Abstract
In a previous study, four Salmonella isolates from humans in the Henan province of China showed reduced susceptibility to ciprofloxacin (MIC, 0.125 to 0.25 μg/ml) but were susceptible to nalidixic acid (MIC, 4 to 8 μg/ml). All isolates were negative for known qnr genes (A, B, and S), aac(6′)Ib-cr, and mutations in gyrA and parC. Plasmid DNA was extracted from all four isolates and transformed into Escherichia coli TG1 and DH10B cells by electroporation, and transformants were selected on 0.06 μg/ml ciprofloxacin containing brain heart infusion agar plates. Resistance to ciprofloxacin could be transferred by electroporation, and a similar 4,270-bp plasmid was found in all transformants. By sequence analysis, the plasmid was found to carry an open reading frame that had similarities to other qnr genes and that encoded a 214-amino-acid pentapeptide repeat protein. This gene, designated qnrD, showed 48% similarity to qnrA1, 61% similarity to qnrB1, and 41% similarity to qnrS1. Further subcloning of the qnrD coding region into the constitutively expressed tetA gene of vector pBR322 showed that the gene conferred an increase in the MIC of ciprofloxacin by a factor of 32 (from an MIC of 0.002 to an MIC of 0.06 μg/ml). For comparison, qnrA1 and qnrS1 were also subcloned into pBR322 and transformed into DH10B cells, conferring MICs of 0.125 and 0.5 μg/ml, respectively. A phylogenetic analysis of all known qnr sequences was performed and showed that qnrD was more closely related to the qnrB variants but formed an independent cluster. To our knowledge, this is the first description of this qnrD gene.
Quinolone resistance in the Enterobacteriaceae is mostly mediated by point mutations in the quinolone resistance-determining regions (QRDR) of the gyrase and topoisomerase IV genes, leading to a target modification. Other resistance mechanisms include efflux pump mechanisms, and more recently, target protection mechanisms encoded by the qnr genes and enzymatic modifications encoded by aac(6′)Ib-cr have also been found to contribute to resistance to drugs belonging to this antimicrobial class. First, qnrA1 from a clinical strain of Klebsiella pneumoniae isolated in Alabama was described by Martinez-Martinez et al. in 1998 (17). This strain carried plasmid pMG252, which contained the gene encoding quinolone resistance, later named qnrA1 (17).
In 2005, Hata et al. described another qnr gene from a Shigella flexneri 2b isolate isolated during an outbreak of food poisoning in Japan. This strain contained a plasmid, designated pAH0376, containing a gene with high similarity to qnr, which was designated qnrS. The QnrS protein was also a 218-amino-acid protein with 59% similarity to QnrA that conferred low-level resistance to fluoroquinolones (11).
In 2006, Jacoby and colleagues described a third gene encoding quinolone resistance, qnrB. This gene was first found in a Klebsiella pneumoniae isolate from India and encoded a 214-amino-acid protein of the pentapeptide repeat family, which had 41% amino acid identity with qnrA and 39% amino acid identity with qnrS (14).
To date, a total of 6 qnrA, 4 qnrS, and 20 qnrB variants have been described in the literature and are listed in the database maintained at the website http://www.lahey.org/qnrStudies (13). Furthermore, other similar chromosomal genes such as Vibrio vulnificus qnr, Vibrio parahaemolyticus qnr, Vibrio cholerae qnr, Photobacterium profundum qnr, Enterococcus faecalis qnr, and Enterococcus faecalis qnr were described previously (1, 3, 9, 13, 19-22).
Later, other transferable resistance genes were found to cause reduced susceptibility to quinolones. The aminoglycoside acetyltransferase variant aac(6′)Ib-cr is able to modify ciprofloxacin and norfloxacin. Moreover, in 2007, in Japan, but also in Belgium, another gene, qepA, was discovered to encode a putative specific efflux pump, which is able to reduce susceptibility to hydrophilic quinolones (18, 26). Furthermore, a second variant, named qepA2, from France was recently described (4).
