Abstract
Voriconazole is an effective antifungal drug, but adverse drug-drug interactions associated with its use are of major clinical concern. To identify the mechanisms of these interactions, we tested the inhibitory potency of voriconazole with eight human cytochrome P450 (CYP) enzymes. Isoform-specific probes were incubated with human liver microsomes (HLMs) (or expressed CYPs) and cofactors in the absence and the presence of voriconazole. Preincubation experiments were performed to test mechanism-based inactivation. In pilot experiments, voriconazole showed inhibition of CYP2B6, CYP2C9, CYP2C19, and CYP3A (half-maximal [50%] inhibitory concentrations, <6 μM); its effect on CYP1A2, CYP2A6, CYP2C8, and CYP2D6 was marginal (<25% inhibition at 100 μM voriconazole). Further detailed experiments with HLMs showed that voriconazole is a potent competitive inhibitor of CYP2B6 (Ki < 0.5), CYP2C9 (Ki = 2.79 μM), and CYP2C19 (Ki = 5.1 μM). The inhibition of CYP3A by voriconazole was explained by noncompetitive (Ki = 2.97 μM) and competitive (Ki = 0.66 μM) modes of inhibition. Prediction of the in vivo interaction of voriconazole from these in vitro data suggests that voriconazole would substantially increase the exposure of drugs metabolized by CYP2B6, CYP2C9, CYP2C19, and CYP3A. Clinicians should be aware of these interactions and monitor patients for adverse effects or failure of therapy.
Voriconazole, a derivative of fluconazole, belongs to the second generation of triazole antifungal drugs and has improved potency and a spectrum of antifungal activity that is expanded compared with the potency and activity of fluconazole (56, 59). Currently, orally or intravenously administered voriconazole is considered the first-line therapy for invasive aspergillosis (39, 56, 59). In addition, voriconazole is widely used for the management of patients infected with a broad range of other fungal pathogens, particularly patients who are intolerant of or who developed resistance to other conventional antifungal therapies (59).
However, despite its proven efficacy, the goal of optimal therapy with voriconazole is made difficult by the occurrence of clinically important drug-drug interactions. Several clinical studies and case reports have documented that voriconazole substantially reduces the clearance of several drugs, including warfarin (49), phenytoin (48), midazolam (53), diazepam (52), immunosuppressant drugs (cyclosporine, sirolimus, and tacrolimus) (46, 47), efavirenz (37), methadone (36), ibuprofen (27), diclofenac (26), fentanyl and alfentanil (54), oxycodone (22), and omeprazole (47). Considering the mechanisms of clearance of the drugs affected (3), many of these drug-drug interactions appear to be attributable to pharmacokinetic changes that can be understood in terms of inhibition of the cytochrome P450 (CYP) system. Indeed, in vitro studies by Niwa et al (41, 43) have documented that voriconazole inhibits CYPs 2C9, 2C19, and 3A, while its effect on the activity of other CYPs (CYPs 1A2, 2D6, and 2E1) was marginal. However, the mechanisms of inhibition were not addressed, and quantitative information that allows accurate prediction of voriconazole drug interactions in vivo was not provided in those studies because 50% inhibitory concentrations (IC50s) obtained by using only a single substrate probe concentration were estimated. In addition, some drug-metabolizing CYPs (e.g., CYP2A6, CYP2C8, and CYP2B6) have not been studied. Comprehensive inhibitory analyses that encompass all major drug-metabolizing CYPs are important because not all pharmacokinetic drug interactions involving voriconazole can be explained by the CYPs studied so far. For example, voriconazole has recently been shown to slow the elimination of efavirenz (37), a drug mainly metabolized by CYP2B6 (9, 66).
The purpose of the present study was to determine the inhibitory potency of voriconazole with eight different major drug-metabolizing CYP isoforms in vitro. Inhibition constants (Ki values) were estimated for those isoforms that were markedly inhibited by voriconazole in pilot experiments to guide predictions of drug interactions in vivo.
MATERIALS AND METHODS
Chemicals.
Voriconazole, efavirenz, 7-hydroxycoumarin, 8-hydroxyefavirenz, bupropion, 4-hydroxybupropion, ritonavir, midazolam, 1′-hydroxymidazolam, and desethylamodiaquine were purchased from Toronto Research Chemicals (North York, Ontario, Canada). Coumarin, glucose-6-phosphate, NADP, glucose-6-phosphate dehydrogenase, 8-methoxypsolaren, dextromethorphan, dextrorphan, chloroquine, desmethyldiazepam, phenacetin, and acetaminophen were purchased from Sigma-Aldrich (St. Louis, MO). Amodiaquine and levallorphan were purchased from the United States Pharmacopeia (Rockville, MD). S-Mephenytoin was purchased from Biomol (Plymouth, PA). All other chemicals and solvents were of high-performance liquid chromatography (HPLC) grade.
HLMs.
Human liver microsomal preparations (lot no. SD101, SD109) were purchased from Cellzdirect (Pittsboro, NC). HLMs (HL 09/14/99) were prepared from liver tissues medically unsuitable for transplantation by ultracentrifugation by standard protocols (8), and protein concentrations were determined by the method of Bradford (5) with bovine serum albumin as a standard. The microsomal pellets were suspended in a reaction buffer to a protein concentration of 10 mg/ml (stock). Insect cells in which human P450s were expressed by baculovirus (with oxidoreductase) were purchased from BD Biosciences (San Jose, CA). All microsomal preparations were stored at −80°C until they were used.
Experiments. (i) General incubation conditions.
The inhibitory effects of voriconazole on the activities of eight different CYP isoforms were studied with HLMs (and expressed CYPs, when required). The isoform-specific probe reactions used were phenacetin O-deethylation (CYP1A2) (58), coumarin 7-hydroxylation (CYP2A6) (45), efavirenz 8-hydroxylation (CYP2B6) (66), bupropion 4-hydroxylation (CYP2B6) (15), amodiquine N-deethylation (CYP2C8) (34), tolbutamide 4-methylhydroxylation (CYP2C9) (50), S-mephenytoin 4′-hydroxylation (CYP2C19) (68), dextromethorphan O-demethylation (CYP2D6) (6), and midazolam 1′-hydroxylation (CYP3A) (18, 62). Clearly, several probes have been used by different laboratories to characterize CYP-mediated in vitro drug interactions for a number of laboratory-specific reasons. The substrate probes used in this study were chosen because they have been well characterized and validated in our laboratory, as documented in our multiple previous publications (8, 10, 31). In addition, all these phenotyping probes except amodiaquine are listed as markers of first choice for the study of drug interactions in vitro in the FDA guidance for industry (17); amodiaquine was listed as an acceptable (second-choice) probe. Traditionally, paclitaxel has been used as the preferred probe for CYP2C8, but we preferred amodiaquine instead to minimize the influence of alternative metabolic pathways of paclitaxel on inhibition. The published assays described below (see “Specific enzyme assays”) were used after slight modification, when necessary, by conducting pilot experiments to optimize the conditions for incubation and/or HPLC.
By using incubation conditions specific to each isoform that were within the linear range for the velocity of the reaction (the incubation time as well as the substrate and protein concentrations), an incubation mixture that consisted of the substrate probe, HLMs, and phosphate reaction buffer (pH 7.4) was prewarmed for 5 min at 37°C without (control) and with multiple concentrations of voriconazole. Voriconazole was dissolved in methanol and was diluted with the same solvent to the required concentrations. Any methanol was removed by drying of the solution in a speed vacuum. The reaction was initiated by addition of an NADPH-generating system (1.3 mM NADP, 3.3 mM glucose-6-phosphate, 3.3 mM MgCl2, and 0.4 U/ml glucose-6-phosphate dehydrogenase) (final incubation volume, 250 μl). The incubations were conducted in duplicate. The reaction was allowed to proceed for the time specific for each isoform and was then terminated by placing the tubes on ice and immediately adding the appropriate reagent. After an internal standard was added, vortex mixed, and then centrifuged at 14,000 rpm for 5 min (Brinkmann Instruments, Westbury, NY), the supernatant was directly injected into an HPLC system (see below) or was first extracted and reconstituted in the mobile phase, from which an aliquot was injected into an HPLC system.
(ii) Analytical procedures.
The concentrations of the metabolites and internal standards were measured by HPLC and UV or fluorescence detection methods specific for each assay. The HPLC system consisted of a Waters (Milford, MA) model 515 pump, model 717 autosampler, and model 490 programmable absorbance UV detectors and a Spectrovision FD-300 dual monochromator fluorescence detector (Groton Technology, Concord, MA). The rates of production of each metabolite from the substrate probes were quantified by using the ratio of the area under the curve (AUC) for the metabolite to the AUC for each internal standard by using an appropriate standard curve generated with authentic metabolite standards.
(iii) Kinetic analysis.
