Abstract
A real-time assay system that allows monitoring of intracellular human enterovirus (HEV) protease activity was established using the principle of fluorescence resonance energy transfer (FRET). It was accomplished by engineering cells to constitutively express a genetically encoded FRET probe. The FRET-based probe was designed to contain an enterovirus 71 3C protease (3Cpro) cleavage motif flanked by the FRET pair composed of green fluorescent protein 2 and red fluorescent protein 2 (DsRed2). Efficient FRET from the stable line was detected in a real-time manner by fluorescence microscopy, and the disruption of FRET was readily monitored upon HEV infection. The level of the repressed FRET was proportional to the input virus titer and the infection duration as measured by the fluorometric method. The FRET biosensor cell line was also responsive to other related HEV serotypes, but not to the phylogenetically distant herpes simplex virus, which was confirmed by Western blot analysis. The FRET biosensor was then utilized to develop a format for the determination of antiviral susceptibility, as the reduced FRET appeared to reflect viral replication. Evaluations of the FRET biosensor system with representative HEV serotypes demonstrated that their susceptibilities to a 3Cpro inhibitor, rupintrivir, were all accurately determined. In summary, this novel FRET-based system is a means for rapid detection, quantification, and drug susceptibility testing for HEVs, with potential for the development of a high-throughput screening assay.
Human enteroviruses (HEVs) belong to the picornavirus family and include coxsackieviruses (CVs), echoviruses, polioviruses, and the numbered enteroviruses (EVs). HEVs are associated with diverse clinical syndromes ranging from minor febrile illness to severe, potentially fatal conditions (e.g., aseptic meningitis, encephalitis, paralysis, myocarditis, and neonatal enteroviral sepsis) (25, 27). Despite sustained effort, treatment of HEV infection remains a significant unmet medical need.
Rapid and sensitive immunological and PCR-based diagnostic assays have been developed for group-specific diagnosis of HEV infections (23, 27). Nevertheless, traditional culture-based assay remains the only method to detect infectious virus particles and to facilitate the analysis of antiviral susceptibility, given that a number of anti-HEV agents are available (1, 32, 34). However, HEV culture is time-consuming, labor-intensive, and relatively insensitive. The development of alternative cell-based methods for rapid and reliable determination of HEV infection remains a high priority.
HEVs are a group of plus-strand RNA viruses with a genome that comprises the untranslated regions at the 5′ and 3′ ends and a long open reading frame encoding a polyprotein (2). The polyprotein is processed into the mature virus proteins through a sequence of cleavages performed by two virus-encoded proteases, 2A (2Apro) and 3C (3Cpro). While 2Apro carries out the initial cleavage to liberate the capsid protein precursor, the majority of subsequent cleavages are accomplished by 3Cpro or its precursor, 3CDpro (2, 29). Since 3Cpro is essential in viral replication and shows a high degree of homology among the members of the picornavirus family, efforts have been made to develop compounds that target 3Cpro (1, 29, 32, 34). Among them, rupintrivir (formerly AG7088) has been shown to potently inhibit the replication of a number of human rhinovirus (HRV) and HEV serotypes tested in cell culture (3, 17, 28).
Fluorescence resonance energy transfer (FRET) has been one of the most promising spectroscopic tools for investigating intracellular protease activity (14, 33, 35). FRET is a nonradioactive process in which energy from an excited donor fluorophore is transferred to an acceptor fluorophore when they have overlapping emission/absorption spectra with suitable orientations and distances (in the range of 1 to 10 nm) (16, 30). It was previously demonstrated that green fluorescent protein 2 (GFP2) and red fluorescent protein 2 (DsRed2) worked as the optimal donor and acceptor for FRET, respectively, permitting sufficient excitation and selective imaging (24). Based on the FRET pair, a genetically engineered biosensor cell line was generated and used for real-time measurement of EV 2Apro activity from infectious viruses (13).
In this study, we developed a stable cell line that expressed a recombinant 3Cpro substrate composed of the GFP2-DsRed2 pair linked by the 3Cpro cleavage motif. The FRET biosensor showed a real-time and quantifiable shift of fluorescence emission from a wavelength of ∼600 nm (red) to ∼510 nm (green) upon infection by the input HEVs. Sole expression of 3Cpro in the fusion substrate-expressing cells was sufficient to substantially abrogate FRET. EV71, the most common nonpoliovirus HEV associated with poliomyelitis-like paralysis (25), was used as a model virus. Furthermore, we have adapted the FRET biosensor for use as a rupintrivir susceptibility assay for the selected HEV serotypes.
MATERIALS AND METHODS
Cells and viruses.
The cell lines HeLa and Vero (African green monkey kidney cells) were cultured in Dulbecco's modified Eagle's medium (Gibco-BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS). EV71 (strain BrCr), herpes simplex virus type 1 (HSV-1) strain KOS, and clinical strains of echovirus serotype 9 (Echo9), CVA16, and CVB1 to -4 were propagated in Vero cells. All these viral stocks were propagated for at least two passages in HeLa cells and titrated on a Vero cell monolayer by a plaque assay.
Plasmids.
