Abstract
The purpose of this study was to determine the effect of a single dose of 300 mg of ritonavir on the plasma pharmacokinetics (PK) of a single dose of 20 mg of elvucitabine when the two drugs were coadministered in healthy subjects. In a three-way crossover design, 30 subjects received 20 mg of elvucitabine, 300 mg of ritonavir, or 20 mg of elvucitabine coadministered with 300 mg of ritonavir. Elvucitabine concentrations were analyzed using a validated liquid chromatography-tandem mass spectrometry assay. The PK of elvucitabine was determined using both noncompartmental and compartmental analyses. Models were developed and tested using ADAPT-II, while a population analysis was performed using IT2S. Comparisons of PK parameters between groups were done with SAS. The pharmacokinetic behavior of elvucitabine was best described by a two-compartment linear model using two absorption rates and a first-order elimination rate. Ritonavir significantly impacted the PK of elvucitabine by reducing elvucitabine's bioavailability, with the most plausible explanation being an inhibition on influx transporters by ritonavir. The decrease in elvucitabine bioavailability when elvucitabine was coadministered with ritonavir may be due to ritonavir's inhibiting influx gut transporters. Continued development of elvucitabine is warranted to better characterize its PK and to determine its in vivo efficacy against human immunodeficiency virus.
The number of people infected with human immunodeficiency virus (HIV) is increasing, and it is estimated that more than seventy million people have been infected so far, of whom thirty million have died (14). Since the mid-1990s, the widespread use of highly active antiretroviral therapy (HAART) has dramatically reduced the incidence of mortality associated with HIV (8, 10, 12). HAART regimens include cocktails composed of at least three medications from at least two classes of medication (15, 16). Two of the main classes of HIV drugs are nucleoside reverse transcriptase inhibitors (NRTIs) and protease inhibitors. NRTIs require intracellular metabolism to the triphosphate metabolite in order to be efficacious in stopping the replication of the virus. The majority of protease inhibitors, on the other hand, do not require metabolism and are cleaved by the virus protease, blocking the normal development of the virus and making it impossible for it to infect other cells.
New HIV drugs with favorable pharmacokinetics (PK) profiles as well as improved safety profiles are being developed. These new drugs, which make innovative dosing regimens possible, could simplify the HAART regimen, potentially increasing compliance.
Elvucitabine (2′,3′-dideoxy-2′,3′-didehydro-β-l-5-fluorocytidine), an investigational l-cytosine NRTI, showed a 5- to 10-fold-improved in vitro activity against wild-type HIV isolates (50% inhibitory concentration of ∼1 ng/ml in peripheral blood mononuclear cells) compared to lamivudine. In addition, elvucitabine also showed potentially greater activity against a variety of nucleoside resistant viral isolates, particularly those that are resistant to zidovudine and tenofovir. Preclinical in vitro data for elvucitabine showed that elvucitabine has a plasma protein binding of less than 10%, is metabolized intracellularly into monophosphate, diphosphate, and triphosphate analytes (with elvucitabine triphosphate having a half-life of at least 20 h), has no other significant metabolites (i.e., was not metabolized by CYP enzymes), and is not an inducer or an inhibitor of CYP enzymes. Additionally, preclinical animal studies demonstrated that elvucitabine has a bioavailability of approximately 50% in dogs and has increasing exposure with increasing doses in mouse, rat, and dog studies. Preliminary phase I PK studies of elvucitabine demonstrated that elvucitabine has a long half-life, which could potentially make innovative dosing regimens possible.
The purpose of this study was to determine the effect of a single dose of 300 mg of ritonavir on the plasma PK of a single dose of 20 mg of elvucitabine when the drugs were coadministered in healthy subjects. As ritonavir is a protease inhibitor often used in boosted protease inhibitor regimens, such as Kaletra, it is important to know the effect that it may have on elvucitabine's profile. Ritonavir is a protease inhibitor that causes multiple drug interactions. Ritonavir inhibits intestinal ABCB1 transporters and CYP3A enzymes. Ritonavir also inhibit hepatic CYP2D6 and possibly CYP2C9, CYP2C19, and CYP1A2 enzymes (4, 7, 9). Ritonavir also has a high plasma protein binding (>98%) that may cause it to displace other medications (6).