Kinetic analysis was performed for each substrate probe reaction before initiation of the inhibition experiments with voriconazole, and the data generated were used as a guide for selection of the appropriate concentrations of the substrate probes in the subsequent inhibition experiments. Thus, the kinetic parameters for the metabolism of each probe substrate were determined by incubating increasing concentrations of the substrate (without the inhibitor) at 37°C in duplicate with HLMs and the NADPH-generating system. Phenacetin (5 to 500 μM), coumarin (0.1 to 50 μM), efavirenz (0.1 to 500 μM), bupropion (1 to 500 μM), amodiaquine (0.1 to 100 μM), tolbutamide (5 to 500 μM), S-mephenytoin (5 to 500 μM), dextromethorphan (1 to 200 μM), and midazolam (1 to 300 μM) were used. The rates of formation of metabolite versus the substrate concentrations were fit to appropriate enzyme kinetic models to estimate the apparent kinetic parameters. Km values and the maximum rate of metabolism (Vmax) were 43.8 μM and 84.9 pmol/min/mg protein, respectively, for CYP1A2 (high affinity component); 10.5 μM and 1,525 pmol/min/mg protein, respectively, for CYP2A6; 29.7 μM and 2,311 pmol/min/mg protein, respectively, for CYP2B6; 3.3 μM and 2,197 pmol/min/mg protein, respectively, for CYP2C8; 154.1 μM and 200 pmol/min/mg protein, respectively, for CYP2C9; 77.9 μM and 640 pmol/min/mg protein, respectively, for CYP2C19; 28.6 μM and 57 pmol/min/mg protein, respectively, for CYP2D6; 2.1 μM and 1,369 pmol/min/mg protein, respectively, for CYP3A-catalyzed midazolam 1′hydroxylation; and 30.6 μM and 789 pmol/min/mg protein, respectively, for CYP3A-catalyzed midazolam 4′-hydroxylation. These Km values broadly agree with values published in the literature: 22 to 46 μM for CYP1A2 (4), 17 to 23 μM for CYP2B6 (66), 2.4 μM for CYP2C8 (34), 164 to 371 μM for CYP2C9 (4), 13.6 to 56.5 μM for CYP2C19 (32), 8.6 to 23 μM for CYP2D6 (4, 32), 2 to 5 μM for CYP3A-catalyzed 1′hydroxylation, and 30 to 40 μM for CYP3A catalyzed 4-hydroxylation (18, 33, 61).
(iv) Inhibition studies for IC50 determination.
A pilot inhibitory analysis of each isoform was performed to determine the potency of inhibition and to select isoforms for further detailed study of their inhibition. A single isoform-specific substrate concentration at about the respective Km value (50 μM phenacetin, 10 μM coumarin, 10 μM efavirenz, 5 μM amodiaquine, 150 μM tolbutamide, 50 μM S-mephenytoin, 25 μM dextromethorphan, and 10 μM midazolam) was incubated at 37°C in duplicate with HLMs and the NADPH-generating system in the absence or the presence of a range of voriconazole concentrations (up to 100 μM). Processing of the incubation mixture and HPLC analysis of the metabolites formed were performed as described above.
Positive control experiments were run in parallel by incubating each probe substrate at 37°C in duplicate with HLMs and the NADPH-generating system in the absence (control) and the presence of the following isoform-specific inhibitors: furafylline (20 μM; specific for CYP1A2), pilocarpine (50 μM; specific for CYP2A6), thiotepa (50 μM; specific for CYP2B6) ticlopidine (5 μM; specific for CYP2B6), quercetin (10 μM; specific for CYP2C8), sulfaphenazole (25 μM; specific for CYP2C9), ticlopidine (5 μM; specific for CYP2C19), quinidine (1 μM; specific for CYP2D6), and ketoconazole (1 μM; specific for CYP3A). The substrate probes and concentrations that were used in the IC50 determination experiments (see above) were used for these positive control experiments. The specific conditions used with these inhibitors have been described in detail in earlier publications (4, 8, 10, 66). The rate of formation of the metabolite in the presence of the isoform-specific inhibitor was compared with that for controls in which the inhibitor was replaced with vehicle.
(v) Determination of Kis.
In pilot experiments (IC50 determination), we noted that voriconazole markedly inhibits CYP2B6, CYP2C9, CYP2C19, and CYP3A, while its effect on the remaining CYPs (CYPs 1A2, 2A6, 2C8, and 2D6) was minimal. Therefore, Dixon plots for the inhibition of CYP2B6, CYP2C9, CYP2C19, and CYP3A were determined by incubating the substrate probe at multiple concentrations without or with the test inhibitor at multiple concentrations with HLMs and cofactors. The inhibition data obtained from the pilot experiments were used as a guide to generate appropriate probe substrate and test inhibitor concentrations for the determination of the Ki values for each isoform. The isoform-specific probe substrate concentrations used were 10 to 75 μM efavirenz and bupropion for CYP2B6, 10 to 500 μM tolbutamide for CYP2C9, 10 to 75 μM S-mephenytoin for CYP2C19, and 5 to 25 μM midazolam for CYP3A. The voriconazole concentrations used were 0 to 25 μM.
Specific enzyme assays. (i) CYP1A2 activity (phenacetin O-deethylation to acetaminophen) (58).
HLMs (1 mg/ml), different concentrations of phenacetin, the NADPH-generating system, and phosphate reaction buffer (pH 7.4) were incubated at 37°C for 30 min in the absence or the presence of voriconazole. Details about the incubation conditions have been described elsewhere (32). Acetonitrile (100 μl) was used to terminate the reaction. Coumarin (100 μM) was used as an internal standard. After centrifugation (14,000 rpm), 100 μl of the supernatant was injected into an HPLC system. The HPLC eluates (acetaminophen and coumarin) were separated with an Alltech Spherisorb ODS-2 column (250 by 4.6 mm; particle size, 5 μm; Waters), a Luna C18 guard column (30 by 4.6 mm; particle size, 5 μm; Phenomenex, Torrance, CA), and a mobile phase composed of 85% 50 mM KH2PO4 (pH 4.5) (flow rate, 0.7 ml/min). Detection was performed with UV detection at 245 nm.
(ii) CYP2A6 activity (coumarin 7-hydroxylation to umbelliferone [7-hydroxycoumarin]) (45).
The rate of coumarin 7-hydroxylation was assayed by methods described previously (13, 20, 45), with minor modifications. Coumarin was dissolved in methanol (stock, 100 mM) and was serially diluted in distilled water to the required concentration (final methanol concentration, ≤0.1%). Coumarin, 0.5 mg/ml HLMs, the NADPH-generating system, and phosphate reaction buffer (pH 7.4) were incubated in the absence or the presence of voriconazole at 37°C for 15 min. The reaction was terminated by adding 100 μl of acetonitrile. After the internal standard, 8-methoxypsolaren (50 μl of 20 μg/ml), was added, the mixture was vortex mixed and centrifuged and an aliquot (150 μl) of the supernatant was transferred to HPLC vials; 100 μl of this aliquot was injected onto the HPLC system. The internal standard and 7-hydroxycoumarin were separated by using a Zorbax SB-C18 column (150 by 4.6 mm; particle size, 3.5 μm; Phenomenex), a Luna C18 guard column (30 by 4.6 mm; particle size, 5 μm; Phenomenex), and a mobile phase composed of 70% 10 mM KH2PO4 (pH 3.0) and 30% (vol/vol) acetonitrile (flow rate, 0.8 ml/min). The column eluate was monitored by UV detection at 280 nm (internal standard) or with a fluorescence detector at an excitation wavelength of 370 nm and an emission wavelength of 450 nm (7-hydroxycoumarin).
(iii) CYP2B6 activity (efavirenz 8-hydroxylation to 8-hydroxyefavirenz) (66).
The assay of efavirenz 8-hydroxylation was performed as described in our earlier publication (66), with a slight modification. A mixture of efavirenz (with or without inhibitors), HLMs (0.5 mg/ml) or expressed CYP2B6 (52 pmol/ml), and the NADPH-generating system was incubated at 37°C for 10 min. The reaction was terminated by addition of 0.5 ml acetonitrile. After an internal standard (ritonavir, 10 μg/ml) was added and the mixture was centrifuged at 14,000 rpm for 5 min, the supernatant was extracted with ethyl acetate under alkaline pH. The organic layer was removed, evaporated to dryness with a speed vacuum, and reconstituted with 150 μl of the mobile phase; 100 μl of the reconstituted organic layer was then injected into an HPLC system. The separation column was the same as that used for the CYP2A6 assay described above. The mobile phase was 55% 10 mM KH2PO4 (pH 2.4) and 45% (vol/vol) acetonitrile (flow rate, 0.8 ml/min). Detection was performed with UV detection at 245 nm.
Evidence exists that drug interactions involving CYP2B6 might be substrate dependent (7). To rule out substrate-specific differences in inhibition, experiments were performed with two additional CYP2B6-catalyzed substrate reactions: bupropion 4-hydroxylation to 4-hydroxybupropion (15) and 8-hydroxyefavirenz 14-hydroxylation to 8,14-dihydroxyefavirenz (66). To determine the IC50s, bupropion (50 μM) or 8-hydroxyefavirenz (5 μM) was incubated with HLMs and the NADPH-generating system in the absence or the presence of voriconazole (0 to 100 μM). Dixon plots were determined by using efavirenz and bupropion, as described above. The methods used for the incubation and assay of 8-hydroxyefavirenz 14-hydroxylase were the same as those described previously for efavirenz metabolism (66). The incubation conditions and sample processing for the bupropion assay were similar to those described above for the efavirenz metabolism assay, except that the incubation period was 15 min and the internal standard was acetaminophen (25 μl of 10 μg/ml). A symmetry C18 column (4.6 by 150 mm; particle size, 3.5 μm) with a guard column, a mobile phase that consisted of 85% 10 mM KH2PO4 (adjusted to pH 3.0 with 85% phosphoric acid) and that was delivered at 1.5 ml/min, and UV detection at 214 nm were used.
(iv) CYP2C8 activity (amodiaquine desethylation to desethylamodiaquine) (34).
A mixture of amodiaquine (with or without voriconazole), HLMs (0.1 mg/ml) or expressed CYP2C8 (52 pmol/ml), and the NADPH-generating system was incubated for 15 min. The enzyme activity assay was performed as described elsewhere (34, 44), with minor modifications. The reaction was terminated by addition of 100 μl acetonitrile, and chloroquine (50 μM) was added as an internal standard. The methods for further processing of the sample and the separation column were the same as those used in the CYP2A6 assay. We used a mobile phase that consisted of water, 16% (vol/vol) methanol, and 0.1% (vol/vol) triethylamine (pH 2.2); a flow rate of 1 ml/min; and UV detection at 340 nm.
(v) CYP2C9 activity (tolbutamide 4-hydroxylation to 4-hydroxytolbutamide) (50).
