Abstract
Infections with Giardia lamblia, Entamoeba histolytica, and Trichomonas vaginalis, which cause diarrhea, dysentery, and vaginitis, respectively, are each treated with metronidazole. Here we show that Giardia, Entamoeba, and Trichomonas have oxygen-insensitive nitroreductase (ntr) genes which are homologous to those genes that have nonsense mutations in metronidazole-resistant Helicobacter pylori isolates. Entamoeba and Trichomonas also have nim genes which are homologous to those genes expressed in metronidazole-resistant Bacteroides fragilis isolates. Recombinant Giardia, Entamoeba, and Trichomonas nitroreductases used NADH rather than the NADPH used by Helicobacter, and two recombinant Entamoeba nitroreductases increased the metronidazole sensitivity of transformed Escherichia coli strains. Conversely, the recombinant nitroimidazole reductases (NIMs) of Entamoeba and Trichmonas conferred very strong metronidazole resistance to transformed bacteria. The Ehntr1 gene of the genome project HM-1:IMSS strain of Entamoeba histolytica had a nonsense mutation, and the same nonsense mutation was present in 3 of 22 clinical isolates of Entamoeba. While ntr and nim mRNAs were variably expressed by cultured Entamoeba and Trichomonas isolates, there was no relationship to metronidazole sensitivity. We conclude that microaerophilic protists have bacterium-like enzymes capable of activating metronidazole (nitroreductases) and inactivating metronidazole (NIMs). While Entamoeba and Trichomonas displayed some of the changes (nonsense mutations and gene overexpression) associated with metronidazole resistance in bacteria, these changes did not confer metronidazole resistance to the microaerophilic protists examined here.
Microaerophilic protists that are important human pathogens include Giardia lamblia, Entamoeba histolytica, and Trichomonas vaginalis, which cause diarrhea, dysentery, and vaginitis, respectively (1, 15, 34). Metronidazole is a nitroimidazole which was originally developed to treat Trichomonas infections but which has subsequently also become a mainstay for the treatment of infections caused by Entamoeba, Giardia, and anaerobic bacteria (17, 33, 38). Metronidazole damages the DNA in target cells when its nitro group is reduced by one electron to form a highly reactive and toxic radical anion (13). In bacteria, metronidazole is reduced and activated by enzymes called nitroreductases, which may be oxygen sensitive if they contain an N-terminal ferredoxin domain (2, 14, 24, 27).
Metronidazole resistance in Helicobacter pylori, which is an important cause of gastritis and gastric cancer, is frequently based upon nonsense mutations (premature stop codons) in oxygen-insensitive NADPH-nitroreductase (the rdxA gene product) and/or NADH-flavin oxidoreductase (the frxA gene product) (2, 14). Metronidazole resistance is widespread among Helicobacter strains in developing countries, such as India.
Strong metronidazole resistance in some strains of Bacteroides occurs by the overexpression of nim genes, which may be located on chromosomes or plasmids (36). A structural study of NimA from Deinococcus showed that metronidazole, in a reaction catalyzed by pyruvate, undergoes a two-electron reduction to form a nitroso group that is eventually reduced to an amine, which is not toxic (18).
Microaerophilic protists, which include Giardia, Entamoeba, and Trichomonas, are sensitive to metronidazole because they share metabolic properties with anaerobic bacteria (25, 29, 33, 38). Each of these protists, which live under anaerobic conditions in the lumen of the bowel or the vagina, is secondarily amitochondriate and lacks the enzymes for oxidative phosphorylation (5, 30, 37). Protist genes encoding fermentation enzymes appear to have been obtained by lateral gene transfer (LGT) from diverse bacteria (6, 8, 21, 22, 31). In Entamoeba and Giardia, these fermentation enzymes are present in the cytosol. In contrast, in Trichomonas, some of these enzymes are present in the hydrogenosome, a modified mitochondrion named for its production of hydrogen (4, 5, 26).
Metronidazole resistance is a major problem in clinical isolates of Trichomonas in the United States and elsewhere, while metronidazole-resistant Entamoeba and Giardia have for the most been prepared by selection in culture (9, 12, 17, 19, 20, 23, 32, 38, 39). Recently, a ferredoxin-nitroreductase fusion protein of Giardia (called GlNR1) which resembles oxygen-sensitive nitroreductases of bacteria and which appears to have been obtained by LGT was shown to have nitroreductase activity when it was expressed as a recombinant protein in bacteria (24, 27).
The goal of the present study was to determine how well two bacterial models for metronidazole resistance (nonsense mutations in Helicobacter genes encoding oxygen-insensitive nitroreductases that activate metronidazole and the overexpression of Bacteroides nim genes encoding enzymes that inactivate metronidazole) apply to microaerophilic protists. The specific questions asked in the present study included the following. Do Entamoeba, Trichomonas, and Giardia have homologs of bacterial nitroreductases and nitroimidazole reductases (NIMs)? Do recombinant protist nitroreductases and NIMs have the expected enzyme activities? Are there nonsense mutations in these protist genes like those that have been described for the ntr genes of metronidazole-resistant Helicobacter strains? Are any of the nim genes overexpressed in metronidazole-resistant protists, as has been described for the nim genes of metronidazole-resistant Bacteroides strains? How do nonsense mutations and/or the overexpression of the ntr and the nim genes relate to the metronidazole sensitivities of axenized Trichomonas and Entamoeba?
MATERIALS AND METHODS
Parasites examined.
