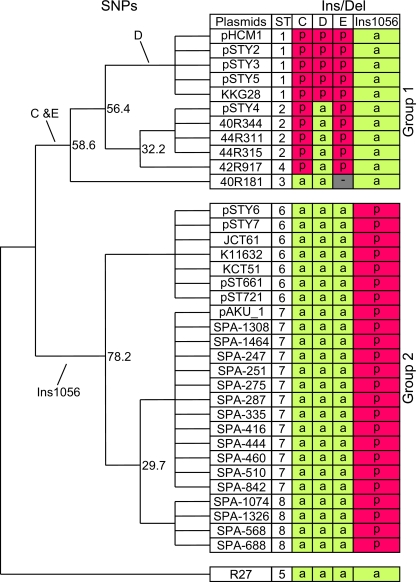
Of the whole plasmid set analyzed, there are three fully sequenced plasmids, each of which represents one group in our PMLST map. The sequence comparison among these three plasmids (pHCM1, R27, and pAKU1) has been described elsewhere (13). We highlight here some features within the core region that we have defined by microarray and long-range PCR (Fig. 1 and 2) and further investigated on our full set of plasmids. Sequence comparison between pHCM1 and R27 defined five variable regions, two of them (regions A and B) encode antibiotic resistance determinants and mobile elements, while the other three (regions C, D, and E) were situated among the conserved regions defined by long-range PCR and do not show any evidence of mobile elements. The genes in regions C, D, and E were present in pHCM1 and absent in both R27 and pAKU1. The sequence contexts surrounding these three regions are similar in R27 and pAKU1. Another feature that lies within the conserved region is a composite transposon containing two IS1 flanking two CDSs (SPAP0105 and SPAP0106). This transposon (hereby called Ins1056) was inserted in the middle of the transfer region of pAKU1 (right after trhC). PCR assays were designed to screen for the presence or absence of these regions in the whole set of plasmids. To avoid false negatives, these assays were designed such that we expect a large PCR product when the region is present and a smaller PCR product when the region is absent.
Figure 4 shows the presence or absence of regions C, D, E, and Ins1056 in each plasmid, along with the phylogenetic tree of plasmids of all STs. Ins1056 was specific to plasmids of group 2, which suggests the singleton plasmid R27 may be more closely related to group 1 than to group 2. Regions C, D, and E were present in all five plasmids of ST1 (group 1). The remaining plasmids of group 1 were negative for regions C and E (except for 40R181). In group 2, regions C, D, and E were generally absent. Region D failed to amplify in the four plasmids of ST8 and two from ST7, suggesting further sequence changes had occurred that affected the binding sites of PCR primers.
FIG. 4.
IncHI1 plasmids phylogenetic tree and insertion/deletion (Ins/Del) events. The phylogenetic tree was built based on the single nucleotide polymorphisms identified within PMLST sequences using parsimony methods for discrete character data (PHYLIP software package), with bootstrap values shown from 1,000 resamplings. The insertion/deletion events were determined by PCR, with “a” for absent, “p” for present, and “-” for negative PCR.
The acquisition or loss of these regions can therefore be inferred within the plasmid lineages represented by the phylogenetic tree. Ins1056 was most likely acquired by a common ancestor of group 2 plasmids, while regions C and E were more likely lost within group 1, after the divergence of ST1. Region D came into a plasmid from group 1 which later on became the predecessor of ST1 plasmids.
It has been suggested previously that R27 and pAKU1 were more closely related to each other than either of them to pHCM1 (13). This is supported by our data showing the absence of regions C, D, and E in 24 plasmids from group 2 (including pAKU1), as well as their absence in R27. Combining the eBURST grouping with the phylogenetic tree and PCR results, we conclude that plasmids of group 2 (post-1993) did not evolve directly from group 1 (pre-1993) but belong to two different lineages.
Replication initiation.
The complete sequence of plasmid pHCM1 revealed five genes that are predicted to encode proteins involved in the initiation of plasmid replication. Two, RepHI1B and RepHI1A, were classified as “core genes” in this analysis since they were present in all 20 serovar Typhi plasmids. However, RepE, RepA, and RepC were variable between isolates and so were classified as accessory genes (Table 5). These replication (Rep) proteins relate to incompatibility, and so this is a possible way in which different plasmid cores (10), or possibly different accessory gene linkage groups, could compete for replication space in the bacterial host. Expression of different Rep proteins, as seen with Neisseria gonorrhoeae (22), may explain the occasional reports of plasmids of different incompatibility groups in serovar Typhi (20). Three possible DNA-binding proteins (HCM1.58c, HCM1.175, and HCM1.286c) clustered within the core genes, as did the plasmid partition proteins parA (HCM1.86) and a putative parB encoding CDS (HCM1.87), with a series of 54-bp repeats upstream that could act as binding sites for the partition proteins.
