Abstract
Salmonella genomic island 1 was identified for the first time in Salmonella enterica serovar Virchow isolated from humans in Taiwan. The complex class 1 integron conferring multidrug resistance was shown to be inserted within open reading frame (ORF) S023 and contains for the first time a partial transpositional module. The 5-bp target duplication flanking the complex integron suggests that its insertion in ORF S023 was by transposition.
Salmonella genomic island 1 (SGI1) is a 43-kb integrative mobilizable element initially characterized in multidrug-resistant Salmonella enterica serovar Typhimurium phage type DT104 strains (hereafter serovar Typhimurium DT104) (1, 4). SGI1 is a site-specific integrative element, which is able to excise itself from or integrate into the last 18 bp of the chromosomal thdF gene (4). Recently, SGI1 was also demonstrated, during in vitro conjugation experiments, to be able to integrate into a secondary specific attachment site in the chromosome of serovar Typhimurium LT2 (6).
SGI1 contains a complex class 1 integron designated In104 responsible for the pentadrug resistance phenotype of serovar Typhimurium DT104 strains (1, 9). The complex integron In104 and variants of it named SGI1-A to SGI1-Q have been described in several other S. enterica serovars and also in Proteus mirabilis (2, 3, 7, 9, 10, 13). All In104 integrons and variants of it are found always at the same position in the SGI1 backbone, i.e., between the res gene (also named tnpR) and open reading frame (ORF) S044 of SGI1 (1, 13).
Recently, Levings et al. have described an unusual SGI1 variant in Salmonella enterica serovar Emek strains isolated between 1999 and 2002 (11). The complex class 1 integron initially named SGI1-J contains the dfrA1-orfC cassette array in the first attI1 site and a deletion at the second attI1 site. Flanked by the two integron-like structures are a new variant of the floR gene called floR2 or cmlA9 and the tetracycline resistance genes tetR and tet(G) (8, 11). For the first time, this complex integron, relative to In104, was found inserted within ORF S023 of the SGI1 backbone. For this major reason, Levings et al. chose to rename SGI1-J as SGI2 (9, 11). This point of nomenclature will be further discussed below.
In the present study, we examined three Salmonella enterica serovar Virchow strains, isolated from human blood in 1993 and 1994 in Taiwan. These strains lacked plasmids and displayed a multidrug resistance profile (Table 1) suggesting the presence of SGI1.
TABLE 1.
Characteristics of multidrug-resistant S. enterica strains
| Serovar and strain(s) | Antibiotic resistance profilea,b | SGI1 integron-borne cassette(s) | Complex integron variant | Integron position in SGI1 (GenBank accession no. AF261825) | Chloramphenicol and florfenicol resistance gene variant | MIC (μg/ml)c
|
|
|---|---|---|---|---|---|---|---|
| Chloramphenicol | Florfenicol | ||||||
| Typhimurium strain BN9181 | AmpChlFloStrSptSulTet | aadA2, blaPSE-1 | In104 | 26636 | floR | 256 | 64 |
| Virchow strains B94, B98, and B100 | ChlFloSulTetTmp-NalFlu | dfrA1-orfC | InSGI1-J | 19747 | floR2 | >256 | 32 |
Abbreviations: Amp, ampicillin; Chl, chloramphenicol; Flo, florfenicol; Str, streptomycin; Spt, spectinomycin; Sul, sulfonamides; Tet, tetracycline; Tmp, trimethoprim; Nal, nalidixic acid; Flu, flumequine.
For antibiotics indicated in boldface, resistance was conferred by SGI1.
The MIC breakpoints for phenicols were defined by the Antibiogram Committee of the French Society of Microbiology, i.e., susceptible (MIC, ≤8 μg/ml) or resistant (MIC, >16 μg/ml) (12).