Tolbutamide (with or without voriconazole), HLMs (1 mg/ml), and the NADPH-generating system were incubated for 60 min at 37°C. Incubation and HPLC analysis were performed as described elsewhere (32), with slight modifications. After the reaction was terminated by addition of 100 μl 10% CClO4 and the internal standard, chlorpropamide (10 μg/ml), the sample was processed as described above for the CYP2A6 assay. The separation column was the same as that used for the CYP1A2 assay. A mobile phase that consisted of 70% 10 mM ammonium acetate (pH 5.4) and 30% (vol/vol) acetonitrile (flow rate, 0.7 ml/min) and UV detection at 230 nm were used.
(vi) CYP2C19 activity (S-mephenytoin 4′-hydroxylation to 4′-hydroxy-S-mephenytoin) (68).
S-Mephenytoin (with or without voriconazole), HLMs (SD101; 0.5 mg/ml), and the NADPH-generating system were incubated for 60 min at 37°C. The incubation and HPLC assay methods described by Ko et al. (32) were used, after slight modification. The reaction was terminated by addition of 100 μl acetonitrile. The sample was centrifuged after addition of phenytoin (5 μg/ml) as an internal standard, and the supernatant was extracted with dichloromethane (2 ml). The organic layer was removed and evaporated to dryness with a speed vacuum. The residue was reconstituted in mobile phase (75% 50 mM KH2PO4 [pH 4.0], 25% [vol/vol] acetonitrile), and 100 μl was injected into an HPLC system. The separation columns were the same as those used for the CYP1A2 assay. The mobile phase was delivered at 0.7 ml/min, and the column eluate was monitored by UV detection at 211 nm.
(vii) CYP2D6 activity (dextromethorphan O-demethylation to dextrorphan) (6).
Dextromethorphan (with or without voriconazole), HLMs (1 mg/ml), and the NADPH-generating system were incubated at 37°C for 30 min. A minor modification of a previously published dextromethorphan O-demethylation assay was used (32). The reaction was terminated by addition of 20 μl 60% CClO4. Levallorphan (40 μl of 16 μM) was added as an internal standard and was processed further as described above for the CYP2A6 assay. An aliquot of the supernatant (100 μl) was injected into the HPLC system, which consisted of the separation column described above for the CYP2A6 assay; the mobile phase was 10 mM KH2PO4 with 0.114% triethylamine (pH 4.0), 23% (vol/vol) acetonitrile, and 20% (vol/vol) methanol (flow rate, 0.5 ml); and fluorescence detection was performed at an excitation wavelength of 200 nm and an emission wavelength of 304 nm.
(viii) CYP3A activity (midazolam 1′-hydroxylation to 1′-hydroxylmidazolam) (18, 62).
The assay method described elsewhere (65) was slightly modified to assess the metabolism of midazolam. A mixture of midazolam (with or without inhibitors), HLMs (SD-109; 0.5 mg/ml), and the NADPH-generating system was incubated for 5 min at 37°C. The reaction was terminated by addition of 100 μl acetonitrile. Desmethyldiazepam (50 μl of 5 μg/ml) was added as an internal standard and was processed further as described above for the CYP2A6 assay. The supernatant (100 μl) was then injected into the HPLC system. Separation of 1′-hydroxymidazolam and the internal standard was performed with the columns described above for the CYP2A6 assay and a mobile phase consisting of 50% 10 mM ammonium acetate, 20% (vol/vol) methanol, and 30% (vol/vol) acetonitrile (pH 7.4) (flow rate, 1 ml/min). A UV detector set at 230 nm was used.
(ix) Mechanism-based inhibition screening.
The potential for voriconazole to cause mechanism-based inhibition was screened by using the following protocols. Voriconazole (10 μM) was preincubated in duplicate with HLM and phosphate reaction buffer (pH 7.4) (without or with the NADPH-generating system) in the absence of a substrate probe for 0 and 15 min at 37°C. The preincubation reaction was started by adding the NADPH-generating system. Controls were preincubated for 0 min without the test inhibitor and without the NADPH-generating system. The total volume of the preincubation mixture was 225 μl. After 0 and 15 min of preincubation, 25 μl of probe substrate was added at a final concentration corresponding to the Vmax, and the mixture was further incubated for the time specific for each assay. The reaction was stopped and processed as described above for the coincubation experiments.
Data analysis and prediction of in vivo data from in vitro data.
The rates of formation of metabolite versus the substrate concentrations were fit to appropriate enzyme kinetic models by using the WinNonlin (version 5.0) program (Pharsight, Mountain View, CA) to estimate the apparent kinetic parameters (Km and Vmax). IC50s were determined by analysis of the plot of the logarithm of the inhibitor concentration versus the percentage of activity remaining after inhibition by using the SigmaPlot (version 10.0) program (Systat Software, Point Richmond, CA). To calculate Ki values, the inhibition data were fit to different models of enzyme inhibition (competitive, noncompetitive, and uncompetitive) by nonlinear least-squares regression analysis with the WinNonlin (version 5.0) program (Pharsight). The appropriate type of model to be used for each data set was selected on the basis of visual inspection of Lineweaver-Burk, Dixon, and Eadie-Scatchard plots, as well as the size of the sum of squares of residuals, the Akaike information criterion, and Schwartz criterion values.
The anticipated in vivo drug interaction potential of voriconazole was estimated by using the following equation: AUCI /AUCUI = 1 + ([I]/Ki), where AUCI/AUCUI is the ratio of the AUC of the substrate after inhibition (AUCI) to the AUC of the uninhibited substrate (AUCUI), Ki is the in vitro inhibition constant, and [I] represents the therapeutic voriconazole concentration. For this purpose, the average maximum steady-state concentration in plasma (Cmax) and the trough concentration in plasma (Cmin) of voriconazole (total and unbound fractions) were used for [I]. Although voriconazole is administered at different doses or by different dosing schedules, we used the concentrations that are achieved after the administration of the most commonly employed therapeutic doses of voriconazole for adults (400 mg twice daily on the first day [loading doses] and then 200 mg twice daily on days 2 to 4). With this dosing schedule, the Cmax and Cmin are approximately 4 and 2 mg/liter, respectively (12), or ∼11.5 to 5.7 μM, respectively, for total concentrations and 4.8 to 2.4 μM, respectively, for the concentrations of the fraction unbound. The fraction unbound was calculated by assuming that voriconazole has a level of plasma protein binding of 58% (51). On the basis of the predicted in vivo changes, voriconazole's ability to alter (inhibit) the pharmacokinetics of the coadministered drugs was classified as strong (≥5-fold increase in AUC), moderate (≥2-fold but <5-fold increase in the AUC), or weak (≥1.25-fold but <2-fold increase in AUC), as proposed by the FDA (17; see http://www.fda.gov/Cder/drug/drugInteractions/, updated October 2008).
RESULTS
Estimation of IC50s.
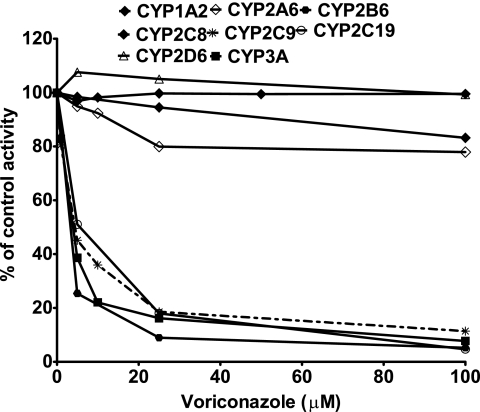
The inhibitory effects of multiple concentrations of voriconazole (0 to 100 μM) on the activity of each CYP isoform determined by the metabolism of a single concentration of isoform-specific probe were tested with HLMs (or expressed CYPs, when needed). Voriconazole showed potent inhibition of CYP2B6 (efavirenz 8-hydroxylation), CYP2C9 (tolbutamide 4′-hydroxylation), CYP2C19 (S-mephenytoin 4′-hydroxylation), and CYP3A (midazolam 1′-hydroxylation), with IC50s of 1.71 μM, 3.62 μM, 5.25 μM, and 2.90 μM, respectively. Relatively higher IC50s were previously reported: for CYP2C9, 8.4 μM when tolbutamide was used as a probe (42); for CYP2C19, 8.7 μM (42) or 32 μM (16) when S-mephenytoin was used as a probe; and for CYP3A, 50 μM when felodipine was used as a probe (16), 10.5 μM when nifedipine was used as a probe (42), 54 μM when testosterone was used as a probe (16), and 10.5 μM when midazolam was used as a probe (42). There is evidence that drug interactions involving CYP3A might be substrate dependent (29). An inhibitor might show inhibition when one substrate is used, while it may not inhibit (or may have a lower level of inhibition) when another substrate is used. Thus, it is often recommended that two structurally unrelated CYP3A4/CYP3A5 substrate probes be used to assess the potential for drug interactions involving this enzyme system (17). Using midazolam (the preferred substrate probe), we have shown potent inhibition at therapeutically relevant concentrations of voriconazole (see also below). Since these data are consistent with in vivo data showing that voriconazole is a potent inhibitor of chemically diverse CYP3A substrates, we do not believe that the differences in IC50s are due to the substrate probes used. Differences in experimental conditions most likely explain the differences in IC50s observed, particularly given that the estimation of IC50s is highly dependent on the substrate concentration used.
The inhibitory effect of voriconazole on the activity of CYP1A2, CYP2A6, CYP2C8, and CYP2D6 was negligible (by less than 25% at 100 μM voriconazole) (Fig. 1). The lack of inhibition of these isoforms by voriconazole is unlikely a result of the substrate probes used. For example, we used dextromethorphan as a probe for CYP2D6 in this study and found no inhibition, consistent with in vitro data obtained with debrisoquin (41) and bufuralol (16) as probes or in a clinical study of the voriconazole-venlafaxine interaction (25). We believe that both probes are good as in vitro probes, but dextromethorphan might be preferred because of its availability for in vivo studies, while special permission for the use of debrisoquin is needed. The extent of inhibition of CYPs by voriconazole was not modified during the preincubation experiments (data not shown). As expected, the activity of each isoform was substantially inhibited by the isoform-specific inhibitor, and the isoform-specific inhibitors served as positive controls (data not shown).