The pG2AwtR plasmid, which encodes the GFP2-DsRed2 fusion protein with the EV71 2Apro-sensitive motif connected in between (13), was used as starting material for molecular cloning. A set of complementary oligonucleotides that encode the EV71 3Cpro cleavage motif (amino acid residues 1435 to 1446 in EV71 strain BrCr) were synthesized as follows: forward, 5′-AGCTTTATAGAAGCACTCTTTCAAGGACCCCCTAAATTCAGGGGGCC-3′, and reverse, 5′-CCCTGAATTTAGGGGGTCCTTGAAAGAGTGCTTCTATAA-3′. Another set of oligomers encoding a mutant 3Cpro cleavage motif was synthesized as follows: forward, 5′-AGCTTTATAGAAAAACTCTTTCAACCTCCCCCTAAATTCAGGGGGCC-3′, and reverse, 5′-CCCTGAATTTAGGGGGAGGTTGAAAGAGTTTTTCTATAA-3′. The engineered HindIII/ApaI restriction sites are underlined. Each set of oligonucleotides was annealed and cloned into the corresponding sites in the pG2AwtR plasmid in frame with the GFP2 and DsRed2 open reading frame. The resulting plasmids were referred to as pG3CwtR and pG3CmutR, harboring a linker region that encodes the wild-type and mutant 3Cpro cleavage motifs, respectively (see Fig. S1 in the supplemental material).
The plasmids pCMV-FLAG-2A and pCMV-FLAG-3C, which bear the 2Apro and 3Cpro coding regions, respectively, were previously described (13).
Stable and transient transfections.
To establish stable FRET cells, subconfluently grown HeLa cells in a 24-well dish were transfected with pG3CwtR or pG3CmutR plasmid at approximately 1.5 μg each using Lipofectamine (Gibco-BRL) according to the manufacturer's instructions. The transfected cells were grown in the presence of zeocin at 50 μg/ml 48 h after transfection. Zeocin-resistant cell lines were isolated further by plating the cells at limiting dilution onto 96-well tissue culture dishes. Each clone was maintained in the presence of 20 μg/ml zeocin.
To transiently express 2Apro or 3Cpro, approximately 70% confluent monolayers of the stable FRET lines grown on 24-well dishes were transfected with the related plasmids for 48 h using the Lipofectamine method.
FRET analysis.
Both imaging and fluorometric methods were used for FRET measurements.
(i) Imaging analysis.
The imaging process was performed under an inverted fluorescence microscope (Nikon TE200). The filters used for observing GFP2 allowed excitation at 390 nm and emission at 500 to 530 nm; for DsRed2, excitation at 540 nm and emission at 575 to 625 nm were used; for FRET of GFP2 and DsRed2, excitation was at 390 nm and emission was at 575 to 625 nm (13).
(ii) Fluorometry.
Cells were washed and resuspended with phosphate-buffered saline and aliquoted into a black, 96-well, flat-bottom microplate (Nunc, Denmark). Fluorescence was measured on a fluorometer apparatus (Fluoroskan Ascent type 374; Labsystems, Rochester, NY) (13). The excitation wavelength was 390 nm (band pass, 20 nm), and the emission wavelengths for the fluorophore donor (GFP2) and acceptor (DsRed2) were 510 nm (band pass, 10 nm) and 590 nm (band pass, 14 nm), respectively.
Western blot analysis.
Cells were harvested at the indicated time points following infection or transfection. Cell extracts were prepared by washing the cells with cold phosphate-buffered saline and scraping them in the lysis buffer (19). The cell extracts were mixed with 2× sample buffer in equal volumes, separated by 13% sodium dodecyl sulfate-gel electrophoresis, and transferred onto nitrocellulose membranes. The blots were incubated with the Living Colors rabbit anti-GFP polyclonal antibody (1:1,000 dilution; no. 632459; BD Bioscience) or an anti-actin monoclonal antibody (1:1,000 dilution; MAB1501; Chemicon) as a loading control, followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit antibody (1:1,000 dilution; sc-2004; Santa Cruz) as the secondary antibody. Proteins were detected using the Enhanced Chemiluminescence Western blotting kit (Amersham).
Antiviral susceptibility testing.
The susceptibilities to rupintrivir (Pfizer Global Research and Development) of EV71, CVA16, CVB1, CVB2, CVB4, and Echo9 strains were measured by the FRET-based method, plaque reduction assay (PRA), and dye reduction assay (DRA) as described below. For each method, the concentrations of rupintrivir tested were twofold serial dilutions ranging from 0.04 to 5.12 μM. Control groups included mock-infected cells and infected cells with no rupintrivir added. The 50% effective concentration (EC50) value from each method was calculated with the PRISM4 program (GraphPad Inc.).
(i) FRET-based assay.
Approximately 80 to 90% confluent monolayers of the stable FRET cells were prepared in 24-well tissue culture plates. The cells were inoculated with each viral strain (100 μl) at a multiplicity of infection (MOI) of 0.04. After 90-min of adsorption, the inoculum was aspirated and 500 liters of medium (Dulbecco's modified Eagle's medium containing 2% FBS) alone or medium with rupintrivir at various concentrations was added to each well. At 36 to 48 h postinfection (p.i.), the infected, rupintrivir-free culture showed 80 to 95% FRET disruptions under the fluorescence microscope; cultures in all conditions were subjected to FRET measurement by fluorometry as described above. The data were expressed as the percentage of the FRET ratio obtained in rupintrivir-treated cells relative to the FRET ratio generated in wells of uninfected, rupintrivir-free cells. The EC50 was defined as the rupintrivir concentration that resulted in 50% of the FRET ratio from the infected cells compared with that from the uninfected, rupintrivir-free cells.