Genome project strains of Giardia lamblia (strain WB), Entamoeba histolytica (strain HM-1:IMSS), and Trichomonas (strain G3) were all grown in axenic culture by standard conditions. For the testing of metronidazole sensitivity and for the determination of ntr and nim gene expression, we grew two other model strains of Entamoeba (strains 200:NIH and Rahman) and Trichomonas (strains B2RC7 and S1). To test for polymorphisms in the ntr and nim genes, we obtained the DNA of 18 Entamoeba clinical isolates from Egbert Tannich of the Bernard Nochte Institute for Tropical Medicine in Hamburg, Germany. We obtained the DNA of four Entamoeba clinical isolates from William Petri of the University of Virginia. We obtained the DNA of six Trichomonas clinical isolates from Evan Secor of the Centers for Disease Control and Prevention.
Bioinformatic methods.
Searches of the NCBI GiardiaDB database or databases managed by The J. Craig Venter Institute for sequences that matched the predicted proteins of Giardia, Entamoeba, and Trichomonas, which were derived by whole-genome sequencing, were done with the BLASTP program and the Giardia ferredoxin-nitroreductase and Bacteroides fragilis NimA (3, 6, 21, 22, 27).
Recombinant expression of protist nitroreductases and NIMs.
Two Entameoba histolytica nitroreductase genes (Ehntr1 and Ehntr2) were amplified from the genomic DNA of strains 200:NIH and HM-1:IMSS, respectively (see Table S1 in the supplemental material for all primers used). A third Entamoeba histolytica nitroreductase gene (Ehntr3) was amplified from the cDNA of HM-1:IMSS with gene-specific primers, as the gene contains an intron. Nitroimidazole resistance genes (nim) from Entamoeba histolytica (Ehnim) and Trichomonas vaginalis (Tvnim) were amplified with gene-specific primers from genomic DNAs. The BamHI and XhoI restriction sites were introduced into the sense and antisense primers, respectively. The amplicons were cloned into the expression vector pQE30 (Qiagen), which produces a recombinant protein with an N-terminal His tag. The protein was expressed in Escherichia coli strain M15 (lacI Kanr on pREP4 F− recA+ uvr+ lon+ lac) with pREP4. Isopropyl-β-d-thiogalactopyranoside (IPTG)-induced soluble recombinant protein was purified on a nickel-nitrilotriacetic acid affinity column and was eluted with 200 mM imidazole. To recover the proteins in the pellet (EhNTR3), the inclusion body was dissolved in 8 M urea and the mixture was briefly centrifuged, and then the supernatant was loaded onto the column. Slow renaturation of the protein was done with a gradient of urea (6 M, 4 M, 2 M, and 1 M), and finally, the protein was eluted with 200 mM imidazole.
The single nitroreductase gene from Giardia (Glntr1) and 1 of 11 nitroreductase genes of Trichomonas (Tvntr8) were amplified from genomic DNAs with specific primers. Amplified Giardia and Trichomonas nitroreductase genes were cloned into the pGEX6P vector (Amersham Biosciences) at the EcoRI-XhoI and BamHI-XhoI sites, respectively (35). E. coli strain BL21 was used for the overexpression of recombinant proteins, which were allowed to bind to glutathione-conjugated beads overnight. The bound protein was eluted with 15 mM reduced glutathione and was dialyzed against phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride to remove the glutathione. The purified protein was immediately used for the enzyme assays or was stored at −20°C in 10% glycerol.
Nitroreductase and NIM enzyme assays.
The nitroreductase assay was performed under the conditions described previously (14). The reaction mixture contained Tris-acetate (100 mM Tris-HCl, 50 mM acetate buffer, pH 7.0), 50 μM metronidazole, 0.3 mM NADPH or NADH, and enzyme in a 1-ml reaction volume. The assay was carried out at 25°C in a UV-visible spectrophotometer (Lambda 25; Perkin-Elmer) in quartz cuvettes with a 1-cm path length. The results of the assay were determined by measurement of the oxidation of NADPH or NADH at 340 nm (E = 6.22 mM−1 cm−1) or by determination of the reduction of metronidazole at 320 nm (E = 9.2 mM−1 cm−1). The NADH concentration was varied while the concentration of metronidazole (50 μM) was kept constant. Alternatively, the metronidazole concentration was varied while the NADH concentration (0.3 mM) was kept constant. The initial velocity of the purified enzyme was measured by determining the amount of change in the substrate concentration at 10-s intervals. The Michaelis-Menten constant (Km) and the maximal velocity (Vmax) were determined by using a Lineweaver-Burk double-reciprocal plot. All the experiments were performed in duplicate or triplicate with a minimum of four substrate concentrations. Enzymatic specific activities are reported as micromoles per minute per milligram of protein.
The assay methods used for the NIMs, which reduce metronidazole in the presence of NADH or NADPH, were similar to those used for the nitroreductases. E. coli cells harboring the empty pQE30 vector exhibited no significant amounts of enzyme activity.
Tests of metronidazole sensitivity of E. coli expressing recombinant nitroreductases and NIMs.
The sensitivity of transformed E. coli cells expressing protist NIMs to metronidazole was determined by a modified broth dilution procedure. The Ehnim and Tvnim genes were cloned into the pQE30 vector and expressed in E. coli strain JM109, which is relatively sensitive to metronidazole. IPTG-treated E. coli cells were incubated at 37°C for 16 h in the presence of metronidazole at concentrations ranging from 0 μg/ml to 2 mg/ml. The bacterial growth was measured in a UV-visible spectrophotometer (DU 500; Beckman) at 600 nm. The experiment was performed in duplicate and was repeated at least three times.