Recently, Wang and colleagues described another qnr gene, qnrC, which was found in Proteus mirabilis; however, its sequence is not yet publicly available, but it was found that qnrC encodes a 221-amino-acid protein with different amino acid identities from qnrD, which indicates that the gene is different from qnrD (25).
Here, we report the finding of a plasmid containing a novel quinolone resistance gene, qnrD, which has been found to cause reduced susceptibility to fluoroquinolones in isolates of Salmonella enterica serovar Bovismorbificans and Kentucky strains isolated from humans in the Henan province of China. The complete plasmid was sequenced, and the novel qnrD gene was cloned along with both the qnrA1 and qnrS1 genes, which were cloned for comparisons of the susceptibility patterns in vitro. The novel qnrD gene shares similarities with the previously described qnr genes and encodes a putative pentapeptide repeat protein that is able to confer reduced susceptibility to fluoroquinolones. A phylogenetic analysis shows that it clusters separately from the known qnr genes and variants.
MATERIALS AND METHODS
Strains.
Salmonella enterica strains HN-GSS2006-142, HN-GSS2007-0.033, HN-GSS2007-0.056, and HN-GSS-2007-057 were isolated in 2006 to 2007 from human infections in China in the Henan province. These strains belonged to serovars Kentucky (HN-GSS2007-033) and Bovismorbificans (remaining strains) and were included in this study based on their resistance pattern (low-level resistance to ciprofloxacin but susceptibility to nalidixic acid).
For the amplification of the qnrA1 and qnrS1 genes for cloning, DNA samples obtained from the control strains Escherichia coli J53 with pMG252 (kindly provided by George Jacoby) and E. coli MT102(pBC H-2.6) (plasmid DNA kindly provided by Mami Hata) were used.
Competent Escherichia coli cells were used in the transformation and cloning experiments: TG1 cells (Stratagene, Cambridge, United Kingdom) were used in the initial transformation experiments with the natural plasmid and cloned plasmid fragment, and DH10B cells (Invitrogen, Cergy Pontoise, France) were used for transformation with the native plasmid and vector pBR322, containing the resistance genes qnrD, qnrA1, and qnrS1. Plasmids pACYC177 (New England Biolabs, Hitchin, United Kingdom), containing resistance to ampicillin and kanamycin, and pBR322 (New England Biolabs, Hitchin, United Kingdom), containing resistance to tetracycline and ampicillin, were used in the cloning of plasmid restriction fragments and PCR products. The strains were grown on brain heart infusion broth or agar (Becton Dickinson, Sparks, MD) at 37°C.
Susceptibility testing.
MICs were determined in a broth microdilution assay for ciprofloxacin, nalidixic acid, norfloxacin, and ofloxacin according to Clinical and Laboratory Standards Institute (CLSI) standards (8) by using 96-well panels (Sensititre; Trek Diagnostics), and self-made panels were used for the wider range of dilutions of ciprofloxacin, nalidixic acid, norfloxacin, and ofloxacin.
DNA preparation and transformation.
Plasmid DNA was extracted from all strains using the Qiagen (Hilden, Germany) Midi kit. Initial transformation experiments were performed by electroporation (Gene Pulser; Bio-Rad) of the plasmid DNA into competent TG1 cells (Stratagene, Cambridge, United Kingdom). Transformants were selected on brain heart infusion agar plates containing 0.06 μg/ml ciprofloxacin. Plasmid DNA from transformants was extracted and restricted with several restriction enzymes (EcoRV, HindIII, EcoRI, and SmaI) (New England BioLabs, Hitchin, United Kingdom) to observe restriction patterns and choose an enzyme for the restriction of fragments before cloning (data not shown).
Cloning.