MATERIALS AND METHODS
Subjects and study design.
Thirty (30) healthy subjects were enrolled in a single-center comparative randomized single-dose three-way crossover drug interaction study. Twenty-two male and eight female subjects, between 19 and 55 years of age, were administered 20 mg of elvucitabine, 300 mg of ritonavir, or 20 mg of elvucitabine coadministered with 300 mg of ritonavir, under fasting conditions. Doses were administered at one clinical phase I unit (Lincoln, NE). Subjects provided written consents prior to participating in the study, and the study was approved by an ethics committee. When elvucitabine was administered, plasma samples were collected for PK determination prior to dosing and at 1, 2, 3, 3.5, 4, 5, 6, 7, 8, 10, 12, 16, 24, 36, 48, 72, and 96 h postdose.
Drug analysis.
Plasma samples were analyzed for elvucitabine concentrations by a sensitive and specific validated liquid chromatography-tandem mass spectrometry assay (11). The plasma analytical range was 0.500 ng/ml to 100 ng/ml. The precision (coefficient of variation) was less than or equal to 5.2%, and accuracy (bias) ranged from 0.3 to 3.3% for concentrations of 1.5, 15, and 75 ng/ml.
Noncompartmental PK analysis.
Standard noncompartmental analyses were performed on the elvucitabine concentration-versus-time data. The following PK parameters were observed or calculated for elvucitabine administered alone and coadministered with a single dose of 300 mg of ritonavir: maximum observed concentration (Cmax), time of maximum observed concentration (Tmax), area under the curve from time zero to 24 h (AUC0-24), area under the curve from time zero to the last measurable concentration (AUC0-t), area under the curve from time zero to infinity (AUC0-∞), elimination rate constant (kel), and half-life (t1/2). Noncompartmental analyses were performed using Kinetica version 4.3 (InnaPhase Corporation).
Population compartmental PK analysis.
Compartmental PK analyses were performed on elvucitabine data from all subjects. Individual analyses were first performed using maximum-likelihood in ADAPT-II release IV (3). The model discrimination process was based on minimization of the values of the Akaike information criterion test, of the minimum value of the objective function, and of the residual variability. An additional criterion considered in the discrimination process was the maximization of the average coefficient of determination. A population PK analysis was then performed on the final model using an iterative two-stage methodology (IT2S) (1) using previously obtained data from the ADAPT-II analysis in order to get the most accurate population PK parameters, variance, residual variability and individual results. All systemic concentrations of elvucitabine were modeled using a weighting procedure of Wj = 1/Sj2, where the variance Sj2 was calculated for each observation (Y) using the formula (a + b · Y)2. The parameters a and b are the intercept and slope of the variance model. The slope is the residual variability proportional to each concentration, and the intercept is the additive component of the error. Variance parameter estimates from the individual PK analysis (ADAPT-II) were used as beginning estimates and were updated iteratively during the population PK analysis until stable values were found.
Statistical analysis.
Statistical analyses were performed using SAS version 9.1.3 for Windows. Elvucitabine PK parameters obtained after a single dose with and without ritonavir were compared using an analysis of variance (ANOVA) using the Proc Mixed procedure as implemented in SAS. An ANOVA was performed on the ln-transformed PK parameters AUC0-24, AUC0-t, AUC0-∞, Cmax, clearance (CL/F), central volume of distribution (Vc/F), distributional clearance (CLd/F), peripheral volume of distribution (Vp/F), and volume of distribution (Vss/F) and included sequence, treatment, and period as fixed effects and subject nested within sequence as a random effect. Similarly, an ANOVA was performed on the PK parameters absorption rate constant (ka), t1/2, and lag time prior to the start of absorption. Tmax was compared using a Wilcoxon signed rank nonparametric analysis on paired data. Statistical significance was set a priori at P < 0.05.
RESULTS
Noncompartmental PK analysis.