Similar methods were used to determine whether the expression of Entamoeba NTR1 or NTR2 increases the sensitivity of transformed E. coli cells to metronidazole. The Ehntr1 and Ehntr2 genes were cloned into the pQE30 vector and were expressed in E. coli strain JM109. Bacterial growth in the presence of metronidazole was measured in liquid culture.
Identification of introns and nonsense mutations in ntr and nim genes of Entamoeba and Trichomonas.
Genome project strains of Entamoeba (strain HM-1:IMSS) and Trichomonas (strain G3) have predicted introns and/or in-frame nonsense mutations in some of the ntr and nim genes (6, 21). We confirmed that these strains had the introns and stop codons, as well as those identified in other isolates, in three different ways. First, to verify the presence of introns, we performed PCR and reverse transcription-PCR (RT-PCR) with genomic DNA and total RNA, respectively, and we compared the sizes of the products by agarose gel electrophoresis. Second, the amplified products from PCR and RT-PCR were cloned and sequenced. Third, Western blots were performed with total protein, which was isolated from exponentially growing Entamoeba (strains HM-1:IMSS, 200:NIH, and Rahman) and separated in a 4 to 20% precast sodium dodecyl sulfate-polyacrylamide gel (Bio-Rad). The proteins were transferred onto polyvinylidene difluoride membranes, and affinity-purified rabbit antibodies to recombinant EhNTR1 were used for hybridization. Bound antibody was detected with a chemiluminescence kit (Pierce).
Tests of metronidazole sensitivity of cultured Entamoeba and Trichomonas.
Trophozoites (10,000 per ml) were grown in TYI-S-33 (tryptone-yeast extract) (Entamoeba) or TYM (tryptone-yeast extract-maltose) (Trichomonas) medium, each of which was supplemented with 10% heat-inactivated fetal bovine serum, at 37°C in the presence of various concentrations of metronidazole dissolved in 100% dimethyl sulfoxide. After 48 h, the number of viable parasites was counted with a hemocytometer and by the use of trypan blue to identify dead organisms. All experiments were run twice in triplicate with protists treated with dimethyl sulfoxide only (controls).
Real-time PCR for measurement of ntr and nim gene expression by cultured Entamoeba and Trichomonas.
Total RNA was isolated from mid-log-phase Entamoeba and Trichomonas with the Trizol reagent (Invitrogen), and the RNA was treated with a DNA-free reagent (Ambion), according to the manufacturer's instructions. The RNA was reverse transcribed with a RETROscript kit (Ambion) by using oligo(dT)18. Real-time PCR was carried out by the SYBR green method in a 96-well plate format with a Stratagene MX4000 cycler. A typical reaction mixture contained 12.5 μl of twice-concentrated Brilliant SYBR green QPCR master mix (Stratagene), primers (100 nM), and template cDNA in a total volume of 25 μl. The thermal profile for amplification was 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 60 s. The primers used for real-time PCR were designed (Table 1) by using OligoPerfect Designer software (Invitrogen). Gel electrophoresis was carried out with representative samples to confirm the product size. The relative quantities of the mRNA species were determined with MX4000 software (version 4.20; Stratagene) by using the Entamoeba or Trichomonas actin gene as a calibrator.
TABLE 1.
Kinetic properties of recombinant nitroreductases of Entamoeba, Giardia, and Trichomonas
| Protein | Substrate | Km (nM) | Vmax (μmol min−1) | Vmax/Km |
|---|---|---|---|---|
| EhNTR1 | NADH | 57 | 0.068 | 1.19 |
| Metronidazole | 67 | 0.027 | 0.40 | |
| EhNTR2 | NADH | 62 | 0.084 | 1.35 |
| Metronidazole | 81 | 0.031 | 0.38 | |
| EhNTR3 | NADH | 66 | 0.033 | 0.50 |
| Metronidazole | 83 | 0.014 | 0.17 | |
| GlNTR | NADH | 24 | 1.698 | 70.75 |
| NADPH | 19 | 2.222 | 116.9 | |
| Metronidazole | 206 | 0.347 | 1.68 | |
| TvNTR8 | NADH | 16 | 1.661 | 106.5 |
| Metronidazole | 78 | 0.329 | 4.21 |
Nucleotide sequence accession number.
The nucleotide sequence of the Ehntr1 gene of the HK9 strain of Entamoeba histolytica has been submitted to the GenBank database and can be found under accession number ABE99820.
RESULTS AND DISCUSSION
Giardia, Entamoeba, and Trichomonas genomes predict different sets of enzymes which might activate (nitroreductases) or inactivate (NIMs) metronidazole.
Genes encoding putative oxygen-insensitive nitroreductases, which lack an N-terminal ferredoxin domain, were present in a single copy in Giardia (Glntr1), 3 copies in Entamoeba (Ehntr1 to Ehntr3), and 11 copies in Trichomonas (Tvntr1 to Tvntr11) (see Table S1 in the supplemental material).
Bacteroides fragilis NimA, which confers metronidazole resistance, was used to identify a single putative nim gene from Entamoeba (Ehnim1) and three putative nim genes from Trichomonas (Tvnim1 to Tvnim3) (see Table S1 in the supplemental material). Nim genes appeared to be absent from Giardia.
Putative oxygen-sensitive nitroreductase (fdntr) genes, which have a ferredoxin domain at the N termini of the predicted enzymes, were present in two copies in Giardia and one copy in Entamoeba (24, 27). In contrast, fdntr genes appeared to be absent from the genome of Trichomonas. These results show that each microaerophilic protist has different combinations of nitroreductases and NIMs.