FIG. 1.
Inhibition of CYP isoforms by voriconazole in HLMs. A substrate probe at a single concentration was incubated with HLMs and cofactors in the absence (control) or the presence of voriconazole (0 to 100 μM) for times and with protein concentrations that were linear for the respective reaction described in detail in Materials and Methods. The specific concentrations of each substrate probe used are detailed in Materials and Methods. Each point represents the average of duplicate incubations.
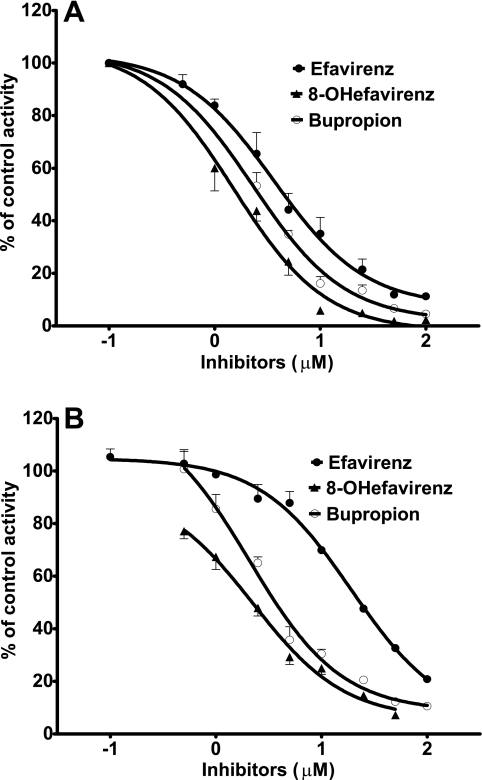
The initial goal of this study was to characterize the ability of voriconazole to inhibit CYP2B6 with the view toward finding an inhibitor probe that can be used in vivo to dissect the role of this enzyme in human drug metabolism. Thus, the interaction of voriconazole with CYP2B6 compared to its interaction with other CYPs was examined in more detail. In particular, since there is evidence that drug interactions involving CYP2B6 might be substrate dependent (7), IC50s were determined by using two additional CYP2B6-catalyzed reactions. Consistent with data obtained with efavirenz as a substrate in HLMs (IC50 = 1.71 μM), voriconazole showed potent inhibition of CYP2B6-mediated bupropion 4-hydroxylation (IC50 = 1.19 μM) and 8-hydroxyefavirenz 14-hydroxylation (IC50 = 0.79 μM) in HLMs (Fig. 2A). In expressed CYP2B6, this inhibitory effect of voriconazole was less pronounced when efavirenz, bupropion, and 8-hydroxyefavirenz were used as probe substrates (IC50s = 17.5 μM, 3.7 μM, and 1.9 μM, respectively) (Fig. 2B).
FIG. 2.
Inhibition of CYP2B6 by voriconazole in HLMs (A) and expressed CYP2B6 (B). Efavirez (10 μM), 8-hydroxyefavirenz (5 μM), or bupropion (50 μM) was incubated with HLMs (0.5 mg/ml) and cofactors in the absence or the presence of voriconazole (0 to 100 μM) for 10 min (efavirenz and 8-hydroxyefavirenz) or 15 min (bupropion). Each point represents the average of duplicate incubations.
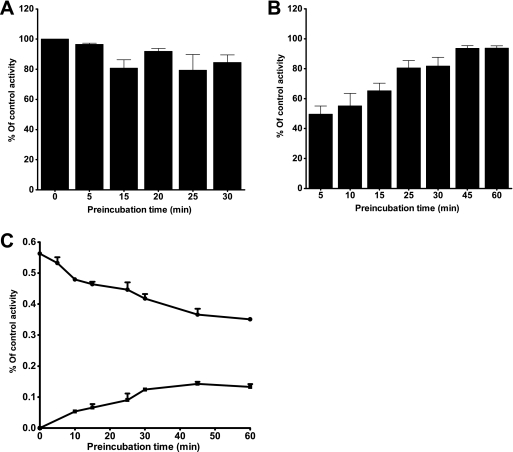
Further experiments were performed to determine the causes for the differential inhibitory potency of voriconazole in HLMs and by CYP2B6, particularly when efavirenz was used as a probe. One possibility is that this difference is due to the conversion of voriconazole in HLMs by CYPs other than CYP2B6 to a metabolite(s) that is a more potent inhibitor of CYP2B6. The main human metabolite of voriconazole is voriconazole N-oxide, which is mainly formed by CYP2C19 (and to some extent by CYP2C9 and CYP3A) (24), while a minor metabolite is hydroxyvoriconazole (40). Since we had no access to these metabolites to directly test their ability to inhibit CYP2B6, we conducted a series of experiments to better understand the difference in susceptibility to voriconazole inhibition between CYP2B6 and HLMs. First, voriconazole was preincubated with HLMs or expressed enzyme for 5, 15, 20, 25, and 30 min in the presence of an NADPH-generating system before efavirenz was added to the incubation mixture, and the activity remaining was compared with that of the control (preincubated for 0 min). We found no marked effect of the preincubation of voriconazole on its ability to inhibit CYP2B6 activity in HLMs (Fig. 3A). Second, the time course (0 min to 60 min) of efavirenz metabolism was determined in the absence or the presence (coincubation) of voriconazole. In this time course study, we measured the amount of 8-hydroxyefavirenz formed and showed that the potency of inhibition of efavirenz 8-hydroxylation by voriconazole was decreased with an increase in the duration of incubation (Fig. 3B). We also evaluated the disappearance of voriconazole and the formation of its main metabolite (voriconazole N-oxide) in the same incubations (Fig. 3C). The amount of voriconazole remaining in incubated microsomes decreased, while the amount of voriconazole formed increased with the incubation time (Fig. 3C). Third, we incubated expressed CYP2B6 in the absence and the presence of expressed CYP2C19. While the presence of expressed CYP2C19 increased the formation of voriconazole N-oxide, the inhibitory potency of voriconazole on CYP2B6 was not modified (IC50 = 12.2 μM). None of the data obtained from these experiments suggest that the more potent inhibition of CYP2B6 in HLMs is because of the voriconazole metabolites that are formed during incubation. Efavirenz 8-hydroxylation is predominantly catalyzed by CYP2B6 (66). However, although it is unlikely, the potent inhibition of efavirenz 8-hydroxylation in HLMs (but less so in expressed CYP2B6) by voriconazole may suggest the inhibition of CYPs in addition to CYP2B6 in efavirenz 8-hydroxylation in vitro.
FIG. 3.
Time-dependent inhibition of CYP2B6-catalyzed efavirenz 8-hydroxylation by voriconazole in HLMs. (A) Voriconazole (10 μM) was preincubated in duplicate with HLM and a phosphate reaction buffer (pH 7.4) (without or with the NADPH-generating system) in the absence or the presence of a substrate probe for 15 min at 37°C. The preincubation reaction was started by adding the NADPH-generating system. The controls were preincubated for 0 min without the test inhibitor and without the NADPH-generating system. The total volume of the preincubation mixture was 225 μl. After 0 and 15 min of preincubation, 25 μl of efavirenz (final concentration, 100 μM) was added and the mixture was further incubated for 10 min with cofactors. The reaction was stopped and processed as described for the coincubation experiments (see Materials and Methods). (B and C) Effects of duration of incubation on the degree of inhibition of efavirenz 8-hydroxylation by voriconazole in HLMs (B) and metabolism of voriconazole, as shown by parent compound depletion and metabolite formation (C). Efavirenz (10 μM) was incubated with HLMs (0.5 mg/ml) and cofactors in the absence or the presence of voriconazole (1 μM) for up to 60 min. Each point represents the average of duplicate incubations.
Estimation of Ki values.
While IC50s are qualitatively informative and help to address whether inhibition has occurred, their values are of limited use because they can be influenced by the substrate concentration selected, and it may not be accurate to use these parameters for the quantitative prediction drug interactions in vivo. Therefore, we performed additional experiments designed to estimate Ki values. The preliminary inhibition data generated from a single probe substrate reaction (Fig. 1) were used to simulate the appropriate range of substrate and inhibitor concentrations for use in the construction of Dixon plots for the inhibition of CYP isoforms by voriconazole in HLMs, from which precise Ki values were estimated.
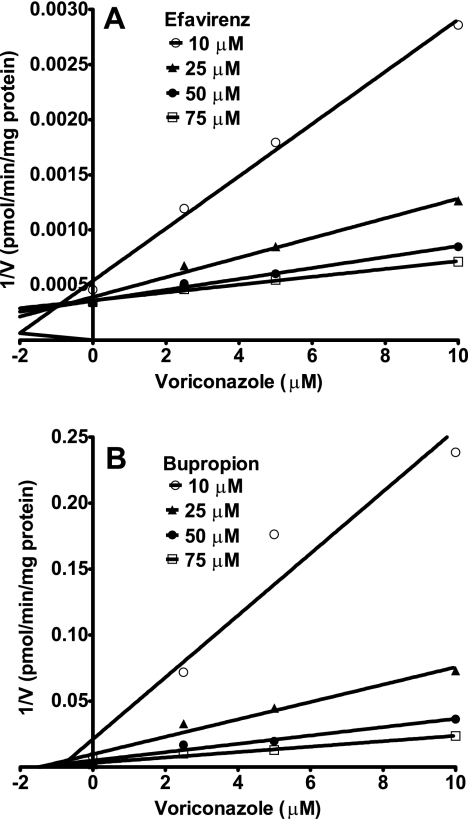
For CYP2B6, Ki values were determined by using efavirenz and bupropion as the probe substrates. Of all the CYPs tested, CYP2B6 was the most sensitive to voriconazole inhibition (Table 1). Representative Dixon plots for the inhibition of CYP2B6 in HLMs are shown in Fig. 4. Visual inspection of the Dixon plots and further analysis of the parameters of the enzyme inhibition models suggested that the inhibition data fit well to a competitive type of inhibition. The Ki values estimated by using a nonlinear regression model for competitive enzyme inhibition of CYP2B6-catalyzed efavirenz 8-hydroxylation and bupropion 4-hydroxylation in HLMs were less than 0.5 μM (Table 1).