Standard PK parameters including AUC0-24, AUC0-t, AUC0-∞, Cmax, kel, and t1/2 were calculated for 20 mg elvucitabine administered alone or coadministered with ritonavir. Results are presented in Table 1. Based on AUC0-∞ results, coadministration of ritonavir reduced the exposure of elvucitabine by approximately 30% (90% confidence interval [CI], 61.7 to 83.3). Elvucitabine's Cmax was reduced by 40.3% (90% CI, 44.8 to 79.6). The t1/2 of approximately 60 h was more than likely underestimated due to the sampling scheme of 96 h.
TABLE 1.
Noncompartmental elvucitabine (ELV) PK parameters in plasma
| ELV regimen | Mean value (CV [%]) [ratio (90% confidence interval)]
|
||||||
|---|---|---|---|---|---|---|---|
| AUC0-24 (ng · h/ml) | AUC0-t (ng · h/ml) | AUC0-∞ (ng · h/ml) | Cmax (ng/ml) | CL (liters/h) | Tmax (h) | t1/2 (h) | |
| Alone | 732 (33.8) | 1,029 (28.7) | 1,243 (29.7) | 136.1 (37.7) | 17.6 (31.2) | 3.78 (35.8) | 59.1 (22.1) |
| With ritonavir | 568 (54.8) [70.6 (60.2-82.9)] | 790 (52.5) [65.9 (53.1-81.7)] | 979.3 (50.0) [71.7 (61.7-83.3)] | 102.3 (62.2) [59.7 (44.8-79.6)] | 28.0 (66.1) [138.1 (119.0-160.3)] | 5.12 (116) [NSa] | 54.2 (23.2) [91.0 (84.2-98.3)] |
Tested using a Wilcoxon signed rank nonparametric analysis on paired data. NS, not statistically significant (P > 0.05).
Population compartmental analyses.
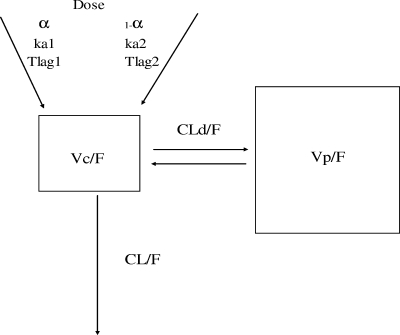
The population analysis was performed in two stages. The first consisted of testing different models in ADAPT-II to determine which was the simplest to explain the observed elvucitabine concentrations. The same analyses were performed for elvucitabine administered alone and for elvucitabine coadministered with ritonavir. For elvucitabine administered alone and with ritonavir, a linear two-compartment model with two first-order absorption rate constants and a first-order elimination process was determined to be the simplest model to explain the elvucitabine data. Results of the discrimination process are presented in Table 2. The final model is presented graphically in Fig. 1. This was consistent with the results found in a multiple-dose study of elvucitabine in HIV-1-infected subjects (2).
TABLE 2.
Discrimination criteria between PK modelsa
| Model | Mean value for ELV
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Alone
|
With ritonavir
|
|||||||
| ECV | AIC | r2 | Residual variability (%) | ECV | AIC | r2 | Residual variability (%) | |
| 2 CPT; 1 ka; 1 lag time per dose | 33.3 | 82.6 | 0.983 | 8.1 | 35.1 | 86.2 | 0.872 | 14.3 |
| 2 CPT; 1 ka; 2 lag time per dose; including alpha | 28.4 | 76.9 | 0.991 | 7.6 | 29.1 | 78.1 | 0.937 | 12.6 |
| 2 CPT; 2kaand 2 lag time per dose; including alpha | 25.2 | 72.3 | 0.992 | 6.2 | 26.9 | 75.9 | 0.932 | 11.3 |
| 3 CPT; 1 ka; 1 lag time per dose | 30.7 | 81.5 | 0.984 | 8.3 | 33.9 | 87.7 | 0.810 | 15.9 |
| 3 CPT; 1 ka; 2 lag time per dose; including alpha | 27.7 | 79.4 | 0.992 | 6.6 | 30.9 | 85.7 | 0.832 | 15.9 |
| 3 CPT; 2 ka and 2 lag time per dose; including alpha | 25.6 | 77.1 | 0.991 | 5.7 | 28.3 | 82.6 | 0.912 | 11.5 |
Bold type indicates the selected model. ELV, elvucitabine; CPT, compartment; ECV, estimator criteria value; AIC, Akaike information criterion.