Phylogenetic reconstructions of protist and bacterial ntr and nim genes strongly suggested that Entamoeba and Giardia received their ntr genes from two different bacteria, while Entamoeba and Trichomonas received their nim genes from two different bacteria (see Fig. S1 and S2 in the supplemental material).
Recombinant nitroreductases of Giardia, Entamoeba, and Trichomonas reduce metronidazole.
All the nitroreductases (i.e., Giardia lamblia NTR1 [GlNTR1]; Entamoeba histolytica NTR1, NTR2, and NTR3 [EhNTR1, EhNTR2, and EhNTR3, respectively]; and Trichomonas vaginalis NTR8 [TvNTR8]) which were expressed as recombinant enzymes in the cytosol of E. coli reduced metronidazole (Table 1). While the Entamoeba and Trichomonas nitroreductases used NADH as the electron donor, the Giardia nitroreductase used either NADH or NADPH. The Kms for NADH and metronidazole were similar for the nitroreductases examined, but the activities of the recombinant Giardia and Trichomonas nitroreductases were much greater than those of the Entamoeba nitroreductases.
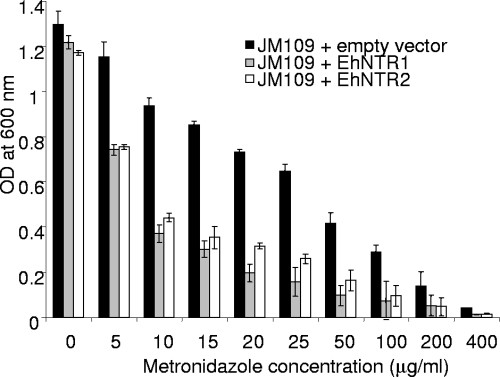
Two Entamoeba nitroreductases increased the metronidazole sensitivity of transformed strain JM109 of E. coli by greater than threefold (Fig. 1). JM109 transformed with an empty vector had a 50% effective concentration (EC50) slightly greater than 25 μg/ml metronidazole, while JM109 transformed with EhNTR1 and JM109 transformed with EhNTR2 had EC50s of 7 to 8 μg/ml metronidazole.
FIG. 1.
Recombinant expression of Entamoeba nitroreductases increased the sensitivity of transformed bacteria to metronidazole. E. coli JM109 was transformed with either the Ehntr1 gene, the Ehntr2 gene, or an empty PQE30 vector. The expression of each recombinant Entamoeba nitroreductase was induced with IPTG, and the bacteria were grown in the presence of serial dilutions of metronidazole for 16 h, at which time the optical density (OD) at 600 nm was measured.
Entamoeba and Trichomonas NIMs confer metronidazole resistance to E. coli.
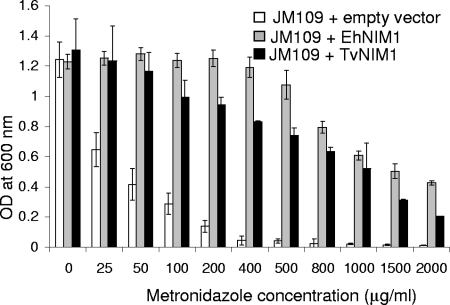
E. coli JM109 cells transformed with either the Entamoeba or the Trichomonas nim gene were 30 and 20 times more resistant to metronidazole, respectively, than E. coli cells transformed with an empty vector (Fig. 2). JM109 transformed with an empty vector had an EC50 of ∼25 μg/ml metronidazole, while JM109 transformed with EhNIM1 and JM109 transformed with TvNIM1 had EC50s of 750 and 500 μg/ml metronidazole, respectively. These results show that EhNIM1 and TvNIM1 confer strong metronidazole resistance to transformed E. coli cells.
FIG. 2.
Recombinant expression of Entamoeba and Trichomonas NIMs conferred metronidazole resistance to transformed bacteria. E. coli JM109 was transformed with either the Ehnim1 gene, the Tvnim1 gene, or an empty PQE30 vector. The expression of either the recombinant Entamoeba or the Trichomonas NIMs was induced with IPTG, and the bacteria were grown in the presence of serial dilutions of metronidazole for 16 h, at which time the optical density (OD) at 600 nm was measured.
The kinetics of the reduction of metronidazole by Entamoeba NIM1 showed that either NADPH or NADH may be an electron donor (Table 2).
TABLE 2.
Kinetic properties of a recombinant EhNIM1 of Entamoeba
| Substrate | Km (nM) | Vmax (μmol min−1) | Vmax/Km |
|---|---|---|---|
| NADH | 57 | 0.042 | 0.736 |
| NADPH | 96 | 0.033 | 0.34 |
| Metronidazole | 8 | 0.057 | 6.25 |
An Entamoeba nitroreductase gene (Ehntr1) has a nonsense mutation in the genome project strain and some clinical isolates.
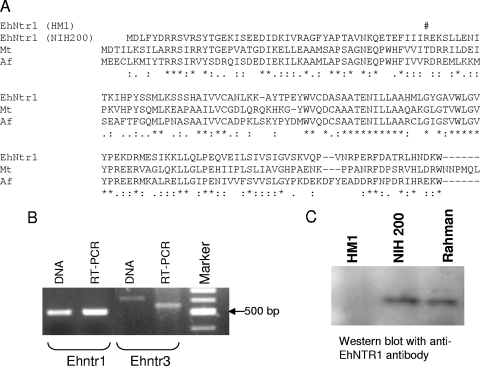
One of the goals of the present study was to determine whether there are any nonsense mutations in protist ntr genes, as have been shown for the ntr genes of metronidazole-resistant Helicobacter (2, 14). No nonsense mutations or introns were identified in the 11 ntr genes of genome project strain G3 of Trichomonas or the single ntr gene of genome project strain WB of Giardia. In contrast, the Ehntr1 gene of genome project strain HM-1:IMSS of Entamoeba, which was previously predicted to have a zero-frame 81-bp intron, appeared to have a nonsense mutation for the following reasons (Fig. 3).