For restriction and cloning experiments, the plasmid DNA of Salmonella enterica serovar Bovismorbificans strain GSS-HN-2007-057 was used. Plasmid DNA of the transformant Tf1-HN-GSS-2007-057 was restricted using EcoRV (New England BioLabs, Hitchin, United Kingdom). The restriction product containing the plasmid fragments was purified using a Gfx purification kit (Amersham Biosciences, Piscataway, NJ) and ligated into pACYC177 vector DNA (New England Biolabs, Hitchin, United Kingdom) digested with SmaI (New England BioLabs, United Kingdom) and dephosphorylated with shrimp alkaline phosphatase (USB Corporation). The ligation products of pACYC177 with the inserts were electroporated into competent E. coli TG1 cells (Stratagene, Cambridge, United Kingdom), selected on ampicillin (50 μg/ml)- and on ciprofloxacin (0.06 μg/ml)-containing plates, and replicated on kanamycin (50 μg/ml)-containing brain heart infusion agar plates (Becton Dickinson, Sparks, MD) to identify inserts.
Total plasmid sequencing.
The cloned fragments were amplified using plasmid DNA as a template for a long PCR (Phusion high-fidelity PCR kit; Finnzymes, Espoo, Finland) with primers designed with the kanamycin resistance gene to amplify the insert and the flanking regions of the SmaI restriction site (Table 1) with HF buffer (Phusion high-fidelity PCR kit; Finnzymes, Espoo, Finland) and under normal conditions as indicated by the manufacturer, at an annealing temperature of 50°C, allowing an extension time of 4 min. Sequencing of the initial fragments in both senses was performed with the primers used for the amplification of the cloned fragment. After the first round of sequencing, further primers were designed based on the first sequences to continue the amplification and sequencing rounds, monitoring the sequence by primer walking. Furthermore, primer walking was also performed for the remaining portion of the isolated plasmid, designing primers directed outwards from the cloned fragment that were used to amplify the remaining sequence using the plasmid DNA extracted from Salmonella isolate GSS-HN-2007-057 as a template, and further primers designed from the previous sequences were used to obtain the total sequence of the plasmid. Primers used for sequencing are shown in Table 1. Amplification conditions for each primer pair were adapted to the primers and sequences to be amplified. Furthermore, PCR mapping was performed on the plasmid DNA obtained from the transformants to observe the similarities between the plasmids that were isolated from the original isolates and p2007057. The fragments obtained in the amplification rounds were sent for sequencing at Macrogen Laboratories (Seoul, South Korea).
TABLE 1.
Primers used for amplification/sequencing and cloning of plasmid p2007057
| Primer | Sequencea | Used for |
|---|---|---|
| PACYC177 SmaI insert fw | 5′-CGTACTCCTGATGATGCATG-3′ | Seq inserted fragment |
| PACYC177 SmaI insert rev | 5′-GCGCATCAACAATATTTTCAC-3′ | Seq inserted fragment |
| P1 | 5′-GCCTTTTCAAATTGTGATTTTTC-3′ | Seq plasmid |
| P2 | 5′-CGTTTCCTGCTTCACAAAAT-3′ | Seq plasmid |
| P3 | 5′-CTGTACGTAATCGTTCGGTTTC-3′ | Seq plasmid |
| P4 | 5′-CCAGCGGTATCGAGGTAAAC-3′ | Seq plasmid |
| P5 | 5′-TTACTGGTTGTGATTTAACGGG-3′ | Seq plasmid |
| P6 | 5′-TCAGTAACGTCGAATGGCTTA-3′ | Seq plasmid |
| P7 | 5′-AGGCCGGAAGTCTCAAAAG-3′ | Seq plasmid |
| P8 | 5′-ATATCAGACAGTGTGGCACAG-3′ | Seq plasmid |
| P9 | 5′-CTGAAACGCGCTCAGG- 3′ | Seq plasmid |
| P10 | 5′-AACTTCTCACACTCCTGCTGTC-3′ | Seq plasmid |
| P11 | 5′-CTTTTGAGACTTCCGGCCT-3′ | Seq plasmid |
| P12 | 5′-CTGTGCCACACTGTCTGATAT-3′ | Seq plasmid |
| qnrD start EcoRV | 5′-GGGGATATCTTAAGGTTGTTCAAATTAATGTAC-3′ | Cloning qnrD |
| qnrD end SalI | 5′-CCCGTCGACTTTGATTAGTACCACATTGG-3′ | Cloning qnrD |
| qnrD fw | 5′-CGAGATCAATTTACGGGGAATA-3′ | Amplification of qnrD gene |
| qnrD rev | 5′-AACAAGCTGAAGCGCCTG-3′ | Amplification of qnrD gene |
| qnrA start EcoRV | 5′-CCCGATATCCTGATTAAAGGAAGCC-3′ | Cloning qnrA1 |
| qnrA end SalI | 5′-GGGGGTCGACAGAGCTAATCCGGCAG-3′ | Cloning qnrA1 |
| qnrS start PvuII | 5′-GGGCAGCTGCCTTTCAACAAGGAGTACTC-3′ | Cloning qnrS1 |
| qnrS end SalI | 5′-CCCGTCGACAATTAGTCAGGATAAACAACA-3′ | Cloning qnrS1 |
The recognition sites for the restriction enzymes are underlined in the nucleotide sequences of the primers used for cloning.