(ii) PRA.
Confluent Vero cells grown in six-well tissue culture plates were infected with each HEV serotype by inoculating approximately 80 to 100 PFU of each virus inoculum. The cells were then overlaid with medium containing 0.8% agar, 2% FBS, and the various concentrations of rupintrivir. The assay cultures were incubated for 4 to 7 days, and the plaques were counted after fixation and staining with a crystal violet formalin solution. The EC50 was defined as the rupintrivir concentration that reduced the number of plaques by 50% compared with those of the untreated controls.
(iii) DRA.
The DRA was used to measure EC50s following the protocol of Patick et al. (28) with some modifications. Approximately 70 to 80% confluent HeLa cells on 96-well dishes were infected with each HEV serotype at an MOI of 0.04 in the presence of the various concentrations of rupintrivir. Between 2 and 4 days p.i., when the cells in the rupintrivir-free wells showed 75 to 90% cytopathic effect (CPE), a cell survival assay (CellTiter 96 AQueous Cell Proliferation Assay; Promega) was performed according to the manufacturer's instructions. The absorbance value of each well was measured spectrophotometrically at 490 nm absorption for the soluble formazan product. The data were expressed as the percentage of formazan produced in the infected, rupintrivir-treated cells compared to the formazan produced in wells of uninfected, rupintrivir-free cells. The EC50 was calculated as the concentration of rupintrivir that reduced formazan production in infected, rupintrivir-treated cells to 50% of that produced by uninfected, rupintrivir-free cells.
RESULTS
Development of stable FRET cells.
A cell-based 3Cpro activity assay was developed based on the GFP2-DsRed2 tags, an optimal FRET pair we previously demonstrated (13). In principle, FRET can be generated due to the close proximity of GFP2-DsRed2, and it should significantly decline following the separation of the tandem fluorophores, which is mediated by 3Cpro activity. To this end, a recombinant construct was generated that encoded the GFP2-DsRed2 fusion protein tethered by amino acid sequences (19 amino acids in length) with the cleavage motif (IEALFQ↓GPPKFR; the arrow indicates the scissile bond) for EV71 3Cpro embedded. The linker contained the boundary region of EV71 2C-3A, the primary cleavage site of 3Cpro in many HEVs (26). Moreover, the importance of residues at the P1′ (mostly Gly) and P4 (mostly Ala) positions of the cleavage motif was previously documented (5, 6, 26); consequently, a recombinant plasmid, pG3CmutR, that bears the mutant cleavage motif (IEKLFQPPPKFR; the underlined residues represent the mutations in the P4 and P1′ positions) was also generated as a control (see Fig. S1 in the supplemental material).
To continuously monitor fluorescence, we isolated two stable clones, the HeLa-G3CwtR and HeLa-G3CmutR lines, that were stably transfected with the pG3CwtR and pG3CmutR plasmids, respectively. Constitutive expression of the fluorescent substrate did not appear to retard the growth of the clones, as they propagated at a rate similar to that of parental HeLa cells (data not shown). The two stable lines showed identical susceptibilities to viral infections because similar levels of CPE were seen upon infection. The expression levels of GFP2 and DsRed2 were verified by inspecting the stable lines under a fluorescence microscope using filter sets with excitation at 390 nm and emission at 515 nm (390ex/515em) and 540ex/600em, respectively. More importantly, detection of FRET was achieved with the 390ex/600em filter set. As an indication of FRET efficiency, the images from 390ex/515em and 390ex/600em were superimposed and could be seen in yellow (Fig. 1A and B, leftmost columns). The two stable clones showed FRET signals that were relatively high among the clones and comparable to each other.
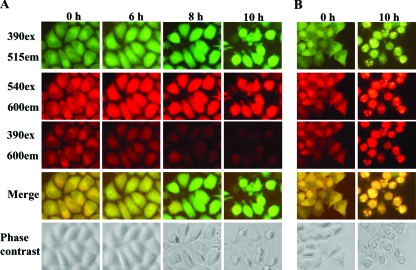
FIG. 1.
FRET image analysis. HeLa-G3CwtR (A) and HeLa-G3CmutR (B) cells grown in a 24-well dish were inoculated with EV71 stock at an MOI of 3 for 10 h. At the indicated time points, the cells were visualized with a fluorescence microscope (Nikon TE200) at 390ex/515em for GFP2, 540ex/600em for DsRed2, and 390ex/600em for FRET. The images from the 390ex/515em and 390ex/600em filter sets were merged. The cells were also observed with a phase-contrast microscope.
Imaging of 3Cpro dynamics in the context of virus infection.