FIG. 3.
A nonsense mutation (in-frame stop codon) was present in the Ehntr1 gene of genome project strain HM-1:IMSS of Entamoeba and three clinical isolates. (A) The nitroreductases of Entamoeba strains HM-1:IMSS and 200:NIH, as well as those Methanothermobacter thermautotrophicus (Mt) and Archaeoglobus fulgidus (Af), were aligned by using the single-letter code. In this alignment, identical amino acids are marked with an asterisk, while similar amino acids are marked with a colon or a period. Within a putative zero-frame 81-bp intron of EhNTR1 of genome project strain HM-1:IMSS, there was a stop codon (#) where there was an Arg (R) in the wild-type EhNTR1 of 200:NIH. The stop codon, which was also present in the Ehntr1 genes of 3 of 22 clinical isolates examined (isolates BM1, CM2, and H22), was present in a region that is conserved in bacterial nitroreductases. (B) PCR and RT-PCR with Ehntr1 gene primers from HM-1:IMSS DNA and RNA, respectively, produced products of the same size, arguing against the presence of an in-frame intron in the Ehntr1 gene. In contrast, the product obtained by RT-PCR with Ehntr3 gene primers from HM-1:IMSS RNA was smaller than the product obtained by PCR from DNA, consistent with the presence of an intron in the Ehntr3 gene of Entamoeba. The sequence of the Ehntr3 RT-PCR product confirmed the presence of the intron at the position predicted by the genome project (data not shown). (C) Western blotting with a rabbit polyclonal antibody to a recombinant EhNTR1 protein showed that strains 200:NIH and Rahman, which had the wild-type Ehntr1 gene, both expressed the NTR1 protein. In contrast, strain HM-1:IMSS, which had a nonsense mutation in the Ehntr1 gene, did not express the NTR1 protein.
First, the Ehntr1 genes from two other axenized strains of Entamoeba (strains 200:NIH and Rahman) had a single base change compared with the Ehntr1 sequence of HM-1:IMSS, so that their Ehntr1 genes had an open reading frame that was not interrupted by an intron (Fig. 3A). The wild-type, intron-less Ehntr1 gene was also present in the PCR products of 19 of 22 clinical isolates of Entamoeba, while 3 clinical isolates had the exact same nonsense mutation in Ehntr1 as genome project strain HM-1:IMSS (see Fig. S3 in the supplemental material). An intron-less ntr1 gene was also predicted from the whole-genome sequence of Entamoeba dispar.
Second, the predicted intron in Ehntr1 of the genome project strain caused a deletion of residues which are conserved in bacterial nitroreductases (Fig. 3A). Third, RT-PCRs with Ehntr1 gene-specific primers from strains HM-1:IMSS, 200:NIH, and Rahman RNAs failed to produce a spliced product (Fig. 3B). In contrast, RT-PCR of the Ehntr3 gene showed removal of its predicted intron (Fig. 3B). Fourth, antibodies to recombinant EhNTR1 bound to a 20-kDa protein band in Western blots of proteins from Entamoeba strains 200:NIH and Rahman, which had the wild-type Ehntr1 gene (Fig. 3C). In contrast, these anti-EhNTR1 antibodies failed to bind to proteins from HM-1:IMSS, which had the nonsense mutation in its Ehntr1gene (Fig. 3C).
To our knowledge, this is the first report of the identification of a polymorphic nonsense mutation in an Entamoeba gene, although pseudogenes for Entamoeba P glycoproteins and Entamoeba dispar cysteine proteinase 5 have been observed (11, 41). We were able to identify just eight other genes with nonsense mutations in genome project strain HM-1:IMSS of Entamoeba (our unpublished data).
Trichomonas nim genes included a nonsense mutation in Tvnim1 in strain S1 and a truncated Tvnim3 gene in all three strains examined (see Fig. S4 in the supplemental material). We were able to identify nonsense mutations in ∼5% of the predicted Trichomonas genes (our unpublished data).
Nitroreductase and NIM mRNAs are variably expressed by cultured Entamoeba and Trichomonas, but there is no relationship to metronidazole sensitivity.
The goal of the evaluation of nitroreductase and NIM mRNA expression was to determine whether there is any relationship between either ntr or nim gene expression by Entamoeba and Trichomonas or the presence of nonsense mutations in their ntr or nim genes and the sensitivity of these microaerophilic protists to metronidazole.