FIG. 1.
Final PK model used in the compartmental analysis.
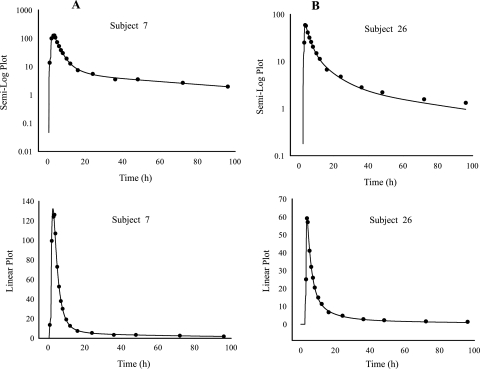
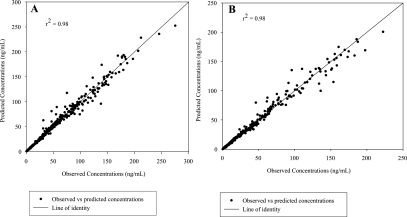
A population analysis was performed on the final model using a mixed-effect modeling approach (IT2S). This analysis was done to obtain better estimates of the population PK parameters, their variance (intersubject variability), the residual variability (also known as intrasubject variability) as well as the subject's individual results. The model contained the following PK parameters: two absorption rate constants (ka1 and ka2), a different lag time for each absorption rate constant, CL/F, Vc/F, Vp/F, and CLd/F. Results of the population PK parameters are presented in Table 3, and a graphical depiction of the predicted and observed concentrations of a representative subject is given in Fig. 2. The population predicted versus observed elvucitabine concentrations when administered alone and coadministered with ritonavir are presented in Fig. 3.
TABLE 3.
Elvucitabine (ELV) PK parameters estimated using IT2S population compartmental analyses
| ELV regimen | Mean (CV [%]), median (range) [ratio (90% confidence interval)]
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peak 1% (%) | Day 1 lag 1 (h) | Day 1 lag 2 (h) | ka1 (h−1) | ka2 (h−1) | CL/F (liters/h) | Vc/F (L) | CLd/F (liters/h) | Vp/F (liters) | Vss/F (liters) | λZ-HL (h) | Residual variability (%) | |
| Alone | 44.2 (47.4), 40.1 (9.9-99.0) | 1.60 (69.2), 1.39 (0.078-5.97) | 2.30 (46.0), 2.05 (0.607-6.38) | 0.183 (32.8), 0.192 (0.049-0.311) | 0.459 (21.0), 0.442 (0.275-0.679) | 17.8 (29.2), 16.1 (10.7-28.0) | 26.1 (67.5), 20.5 (6.9-73.8) | 16.3 (33.9), 15.5 (7.4-26.0) | 703 (30.5), 649 (336-1247) | 729 (29.9), 683 (376-1286) | 58.6 (12.0), 57.3 (46.8-75.7) | 9.1 |
| With ritonavir | 53.9 (18.9), 56.2 (30.2-73.4) [134.7 (112.3-161.8)] | 2.96 (141.0), 2.43 (0.623-24.08) [166.2 (123.7-223.2)] | 3.54 (116.2), 2.85 (0.96-24.3) [126.3 (105.1-151.8)] | 0.127 (56.5), 0.141 (0.004-0.246) [61.4 (45.9-82.1)] | 0.483 (37.7), 0.450 (0.194-0.809) [102.3 (91.3-114.7)] | 29.2 (64.8), 20.2 (11.0-66.8) [135.0 (115.8-157.4)] | 24.8 (53.0), 26.8 (4.9-51.6) [96.6 (75.6-123.5)] | 19.3 (35.6), 18.1 (8.2-33.6) [116.0 (103.3-130.4)] | 897 (31.6), 825 (352-1435) [124.6 (111.9-138.8)] | 921 (30.3), 859 (375-1451) [123.7 (111.4-137.4)] | 60.2 (26.1), 58.9 (33.6-96.0) [101.3 (92.9-110.5)] | 9.2 |
FIG. 2.