Sequence analysis.
Sequence analysis was performed using the Vector NTI program tools Contig Express and AlignX (VectorNTI Suite Informax, Inc.). BLAST searches were performed using the National Center for Biotechnology Information website (http.//www.ncbi.nlm.nih.gov) using both the blastn and the tblastx algorithms.
For the plasmid sequence analysis, the Vector NTI ORF Finder tool was used, and the open reading frame (ORF) sequences were compared against GenBank sequences using BLAST algorithms.
Phylogenetic analysis.
All representative sequences of the known qnr genes described above were obtained from GenBank in FASTA format. Phylogenetic analysis was conducted using the neighbor-joining method using MEGA 4 software, version 4 (23).
Cloning of the qnrD, qnrA1, and qnrS1 genes.
Subcloning of the gene and flanking regions was performed to confirm if it conferred the observed quinolone resistance phenotype. Primers were designed to amplify the entire ORF and some of the flanking region (62 nucleotides upstream and 46 nucleotides downstream) carrying an EcoRV restriction site in the forward primer and a SalI site in the reverse primer (Table 1). PCR was performed in a 50-μl PCR mixture including 5 μl 10× reaction buffer (Ampliqon), 0.5 μl forward and reverse primer, 0.5 μl deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.2 μl Taq polymerase (Ampliqon), and 2 μl DNA template. PCR was run with the same program using a Trio thermocycler (Biometra): 94°C for 5 min; 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min for 30 cycles; and 72°C for 10 min. The amplified fragment digested with EcoRV and SalI (New England BioLabs, Hitchin, United Kingdom) was then ligated into the tetA gene of vector pBR322 (New England Biolabs, Hitchin, United Kingdom) downstream of the tetA promoter digested with EcoRV and SalI (New England BioLabs, Hitchin, United Kingdom). The ligation product was electroporated into competent E. coli DH10B cells (Invitrogen, Cergy Pontoise, France). Transformants were selected on ampicillin (50 μg/ml)-containing brain heart infusion agar plates (Becton Dickinson) and replicated on plates containing tetracycline to identify inserts. The presence of the qnr-like (qnrD) gene was confirmed by PCR amplification using primers qnrD fw and qnrD reverse (Table 1), and the integrity of the sequence was confirmed by amplification and sequencing of the inserted gene. Further constructs were constructed in the same way from plasmids containing the other quinolone resistance genes qnrA1 and qnrS1, for comparison, using amplified fragments from the control strains E. coli J53(pMG252) and E. coli MT102 (pBC H-2.6). Cloning of qnrA1 and qnrS1 was performed using either EcoRV or PvuII restriction site in the forward primer and SalI in the reverse primer, and consequently, the restriction of the PCR product was performed with either EcoRV or PvuII and SalI (New England BioLabs, Hitchin, United Kingdom).