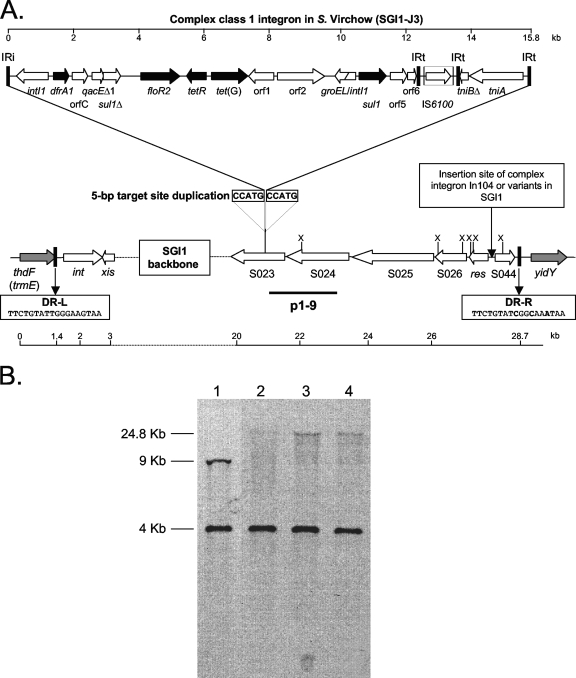
Detection of SGI1 and its location in the chromosome were performed by PCR as previously described (5). PCR results were positive for the left and right junctions, indicating that the three serovar Virchow strains harbor SGI1 at the same chromosomal location, i.e., between the chromosomal thdF and yidY genes as in other S. enterica serovars (Fig. 1A). The junction PCR products were sequenced to analyze the left and right direct repeats DR-L and DR-R, respectively. The 18-bp DR-L sequence is almost identical to the 18-bp attP sequence of SGI1 previously described (4). However, the 18-bp DR-R sequence showed three substitutions compared to DR-L (Fig. 1A). PCR mapping of the In104 integron of SGI1 was performed as previously described (5). This mapping revealed only the presence of the tetracycline resistance genes tetR and tet(G) (5). Thus, PCR of the integron-borne cassettes was undertaken and revealed only a 1.3-kb fragment in the three serovar Virchow strains. Sequence analysis showed the presence of the trimethoprim resistance gene cassette dfrA1 and a gene cassette of unknown function, orfC. These results suggested the occurrence of the SGI1-J complex integron variant recently described in serovar Emek strains (11). Then, PCRs were carried out from the sul1Δ gene to the tetR gene and the resulting product was sequenced. Sequence analysis confirmed the occurrence of the floR2 resistance gene (Fig. 1A). Moreover, chloramphenicol and florfenicol MICs suggested that this variant of the floR gene conferred a resistant phenotype on both antibiotics (Table 1) according to susceptibility breakpoints (12). Thus, the name floR2 may be much more appropriate than cmlA9 as proposed by Levings et al. (11), as all cmlA genes confer only chloramphenicol resistance and not florfenicol resistance (16). Interestingly, the region extending from IRi to the groEL-intI1 fusion gene was also found in the antibiotic resistance gene cluster from an epidemic multidrug-resistant Acinetobacter baumannii strain isolated in France (8). The remaining part of the complex integron was mapped by PCR from the tetR gene to the IS6100 element by using primers previously described (7).
FIG. 1.
(A) Genetic organization of the complex class 1 integron in SGI1-J3 from serovar Virchow isolates. Black and gray arrows correspond to SGI1 antibiotic resistance genes and chromosomal genes flanking SGI1, respectively. DR-L and DR-R are the 18-bp left and right direct repeats, respectively, bracketing SGI1. IRi and IRt are 25-bp imperfect inverted repeats defining the left and right end of complex class 1 integrons. The usual insertion point of the complex integron In104 or variants of it in SGI1 and the 5-bp target site duplication flanking the complex integron (SGI1-J3) in ORF S023 are indicated (GenBank accession number EU924797). The p1-9 probe and XbaI restriction sites (X) are indicated. (B) Southern blot hybridization with the p1-9 probe of XbaI-digested genomic DNAs of S. enterica serovar Typhimurium DT104 control strain BN9181 carrying SGI1 (lane 1), serovar Virchow strain B94 (lane 2), serovar Virchow strain B98 (lane 3), and serovar Virchow strain B100 (lane 4).
To detect the boundaries of the complex integron with the SGI1 backbone, PCRs were applied using a forward primer in S023 with a reverse primer in the intI1 gene and an IS6100 forward primer with a reverse primer in S024. Sequence analysis of the left boundary of the complex integron showed that the complex integron of serovar Virchow is inserted in SGI1 exactly at the same position within ORF S023 as in serovar Emek (Table 1; Fig. 1A). However, the right boundary PCR product was 2.1 kb larger than the size expected from the SGI1-J (SGI2) sequence with the GenBank accession number AY963803 (11). Sequence analysis revealed that, adjacent to the short segment of 152 bp derived from the IRt outer end of Tn402, the complex integron of serovar Virchow strains harbored the right outer end of the mercury resistance transposon Tn5058 (GenBank accession no. Y17897) (14, 15). This fragment contains a short part of the tniB gene (tniBΔ) coding for an ATPase DNA binding protein, a transposase tniA gene, and the IRt outer end of Tn5058 (Fig. 1A). The 5-bp duplication target is found downstream of this third IRt copy of Tn5058. The location of the complex integron within S023 was confirmed by XbaI-Southern blot hybridization using the p1-9 probe (Fig. 1B) (4, 6). The p1-9 probe showed two XbaI fragments, one fragment of the expected 4-kb size as in the SGI1-carrying control strains and another less-visible fragment larger than 20 kb due to the insertion of the complex integron in S023 (Fig. 1A and B). The 5′ region of SGI1 of approximately 15 kb was mapped by PCR and did not reveal differences of genetic organization (data not shown).