TABLE 1.
Ki values of voriconazole for the inhibition of CYPs in HLMs and prediction of changes in AUC in vivo
| CYP | Ki value (μM) | Predicted fold change in AUC ata:
|
Experimental conditions and observed fold change in AUC
|
Reference | |||
|---|---|---|---|---|---|---|---|
| Total [I] (Cmin-Cmax) | Free (fu) [I] (Cmin-Cmax)b | Voriconazole dose | Substrate (dose) | Fold change | |||
| CYP2B6 | 0.40 ± 0.1e | 16-32 | 7.5-14 | 400 mg BIDc on day 1, 200 mg BID on days 2-3 | Efavirenz (400 mg/day) | 1.44 | 37 |
| 0.34 ± 0.1f | 400 mg BID on day 1, 200 mg BID on days 2-3 | S-Methadone (30-100 mg/day racemic methadone orally) | 2.2 | 36 | |||
| CYP2C9 | 2.8 ± 0.2 | 3.0-5.1 | 1.9-2.71 | 400 mg/day for 10 days | Phenytoin (300 mg/day orally) | 1.9 | 48 |
| 400 mg BID on day 1, 200 mg BID on day 2 | Diclofenac (50 mg, single dose) | 1.8 | 26 | ||||
| 400 mg BID on day 1, 200 mg BID on day 2 | S-Ibuprofen (400 mg, single oral dose) | 2.0 | 27 | ||||
| 300 mg/days for 12 days | Warfarin (30 mg, single dose) | ∼2d | 49 | ||||
| CYP2C19 | 5.1 ± 0.3 | 2.1-3.3 | 1.5-1.9 | 400 mg BID on day 1, 200 mg BID on days 2-6 | Omeprazole (40 mg/day for 7 days) | 4.0 | 47 |
| 200 mg BID | Omeprazole (40 mg/day) | 2.8 | 16 | ||||
| CYP3A | 2.97 ± 0.2g | 2.9-4.8 | 1.8-2.6 | 400 mg BID on day 1, 200 mg BID on day 2 | Midazolam (7.5 mg orally) | 10.3 | 53 |
| 0.66 ± 0.2h | 9.6-18.4 | 4.6-8.3 | 400 mg BID on day 1, 200 mg BID on day 2 | Midazolam (0.05 mg/kg of body weight intravenously) | 3.8 | 53 | |
The average plasma voriconazole concentrations after administration of the usual therapeutic dose (400 mg every 12 h on day 1 and then 200 mg every 12 h for days 2 and 4) were approximately 4 μg/ml (11.5 μM) at Cmax and 2 μg/ml (5.7 μM) at Cmin (12), and these concentrations were used. Cmax and Cmin values are in micromolar.
The fraction unbound (fu) (2.4 to 4.8 μM for Cmin and Cmax, respectively) was calculated on the basis of 58% plasma protein binding of voriconazole, which was independent of the dose or the plasma concentrations (51). The change in the AUC was calculated by using the formula described in the text (see “Data analysis and prediction of in vivo data from in vitro data”), assuming competitive and noncompetitive modes of inhibition.
BID, twice a day.
Indicates the fold change in pharmacodynamics.
Efavirenz as a substrate.
Bupropion as a substrate.
Ki for inhibition of midazolam 1′-hydroxylation derived from noncompetitive model.
Ki for inhibition of midazolam 1′-hydroxylation derived from competitive model.
FIG. 4.
Dixon plots for the inhibition of efavirenz 8-hydroxylation (A) and bupropion 4-hydroxylation (B) by voriconazole in HLMs. Efavirnez (10 to 75 μM) or bupropion (10 to 75 μM) was incubated with HLMs (0.5 mg/ml; HL-09/14/99) and the NADPH-generating system at 37°C for 10 min (efavirenz) or 15 min (bupropion) without or with voriconazole (2.5 to 10 μM). Each point represents the mean of duplicate incubations.
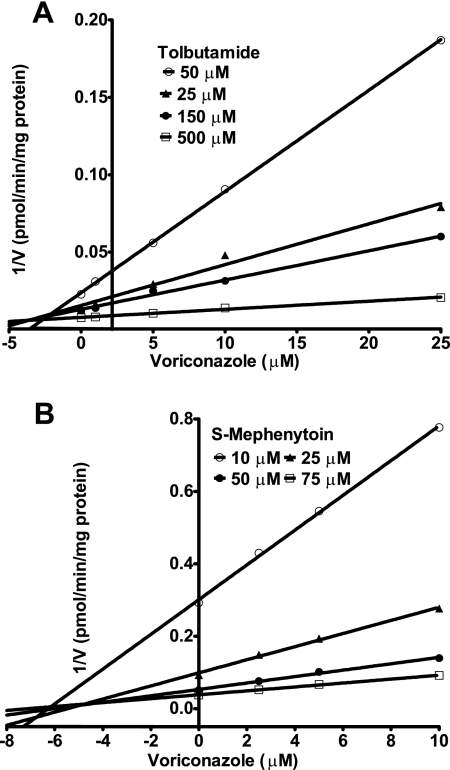
Figure 5 shows Dixon plots for the inhibition of CYP2C9 (Fig. 5A) and CYP2C19 (Fig. 5B) by voriconazole in HLMs. Voriconazole inhibited CYP2C9 and CYP2C19 competitively, with estimated Ki values of 2.79 μM and 5.07 μM, respectively (Table 1).
FIG. 5.
Dixon plots for the inhibition of CYP2C9-catalyzed tolbutamide 4-methylhydroxylation (A) and CYP2C19-catalyzed S-mephenytoin 4′-hydroxylation (B) by voriconazole in HLMs. Each substrate probe was incubated with HLMs and cofactors without or with a range of voriconazole concentrations at 37°C for 60 min. Tolbutamide (50 to 500 μM), HLMs (1 mg/ml; HL-09/14/99), and voriconazole (1 to 25 μM) were used for the CYP2C9 assay; and S-mephenytoin (10 to 75 μM), HLMs (0.5 mg/ml; SD-101), and voriconazole (2.5 to 10 μM) were used for the CYP2C19 assay. Each point represents the mean of duplicate measurements.
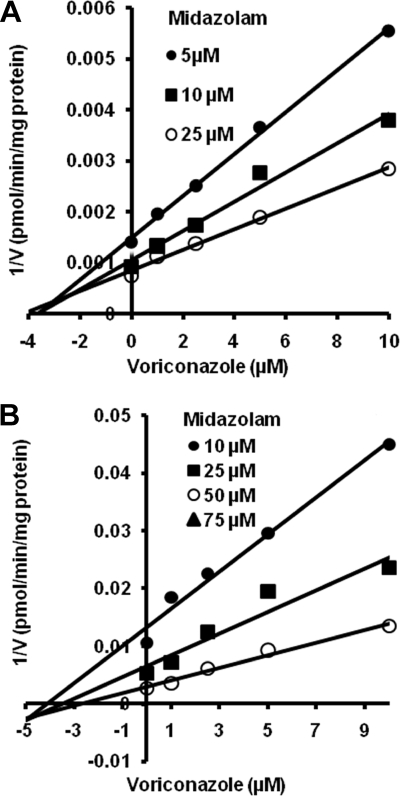
As shown in Fig. 6, the inhibition of CYP3A-catalyzed midazolam 1′-hydroxylation (Fig. 6A) and 4′-hydroxylation (Fig. 6B) by voriconazole fit well to both noncompetitive and competitive enzyme inhibition models. The Ki values estimated from a noncompetitive inhibition equation and a competitive inhibition equation were less than 3 μM and 1 μM, respectively (Table 1), while the Ki value derived for inhibition of midazolam 4-hydroxylation by noncompetitive inhibition model was 2.9 ± 1.3 μM. To further understand the mechanism by which voriconazole inhibits the activity of CYP3A, voriconazole was preincubated in the presence of an NADPH-generating system and HLMs before initiation of the reaction by the addition of midazolam. Preincubation with voriconazole did not enhance its potency toward the inhibition of CYP3A activity (data not shown), suggesting that the noncompetitive type of inhibition observed during coincubation is unlikely due to a time-dependent inactivation of CYP3A by the inhibitor.
FIG. 6.
Dixon plots for the inhibition of midazolam 1′-hydroxylation (A) and midazolam 4-hydroxylation (B) by voriconazole in HLMs. Midazolam (5 to 25 μM) was incubated without or with voriconazole (1 to 10 μM) with HLMs (0.5 mg/ml, SD-109) and the NADPH-generating system at 37°C for 5 min. Each point represents the mean of duplicate incubations.
DISCUSSION
In the present study, (i) we have shown for the first time that voriconazole is a highly potent inhibitor of CYP2B6 and that its effect on CYP2A6 and CYP2C8 activity is marginal; and (ii) we have confirmed previous findings (16, 42) that voriconazole is a potent inhibitor of CYP2C9, CYP2C19, and CYP3A and also reported Ki values that can be used to predict the magnitude of drug interactions in vivo. On the basis of our in vitro data, voriconazole would be predicted to slow the elimination of drugs predominantly cleared by CYP2B6, CYP2C9, CYP2C19, and CYP3A or by a combination of these CYPs (Table 1). These data help to explain the multiple voriconazole drug interactions that have been reported in the literature and can be used to predict and to identify new drug-drug interactions. Our data and findings from other in vitro studies (16, 41) and in vivo studies (25, 27) suggest that it is unlikely that voriconazole alters the pharmacokinetics of drugs metabolized by CYP1A2, CYP2A6, CYP2C8, CYP2D6, and CYP2E1.