Susceptibility testing was performed for all the constructed clones and for E. coli DH10B cells and DH10B cells containing pBR322 and the plasmid extracted from the transformant of Salmonella enterica serovar Bovismorbificans GSS-HN-2007-057 for comparison.
Nucleotide sequence accession numbers.
The nucleotide sequence of the qnrD gene has been deposited in the GenBank data library under accession number EU692908. The nucleotide sequence of plasmid p2007057 has been deposited in the GenBank data library under accession number FJ228229.
RESULTS
Susceptibility testing.
The Salmonella enterica serovar Kentucky (n = 1) and Bovismorbificans (n = 3) isolates included in this study showed reduced susceptibility to ciprofloxacin (MIC, 0.125 or 0.25 μg/ml) but were susceptible to nalidixic acid (MIC = 8 μg/ml). All four isolates were susceptible to the remaining drugs tested.
All PCR screenings performed previously for qnrA, qnrB, qnrS, aac(6′)Ib-cr, and qepA were negative for all four isolates.
No amino acid substitutions were found in the QRDR of the gyrA or parC topoisomerase gene. One amino acid substitution outside of the QRDR in the parC gene (T57S) was detected in all but one isolate (GSS-HN-2007-033). This mutation was previously described but not likely related to quinolone resistance since it was found in susceptible strains previously (2).
Plasmid extraction and transfer of resistance.
All four strains contained one or several small plasmids as observed by plasmid extraction. The transfer of resistance by the transformation of electrocompetent TG1 cells with the extracted plasmid DNA was successful for all the strains. The transformants obtained were able to grow on plates containing 0.06 μg/ml ciprofloxacin. Electrophoresis of plasmid DNA extracts from the transformants showed the presence of a small plasmid of about 4.3 kb in all transformants obtained and an additional small-sized plasmid that was cotransferred to a transformant obtained from isolate GSS-HN-2007-033, which was considered not to be related to the resistance phenotype observed since it was present in only one of the transformants, and no difference in their phenotypes was noted for the other transformants containing only the 4.3-kb plasmid. Further cloning and sequencing were proceeded on the 4.3-kb plasmid, which was suspected to contain the quinolone resistance determinant.
Restriction, cloning, and sequencing of plasmid.
Restriction of plasmid DNA with EcoRV resulted in two distinct fragments of about 3.2 kb and 1.1 kb. All isolates carried a plasmid with a similar restriction pattern using the EcoRV enzyme, and due to subsequent PCR mapping, we believe that the plasmids carried by the other isolate were similar to the plasmid that was cloned and sequenced.
Cloning of the digested plasmid DNA from strain HN-GSS-2007-057 into the SmaI site located in the aph(3′)-Ia kanamycin resistance gene of vector plasmid pACYC177 allowed us to select two ampicillin-resistant and kanamycin-susceptible clones carrying a fragment of about 3.2 kb.
Amplification and sequencing of the cloned fragment in both directions showed that the same fragment (although in inverted positions) was present in both selected clones. Further sequencing resulted in the full sequencing of the 3.2-kb fragment, which was suspected to contain the quinolone resistance determinant, but also of the remaining 1.1 kb-portion of the plasmid, which was totally sequenced and assembled and was deposited in the GenBank library under accession number FJ228229. The sequence of the cloned 3.2-kb fragment contained an ORF encoding a 214-amino-acid protein that showed similarity to previously identified qnr genes. Its nucleotide sequence showed 45% similarity to qnrA1, 65% similarity to qnrB1, and 38% similarity to qnrS1. The nucleotide and translated amino acid sequences were compared to the those of qnr genes known and aligned by using Clustal W for comparisons (Fig. 1).
FIG. 1.
QnrD amino acid sequence. (a) Alignment of amino acid sequences encoded by the qnrA1, qnrB1, qnrS1, and qnrD genes obtained using AlignX with Vector NTI software (InformaxVector NTI Suite 8). (b) Hypothetical structure of the QnrD protein. The amino acid sequence was represented and divided into pentapeptide repeats. The conserved amino acids according to the consensus sequence (A/C/S/T/V)(D/N)(L/F)(S/T/R)(G/R) (22) are in boldface type, and the most characteristic pentapeptide units are underlined.