The three serovar Virchow strains studied here harbored an SGI1 variant very similar to that recently described for serovar Emek (9, 11). Levings et al. sequenced 20% of the SGI1 backbone in serovar Emek and revealed more than 99.7% nucleotide identity to the sequence of SGI1 (GenBank accession no. AF261825) (11). Except the complex integron variant, the only major change from other SGI1 variants is the insertion point within the SGI1 backbone. Thus, according to the previously proposed SGI1 nomenclature, it would be preferable to keep the name SGI1 and to add a letter and a number if necessary to classify variants of SGI1 (7). We propose to maintain the name SGI1-J for the variant described in serovar Emek strains (SGI1-J corresponds to SGI2 and SGI1-J2 corresponds to SGI2-A) and to name the variant described here in serovar Virchow as SGI1-J3 according to the few differences between these two variants.
In conclusion, serovar Virchow represents the15th serovar of S. enterica harboring a variant of SGI1 named SGI1-J3 in which the integron transposition occurred in a different location than those in the other SGI1 variants. The presence of a large part of the tni module of Tn5058 containing the transposase tniA gene could facilitate the transposition event of this complex integron. SGI1 seems to be a “hot spot” of acquisition of complex In4-type integrons or transposon structures as recently described in variants SGI1-K, -P, and -Q (7) and thus serves as a vehicle to transfer multidrug resistance between different bacterial genera.
Nucleotide sequence accession number.
The partial sequence of the SGI1-J3 variant has been deposited in GenBank under accession number EU924797.
Acknowledgments
We gratefully acknowledge François-Xavier Weill for serotyping of the strains and the expert technical assistance of Karine Praud and Raquel Bes Torres.
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Boyd, D. A., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, D. A., X. Shi, Q. H. Hu, L. K. Ng, B. Doublet, A. Cloeckaert, and M. R. Mulvey. 2008. Salmonella genomic island 1 (SGI1), variant SGI1-I, and new variant SGI1-O in Proteus mirabilis clinical and food isolates from China. Antimicrob. Agents Chemother. 52:340-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cloeckaert, A., K. Praud, B. Doublet, M. Demartin, and F.-X. Weill. 2006. Variant Salmonella genomic island 1-L antibiotic resistance cluster in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 50:3944-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doublet, B., D. A. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 5.Doublet, B., F.-X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doublet, B., G. R. Golding, M. R. Mulvey, and A. Cloeckaert. 2008. Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1. PLoS ONE 3:e2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doublet, B., K. Praud, S. Bertrand, J.-M. Collard, F.-X. Weill, and A. Cloeckaert. 2008. Novel insertion sequence- and transposon-mediated genetic rearrangements in the genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52:3745-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fournier, P.-E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J.-M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumanii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levings, R. S., S. R. Partridge, S. P. Djordjevic, and R. M. Hall. 2007. SGI1-K, a variant of the SGI1 genomic island carrying a mercury resistance region, in Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 51:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levings, R. S., S. P. Djordjevic, and R. M. Hall. 2008. SGI2, a relative of the Salmonella genomic island SGI1 with an independent origin. Antimicrob. Agents Chemother. 52:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Members of the SFM Antibiogram Committee. 2003. Comité de l'Antibiogramme de la Société Française de Microbiologie report 2003. Int. J. Antimicrob. Agents 21:364-391. [DOI] [PubMed] [Google Scholar]
- 13.Mulvey, M. R., D. A. Boyd, A. B. Olson, B. Doublet, and A. Cloeckaert. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915-1922. [DOI] [PubMed] [Google Scholar]
- 14.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz, S., C. Kehrenberg, B. Doublet, and A. Cloeckaert. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519-542. [DOI] [PubMed] [Google Scholar]