Both HeLa-G3CwtR and HeLa-G3CmutR cells were subjected to infections by EV71 and analyzed by fluorescence microscopy within 10 h p.i. For the infected HeLa-G3CwtR cells, a gradual increase in GFP2 intensities (Fig. 1A, top row) but a progressive decline of FRET (Fig. 1A, third row from top), with color shifting from yellow to green in the merged images (Fig. 1A, second row from bottom), were observed as the infection proceeded; this was presumably due to the cleavage of the fluorescent substrate in vivo by 3Cpro, thereby decreasing FRET between the two fluorophores. In contrast, no appreciable FRET abrogation was shown for HeLa-G3CmutR cells at 10 h p.i., suggesting high specificity of cleavage by 3Cpro activity in vivo (Fig. 1B). For both stable FRET cell lines, the levels of DsRed2 intensity remained constant throughout the infections (Fig. 1A and B, second rows from top), arguing against the possibility that host protein shutoff due to picornavirus infection could account for the FRET disruptions (7, 18). Of note, concomitant inspection of the infected cells by phase-contrast microscopy did not reveal characteristic CPE until 10 h p.i. (Fig. 1A and B, bottom rows), while distinct FRET alteration was readily appreciable at 6 to 8 h p.i.
Quantitative measurements of FRET in vivo.
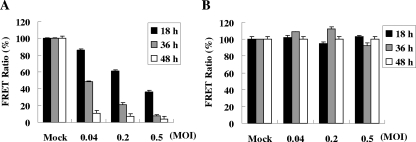
We then quantitatively measured FRET efficiency by the fluorometric method (13, 33). Both stable FRET lines were inoculated with EV71 stocks at various MOIs ranging from 0.04 to 0.5 for up to 48 h. The infected HeLa-G3CwtR cells exhibited a progressive decline in the FRET ratio in a dose- and time-dependent fashion (Fig. 2A), while infection of HeLa-G3CmutR did not cause detectable FRET abrogation regardless of the titers and durations tested (Fig. 2B), consistent with the imaging analyses (Fig. 1). The quantitative data indicated that FRET from HeLa-G3CwtR cells was inversely correlated with EV71 multiplication.
FIG. 2.
Quantitative fluorometry measurement of EV71-infected FRET cells. HeLa-G3CwtR (A) and HeLa-G3CmutR (B) cells on 24-well dishes were infected with EV71 stocks at MOIs of 0.04, 0.2, and 0.5 or mock infected for 18, 36, and 48 h. Cells were harvested and subjected to measurement by a fluorescent-plate reader with the excitation wavelength at 390/20 nm and the emission wavelength at 510/10 nm (for GFP2) or 590/14 nm (for DsRed2). The FRET ratio was defined as the intensity of emission at 590/14 nm divided by that at 510/10 nm. For each infection, the percent FRET ratio was determined by comparing it with that of the mock-infected control. The data presented are mean values of experiments in triplicate, and the error bars represent the standard deviations.
Physical separation of the tandem fluorophores in the infected cells.
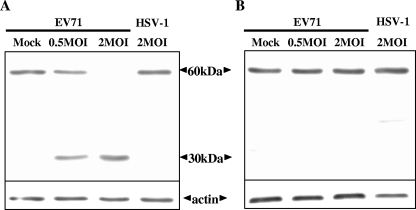
Western blotting was performed to examine if the in vivo fusion substrate was specifically cleaved upon EV71 infection. A phylogenetically distant virus, HSV-1, which carries an irrelevant protease activity (10), was used as a negative control. Only full-length fusion substrate (∼60 kDa) was detected in the extracts of the mock- and HSV-1-infected HeLa-G3wtR cells (Fig. 3A), as well as those of the infected HeLa-G3CmutR cells (Fig. 3B). Conversely, for HeLa-G3CwtR cells infected by EV71, the full fusion substrate was strikingly lost, with concurrent appearance of the cleavage product (∼30 kDa) in a dose-dependent manner (Fig. 3A).
FIG. 3.
Western blot analysis of the fluorogenic fusion protein in the FRET cells. HeLa-G3CwtR (A) and HeLa-G3CmutR (B) cells were mock infected, infected with EV71 stock at MOIs of 0.5 and 2, or infected with HSV-1 stock at an MOI of 2 for 12 h. Total-protein extracts were prepared and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 13% gels and then probed with an anti-GFP polyclonal antibody (BD Bioscience). The bands corresponding to the full-size fusion protein (60 kDa) or its cleavage product (30 kDa) are indicated. The actin level was used as the loading control.
3Cpro alone causes FRET disruptions.
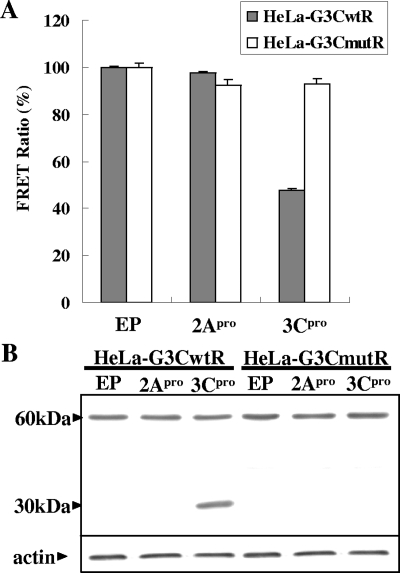
EVs carry two protease activities, namely, 2Apro and 3Cpro (2). To confirm that the FRET disruptions were specifically mediated by the 3Cpro activity, the 2Apro or 3Cpro expression plasmid was used to transfect the stable FRET lines. FRET was significantly impaired in the 3Cpro-expressing HeLa-G3CwtR cells to the level of ∼50%, while FRET remained unaffected for the remaining transfected cells (Fig. 4A). These findings were verified by a Western blot analysis in which the cleavage product (∼30 kDa) was seen only in the extract of 3Cpro-expressing HeLa-G3CwtR cells and not in those of other transfected cells (Fig. 4B). Transient expression of EV71 2Apro or 3Cpro was reported to elicit apoptotic cell death (19, 20); based on that, the transfection efficiencies in this study for the 2Apro- and 3Cpro-transfected cells appeared to be comparable (at a level of ∼50%), since they induced similar levels of CPE (data not shown). The data indicated that 3Cpro alone, rather than 2Apro or any redundant proteolytic activity associated with EV71, was responsible for the FRET disruption.