Phylogenetic analysis.
The gene found was, to our knowledge, unknown but was related to the known qnr genes and encoded a putative protein of the pentapeptide repeat family, and therefore, we have conducted a molecular evolutionary genetic analysis using MEGA 4 software according to methods described previously by Tamura et al. (23), which is represented as a phylogenetic tree (Fig. 2).
FIG. 2.
Distance tree between the known qnr genes and variants at the nucleotide level. The evolutionary history was inferred using the neighbor-joining method. Phylogenetic analyses were conducted using MEGA 4 software (23).
The qnr gene variants clustered in the different major gene groups. However, the novel gene clustered separately from the major groups but showed a closer similarity to the qnrB group.
Total plasmid sequencing.
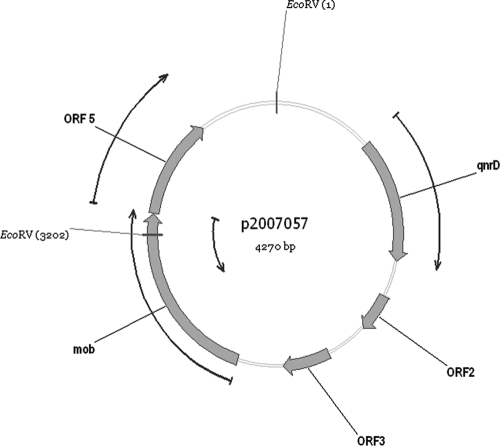
In the plasmid sequence analysis, we observed that it contained five ORFs, as shown in Fig. 3. Apart from the qnrD gene, no other known structures and no further resistance genes have been found in the plasmid sequence (Fig. 3). Surprisingly, we were also unable to locate any similarity to replication proteins or repeat sequences previously deposited in the GenBank database.
FIG. 3.
Graphical map of plasmid p2007057 (GenBank accession number FJ228229). qnrD (positions 554 to 1198) encodes a pentapeptide repeat protein related to previously described qnr genes, ORF2 (positions 1397 to 1597) is a hypothetical protein with 88% similarity to a hypothetical protein described previously (accession number AF448250), ORF3 (positions 1828 to 2091) is a hypothetical protein with no significant matches, ORF4 (positions 2347 to 3309) is a hypothetical protein with 83% similarity to a partial sequence of a mob gene described previously (accession number EU90225), and ORF5 (positions 3312 to 3842) is a hypothetical protein with 73% similarity to a partial sequence of a hypothetical protein described previously (accession number DQ995354). Possible ORFs were searched using Vector NTI software. BLAST searches were performed using the blastn algorithm at the NCBI server (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Susceptibility testing of transformants with natural and cloned plasmid clones.
The MIC of ciprofloxacin in E. coli DH10B cells increased in the transformant carrying the plasmid from strain GSS-HN-2007-057 and also in the strain carrying plasmid pBR322 with the cloned qnrD gene from 0.002 μg/ml to 0.06 μg/ml (by a factor of 32). The increase in the MIC of ciprofloxacin was slightly higher for E. coli DH10B cells carrying pBR322 with qnrA1 (MIC = 0.125 μg/ml) and E. coli DH10B cells carrying pBR322 with qnrS1 (MIC = 0.5 μg/ml). The MIC of norfloxacin increased from 0.015 μg/ml to 0.25, 0.5, and 0.06 μg/ml for qnrA, qnrS, and qnrD, respectively, and the MIC of ofloxacin increased from 0.015 μg/ml to 0.25, 0.5, and 0.125 μg/ml, respectively. The changes in the MIC of nalidixic were smaller than those observed for the fluoroquinolones (only 1 to 2 dilution steps) in all strains carrying qnr genes (Table 2).
TABLE 2.