Of the eight CYPs studied, CYP2B6 was most sensitive to inhibition by voriconazole (Ki < 0.5 μM) (Table 1). Although findings from in vivo studies are generally limited, our in vitro data are consistent with the findings of in vivo studies that showed that voriconazole interacts with certain CYP2B6 substrates. Voriconazole has recently been reported to significantly slow the elimination of efavirenz in healthy volunteers (37). Although the authors of that study (37) suggested that CYP3A was the mechanism for this interaction, the available in vitro evidence (9, 66) and in vivo evidence (21, 38, 69; Bristol-Myers Squibb Company, efavirenz [Sustiva] package insert, updated March 2008) strongly support the conclusion that CYP2B6 (but not CYP3A) is important in the clearance of efavirenz and that inhibition of CYP2B6 is the main mechanism by which voriconazole increases efavirenz exposure in humans (37). Voriconazole has also been shown to increase the level of exposure to methadone, particularly that to S-methadone (36), an enantiomer whose clearance appears to be more dependent on CYP2B6 activity than the clearance of R-methadone is (63). On the basis of the in vitro Ki values, voriconazole is predicted to be a strong inhibitor of CYP2B6 and is expected to substantially alter the pharmacokinetics of drugs predominantly metabolized by this enzyme (16- to 32-fold and 7.5- to 14-fold changes in the AUC when the total and unbound steady-state Cmin and Cmax of voriconazole, respectively, are considered) (Table 1). However, in the steady-state efavirenz-voriconazole interaction (37), the actual change in efavirenz exposure observed after voriconazole was much lower than what has been predicted from our in vitro data (Table 1). This discrepancy may be due to a bidirectional drug interaction, because efavirenz was shown in the same study (37) to substantially decrease the level of voriconazole exposure; the Cmin and the Cmax of voriconazole were reduced to ∼0.2 to 1.15 mg/ml, respectively, which are equivalent to 0.33 to 1.99 μM, respectively (unbound concentrations, 0.139 to 0.836 μM, respectively). If the concentrations of voriconazole obtained after induction are taken into account in the determination of [I], the predicted AUC changes (1.4- to 6.4-fold) will be within the clinically observed range (37) (Table 1). These data emphasize that the estimation of [I] should take bidirectional drug interactions into account, particularly when those interactions involve the induction of voriconazole metabolism, and that the intrinsic ability of voriconazole to alter the pharmacokinetics of substrate drugs primarily metabolized by CYP2B6 in a noninduced state is likely to be greater than what has been reported for efavirenz (37) and methadone (36) because of the induction of voriconazole metabolism by the former and the involvement of multiple enzymes in methadone elimination (14, 63). Although CYP2B6 has been studied less than the other CYPs, a growing list of drugs have been identified as substrates of CYP2B6 (64). Therefore, the inhibition of CYP2B6 by voriconazole is likely to be clinically important.
The ability of voriconazole to slow the elimination of CYP3A substrates in vivo, e.g., immunosuppressant drugs (cyclosporine, sirolimus, and tacrolimus) (46, 47), fentanyl and alfentanil (55), oxycodone (22), etoricoxib (28), and midazolam (53), has been well documented. Using the most reliable standard substrate for evaluation of the in vivo inhibition of CYP3A, midazolam (19), we found that voriconazole is a strong inhibitor of CYP3A (IC50 = 2.90 μM; Ki = 0.66 μM for the competitive enzyme inhibition model and Ki = 2.97 μM for the noncompetitive enzyme inhibition model) (Table 1). In other studies, IC50s for the inhibition of the CYP3A-catalyzed metabolism of nifedipine, felodipine, and testosterone by voriconazole were reported to be relatively high (10.5, 50, and 54 μM, respectively) (16, 42). Although the difference in inhibition constants may reflect in part the substrate-dependent interaction of CYP3A substrates with voriconazole, as this possibility has been reported previously (29), this seems unlikely given that voriconazole alters the in vivo elimination of several structurally diverse CYP3A substrates. The discrepancy could simply reflect differences in experimental conditions. In clinical studies, voriconazole increases the levels of exposure to midazolam administered orally and intravenously by 10.3-fold and 3.7-fold, respectively (53). When the Ki values derived by using the noncompetitive model were used to predict the change in AUC in vivo, the changes in AUC observed after the oral administration of midazolam were much higher than those predicted from our in vitro data (2.9- to 4.9-fold changes in the midazolam AUC when the total Cmin and Cmax of voriconazole were used and 1.8- to 2.6-fold changes in the midazolam AUC when the fraction unbound is considered); the values observed after the intravenous administration of midazolam (53) were consistent with our predicted values (Table 1). As no differences in the potency of inhibition between the preincubation and the coincubation experiments were observed, this observation is not due to the contribution of time-dependent inhibition. The possibility that circulating oxidative metabolites of voriconazole (24, 40) might contribute to the in vivo inhibition of CYP3A cannot be excluded. Also, as CYP3A is the main enzyme catalyzing midazolam not only in the liver but also in the gut wall (19, 53, 60), it is likely that the greater interaction observed in vivo after the oral administration of midazolam (53) can be attributed to the inhibition of CYP3A by voriconazole at both of these sites (53). Alternatively, in vivo inhibition might occur predominantly through competitive inhibition. Using the competitive inhibition model, we predicted 9.6- to 18.4-fold changes in the AUC when total Cmin and Cmax were considered and 4.6- to 8.3-fold when the Cmin and the Cmax for the fraction unbound were used. These values are broadly consistent with the AUC changes that have been observed previously (53). Together, these data indicate that voriconazole can be considered a strong inhibitor of CYP3A, an enzyme system that is abundant in the gut wall and the liver and that is involved in the metabolism of more than 50% of those clinically used drugs cleared by oxidation (67).
The Ki value for the inhibition of CYP2C9 by voriconazole in HLMs was 2.79 μM. On the basis of these data, we would expect changes in the AUC of 3- to 5-fold for the CYP2C9 substrate during voriconazole administration when the total Cmin and the Cmax of voriconazole are considered and 1.9- to 2.7-fold when the Cmin and the Cmax of the fraction unbound are considered. Consistent with our in vitro data and the predicted values, voriconazole has been shown to increase (1.8- to 2-fold) the AUCs of known CYP2C9 substrates (30) (Table 1). Approximately 100 clinically used drugs are metabolized by CYP2C9 (30). Although voriconazole can be categorized as a weak inhibitor of CYP2C9, it may have clinically important consequences for some substrates with narrow therapeutic ranges (e.g., warfarin, phenytoin, and sulfonylureas).
CYP2C19 is responsible for the metabolic detoxification or activation of a number of clinically important drugs (11). Our data show that voriconazole is a moderate inhibitor of CYP2C19 (IC50 = 5.25 μM, Ki = 5.07 μM), consistent with the findings of a previous in vitro study, which reported an IC50 of 8.7 μM (42); in contrast, another study (16) reported a higher IC50 (32 μM) when the same substrate probe was used. We predicted changes in the AUC of 2.1- to 3.3-fold for CYP2C19 substrates predominantly metabolized by the enzyme when the total Cmin and Cmax of voriconazole were used and 1.5- to 2-fold when the Cmin and the Cmax of the fraction unbound were considered (Table 1). According to information provided in the package insert, the level of omeprazole exposure was increased fourfold by voriconazole. Since omeprazole is predominantly cleared by CYP2C19 and to some extent by CYP3A (1, 2), the fact that the observed effect of voriconazole was relatively higher than our predicted effect probably reflects a net effect of voriconazole on both of these enzymes. While voriconazole is expected to moderately alter the exposure of drugs primarily cleared by CYP2C19 (11), the magnitude of the interaction may be greater if alternative metabolic pathways are inhibited simultaneously. In addition, the clinical impact of voriconazole might be greater when it is coprescribed with prodrugs requiring CYP2C19-mediated metabolic activation, e.g., clopidogrel (23), cyclophosphamide (57), and thalidomide (35).
In summary, we have shown that voriconazole is a potent inhibitor of CYP2B6, CYP2C9, CYP2C19, and CYP3A at clinically relevant concentrations. Our in vitro data broadly predict clinically important drug interactions of voriconazole with substrates of these enzymes, and there is convincing evidence that this is the case in clinical studies. Together, these four enzymes are the clearance mechanisms for ∼67% of the drugs currently marketed (70). Given that the number of patients requiring voriconazole is on the increase and that these patients are often seriously ill, the likelihood that voriconazole will be coprescribed with drugs that interact and thus elicit frequent and severe adverse drug interactions in the population is very high. However, it is important to note that the extent of drug interactions with this drug varies greatly among individuals, because the level of voriconazole exposure after the administration of therapeutic doses exhibits large intersubject variability due to factors that include CYP2C19 genetic polymorphisms, nonlinear pharmacokinetics at different doses, the duration of treatment, the routes of administration, the level of hepatic impairment, the presence of inflammatory conditions, age, and the concurrent administration of substrate drugs that reduce or enhance the clearance of voriconazole (e.g., efavirenz, carbamazepine, ritonavir) (37, 47). In addition, the simultaneous inhibition of alternative routes of metabolism of substrates (e.g., omeprazole) (47) or the simultaneous inhibition of gut wall and hepatic metabolism (e.g., CYP3A substrates) versus the inhibition of hepatic metabolism after drug administration by the intravenous route (inhibition only at the hepatic site) (e.g., midazolam) (53) could influence the extent of the interaction with voriconazole.
Acknowledgments
The study was supported by an R01 grant (grant 5R01GM078501-02), an R56 grant (grant 2R56GM067308-09A1), and a clinical pharmacology training grant (grant T32GM008425) from the National Institute of General Medical Sciences, National Institutes of Health (Bethesda, MD). S. Jeong received a fellowship award from the Research Institute of Medical Science of Chonnam National University, Gwangju, South Korea.
Footnotes
Published ahead of print on 24 November 2008.