FIG. 4.
Expression of 3Cpro alone specifically causes FRET disruption. HeLa-G3CwtR and HeLa-G3CmutR cells were transfected with empty plasmid (pFLAG-CMV-2), pCMV-FLAG-2A (2Apro expression plasmid), or pCMV-FLAG-3C (3Cpro expression plasmid) at 1.5 μg each for 48 h. The percent FRET ratio (A) and Western analysis of the intracellular fusion substrate (B) were determined and demonstrated as described in the legends to Fig. 2 and 3, respectively. EP, empty plasmid.
Functional conservation of HEV 3Cpros.
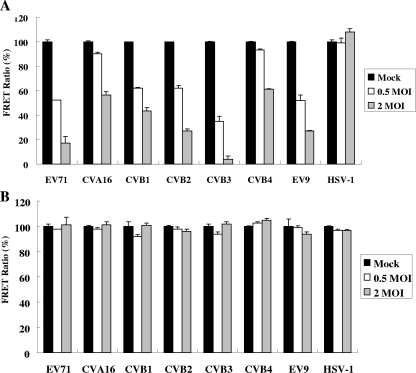
Previous sequence analyses revealed the high identity of 3Cpros among various picornaviruses and strict conservation of the catalytic triad of the 3Cpros (3). We then assessed if the fluorogenic substrate was responsive to 3Cpro activities from other HEV serotypes by inoculation of the stable FRET lines with stocks of CVA16, Echo9, CVB1 to -4, and the distantly related HSV-1. Analysis by fluorometry revealed that infections by all the HEVs at MOIs of 0.5 and 2 appeared to result in dose-dependent declines in FRETs from HeLa-G3CwtR cells, albeit to moderately varied extents; infection by CVB3 was the most sensitive to FRET changes, whereas infection by HSV-1 did not cause a measurable decline in FRET (Fig. 5A). On the other hand, infection of HeLa-G3CmutR cells by any of the viral stocks did not lead to detectable FRET disruptions (Fig. 5B). The data indicated that the fluorogenic substrate in HeLa-G3CwtR cells appeared to be a universal one for 3Cpros from the input HEVs examined, supporting the general use of the FRET biosensor for quantitative measurement of the activity of HEV 3Cpros and detection of several HEV serotypes.
FIG. 5.
Quantitative FRET detection of various HEV serotypes. HeLa-G3CwtR (A) and HeLa-G3CmutR (B) cells were mock infected or infected with stocks of EV71, CVA16, CVB1, CVB2, CVB3, CVB4, Echo9 (EV9), and HSV-1 at MOIs of 0.5 and 2 for 12 h. Analysis was conducted by the fluorometer assay, and the data are presented as described in the legend to Fig. 2.
FRET cell-based antiviral susceptibility testing.
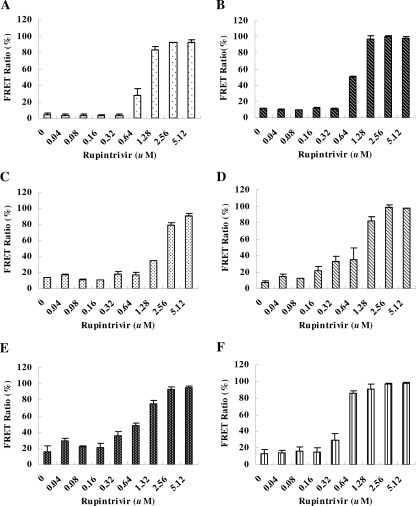
Since the levels of the depressed FRET in HeLa-G3CwtR cells were proportional to the input virus titer (Fig. 2) and indicative of the 3Cpro activity (Fig. 4), we next explored the feasibility of using the HeLa-G3CwtR line as an alternative anti-3Cpro susceptibility assay. For this purpose, we employed rupintrivir, an irreversible inhibitor of 3Cpro that showed broad-spectrum potency against a number of HRV and HEV serotypes tested in vitro (3, 17, 28). The tested viruses at an MOI of 0.04, the minimal titer that resulted in a FRET ratio of ∼10% (Fig. 2A), were used to infect HeLa-G3CwtR cells for 36 to 48 h. Representative HEV strains, including EV71, CVA16, CVB1, CVB2, CVB4, and Echo9, were used to assess the novel antiviral susceptibility testing system (Fig. 6). Moreover, two reference methods, namely, PRA and DRA, were conducted in parallel for comparison (Table 1). Good correlation was obtained for the EC50s between the FRET biosensor system and PRA (R2 = 0.985), higher than that between the FRET assay and DRA (R2 = 0.848). It was noted that while PRA and DRA took 4 to 7 days and 2 to 4 days, respectively, the FRET-based assay took only 36 to 48 h. This study underscores the usefulness of the FRET biosensor for rapid and quantitative determination of antiviral susceptibility.