Results of susceptibility testing by broth microdilution of Escherichia coli DH10B and isogenic strains carrying qnr genes
| Antimicrobial drug | MIC (μg/ml) (fold increase)a
|
|||||
|---|---|---|---|---|---|---|
| E. coli DH10B | E. coli DH10B(pBR322) | E. coli DH10B+pBR322) qnrA1 | E. coli DH10B(pBR322) qnrS1 | E. coli DH10B(pBR322) qnrD | E. coli DH10B(p2007057) | |
| Ciprofloxacin | 0.002 | 0.004 | 0.125 (64) | 0.5 (256) | 0.06 (32) | 0.125 (64) |
| Norfloxacin | 0.015 | 0.015 | 0.25 (16) | 0.5 (32) | 0.06 (4) | 0.125 (64) |
| Ofloxacin | 0.015 | 0.032 | 0.25 (16) | 0.5 (32) | 0.125 (8) | 0.25 (16) |
| Nalidixic acid | 2 | 2 | 8 (4) | 8 (4) | 4 (2) | 8 (4) |
All MIC determinations were performed in broth microdilution assays according to CLSI standards (8).
DISCUSSION
We have identified and cloned a novel gene conferring resistance to ciprofloxacin in four Salmonella enterica serovar Bovismorbificans and Kentucky isolates obtained from human infections in the Henan province in China.
Resistance to quinolones was observed by low-level resistance to ciprofloxacin and susceptibility to nalidixic acid, a phenotype previously associated with the presence of transferable quinolone resistance determinants (6, 24). However, none of the known quinolone resistance genes was detected by PCR amplification, nor were mutations detected in the QRDR of the gyrA and parC topoisomerase genes. Furthermore, it was observed that resistance could be transferred by transformation, and a novel qnr-like gene responsible for the resistance phenotype was identified in a 4,270-bp plasmid, which was totally sequenced. No further resistance determinants were detected in these strains, as they were susceptible to all other drugs tested.
The qnr-like gene found was designated qnrD due to the recent finding of a gene called qnrC in a Proteus isolate from China (25). qnrD showed similarities to qnrA, qnrB, and qnrS genes and encoded a 214-amino-acid pentapeptide repeat protein. The phylogenetic analysis showed that it clustered independently from the known qnr gene variants but shared the highest similarity with the qnrB variants.
It was demonstrated that the plasmid and also the cloned gene were able to confer an increase in the MIC of ciprofloxacin by a factor of about 32 without a major increase in the MIC of nalidixic acid, which is within the expected phenotype of a qnr-related gene. However, the comparison with the cloned qnrA and qnrS genes showed that qnrD resulted in slightly lower increases in the MIC.
Previously known qnr genes have been found in Salmonella isolates of different origins; however, these genes seem to be carried by different genetic elements (5, 12, 15). Previously sequenced plasmids carrying qnr genes showed different backbones and possibilities of origin (7, 10, 15, 16). Furthermore, the qnr genes have been related to integron structures; however, the plasmid found to carry this novel gene showed a small size and carried no integron structures or further resistance determinants.
Until recently, qnr genes have been observed at low prevalences in most reported screenings; however, further genes and variants might still be discovered, increasing the pool of determinants conferring resistance to quinolones. Although their clinical implications are still unknown, the spread of such resistance determinants is concerning.
Acknowledgments
The study was supported by a grant from the European Union Marie Curie Programme (MEST-CT-2004-007819) and grant 274-05-0117 from the Danish Research Agency.
We thank George Jacoby and Mami Hata for the positive control strains for qnrA1 and qnrS1, respectively. We also thank Jacob Dyring Jensen for excellent technical assistance.
Footnotes
Published ahead of print on 24 November 2008.