TABLE 5.
Variation in pHCM1 CDSs associated with plasmid replication
| Plasmid | Location, date (yr) | Presence (+) or absence (−)a of:
|
||||
|---|---|---|---|---|---|---|
| RepA (HI1B), HCM1.54 | RepA2 (HI1A), HCM1.64 | RepE, F plasmid HCM1.137 | RepA, IncQ HCM1.220 | RepC, IncQ HCM1.221 | ||
| R27 | United Kingdom, 1961 | + | + | + | - | - |
| 40R181 | Mexico, 1972 | + | + | + | - | - |
| 40R344 | India, 1972 | + | + | + | - | - |
| 42R917 | Vietnam, 1970s | + | + | + | - | - |
| 44R311 | Thailand, 1970s | + | + | + | - | - |
| 44R315 | Thailand, 1970s | + | + | + | - | - |
| pPB36 | Peru, 1981 | + | + | + | - | - |
| pSTY2 | Vietnam, 1992 | + | + | + | + | + |
| pHCM1 | Vietnam, 1993 | + | + | + | + | + |
| pSTY1 | Vietnam, 1993 | + | + | + | + | + |
| pTy7 | Vietnam, 1993 | + | + | + | + | + |
| pTy5 | Vietnam, 1993 | + | + | + | + | + |
| pSTY3 | Vietnam, 1993 | + | + | + | + | + |
| pSTY4 | Vietnam, 1993 | + | + | + | - | - |
| pSTY5 | Vietnam, 1993 | + | + | + | + | + |
| pSTY6 | Vietnam, 1993 | + | + | + | - | - |
| pSTY7 | Vietnam, 1996 | + | + | - | + | + |
| p61-57-85 | Pakistan, 2000 | + | + | - | + | + |
+, A normalized ratio indicates the presence of this CDS; -, normalized ratios are below the lower cutoff ratio, indicating that the CDS is absent or highly divergent.
DISCUSSION
Plasmids of incompatibility group HI1 have been shown to be strongly associated with drug resistance in serovar Typhi (40) since the first report of a chloramphenicol resistant typhoid outbreak in 1972 (23) and have now spread globally (12). The RFLP typing of IncHI1 plasmids from Vietnam in a previous study revealed seven RFLP types and a change from RFLP pattern 1 to pattern 7 after 1993 (38). This raised a question about the evolutionary process of this particular group of plasmids. It was hypothesized that R27 was an ancestor of pHCM1 (RFLP pattern 1), which in turn gave rise to plasmids of RFLP pattern 7 (including pSTY7, discussed in the present study), which predominate after 1993, presumably due to increased fitness over their progenitors. In order to test this hypothesis of plasmid evolution, we collected IncHI1 plasmids isolated between 1970 and 2004 and analyzed their variation in much greater detail. Through investigation by DNA microarray, overlapping PCR and sequence comparison we identified both core conserved genes and highly variable resistance-associated regions.
In an attempt to elucidate phylogenetic relationships between the IncHI1 plasmids of serovar Typhi, we sequenced six core genes from each of the IncHI1 plasmids, an approach based on MLST (17). All plasmids from the 1970s, and those circulating in serovar Typhi in Vietnam in the early 1990s, were grouped together (PMLST group 1). We therefore infer that they are directly related and represent the expansion of a successful plasmid backbone in the serovar Typhi population over a period of 20 years. A recognizably distinct plasmid core, PMLST group 2, was consistently present after 1993, suggesting successful competition with those already present in the serovar Typhi population.
Although the earlier RFLP study suggested a new plasmid type arose and spread in Vietnam in the mid 1990s, the origin of the plasmid could not be determined. In contrast to our initial hypothesis of stepwise evolution from R27 to pHCM1 to pSTY7, PMLST analysis suggests that pSTY7 (PMLST group 2) and pHCM1 (PMLST group 1) belong to distinct plasmid lineages. This is also supported by whole-plasmid sequence comparisons (13). Thus, the change in the predominant plasmid type in Vietnam is best explained by the acquisition and spread of a distinct plasmid type (group 2) rather than the clonal expansion of a particular variant already present in the serovar Typhi population. Comparison to the broader collection of plasmids suggests that this replacement of plasmid types in 1993 was not unique in Vietnam but was a global phenomenon.