FIG. 6.
Evaluation of the EC50s of the HEV serotypes by the FRET-based assay. The HEV strains, EV71 (A), CVA16 (B), CVB1 (C), CVB2 (D), CVB4 (E), and Echo9 (F), were used to infect HeLa-G3CwtR cells at an MOI of 0.04 each for 36 to 48 h. Twofold serially diluted rupintrivir at the concentrations indicated was added to the cultures. For each rupintrivir concentration, the percent FRET ratio was determined by comparing it with the control devoid of the drug. For each viral serotype, a plot of the percentage of control versus the rupintrivir concentration was established, and the EC50 was thus deduced. The data represent the mean values of triplicate samples, with the error bars representing the standard deviations.
TABLE 1.
In vitro antiviral activities of rupintrivir against HEV serotypes by three different methods
| HEV serotype | EC50 (μM)a
|
||
|---|---|---|---|
| FRET assay | PRA | DRA | |
| EV71 | 0.781 ± 0.028 | 0.791 ± 0.167 | 0.561 ± 0.083 |
| CVA16 | 0.331 ± 0.007 | 0.419 ± 0.032 | 0.270 ± 0.054 |
| CVB1 | 1.641 ± 0.216 | 1.725 ± 0.187 | 1.247 ± 0.177 |
| CVB2 | 0.857 ± 0.023 | 0.982 ± 0.290 | 0.783 ± 0.021 |
| CVB4 | 0.877 ± 0.100 | 0.843 ± 0.313 | 0.372 ± 0.023 |
| Echo9 | 0.434 ± 0.028 | 0.481 ± 0.105 | 0.360 ± 0.019 |
The results represent the mean ± standard deviation for triplicate experiments.
DISCUSSION
This study addressed the development and characterization of a novel FRET-based biosensor system for HEV infection, as well as its potential application as an innovative antiviral susceptibility assay. The HeLa-G3CwtR cell line allows direct imaging of HEV infection in culture during the infectious cycle (Fig. 1). The level of FRET decay in the biosensor cells is a function of the amount and infection duration of the input virus (Fig. 2), and the protease-mediated cleavage was verified by Western blot analysis (Fig. 3 and 4). The biosensor cells appeared to specifically respond to the infections of HEVs as a group, but not to a phylogenetically distant virus, such as HSV-1 (Fig. 5). Moreover, an effort has been made to adapt it for determination of antiviral susceptibility. Analyses with the representative HEV strains showed that the results from the FRET-based assay closely approximated those from the reference PRA and DRA (Fig. 6 and Table 1).
Several elements in the design of the FRET biosensor assay have contributed to its characteristics. First, FRET technology was applied by developing an intracellular fluorogenic substrate that, when processed by viral 3Cpro, resulted in FRET conversion that can be measured with the common detection modalities, fluorescence microscopy for imaging and a fluorescent-plate reader for quantitation. In this regard, the imaging method is sufficiently sensitive to detect FRET alteration early in the very first cycle of infection (6 to 8 h p.i.), unlike the routine method of inspecting the CPE, which normally takes more than one replication cycle to develop. Second, the HEV 2C-3A junction (12 amino acids in length) was utilized as the cleavage motif, since it was ranked the most efficient among the multiple cleavage sites in the viral polyprotein (26). Also, the primary sequence of this cleavage motif appears to be recognized by the 3Cpros of the HEVs tested but is distinct from the one recognized by the protease of HSV-1. Both features of the linker have contributed to the sensitivity and specificity of the biosensor system. Finally, the choice of HeLa cells as the parental cells permitted infections by several HEV serotypes, including those tested in this study. However, the HeLa-based biosensor cells were refractory to infections by some CVA and echovirus serotypes, as a prolonged infection led to neither the characteristic CPE nor FRET reduction (unpublished data). Future improvements to our system will include the generation of a stable cell line exhibiting susceptibility to other HEVs. Moreover, it was noted that infection by CVB3, among others, caused the most severe disruption of FRETs (Fig. 5A), a scenario that could be at least partly explained by its higher efficiency of replication, as it caused more extensive CPE in a certain period (data not shown), and consequently, potentially more 3Cpro was generated.
