Abstract
The potency of antisense peptide-phosphorodiamidate morpholino oligomers (PPMOs) was improved by varying the peptide composition. An antisense phosphorodiamidate morpholino oligomer (PMO) complementary to the mRNA of the essential gene acpP (which encodes the acyl carrier protein required for lipid biosynthesis) in Escherichia coli was conjugated to the 5′ ends of various cationic membrane-penetrating peptides. Each peptide had one of three repeating sequence motifs: C-N-N (motif 1), C-N (motif 2), or C-N-C (motif 3), where C is a cationic residue and N is a nonpolar residue. Variations in the cationic residues included arginine, lysine, and ornithine (O). Variations in the nonpolar residues included phenylalanine, valine, β-alanine (B), and 6-aminohexanoic acid (X). The MICs of the PPMOs varied from 0.625 to >80 μM (about 3 to 480 μg/ml). Three of the most potent were the (RX)6B-, (RXR)4XB-, and (RFR)4XB-AcpP PMOs, which were further tested in mice infected with E. coli. The (RXR)4XB-AcpP PMO was the most potent of the three conjugates tested in mice. The administration of 30 μg (1.5 mg/kg of body weight) (RXR)4XB-AcpP PMO at 15 min postinfection reduced CFU/ml in blood by 102 to 103 within 2 to 12 h compared to the numbers in water-treated controls. All mice treated with 30 μg/dose of (RXR)4XB-AcpP PMO survived infection, whereas all water-treated mice died 12 h postinfection. The reduction in CFU/ml in blood was proportional to the dose of PPMO from 30 to 300 μg/ml. In summary, the C-N-C motif was more effective than the other two motifs, arginine was more effective than lysine or ornithine, phenylalanine was more effective than 6-aminohexanoic acid in vitro but not necessarily in vivo, and (RXR)4XB-AcpP PMO reduced bacterial infection and promoted survival at clinically relevant doses.
Antibiotic resistance in bacteria, its increasing incidence, and the decreasing rate of discovery of new antibiotics create a need for new strategies of targeting bacterial components for antibiotic therapy. Genomics has created an attractive opportunity to target specific genes for inhibition by the use of antisense oligomers to silence essential genes.
Synthetic antisense oligomers are DNA mimics that bind to target RNA through complementary base pairing. When antisense oligomers are added to pure bacterial cultures, they have been shown to decrease the levels of expression of reporter genes such as luciferase (5, 7), activate endogenous genes such as β-galactosidase (7), or inhibit growth by targeting essential genes or RNA (9, 10, 11, 14, 23, 28). In addition, antisense oligomers reduce infection and increase the survival of mice infected with Escherichia coli (8, 21, 22).
Antisense oligomers require assistance to gain entry into bacterial cells because of their molecular weight and polar characteristics. Recently, antisense oligomers have been conjugated to membrane-penetrating peptides. Peptide-oligomer conjugates are significantly more effective in inhibiting the expression of their specific targets than their nonconjugated counterparts (7, 10). Apparently, the membrane-penetrating peptide carries its cargo (the antisense oligomer) across the outer membrane of gram-negative bacteria, after which it traverses the plasma membrane by an unknown mechanism.
Membrane-penetrating peptides have diverse sequences, but many are cationic and amphipathic. Previous investigations suggest that a repeated peptide motif with one cationic residue followed by either one or two nonpolar residues may be an important feature for efficient membrane penetration (25).
Antisense oligomers conjugated to peptides with either (KFF)3 or (RFF)3 and targeted to essential gene products are potent growth inhibitors of E. coli, Salmonella enterica serovar Typhimurium, Staphylococcus aureus, Mycobacterium smegmatis, and Klebsiella pneumoniae (10, 13, 14, 23). Although antisense peptide-oligomers with variations in their peptide sequences have been screened for their efficacies in the gram-positive bacterium S. aureus (17), the effects of only a few variations on the gram-negative bacterium E. coli have been reported (10, 23). Moreover, there have been few if any reports on the effects of variations in the peptide sequences of antisense peptide-oligomers in animal models of bacterial infection.
In the study described in this report, E. coli was used to compare the efficacies in vitro and in vivo of antisense peptide-phosphorodiamidate morpholino oligomers (PPMOs) that vary in their amino acid sequences.
MATERIALS AND METHODS
Bacterial strains.
Wild-type strain E. coli K-12 W3110 was used in all experiments. Liquid cultures were grown in either Mueller-Hinton II broth (for MIC assays) or LB broth (for preparation of the inoculum for mice).
MICs.
MICs were determined by the microdilution method (4). To control for nonspecific toxicity, each peptide was also conjugated to a second phosphorodiamidate morpholino oligomer (PMO) with a base sequence that was not complementary to any gene in E. coli. In all cases, the MIC of the control PPMO was above the limit of measurement, which was 80 μM.