The typing data for both serovar Typhi host and its plasmid are needed to investigate whether the predomination of group 2 plasmids is due to the spread of the strain carrying the plasmid or the spread of plasmid itself. There is limited data on the background strains of serovar Typhi circulating in Vietnam during these periods. Some authors have shown that multiple RFLP types of serovar Typhi had become MDR between 1993 and 1997 (5), while other suggest clonal expansion of single serovar Typhi strain between 1995 and 2002 (15). Unfortunately, several of the plasmids analyzed in the present study were provided as transconjugants in Escherichia coli, and so it was not possible to type the background serovar Typhi hosts.
We also investigated the accessory gene content of different PMLST groups. The location and date of isolation were the most important factors governing the accessory gene content (Fig. 1b), while date of isolation alone had the greatest influence on core gene PMLST. This suggests that IncHI1 plasmids circulate globally but pick up different accessory genes depending on the environment they inhabit.
These plasmids conjugate with a higher frequency (3 to 4 logs) at ambient temperatures (27°C and below) than at an in vivo temperature (37°C) (R. Curtiss III, unpublished data). This, combined with the fact that contaminated water is the main route for serovar Typhi transmission (2), suggests that the plasmids are most likely to spread in contaminated water at ambient temperature. Accessory genes may be exchanged in this environment.
RFLP is a useful method for plasmid typing that can be applied to most groups of plasmids. Several plasmids in the present study gave the same PMLST and had very similar accessory gene content but demonstrated different RFLP patterns (38). The variation detected by RFLP indicates that the diversity of IncHI1 plasmids in serovar Typhi decreased after 1994 (38). Attempts to draw trees to describe phylogenetic relationships between the different RFLP types, however, failed. This is because the variation detected by RFLP is generated by several genetic mechanisms with different effects: point mutation, rearrangement, restriction protection mechanisms, and insertion/deletion. For the present study PMLST, while only applicable to IncHI1 plasmids, was used to define plasmid groups based on the accumulation of mutations in nonselected regions of the plasmid core. The accumulation of mutations is a function of time, and this typing scheme therefore represents a quantitative estimate of variation which can be related to the ancestry of the plasmids.
Since microarray data gave us no information on gene order, we screened for rearrangements and then mapped the regions of variation using specifically designed PCR primers to amplify 5- to 10-kb fragments around the entire plasmid. The results of these long-range PCRs are presented in Fig. 3. The core genes were shown to share the same arrangement in all plasmids analyzed.
It is intriguing that IncHI1 plasmids, which can be easily transferred to E. coli at high frequency in vitro, are found almost exclusively in two S. enterica serovars, Typhi and Paratyphi A. Plasmid fitness cost must play a role in maintaining IncHI1 plasmids in Salmonella. This involves the integration of plasmid and chromosomal regulatory networks, such as H-NS (6). In serovar Typhimurium there is a detectable cost of having IncHI1 plasmid (6), and so it is possible that in E. coli there is also a cost. Advantages to the bacterial host harboring IncHI1 plasmids have also been demonstrated: an increased level of survival inside monocytic cell lines in serovar Typhimurium (6) or higher levels of serovar Typhi bacteremia during typhoid fever (39) or possibly enhanced tolerance to the conditions faced during the infection cycle.
Taken together, these analyses show that the IncHI1 plasmids of serovar Typhi contain truly conserved regions, as well as truly dynamic regions. This is reminiscent of the situation in E. coli K1 F-like plasmids (19). All of the IncHI1 plasmids studied shared a conserved backbone encoding a set of 163 core genes, into which several acquisitions of different antibiotic resistance and other accessory genes have occurred. These plasmids are thus sampling the mobile gene pool and picking up different alleles of the same resistance genes, which are presumably selected for during treatment. It can therefore be hypothesized that variation in the accessory gene content is driven by selection acting on a phenotype encoded by several distinct genotypes.