Compared with the conventional methods available for testing antiviral susceptibility, the HeLa-G3CwtR cell-based assay described here may represent a novel method with remarkable properties. The biosensor assay eliminates the tedium of manual counting and the variability in the resulting PRA data. It is analogous to the DRA used in this study in that objective quantitation is feasible. Unlike the DRA, however, the developed format of the biosensor assay can be performed in a microplate directly on cells without a time-consuming lysate preparation. It was noted that the DRA EC50s of this study were somewhat higher than those reported by Binford et al. (3), possibly owing to the differences in the dye used, viral-titer measurement (PFU versus 50% tissue culture infective dose), and relative viral replication in different cell types. Moreover, both conventional antiviral susceptibility assays are primarily limited to analyzing downstream gross cell damage caused by virus infection, possibly complicating the assessment of an antiviral compound that itself causes CPE. On the other hand, the gross cell damage did not appear to result in FRET decline, as evidenced by the control study with HSV-1 infection (Fig. 5). Finally, the availability of results in 36 to 48 h with the biosensor assay, compared to 4 to 7 days and 2 to 4 days for PRA and DRA, respectively, and high levels of concordance with the results from both conventional methods should make the HeLa-G3CwtR cell-based assay a valuable tool for determining the potencies of 3Cpro inhibitors against many HEV serotypes.
Proteolysis of a precursor protein carried out by viral protease is pivotal in the multiplication cycles of many viruses (12, 29). Picornavirus 3Cpro, the viral protease selected in this study, was originally employed as a target for anti-HRV compounds, and the application was extended to some HEV serotypes based on the three-dimensional structure (22) and the high degree of homology at the amino acid level (3, 34). Clinically, there is considerable interest in a broad-spectrum antiviral compound for the treatment of picornaviral diseases because no single serotype is exclusively associated with any particular disease and the treatment is primarily symptomatic. Furthermore, recent study has revealed that the presence of increasing concentrations of rupintrivir in the HRV cultures led to only little or moderate reduction in susceptibility (4), whereas in vitro selection with a capsid-binding inhibitor (pleconaril) conferred >200-fold reduction in susceptibility (11, 31). Therefore, picornaviral 3Cpro is considered an attractive molecular target despite the fact that rupintrivir was not licensed following the clinical trials (8). With the development of high-throughput fluorescence microscopy and its associated image analysis system (9, 15, 21), this FRET-based biosensor assay represents an ideal platform adaptable for anti-3Cpro drug screening. Any candidate compounds from the screening can be further validated by assaying them in HeLa-G3CwtR cells transfected with the 3Cpro expression plasmid to rule out those that inhibit virus replication due to their cytotoxic properties.
Supplementary Material
Acknowledgments
We thank Wu-Tse Liu for offering the clinical isolates of HEVs. We are also grateful to Pfizer Global Research and Development for rupintrivir.
This study was supported in part by grant 97QC016 from the Ministry of Education, Taiwan, Republic of China.
Footnotes
Published ahead of print on 17 November 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Barnard, D. L. 2006. Current status of anti-picornavirus therapies. Curr. Pharm. Des. 12:1379-1390. [DOI] [PubMed] [Google Scholar]
- 2.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702-713. [DOI] [PubMed] [Google Scholar]
- 3.Binford, S. L., F. Maldonado, M. A. Brothers, P. T. Weady, L. S. Zalman, J. W. Meador III, D. A. Matthews, and A. K. Patick. 2005. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 49:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binford, S. L., P. T. Weady, F. Maldonado, M. A. Brothers, D. A. Matthews, and A. K. Patick. 2007. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 51:4366-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair, W. S., and B. L. Semler. 1991. Role for the P4 amino acid residue in substrate utilization by the poliovirus 3CD proteinase. J. Virol. 65:6111-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blom, N., J. Hansen, D. Blaas, and S. Brunak. 1996. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 5:2203-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buenz, E. J., and C. L. Howe. 2006. Picornaviruses and cell death. Trends Microbiol. 14:28-36. [DOI] [PubMed] [Google Scholar]
- 8.De Palma, A. M., I. Vliegen, E. De Clercq, and J. Neyts. 2008. Selective inhibitors of picornavirus replication. Med. Res. Rev. doi: 10.1002/med.20125. [DOI] [PubMed]
- 9.Erfle, H., B. Neumann, U. Liebel, P. Rogers, M. Held, T. Walter, J. Ellenberg, and R. Pepperkok. 2007. Reverse transfection on cell arrays for high content screening microscopy. Nat. Protocols. 2:392-399. [DOI] [PubMed] [Google Scholar]
- 10.Flynn, D. L., N. A. Abood, and B. C. Holwerda. 1997. Recent advances in antiviral research: identification of inhibitors of the herpesvirus proteases. Curr. Opin. Chem. Biol. 1:190-196. [DOI] [PubMed] [Google Scholar]