REFERENCES
- 1.Arsene, S., and R. Leclercq. 2007. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob. Agents Chemother. 51:3254-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baucheron, S., E. Chaslus-Dancla, A. Cloeckaert, C. H. Chiu, and P. Butaye. 2005. High-level resistance to fluoroquinolones linked to mutations in gyrA, parC, and parE in Salmonella enterica serovar Schwarzengrund isolates from humans in Taiwan. Antimicrob. Agents Chemother. 49:862-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cattoir, V., L. Poirel, D. Mazel, C. J. Soussy, and P. Nordmann. 2007. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 51:2650-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattoir, V., L. Poirel, and P. Nordmann. 2008. Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob. Agents Chemother. 52:3801-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattoir, V., F. X. Weill, L. Poirel, L. Fabre, C. J. Soussy, and P. Nordmann. 2007. Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 59:751-754. [DOI] [PubMed] [Google Scholar]
- 6.Cavaco, L. M., R. S. Hendriksen, and F. M. Aarestrup. 2007. Plasmid-mediated quinolone resistance determinant qnrS1 detected in Salmonella enterica serovar Corvallis strains isolated in Denmark and Thailand. J. Antimicrob. Chemother. 60:704-706. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y.-T., H.-Y. Shu, L.-H. Li, T.-L. Liao, K.-M. Wu, Y.-R. Shiau, J.-J. Yan, I.-J. Su, S.-F. Tsai, and T.-L. Lauderdale. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 50:3861-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th ed. M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Fonseca, E. L., F. F. Dos Santos, V. V. Vieira, and A. C. Vicente. 2008. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg. Infect. Dis. 14:1129-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay, K., A. Robicsek, J. Strahilevitz, C. H. Park, G. Jacoby, T. J. Barrett, F. Medalla, T. M. Chiller, and D. C. Hooper. 2006. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin. Infect. Dis. 43:297-304. [DOI] [PubMed] [Google Scholar]
- 11.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins, K. L., L. Wootton, M. R. Day, and E. J. Threlfall. 2007. Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J. Antimicrob. Chemother. 59:1071-1075. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby, G., V. Cattoir, D. Hooper, L. Martinez-Martinez, P. Nordmann, A. Pascual, L. Poirel, and M. Wang. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 52:2297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehrenberg, C., S. Friederichs, A. de Jong, G. B. Michael, and S. Schwarz. 2006. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J. Antimicrob. Chemother. 58:18-22. [DOI] [PubMed] [Google Scholar]
- 16.Kehrenberg, C., K. L. Hopkins, E. J. Threlfall, and S. Schwarz. 2007. Complete nucleotide sequence of a small qnrS1-carrying plasmid from Salmonella enterica subsp. enterica Typhimurium DT193. J. Antimicrob. Chemother. 60:903-905. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 18.Perichon, B., P. Courvalin, and M. Galimand. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., A. Liard, J. M. Rodriguez-Martinez, and P. Nordmann. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 56:1118-1121. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, L., J. M. Rodriguez-Martinez, H. Mammeri, A. Liard, and P. Nordmann. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saga, T., M. Kaku, Y. Onodera, S. Yamachika, K. Sato, and H. Takase. 2005. Vibrio parahaemolyticus chromosomal qnr homologue VPA0095: demonstration by transformation with a mutated gene of its potential to reduce quinolone susceptibility in Escherichia coli. Antimicrob. Agents Chemother. 49:2144-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez, M., A. Hernandez, J. Rodriguez-Martinez, L. Martinez-Martinez, and J. Martinez. 2008. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol. 8:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 24.Veldman, K., W. van Pelt, and D. Mevius. 2008. First report of qnr genes in Salmonella in The Netherlands. J. Antimicrob. Chemother. 61:452-453. [DOI] [PubMed] [Google Scholar]
- 25.Wang, M. H., X. Xu, S. Wu, D. Zhu, and M. G. Wang. 2008. A new plasmid-mediated gene for quinolone resistance, qnrC, abstr. O207. Abstr. 18th Eur. Congr. Clin. Microbiol. Infect. Dis., Barcelona, Spain.
- 26.Yamane, K., J. I. Wachino, S. Suzuki, K. Kimura, N. Shibata, H. Kato, K. Shibayama, T. Konda, and Y. Arakawa. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51:3354-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]