The adaptation of the serovar Typhi bacterial host to acquire and retain resistance plasmids, and the subsequent evolution of this bacterium-plasmid combination, has occurred over a 20- to 30-year time frame. It is likely that pressure from chloramphenicol treatment in the 1970s drove the selection of a linkage group of genes capable of survival in the serovar Typhi host. The presence of such linkage group was reported by our results to be the core gene set of IncHI1 plasmids. This core has evolved little, but point mutations accumulated in these conserved genes can be used to discern subtly different plasmid lineages, as demonstrated by our newly developed PMLST scheme. By PMLST we showed for the first time the population structure and the dynamic evolutionary process of IncHI1 plasmids. The two lineages defined by PMLST corresponded perfectly to time of plasmid isolation, suggesting that competition between lineages resulted in a global replacement of group 1 by group 2 plasmids.
Antibiotic pressure concurrently drove the evolution of IncHI1 plasmids by acquisition of accessory genes into this core at certain regions where disruption of DNA can occur without effecting core plasmid functions. Genes conferring the same resistance phenotype appear to have been acquired on multiple occasions. Genetic components that are not involved in resistance to toxic substances have also been acquired.
In summary, the selective pressure from antimicrobial chemotherapy has forced serovar Typhi to gain resistance plasmids that compete to colonize the bacterial host population by allowing this major pathogen access to a wide variety of accessory genes.
Further investigation is required in order to identify the molecular mechanisms responsible for the success of PMLST group 2 plasmids. The spread of these plasmids is also of interest since it may shed light upon the transfer route of antibiotic resistance, whether it is the synergistic spread of a bacterium-plasmid clone or the emergence of a more competitive plasmid. Interrogation of the association between serovar Typhi strains and IncHI1 plasmid types is vital to answer this question. The findings suggest a possible approach to destabilize IncHI1 plasmids in serovar Typhi, rendering them susceptible once again to first-line antibiotics.
Supplementary Material
Acknowledgments
This study was funded by the Wellcome Trust of Great Britain and the Defence Science and Technology Laboratory.
We thank Henry Smith from Colindale, Christiane Dolecek and Chau Tran Thuy from Oxford Clinical Research Unit in the HCMC (Vietnam), To Song Diep, Microbiology, HTD, HCMC (Vietnam), and Bianca Paglietti from Università degli Studi di Sassari (Italy) for supplying IncHI1 plasmids.
Supplemental material for this article may be found at http://aac.asm.org/.
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Ausable, F. M., R. Brent, and R. E. Kingston. 1996. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 2.Bhan, M. K., R. Bahl, and S. Bhatnagar. 2005. Typhoid and paratyphoid fever. Lancet 366:749-762. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colquhoun, J., and R. Weetch. 1950. Resistance to chloramphenicol developing during treatment of typhoid fever. Lancet i:869. [DOI] [PubMed] [Google Scholar]
- 5.Connerton, P., J. Wain, T. T. Hien, T. Ali, C. Parry, N. T. Chinh, H. Vinh, V. A. Ho, T. S. Diep, N. P. Day, N. J. White, G. Dougan, and J. J. Farrar. 2000. Epidemic typhoid in Vietnam: molecular typing of multiple-antibiotic-resistant Salmonella enterica serotype Typhi from four outbreaks. J. Clin. Microbiol. 38:895-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle, M., M. Fookes, A. Ivens, M. W. Mangan, J. Wain, and C. J. Dorman. 2007. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315:251-252. [DOI] [PubMed] [Google Scholar]
- 7.Dutta, S., D. Sur, B. Manna, S. K. Bhattacharya, J. L. Deen, and J. D. Clemens. 2005. Rollback of Salmonella enterica serotype Typhi resistance to chloramphenicol and other antimicrobials in Kolkata, India. Antimicrob. Agents Chemother. 49:1662-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essa, A. M., D. J. Julian, S. P. Kidd, N. L. Brown, and J. L. Hobman. 2003. Mercury resistance determinants related to Tn21, Tn1696, and Tn5053 in enterobacteria from the preantibiotic era. Antimicrob. Agents Chemother. 47:1115-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabant, P., P. Newnham, D. Taylor, and M. Couturier. 1993. Isolation and location on the R27 map of two replicons and an incompatibility determinant specific for IncHI1 plasmids. 175:7697-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, R. M., and H. W. Stokes. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115-132. [DOI] [PubMed] [Google Scholar]