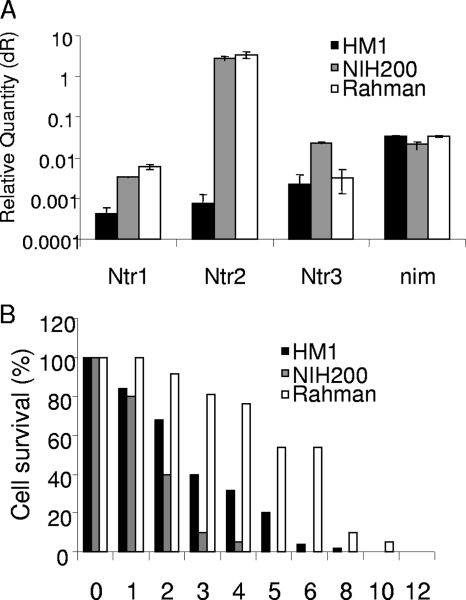
Entamoeba genome project strain HM-1:IMSS strain had an Ehntr1 gene with a nonsense mutation, and the Ehntr2 gene of HM-1:IMSS was expressed at a level 3 orders of magnitude less than the level of Ehntr2 expression by strains 200:NIH and Rahman. In addition, the level of expression of the Ehntr3 gene of HM-1:IMSS was 1 order of magnitude less than that of the Ehntr3 gene of 200:NIH, while all three Entamoeba strains examined had similar levels of expression of the Ehnim1 gene (Fig. 4A; note that the plots of gene expression are on a log scale). These results predicted that strain HM-1:IMSS amebae would be much less sensitive to metronidazole than the other Entamoeba strains because HM-1:IMSS has fewer nitroreductases that activate metronidazole. While strain HM-1:IMSS was slightly less sensitive to metronidazole than strain 200:NIH, strain Rahman was the least sensitive to metronidazole (Fig. 4B). These results suggest that there is no relationship between metronidazole sensitivity and Ehntr gene expression and/or the presence of nonsense mutations in the Ehntr genes of the three axenized Entamoeba strains examined here.
FIG. 4.
There was no correlation between the expression of nitroreductase and NIM mRNAs and the sensitivities of three axenized strains of Entamoeba to metronidazole. (A) Results of real-time PCR with actin as a calibrator plotted on a log scale showing that the level of expression of nitroreductase mRNAs is markedly decreased in strain HM-1:IMSS compared with the levels of expression in strains 200:NIH and Rahman, while the levels of NIM expression are similar. These results predict that strain HM-1:IMSS would be the least sensitive to metronidazole, while the other two strains would have similar sensitivities to metronidazole. dR, baseline subtracted fluorescent reading. (B) In contrast, strain Rahman was the least sensitive to metronidazole, strain HM-1:IMSS was intermediate in its sensitivity to metronidazole, and strain 200:NIH was the most sensitive to metronidazole.
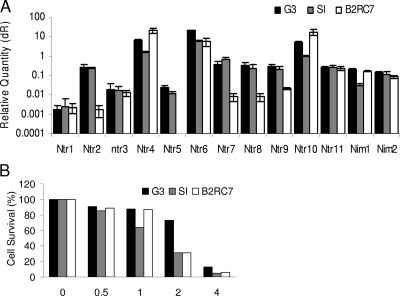
The levels of expression of the 11 Tvntr genes varied by 4 orders of magnitude among the three Trichomonas strains examined here (Fig. 5A; note that the plots of gene expression are on a log scale). Genome project strain G3 and the S1 strain of Trichomonas had similar levels of Tvntr and Tvnim gene expression, suggesting that they would have similar sensitivities to metronidazole. In contrast, strain B2RC7 expressed some Tvntr genes at higher levels and some Tvntr genes at lower levels than those by strains G3 and the S1, so it was difficult to predict the metronidazole sensitivity of B2RC7 compared with the sensitivities of the other strains. As it turns out, all three Trichomonas strains were relatively sensitive to metronidazole (Fig. 5B). These results suggest that there is no relationship between metronidazole sensitivity and Tvntr gene expression and/or the presence of nonsense mutations in the Tvntr genes of the three axenized Trichomonas strains examined here.
FIG. 5.
There was no correlation between the expression of nitroreductase and NIM mRNAs and the sensitivities of three model strains of Trichomonas to metronidazole. (A) The levels of expression of nitroreductases, as measured by real-time PCR by using actin as a calibrator, by 11 Trichomonas varied by multiple log units. In contrast, they showed similar levels of expression of NIMs. These results were so complex that it was not possible to predict which of the three model strains of Trichomonas would be the most sensitive or the most resistant to metronidazole. dR, baseline subtracted fluorescent reading. (B) Indeed, none of these axenized strains of Trichomonas were resistant to metronidazole.
Major conclusions and unanswered questions.
The results presented here show that microaerophilic protists have different combinations of enzymes which activate metronidazole (nitroreductases and ferredoxin-nitroreductase fusions) or inactivate metronidazole (NIMs) in bacteria (2, 14, 18, 24, 27, 36). While it is likely that these ntr and nim genes, which are absent from the vast majority of eukaryotes, were obtained from anaerobic bacteria by LGT, the phylogenetic analyses were not conclusive for some of the protist genes (see Fig. S1 and S2 in the supplemental material) (8, 21). In addition, there was no evidence that LGT directly contributes to metronidazole resistance in these protists.
The most important results were that all of the Giardia, Entamoeba, and Trichomonas enzymes examined here activate metronidazole (nitroreductases) or inactivate metronidazole (NIMs) when they are expressed in E. coli. These results, as well as the demonstration of nitroimidazole activation by a Giardia ferredoxin-nitroreductase fusion enzyme (24), strongly suggest that these enzymes may contribute to metronidazole activation or inactivation in these microaerophilic protists, as has been demonstrated in bacteria (2, 14, 18, 36). A recombinant Trichomonas ferredoxin:NADH was also shown to reduce metronidazole, although the kinetics were not determined (16). Similarly, a recent report showed that the knockout of a Trypanosoma nitroreductase confers cross-resistance to nifurtimox and benznidazole (40).