- 11.Groarke, J. M., and D. C. Pevear. 1999. Attenuated virulence of pleconaril-resistant coxsackievirus B3 variants. J. Infect. Dis. 179:1538-1541. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, J. T., H. C. Wang, G. W. Chen, and S. R. Shih. 2006. Antiviral drug discovery targeting to viral proteases. Curr. Pharm. Des. 12:1301-1314. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, Y.-Y., Y.-N. Liu, W. Wang, F.-J. Kao, and S.-H. Kung. 2007. In vivo dynamics of enterovirus protease revealed by fluorescence resonance emission transfer (FRET) based on a novel FRET pair. Biochem. Biophys. Res. Commun. 353:939-945. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, Y.-C., W. Chen, and M. V. Yates. 2006. Use of fluorescence resonance energy transfer for rapid detection of enteroviral infection in vivo. Appl. Environ. Microbiol. 72:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang, Y. C., J. J. Chu, P. L. Yang, W. Chen, and M. V. Yates. 2008. Rapid identification of inhibitors that interfere with poliovirus replication using a cell-based assay. Antivir. Res. 77:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jares-Erijman, E. A., and T. M. Jovin. 2006. Imaging molecular interactions in living cells by FRET microscopy. Curr. Opin. Chem. Biol. 10:409-416. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser, L., C. E. Crump, and F. G. Hayden. 2000. In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antivir. Res. 47:215-220. [DOI] [PubMed] [Google Scholar]
- 18.Kuechler, E., J. Seipelt, D. D. Liebig, and W. Sommergruber. 2002. Picornavirus proteinase-mediated shutoff of host cell translation: direct cleavage of a cellular initiation factor, p. 301-311. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 19.Kuo, R.-L., S.-H. Kung, Y.-Y. Hsu, and W.-T. Liu. 2002. Infection with enterovirus 71 or expression of its 2A protease induces apoptotic cell death. J. Gen. Virol. 83:1367-1376. [DOI] [PubMed] [Google Scholar]
- 20.Li, M.-L., T.-A. Hsu, T.-C. Chen, S.-C. Chang, J.-C. Lee, C.-C. Chen, V. Stollar, and S.-R. Shih. 2002. The 3C protease activity of enterovirus 71 induces human neural cell apoptosis. Virology 293:386-395. [DOI] [PubMed] [Google Scholar]
- 21.Liebel, U., V. Starkuviene, H. Erfle, J. C. Simpson, A. Poustka, S. Wiemann, and R. Pepperkok. 2003. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 554:394-398. [DOI] [PubMed] [Google Scholar]
- 22.Matthews, D. A., P. S. Dragovich, S. E. Webber, S. A. Fuhrman, A. K. Patick, L. S. Zalman, T. F. Hendrickson, R. A. Love, T. J. Prins, J. T. Marakovits, R. Zhou, J. Tikhe, C. E. Ford, J. W. Meador, R. A. Ferre, E. L. Brown, S. L. Binford, M. A. Brothers, D. M. DeLisle, and S. T. Worland. 1999. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. USA 96:11000-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muir, P., U. Kammerer, K. Korn, M. N. Mulders, T. Poyry, B. Weissbrich, R. Kandolf, G. M. Cleator, and A. M. van Loon. 1998. Molecular typing of enteroviruses: current status and future requirements. Clin. Microbiol. Rev. 11:202-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller-Taubenberger, A., and K. Anderson. 2007. Recent advances using green and red fluorescent protein variants. Appl. Microbiol. Biotechnol. 77:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Palacios, G., and M. S. Oberste. 2005. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11:424-433. [DOI] [PubMed] [Google Scholar]
- 26.Pallai, P. V., F. Burkhardt, M. Skoog, K. Schreiner, P. Bax, K. A. Cohen, G. Hansen, D. E. Palladino, K. S. Harris, and M. J. Nicklin. 1989. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J. Biol. Chem. 264:9738-9741. [PubMed] [Google Scholar]
- 27.Pallansch, M., and R. Roos. 2007. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 839-893. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 28.Patick, A. K., S. L. Binford, M. A. Brothers, R. L. Jackson, C. E. Ford, M. D. Diem, F. Maldonado, P. S. Dragovich, R. Zhou, T. J. Prins, S. A. Fuhrman, J. W. Meador, L. S. Zalman, D. A. Matthews, and S. T. Worland. 1999. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 43:2444-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patick, A. K., and K. E. Potts. 1998. Protease inhibitors as antiviral agents. Clin. Microbiol. Rev. 11:614-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piston, D. W., and G.-J. Kremers. 2007. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 32:407-414. [DOI] [PubMed] [Google Scholar]
- 31.Schmidtke, M., E. Hammerschmidt, S. Schuler, R. Zell, E. Birch-Hirschfeld, V. A. Makarov, O. B. Riabova, and P. Wutzler. 2005. Susceptibility of coxsackievirus B3 laboratory strains and clinical isolates to the capsid function inhibitor pleconaril: antiviral studies with virus chimeras demonstrate the crucial role of amino acid 1092 in treatment. J. Antimicrob. Chemother. 56:648-656. [DOI] [PubMed] [Google Scholar]
- 32.Shih, S.-R., S.-J. Chen, G. H. Hakimelahi, H.-J. Liu, C.-T. Tseng, and K.-S. Shia. 2004. Selective human enterovirus and rhinovirus inhibitors: an overview of capsid-binding and protease-inhibiting molecules. Med. Res. Rev. 24:449-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tawa, P., J. Tam, R. Cassady, D. W. Nicholson, and S. Xanthoudakis. 2001. Quantitative analysis of fluorescent caspase substrate cleavage in intact cells and identification of novel inhibitors of apoptosis. Cell Death Differ. 8:30-37. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Q. M., and S. H. Chen. 2007. Human rhinovirus 3C protease as a potential target for the development of antiviral agents. Curr. Protein Pept. Sci. 8:19-27. [DOI] [PubMed] [Google Scholar]
- 35.Yang, J., Z. Zhang, J. Lin, J. Lu, B.-F. Liu, S. Zeng, and Q. Luo. 2007. Detection of MMP activity in living cells by a genetically encoded surface-displayed FRET sensor. Biochim. Biophys. Acta 1773:400-407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.