Predicted (·) versus observed (-) concentrations of elvucitabine (20 mg) administered alone (A) or with ritonavir (B). Data are from a representative subject.
FIG. 3.
Predicted versus observed concentrations for elvucitabine (20 mg) administered alone (A) or with ritonavir (B).
Results from the population analysis show that elvucitabine's half-life remained the same, while both CL/F and Vss/F increased by approximately 30% when coadministered with ritonavir. This indicates that the bioavailability of elvucitabine was significantly decreased by ritonavir. Results also show that ritonavir delayed and slowed elvucitabine's rate of absorption.
DISCUSSION
The PK behavior of elvucitabine was best described by a two-compartment linear model using two absorption rates with different lag times. Results indicated that the quality of fit was consistently better when two absorption rates were included in the model regardless of whether elvucitabine was administered alone or with ritonavir (Table 2). This is not surprising, as elvucitabine is a delayed-release formulation, and multiple absorption rates are often required when modified-release formulations are modeled (5).
The residual variability left from the population analyses was low at approximately 9% for elvucitabine administered alone or with ritonavir. This suggests that the model chosen was appropriate. In addition, the predicted versus observed concentrations (Fig. 3) were close to the line of identity, and no trend was detected on the plot (absence of bias), which further demonstrates the validity of the model.
Comparing the noncompartmental and compartmental analyses can be useful in determining consistency between PK methods. Both methods provided similar clearance values for elvucitabine administered alone (17.8 versus 17.6 liters/h) and coadministered with ritonavir (29.2 versus 28.0). Results from the compartmental analysis indicate that ritonavir delayed the start of absorption of elvucitabine by 1 h and decreased one of its rates of absorption by nearly 40%. These results are in agreement with the observed 1.3 h shift in Tmax and lower Cmax of elvucitabine.
This in vivo drug-drug interaction study indicated a clinically significant PK interaction of ritonavir with elvucitabine. Elvucitabine's AUC0-∞ and Cmax decreased by 28.3% (61.7 to 83.3%) and 40.3% (44.8 to 79.6%), respectively, when it was coadministered with ritonavir. This clinically significant PK interaction with ritonavir appears to be due to a decrease in the bioavailability and not in a change in the elimination rate. As elvucitabine is not metabolized by CYP enzymes, any impact by ritonavir on the hepatic and intestinal CYP enzymes would be negligible. As elvucitabine is renally eliminated unchanged and the half-life for elvucitabine was not affected, it can be assumed that the elimination process (renal transporters) was not affected. Therefore, a plausible cause for the decrease in elvucitabine exposure by ritonavir would be an alteration in the activity of gut transporters. We can assume that some transporters other than the efflux ABCB1 are affected, as F decreased instead of increasing. This large dose of ritonavir (300 mg) therefore likely affected some influx gut transporters. Although the effect of ritonavir on influx transporters has not been reported to our knowledge, a closely related compound, saquinavir, is a substrate for OATP-A transporters (13). It is therefore possible that ritonavir may affect the activity of OATP transporters.
Conclusion.
Elvucitabine PK behavior was well described by a linear two-compartment model with two first-order absorption rates and a first-order elimination rate.
Ritonavir significantly decreased the exposure of elvucitabine by decreasing its bioavailability. A probable cause for this decrease may be an inhibition of absorption influx transporters by ritonavir.
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Collins, D., and A. Forrest. 1995. IT2S user's guide. State University of New York at Buffalo, Buffalo, NY.
- 2.Colucci, P., J. C. Pottage, H. Robison, J. Turgeon, D. Schürmann, I. M. Hoepelman, and M. P. Ducharme. 2009. Multiple-dose pharmacokinetic behavior of elvucitabine, a nucleoside reverse transcriptase inhibitor, administered over 21 days with lopinavir-ritonavir in human immunodeficiency virus type 1-infected subjects. Antimicrob. Agents Chemother. 53:662-669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Argenio, D., and A. Schumitzky. 1997. ADAPT-II user's manual. Biomedical Simulations Resource, University of Southern California, Los Angeles.