The nitroreductases of Giardia, Entamoeba, and Trichomonas, as well as the NIM of Entamoeba, which lacked targeting sequences, are likely present in the cytosol. In contrast, Trichomonas NIMs, which contained organelle-targeting sequences, are likely present in the hydrogenosome (4). The results for Trichomonas, which suggest that metronidazole might be activated by nitroreductases in the cytosol and inactivated by NIMs in the hydrogenosome, are somewhat surprising, because previous reports suggested that metronidazole is activated in the hydrogenosome (7, 16, 17, 28). It is possible, then, that there are multiple locations for metronidazole activation in Trichomonas and that there are competing reactions with metronidazole in the hydrogenosome.
As is the case in bacteria (2, 14, 18, 36), there were nonsense mutations in protist nitroreductase and nim genes (Ehntr1 in strain HM-1:IMSS, Tvnim1 in strain S1, and Tvnim3 in all Trichomonas strains examined) and marked differences in the mRNA expression of numerous ntr and nim genes in these microaerophilic protists. However, in contrast to Helicobacter and Bacteroides, the presence of nonsense mutations in ntr genes and the overexpression of nim mRNAs did not accurately predict the metronidazole sensitivity of the axenized Entamoeba and Trichomonas.
While the present studies have focused on the nitroreductases and NIMs of Giardia, Entamoeba, and Trichomonas, previous observations show that the metronidazole sensitivities of these microaerophilic protists may also be determined by (i) the availability of reduced ferredoxin or NAD(P)H to activate or inactivate metronidazole (7, 10, 16, 17, 20, 28) or (ii) the ability of these organisms to quench the reactive species generated by metronidazole activation (19, 23, 32, 39). In Trichomonas, low-level, microaerophilic resistance to metronidazole is associated with impaired oxygen scavenging, which results in the reoxidation of metronidazole or the removal of electrons by oxygen (17). In contrast, high-level, anaerobic resistance to metronidazole is associated with marked changes in hydrogenosomal enzymes that metabolize pyruvate (PFOR and hydrogenase) or metabolize malate (malic enzyme and NADH:ferredoxin oxidoreductase) (the so-called alternative pathway) (7, 16, 28).
Supplementary Material
Acknowledgments
We thank Egbert Tannich, William Petri, Patricia Johnson, B. N. Singh, and Evan Secor for supplying parasites and/or parasite DNA.
This work was supported in part by NIH grant AI48082 to J.S. and by CSIR and ICMR grants from the government of India to S.K.G.
Footnotes
Published ahead of print on 17 November 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldana, L. P., M. Kato, T. Kondo, S. Nakagawa, R. Zheng, T. Sugiyama, M. Asaka, and D. H. Kwon. 2005. In vitro induction of resistance to metronidazole, and analysis of mutations in rdxA and frxA genes from Helicobacter pylori isolates. J. Infect. Chemother. 11:59-63. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley, P. J., C. J. Lahti, E. Plümper, and P. J. Johnson. 1997. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 16:3484-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui, E. T., P. J. Bradley, and P. J. Johnson. 1996. A common evolutionary origin for mitochondria and hydrogenosomes. Proc. Natl. Acad. Sci. USA 93:9651-9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlton, J. M., R. P. Hirt, J. C. Silva, A. L. Delcher, M. Schatz, Q. Zhao, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, A., R. Cammack, D. Linstead, and D. Lloyd. 1985. The generation of metronidazole radicals in hydrogenosomes isolated from Trichomonas vaginalis. J. Gen. Microbiol. 131:2141-2144. [DOI] [PubMed] [Google Scholar]
- 8.Clark, C. G., U. C. Alsmark, M. Tazreiter, Y. Saito-Nakano, V. Ali, S. Marion, C. Weber, C. Mukherjee, et al. 2007. Structure and content of the Entamoeba histolytica genome. Adv. Parasitol. 65:51-190. [DOI] [PubMed] [Google Scholar]
- 9.Crowell, A. L., K. A. Sanders-Lewis, and W. E. Secor. 2003. In vitro metronidazole and tinidazole activities against metronidazole-resistant strains of Trichomonas vaginalis. Antimicrob. Agents Chemother. 47:1407-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dan, M., A. L. Wang, and C. C. Wang. 2000. Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme. Mol. Microbiol. 36:447-456. [DOI] [PubMed] [Google Scholar]
- 11.Descoteaux, S., P. Ayala, E. Orozco, and J. Samuelson. 1992. Primary sequences of two P-glycoprotein genes of Entamoeba histolytica. Mol. Biochem. Parasitol. 54:201-211. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh, S., M. Frisardi, L. Ramirez-Avila, S. Descoteaux, K. Sturm-Ramirez, O. A. Newton-Sanchez, J. I. Santos-Preciado, C. Ganguly, A. Lohia, S. Reed, and J. Samuelson. 2000. Molecular epidemiology of Entamoeba spp.: evidence of a bottleneck (demographic sweep) and transcontinental spread of diploid parasites. J. Clin. Microbiol. 38:3815-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman, P., R. L. Koch, T. C. Yeung, E. J. Chrystal, B. B. Beaulieu, Jr., M. A. McLafferty, and G. Sudlow. 1986. Comparing the reduction of nitroimidazoles in bacteria and mammalian tissues and relating it to biological activity. Biochem. Pharmacol. 35:43-51. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin, A., D. Kersulyte, G. Sisson, S. J. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 15.Haque, R., C. D. Huston, M. Hughes, E. Houpt, and W. A. Petri, Jr. 2003. Amebiasis. N. Engl. J. Med. 348:1565-1573. [DOI] [PubMed] [Google Scholar]