- 12.Hampton, M. D., L. R. Ward, B. Rowe, and E. J. Threlfall. 1998. Molecular fingerprinting of multidrug-resistant Salmonella enterica serotype Typhi. Emerg. Infect. Dis. 4:317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt, K. E., N. R. Thomson, J. Wain, M. D. Phan, S. Nair, R. Hasan, Z. A. Bhutta, M. A. Quail, H. Norbertczak, D. Walker, G. Dougan, and J. Parkhill. 2007. Multidrug-resistant Salmonella enterica serovar Paratyphi A harbors IncHI1 plasmids similar to those found in serovar Typhi. J. Bacteriol. 189:4257-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 15.Le, T. A., M. Lejay-Collin, P. A. Grimont, T. L. Hoang, T. V. Nguyen, F. Grimont, and M. R. Scavizzi. 2004. Endemic, epidemic clone of Salmonella enterica serovar Typhi harboring a single multidrug-resistant plasmid in Vietnam between 1995 and 2002. J. Clin. Microbiol. 42:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer, A. A., G. Morelli, M. Heuzenroeder, M. Kamke, and M. Achtman. 1984. Conservation of plasmids among Escherichia coli K1 isolates of diverse origins. Infect. Immun. 46:649-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirza, S., S. Kariuki, K. Z. Mamun, N. J. Beeching, and C. A. Hart. 2000. Analysis of plasmid and chromosomal DNA of multidrug-resistant Salmonella enterica serovar Typhi from Asia. J. Clin. Microbiol. 38:1449-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olarte, J. 1981. R factors present in epidemic strains of shigella and salmonella species found in Mexico, p. 11-19. In S. Levy (ed.), Molecular biology, pathology, and ecology of bacterial plasmids. Plenum Press, Inc., New York, NY.
- 22.Pagotto, F., and J. A. Dillon. 2001. Multiple origins and replication proteins influence biological properties of beta-lactamase-producing plasmids from Neisseria gonorrhoeae. J. Bacteriol. 183:5472-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paniker, C. K., and K. N. Vimala. 1972. Transferable chloramphenicol resistance in Salmonella typhi. Nature 239:109-110. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 25.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 28.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 30.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177-181. [DOI] [PubMed] [Google Scholar]
- 31.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 33.Summers, A., J. Wireman, M. Vimy, F. Lorscheider, B. Marshall, S. Levy, S. Bennett, and L. Billard. 1993. Mercury released from dental “silver” fillings provokes an increase in mercury- and antibiotic-resistant bacteria in oral and intestinal floras of primates. Antimicrob. Agents Chemother. 37:825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thisted, T., A. K. Nielsen, and K. Gerdes. 1994. Mechanism of post-segregational killing: translation of Hok, SrnB, and Pnd mRNAs of plasmids R1, F, and R483 is activated by 3′-end processing. EMBO J. 13:1950-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thisted, T., N. S. Sorensen, E. G. Wagner, and K. Gerdes. 1994. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 13:1960-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, C. M. 2000. Paradigms of plasmid organization. Mol. Microbiol. 37:485-491. [DOI] [PubMed] [Google Scholar]
- 37.Threlfall, E. J. 2002. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and waterborne infections. FEMS Microbiol. Rev. 26:141-148. [DOI] [PubMed] [Google Scholar]
- 38.Wain, J., L. T. Diem Nga, C. Kidgell, K. James, S. Fortune, T. Song Diep, T. Ali, O. G. P., C. Parry, J. Parkhill, J. Farrar, N. J. White, and G. Dougan. 2003. Molecular analysis of incHI1 antimicrobial resistance plasmids from Salmonella serovar Typhi strains associated with typhoid fever. Antimicrob. Agents Chemother. 47:2732-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wain, J., T. S. Diep, V. A. Ho, A. M. Walsh, T. T. Nguyen, C. M. Parry, and N. J. White. 1998. Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. J. Clin. Microbiol. 36:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wain, J., and C. Kidgell. 2004. The emergence of multidrug resistance to antimicrobial agents for the treatment of typhoid fever. Trans. R. Soc. Trop. Med. Hyg. 98:423-430. [DOI] [PubMed] [Google Scholar]
- 41.Whelan, K. F., D. Maher, E. Colleran, and D. E. Taylor. 1994. Genetic and nucleotide sequence analysis of the gene htdA, which regulates conjugal transfer of IncHI plasmids. J. Bacteriol. 176:2242-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodward, T. E., and J. E. Smadel. 1948. Preliminary report on the beneficial effect of chloromycetin in the treatment of typhoid fever. Ann. Intern. Med. 29:131-134. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, R., K. Eggleston, V. Rotimi, and R. J. Zeckhauser. 2006. Antibiotic resistance as a global threat: evidence from China, Kuwait, and the United States. Global Health 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.