REFERENCES
- 1.Abelo, A., T. B. Andersson, M. Antonsson, A. K. Naudot, I. Skanberg, and L. Weidolf. 2000. Stereoselective metabolism of omeprazole by human cytochrome P450 enzymes. Drug Metab. Dispos. 28:966-972. [PubMed] [Google Scholar]
- 2.Andersson, T. 1996. Pharmacokinetics, metabolism and interactions of acid pump inhibitors. Focus on omeprazole, lansoprazole and pantoprazole. Clin. Pharmacokinet. 31:9-28. [DOI] [PubMed] [Google Scholar]
- 3.Bertz, R. J., and G. R. Granneman. 1997. Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin. Pharmacokinet. 32:210-258. [DOI] [PubMed] [Google Scholar]
- 4.Bourrie, M., V. Meunier, Y. Berger, and G. Fabre. 1996. Cytochrome P450 isoform inhibitors as a tool for the investigation of metabolic reactions catalyzed by human liver microsomes. J. Pharmacol. Exp. Ther. 277:321-332. [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Broly, F., C. Libersa, M. Lhermitte, P. Bechtel, and B. Dupuis. 1989. Effect of quinidine on the dextromethorphan O-demethylase activity of microsomal fractions from human liver. Br. J. Clin. Pharmacol. 28:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bumpus, N. N., C. Sridar, U. M. Kent, and P. F. Hollenberg. 2005. The naturally occurring cytochrome P450 (P450) 2B6 K262R mutant of P450 2B6 exhibits alterations in substrate metabolism and inactivation. Drug Metab. Dispos. 33:795-802. [DOI] [PubMed] [Google Scholar]
- 8.Desta, Z., T. Kerbusch, N. Soukhova, E. Richard, J. W. Ko, and D. A. Flockhart. 1998. Identification and characterization of human cytochrome P450 isoforms interacting with pimozide. J. Pharmacol. Exp. Ther. 285:428-437. [PubMed] [Google Scholar]
- 9.Desta, Z., T. Saussele, B. Ward, J. Blievernicht, L. Li, K. Klein, D. A. Flockhart, and U. M. Zanger. 2007. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8:547-558. [DOI] [PubMed] [Google Scholar]
- 10.Desta, Z., N. V. Soukhova, and D. A. Flockhart. 2001. Inhibition of cytochrome P450 (CYP450) isoforms by isoniazid: potent inhibition of CYP2C19 and CYP3A. Antimicrob. Agents Chemother. 45:382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desta, Z., X. Zhao, J. G. Shin, and D. A. Flockhart. 2002. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin. Pharmacokinet. 41:913-958. [DOI] [PubMed] [Google Scholar]
- 12.Dowell, J. A., J. Schranz, A. Baruch, and G. Foster. 2005. Safety and pharmacokinetics of coadministered voriconazole and anidulafungin. J. Clin. Pharmacol. 45:1373-1382. [DOI] [PubMed] [Google Scholar]
- 13.Draper, A. J., A. Madan, and A. Parkinson. 1997. Inhibition of coumarin 7-hydroxylase activity in human liver microsomes. Arch. Biochem. Biophys. 341:47-61. [DOI] [PubMed] [Google Scholar]
- 14.Eap, C. B., T. Buclin, and P. Baumann. 2002. Interindividual variability of the clinical pharmacokinetics of methadone: implications for the treatment of opioid dependence. Clin. Pharmacokinet. 41:1153-1193. [DOI] [PubMed] [Google Scholar]
- 15.Faucette, S. R., R. L. Hawke, E. L. Lecluyse, S. S. Shord, B. Yan, R. M. Laethem, and C. M. Lindley. 2000. Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab. Dispos. 28:1222-1230. [PubMed] [Google Scholar]
- 16.FDA Antiviral Drugs Advisory Committee. 2001. Briefing document for Voriconazole. 4 October 2001. FDA, Rockville, MD.
- 17.FDA (Clinical Pharmacology). 2006. Guidance for industry: drug interaction studies—study design, data analysis, and implications for dosing and labeling. FDA, Rockville, MD. [DOI] [PubMed]
- 18.Gorski, J. C., S. D. Hall, D. R. Jones, M. Vandenbranden, and S. A. Wrighton. 1994. Regioselective biotransformation of midazolam by members of the human cytochrome P450 3A (CYP3A) subfamily. Biochem. Pharmacol. 47:1643-1653. [DOI] [PubMed] [Google Scholar]
- 19.Gorski, J. C., S. Vannaprasaht, M. A. Hamman, W. T. Ambrosius, M. A. Bruce, B. Haehner-Daniels, and S. D. Hall. 2003. The effect of age, sex, and rifampin administration on intestinal and hepatic cytochrome P450 3A activity. Clin. Pharmacol. Ther. 74:275-287. [DOI] [PubMed] [Google Scholar]
- 20.Greenlee, W. F., and A. Poland. 1978. An improved assay of 7-ethoxycoumarin O-deethylase activity: induction of hepatic enzyme activity in C57BL/6J and DBA/2J mice by phenobarbital, 3-methylcholanthrene and 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Pharmacol. Exp. Ther. 205:596-605. [PubMed] [Google Scholar]
- 21.Haas, D. W., H. J. Ribaudo, R. B. Kim, C. Tierney, G. R. Wilkinson, R. M. Gulick, D. B. Clifford, T. Hulgan, C. Marzolini, and E. P. Acosta. 2004. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS 18:2391-2400. [PubMed] [Google Scholar]
- 22.Hagelberg, N. M., T. H. Nieminen, T. I. Saari, M. Neuvonen, P. J. Neuvonen, K. Laine, and K. T. Olkkola. 3 October 2008. Voriconazole drastically increases exposure to oral oxycodone. Eur. J. Clin. Pharmacol. [Epub ahead of print.] [DOI] [PubMed]
- 23.Hulot, J. S., A. Bura, E. Villard, M. Azizi, V. Remones, C. Goyenvalle, M. Aiach, P. Lechat, and P. Gaussem. 2006. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood 108:2244-2247. [DOI] [PubMed] [Google Scholar]
- 24.Hyland, R., B. C. Jones, and D. A. Smith. 2003. Identification of the cytochrome P450 enzymes involved in the N-oxidation of voriconazole. Drug Metab. Dispos. 31:540-547. [DOI] [PubMed] [Google Scholar]
- 25.Hynninen, V. V., K. T. Olkkola, L. Bertilsson, K. Kurkinen, P. J. Neuvonen, and K. Laine. 2008. Effect of terbinafine and voriconazole on the pharmacokinetics of the antidepressant venlafaxine. Clin. Pharmacol. Ther. 83:342-348. [DOI] [PubMed] [Google Scholar]
- 26.Hynninen, V. V., K. T. Olkkola, K. Leino, S. Lundgren, P. J. Neuvonen, A. Rane, M. Valtonen, and K. Laine. 2007. Effect of voriconazole on the pharmacokinetics of diclofenac. Fundam. Clin. Pharmacol. 21:651-656. [DOI] [PubMed] [Google Scholar]
- 27.Hynninen, V. V., K. T. Olkkola, K. Leino, S. Lundgren, P. J. Neuvonen, A. Rane, M. Valtonen, H. Vyyrylainen, and K. Laine. 2006. Effects of the antifungals voriconazole and fluconazole on the pharmacokinetics of S-(+)- and R-(−)-ibuprofen. Antimicrob. Agents Chemother. 50:1967-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hynninen, V. V., K. T. Olkkola, P. J. Neuvonen, and K. Laine. 9 September 2008. Oral voriconazole and miconazole oral gel produce comparable effects on the pharmacokinetics and pharmacodynamics of etoricoxib. Eur. J. Clin. Pharmacol. [Epub ahead of print.] [DOI] [PubMed]
- 29.Kenworthy, K. E., J. C. Bloomer, S. E. Clarke, and J. B. Houston. 1999. CYP3A4 drug interactions: correlation of 10 in vitro probe substrates. Br. J. Clin. Pharmacol. 48:716-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirchheiner, J., and J. Brockmoller. 2005. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin. Pharmacol. Ther. 77:1-16. [DOI] [PubMed] [Google Scholar]
- 31.Ko, J. W., Z. Desta, N. V. Soukhova, T. Tracy, and D. A. Flockhart. 2000. In vitro inhibition of the cytochrome P450 (CYP450) system by the antiplatelet drug ticlopidine: potent effect on CYP2C19 and CYP2D6. Br. J. Clin. Pharmacol. 49:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko, J. W., N. Sukhova, D. Thacker, P. Chen, and D. A. Flockhart. 1997. Evaluation of omeprazole and lansoprazole as inhibitors of cytochrome P450 isoforms. Drug Metab. Dispos. 25:853-862. [PubMed] [Google Scholar]
- 33.Kronbach, T., D. Mathys, M. Umeno, F. J. Gonzalez, and U. A. Meyer. 1989. Oxidation of midazolam and triazolam by human liver cytochrome P450IIIA4. Mol. Pharmacol. 36:89-96. [PubMed] [Google Scholar]
- 34.Li, X. Q., A. Bjorkman, T. B. Andersson, M. Ridderstrom, and C. M. Masimirembwa. 2002. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8: a new high affinity and turnover enzyme-specific probe substrate. J. Pharmacol. Exp. Ther. 300:399-407. [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., J. Hou, H. Jiang, D. Wang, W. Fu, Z. Yuan, Y. Chen, and L. Zhou. 2007. Polymorphisms of CYP2C19 gene are associated with the efficacy of thalidomide based regimens in multiple myeloma. Haematologica 92:1246-1249. [DOI] [PubMed] [Google Scholar]
- 36.Liu, P., G. Foster, R. Labadie, E. Somoza, and A. Sharma. 2007. Pharmacokinetic interaction between voriconazole and methadone at steady state in patients on methadone therapy. Antimicrob. Agents Chemother. 51:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, P., G. Foster, R. R. LaBadie, M. J. Gutierrez, and A. Sharma. 2008. Pharmacokinetic interaction between voriconazole and efavirenz at steady state in healthy male subjects. J. Clin. Pharmacol. 48:73-84. [DOI] [PubMed] [Google Scholar]
- 38.Mouly, S., K. S. Lown, D. Kornhauser, J. L. Joseph, W. D. Fiske, I. H. Benedek, and P. B. Watkins. 2002. Hepatic but not intestinal CYP3A4 displays dose-dependent induction by efavirenz in humans. Clin. Pharmacol. Ther. 72:1-9. [DOI] [PubMed] [Google Scholar]
- 39.Muijsers, R. B., K. L. Goa, and L. J. Scott. 2002. Voriconazole: in the treatment of invasive aspergillosis. Drugs 62:2655-2664. [DOI] [PubMed] [Google Scholar]
- 40.Murayama, N., N. Imai, T. Nakane, M. Shimizu, and H. Yamazaki. 2007. Roles of CYP3A4 and CYP2C19 in methyl hydroxylated and N-oxidized metabolite formation from voriconazole, a new anti-fungal agent, in human liver microsomes. Biochem. Pharmacol. 73:2020-2026. [DOI] [PubMed] [Google Scholar]
- 41.Niwa, T., S. Inoue-Yamamoto, T. Shiraga, and A. Takagi. 2005. Effect of antifungal drugs on cytochrome P450 (CYP) 1A2, CYP2D6, and CYP2E1 activities in human liver microsomes. Biol. Pharm. Bull. 28:1813-1816. [DOI] [PubMed] [Google Scholar]
- 42.Niwa, T., T. Shiraga, and A. Takagi. 2005. Effect of antifungal drugs on cytochrome P450 (CYP) 2C9, CYP2C19, and CYP3A4 activities in human liver microsomes. Biol. Pharm. Bull. 28:1805-1808. [DOI] [PubMed] [Google Scholar]
- 43.Niwa, T., T. Shiraga, and A. Takagi. 2005. Drug-drug interaction of antifungal drugs. Yakugaku Zasshi 125:795-805. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 44.Parikh, S., J. B. Ouedraogo, J. A. Goldstein, P. J. Rosenthal, and D. L. Kroetz. 2007. Amodiaquine metabolism is impaired by common polymorphisms in CYP2C8: implications for malaria treatment in Africa. Clin. Pharmacol. Ther. 82:197-203. [DOI] [PubMed] [Google Scholar]
- 45.Pearce, R., D. Greenway, and A. Parkinson. 1992. Species differences and interindividual variation in liver microsomal cytochrome P450 2A enzymes: effects on coumarin, dicumarol, and testosterone oxidation. Arch. Biochem. Biophys. 298:211-225. [DOI] [PubMed] [Google Scholar]
- 46.Pearson, M. M., P. D. Rogers, J. D. Cleary, and S. W. Chapman. 2003. Voriconazole: a new triazole antifungal agent. Ann. Pharmacother. 37:420-432. [DOI] [PubMed] [Google Scholar]
- 47.Pfizer Inc. 2008. Voriconazole prescribing information, March 2008 revision. Pfizer Inc., New York, NY.