- 16.Hrdý, I., R. Cammack, P. Stopka, J. Kulda, and J. Tachezy. 2005. Alternative pathway of metronidazole activation in Trichomonas vaginalis hydrogenosomes. Antimicrob. Agents Chemother. 49:5033-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulda, J. 1999. Trichomonads, hydrogenosomes and drug resistance. Int. J. Parasitol. 29:199-212. [DOI] [PubMed] [Google Scholar]
- 18.Leiros, H. K., S. Kozielski-Stuhrmann, U. Kapp, L. Terradot, G. A. Leonard, and S. M. McSweeney. 2004. Structural basis of 5-nitroimidazole antibiotic resistance: the crystal structure of NimA from Deinococcus radiodurans. J. Biol. Chem. 279:55840-55849. [DOI] [PubMed] [Google Scholar]
- 19.Leitsch, D., D. Kolarich, I. B. Wilson, F. Altmann, and M. Duchêne. 2007. Nitroimidazole action in Entamoeba histolytica: a central role for thioredoxin reductase. PLoS Biol. 5:e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S. M., D. M. Brown, P. O'Donoghue, P. Upcroft, and J. A. Upcroft. 2000. Ferredoxin involvement in metronidazole resistance of Giardia duodenalis. Mol. Biochem. Parasitol. 108:137-140. [DOI] [PubMed] [Google Scholar]
- 21.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, et al. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865-868. [DOI] [PubMed] [Google Scholar]
- 22.Morrison, H. G., A. G. McArthur, F. D. Gillin, S. B. Aley, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921-1926. [DOI] [PubMed] [Google Scholar]
- 23.Müller, J., S. Ley, I. Felger, A. Hemphill, and N. Müller. 2008. Identification of differentially expressed genes in a Giardia lamblia WB C6 clone resistant to nitazoxanide and metronidazole. J. Antimicrob. Chemother. 62:72-82. [DOI] [PubMed] [Google Scholar]
- 24.Müller, J., J. Wastling, S. Sanderson, N. Müller, and A. Hemphill. 2007. A novel Giardia lamblia nitroreductase, GlNR1, interacts with nitazoxanide and other thiazolides. Antimicrob. Agents Chemother. 51:1979-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller, M. 1988. Energy metabolism of protozoa without mitochondria. Annu. Rev. Microbiol. 42:465-488. [DOI] [PubMed] [Google Scholar]
- 26.Müller, M. 1993. The hydrogenosome. J. Gen. Microbiol. 139:2879-2889. [DOI] [PubMed] [Google Scholar]
- 27.Nixon, J. E. J., A. Wang, J. Field, H. G. Morrison, A. G. McArthur, M. L. Sogin, B. Loftus, and J. Samuelson. 2002. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukarot. Cell 1:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasoloson, D., S. Vanácová, E. Tomková, J. Rázga, I. Hrdy, J. Tachezý, and J. Kulda. 2002. Mechanisms of in vitro development of resistance to metronidazole in Trichomonas vaginalis. Microbiology 148:2467-2477. [DOI] [PubMed] [Google Scholar]
- 29.Reeves, R. E. 1984. Metabolism of Entamoeba histolytica Scaudinn, 1903. Adv. Parasitol. 23:105-142. [DOI] [PubMed] [Google Scholar]
- 30.Roger, A. J., S. G. Svard, J. Tovar, C. G. Clark, M. W. Smith, F. D. Gillin, and M. L. Sogin. 1998. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc. Natl. Acad. Sci. USA 95:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenthal, B., Z. Mai, D. Caplivski, S. Ghosh, H. de la Vega, T. Graf, and J. Samuelson. 1997. Evidence for the bacterial origin of genes encoding fermentation enzymes of the amitochondriate protozoan parasite Entamoeba histolytica. J. Bacteriol. 179:3736-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samarawickrema, N. A., D. M. Brown, J. A. Upcroft, N. Thammapalerd, and P. Upcroft. 1997. Involvement of superoxide dismutase and pyruvate:ferredoxin oxidoreductase in mechanisms of metronidazole resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 40:833-840. [DOI] [PubMed] [Google Scholar]
- 33.Samuelson, J. 1999. Why metronidazole is active against both bacteria and parasites. Antimicrob. Agents Chemother. 43:1533-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwebke, J. R., and D. Burgess. 2004. Trichomoniasis. Clin. Microbiol. Rev. 17:794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 36.Sóki, J., M. Gal, J. S. Brazier, V. O. Rotimi, E. Urbán, E. Nagy, and B. I. Duerden. 2006. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteroides strains. J. Antimicrob. Chemother. 57:212-220. [DOI] [PubMed] [Google Scholar]
- 37.Tovar, J., A. Fischer, and C. G. Clark. 1999. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol. 32:1013-1021. [DOI] [PubMed] [Google Scholar]
- 38.Upcroft, P., and J. A. Upcroft. 2001. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin. Microbiol. Rev. 14:150-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassman, C., A. Hellberg, E. Tannich, and I. Bruchhaus. 1999. Metronidazole resistance in the protozoan parasite Entamoeba histolytica is associated with increased expression of iron-containing superoxide dismutase and peroxiredoxin and decreased expression of ferredoxin 1 and flavin reductase. J. Biol. Chem. 274:26051-26056. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson, S. R., M. C. Taylor, D. Horn, J. M. Kelly, and I. Cheeseman. 2008. A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proc. Natl. Acad. Sci. USA 105:5022-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willhoeft, U., L. Hamann, and E. Tannich. 1999. A DNA sequence corresponding to the gene encoding cysteine proteinase 5 in Entamoeba histolytica is present and positionally conserved but highly degenerated in Entamoeba dispar. Infect. Immun. 67:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.