- 4.Drewe, J., H. Gutmann, G. Fricker, M. Torok, C. Beglinger, and J. Huwyler. 1999. HIV protease inhibitor ritonavir: a more potent inhibitor of P-glycoprotein than the cyclosporine analog SDZ PSC 833. Biochem. Pharmacol. 57:1147-1152. [DOI] [PubMed] [Google Scholar]
- 5.Fradette, C., J. Lavigne, D. Waters, and M. P. Ducharme. 2005. The utility of the population approach applied to bioequivalence in patients: comparison of 2 formulations of cyclosporine. Ther. Drug Monit. 27:592-600. [DOI] [PubMed] [Google Scholar]
- 6.Gerber, J. G., S. Rosenkranz, Y. Segal, J. Aberg, R. D'Amico, D. Mildvan, R. Gulick, V. Hughes, C. Flexner, F. Aweeka, A. Hsu, and J. Gal. 2001. Effect of ritonavir/saquinavir on stereoselective pharmacokinetics of methadone: results of AIDS Clinical Trials Group (ACTG) 401. J. Acquir. Immune Defic. Syndr. 27:153-160. [DOI] [PubMed] [Google Scholar]
- 7.Kumar, G. N., A. D. Rodrigues, A. M. Buko, and J. F. Denissen. 1996. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J. Pharmacol. Exp. Ther. 277:423-431. [PubMed] [Google Scholar]
- 8.Messeri, P., G. Lee, D. M. Abramson, A. Aidala, M. A. Chiasson, and D. J. Jessop. 2003. Antiretroviral therapy and declining AIDS mortality in New York City. Med. Care 41:512-521. [DOI] [PubMed] [Google Scholar]
- 9.Olson, D. P., D. T. Scadden, R. T. D'Aquila, and M. P. De Pasquale. 2002. The protease inhibitor ritonavir inhibits the functional activity of the multidrug resistance related-protein 1 (MRP-1). AIDS 16:1743-1747. [DOI] [PubMed] [Google Scholar]
- 10.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 11.Sheldon, C., R. Sukovaty, and V. Andaloro. 2004. Validation of an LC-MS/MS method for the quantitation of ACH-126,443 in human EDTA plasma. Project 27427_1. MDS Pharma Services, Lincoln, NE.
- 12.Smit, C., R. Geskus, D. Uitenbroek, D. Mulder, A. Van Den Hoek, R. A. Coutinho, and M. Prins. 2004. Declining AIDS mortality in Amsterdam: contributions of declining HIV incidence and effective therapy. Epidemiology 15:536-542. [DOI] [PubMed] [Google Scholar]
- 13.Su, Y., X. Zhang, and P. J. Sinko. 2004. Human organic anion-transporting polypeptide OATP-A (SLC21A3) acts in concert with P-glycoprotein and multidrug resistance protein 2 in the vectorial transport of saquinavir in Hep G2 cells. Mol. Pharm. 1:49-56. [DOI] [PubMed] [Google Scholar]
- 14.WHO. 2006. Report on the global AIDS epidemic. World Health Organization, Geneva, Switzerland.
- 15.Yeni, P. G., S. M. Hammer, C. C. Carpenter, D. A. Cooper, M. A. Fischl, J. M. Gatell, B. G. Gazzard, M. S. Hirsch, D. M. Jacobsen, D. A. Katzenstein, J. S. Montaner, D. D. Richman, M. S. Saag, M. Schechter, R. T. Schooley, M. A. Thompson, S. Vella, and P. A. Volberding. 2002. Antiretroviral treatment for adult HIV infection in 2002: updated recommendations of the International AIDS Society—USA Panel. JAMA 288:222-235. [DOI] [PubMed] [Google Scholar]
- 16.Yeni, P. 2006. Update on HAART in HIV. J. Hepatol. 44(Suppl. 1):S100-S103. [DOI] [PubMed] [Google Scholar]