- 48.Purkins, L., N. Wood, P. Ghahramani, E. R. Love, M. D. Eve, and A. Fielding. 2003. Coadministration of voriconazole and phenytoin: pharmacokinetic interaction, safety, and toleration. Br. J. Clin. Pharmacol. 56(Suppl. 1):37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purkins, L., N. Wood, D. Kleinermans, and D. Nichols. 2003. Voriconazole potentiates warfarin-induced prothrombin time prolongation. Br. J. Clin. Pharmacol. 56(Suppl. 1):24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Relling, M. V., T. Aoyama, F. J. Gonzalez, and U. A. Meyer. 1990. Tolbutamide and mephenytoin hydroxylation by human cytochrome P450s in the CYP2C subfamily. J. Pharmacol. Exp. Ther. 252:442-447. [PubMed] [Google Scholar]
- 51.Roffey, S. J., S. Cole, P. Comby, D. Gibson, S. G. Jezequel, A. N. Nedderman, D. A. Smith, D. K. Walker, and N. Wood. 2003. The disposition of voriconazole in mouse, rat, rabbit, guinea pig, dog, and human. Drug Metab. Dispos. 31:731-741. [DOI] [PubMed] [Google Scholar]
- 52.Saari, T. I., K. Laine, L. Bertilsson, P. J. Neuvonen, and K. T. Olkkola. 2007. Voriconazole and fluconazole increase the exposure to oral diazepam. Eur. J. Clin. Pharmacol. 63:941-949. [DOI] [PubMed] [Google Scholar]
- 53.Saari, T. I., K. Laine, K. Leino, M. Valtonen, P. J. Neuvonen, and K. T. Olkkola. 2006. Effect of voriconazole on the pharmacokinetics and pharmacodynamics of intravenous and oral midazolam. Clin. Pharmacol. Ther. 79:362-370. [DOI] [PubMed] [Google Scholar]
- 54.Saari, T. I., K. Laine, K. Leino, M. Valtonen, P. J. Neuvonen, and K. T. Olkkola. 2006. Voriconazole, but not terbinafine, markedly reduces alfentanil clearance and prolongs its half-life. Clin. Pharmacol. Ther. 80:502-508. [DOI] [PubMed] [Google Scholar]
- 55.Saari, T. I., K. Laine, M. Neuvonen, P. J. Neuvonen, and K. T. Olkkola. 2008. Effect of voriconazole and fluconazole on the pharmacokinetics of intravenous fentanyl. Eur. J. Clin. Pharmacol. 64:25-30. [DOI] [PubMed] [Google Scholar]
- 56.Scott, L. J., and D. Simpson. 2007. Voriconazole: a review of its use in the management of invasive fungal infections. Drugs 67:269-298. [DOI] [PubMed] [Google Scholar]
- 57.Takada, K., M. Arefayene, Z. Desta, C. H. Yarboro, D. T. Boumpas, J. E. Balow, D. A. Flockhart, and G. G. Illei. 2004. Cytochrome P450 pharmacogenetics as a predictor of toxicity and clinical response to pulse cyclophosphamide in lupus nephritis. Arthritis Rheum. 50:2202-2210. [DOI] [PubMed] [Google Scholar]
- 58.Tassaneeyakul, W., D. J. Birkett, M. E. Veronese, M. E. McManus, R. H. Tukey, L. C. Quattrochi, H. V. Gelboin, and J. O. Miners. 1993. Specificity of substrate and inhibitor probes for human cytochromes P450 1A1 and 1A2. J. Pharmacol. Exp. Ther. 265:401-407. [PubMed] [Google Scholar]
- 59.Theuretzbacher, U., F. Ihle, and H. Derendorf. 2006. Pharmacokinetic/pharmacodynamic profile of voriconazole. Clin. Pharmacokinet. 45:649-663. [DOI] [PubMed] [Google Scholar]
- 60.Thummel, K. E., D. O'Shea, M. F. Paine, D. D. Shen, K. L. Kunze, J. D. Perkins, and G. R. Wilkinson. 1996. Oral first-pass elimination of midazolam involves both gastrointestinal and hepatic CYP3A-mediated metabolism. Clin. Pharmacol. Ther. 59:491-502. [DOI] [PubMed] [Google Scholar]
- 61.Thummel, K. E., D. D. Shen, T. D. Podoll, K. L. Kunze, W. F. Trager, C. E. Bacchi, C. L. Marsh, J. P. McVicar, D. M. Barr, and J. D. Perkins. 1994. Use of midazolam as a human cytochrome P450 3A probe. II. Characterization of inter- and intraindividual hepatic CYP3A variability after liver transplantation. J. Pharmacol. Exp. Ther. 271:557-566. [PubMed] [Google Scholar]
- 62.Thummel, K. E., D. D. Shen, T. D. Podoll, K. L. Kunze, W. F. Trager, P. S. Hartwell, V. A. Raisys, C. L. Marsh, J. P. McVicar, and D. M. Barr. 1994. Use of midazolam as a human cytochrome P450 3A probe. I. In vitro-in vivo correlations in liver transplant patients. J. Pharmacol. Exp. Ther. 271:549-556. [PubMed] [Google Scholar]
- 63.Totah, R. A., P. Sheffels, T. Roberts, D. Whittington, K. Thummel, and E. D. Kharasch. 2008. Role of CYP2B6 in stereoselective human methadone metabolism. Anesthesiology 108:363-374. [DOI] [PubMed] [Google Scholar]
- 64.Turpeinen, M., H. Raunio, and O. Pelkonen. 2006. The functional role of CYP2B6 in human drug metabolism: substrates and inhibitors in vitro, in vivo and in silico. Curr. Drug Metab. 7:705-714. [DOI] [PubMed] [Google Scholar]
- 65.Wang, Y. H., D. R. Jones, and S. D. Hall. 2005. Differential mechanism-based inhibition of CYP3A4 and CYP3A5 by verapamil. Drug Metab. Dispos. 33:664-671. [DOI] [PubMed] [Google Scholar]
- 66.Ward, B. A., J. C. Gorski, D. R. Jones, S. D. Hall, D. A. Flockhart, and Z. Desta. 2003. The cytochrome P4502B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J. Pharmacol. Exp. Ther. 306:287-300. [DOI] [PubMed] [Google Scholar]
- 67.Wilkinson, G. R. 2005. Drug metabolism and variability among patients in drug response. N. Engl. J. Med. 352:2211-2221. [DOI] [PubMed] [Google Scholar]
- 68.Wrighton, S. A., J. C. Stevens, G. W. Becker, and M. Vandenbranden. 1993. Isolation and characterization of human liver cytochrome P450 2C19: correlation between 2C19 and S-mephenytoin 4′-hydroxylation. Arch. Biochem. Biophys. 306:240-245. [DOI] [PubMed] [Google Scholar]
- 69.Zanger, U. M., K. Klein, T. Saussele, J. Blievernicht, H. Hofmann, and M. Schwab. 2007. Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics 8:743-759. [DOI] [PubMed] [Google Scholar]
- 70.Zanger, U. M., M. Turpeinen, K. Klein, and M. Schwab. 2008. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 392:1093-1108. [DOI] [PubMed] [Google Scholar]