PMO.
PPMOs were synthesized at AVI BioPharma (Corvallis, OR) as described previously (23). The base sequence of the AcpP PMO is 5′-CTTCGATAGTG-3′, which is complementary to bases 1153197 to 1153207 on the E. coli W3110 genomic map (NCBI accession number AP009048). The target sequence is immediately 3′ to the start codon. The rationale for the length and position of the PMO on the target mRNA has been characterized previously (5). This PMO sequence has been shown to inhibit the growth of E. coli in pure cultures (5, 8, 23), infected tissue culture (23), and mouse models of infection (8, 22). The base sequence of the nonsensical control PMOs is 5′-TCTCAGATGGT-3′. The nonsensical (scrambled) sequence is a random sequence of the same 11 bases used in the AcpP PMO and has previously been shown not to inhibit the growth of E. coli (5, 8, 23, 24).
Mouse experiments.
All mice were infected and treated with the PPMOs as described previously (22). Briefly, groups of three to four mice were infected by the intraperitoneal injection of E. coli W3110. The mice were treated by intraperitoneal injection at 15 min and 12 h postinfection. Blood was collected at 2, 6, 12, 24, and 48 h postinfection; diluted; and plated to determine the numbers of viable bacteria.
All animal procedures were approved by the Oregon State University Animal Care and Use Committee and complied with all state and federal laws.
Statistical analysis.
The mean numbers of CFU/ml blood and standard deviation were calculated for each treatment group by using Prism (version 4.0) software (GraphPad Software, San Diego, CA). Survival curves were analyzed by the Kaplan-Meier method with Prism (version 4.0) software. Group means were compared by transforming the numbers of CFU/ml to log CFU/ml and then using one-way analysis of variance, parametric methods, the Kolmogorov and Smirnov normality test, and the Tukey-Kramer multiple comparison post test (InStat, version 3.05, software; GraphPad Software).
RESULTS
Efficacies of various PPMOs in pure culture.
Various PPMOs were synthesized with the same base sequence targeting a region near the start codon of mRNA of acpP, but they had different peptides attached to the 5′ end (Table 1). Each peptide was designed with one of three repeated sequence motifs: (i) C-N-N (motif 1), (ii) C-N (motif 2), or (iii) C-N-C (motif 3), where C is a cationic residue and N is a nonpolar residue. The cationic amino acids that were used were arginine, lysine, and ornithine (O); and the nonpolar amino acids that were used were phenylalanine, valine, β-alanine (B), and 6-aminohexanoic acid (X).
TABLE 1.
MICs of peptide-AcpP11 PMO conjugates in pure cultures of E. coli
| Motif and PMO no. | Conjugated peptide | MIC
|
|
|---|---|---|---|
| μM | μg/ml | ||
| Motif 1 (C-N-N) | |||
| 05-0198 | RFFXB | >80 | >360 |
| 05-0197 | RFFRFFXB | 20 | 99 |
| 05-0200 | RFFRFFRFFRXB | 2.5 | 14 |
| 05-0653 | dRFdFdRdFdFdRdFdFdRXBa | 5 | 28 |
| 23-248 | RXXRXXRXXB | 20 | 102 |
| 06-0199 | KFFKFFKFFKXB | 10 | 54 |
| 06-0064 | RBBRBBRBBRXB | >80 | >408 |
| 06-0079 | RVVRVVRVVRXB | >80 | >422 |
| 06-0082 | OFFOFFOFFOXB | >80 | >431 |
| Motif 2 (C-N) | |||
| 06-0067 | RXRXB | >80 | >358 |
| 06-0070 | RXRXRXRXB | 80 | 401 |
| 06-0073 | RXRXRXRXRXRXB | 1.25 | 6.9 |
| 06-0761 | KXKXKXKXKXKXB | >80 | >430 |
| 06-0762 | OXOXOXOXOXOXB | >80 | >424 |
| Motif 3 (C-N-C) | |||
| 06-0076 | RXRRXRRXRRXRXB | 1.25 | 7.2 |
| 06-0824 | KXKKXKKXKKXKXB | 20 | 111 |
| 06-0763 | OXOOXOOXOOXOXB | 10 | 54 |
| 07-0795 | RFRRFRRFRRFRXB | 0.625 | 3.7 |
| 07-1119 | RXRRBRRXRRBRXB | 2.5 | 14 |
| Control (ampicillin) | 6.7 | 2.5 | |
d, the residue that follows is of the d-isomeric form.
The MIC of each PPMO with motif 1 (C-N-N) was measured and is shown in Table 1. Control PPMOs with the same peptides attached to a PMO without a complementary sequence had MICs above 80 μM (data not shown). The results indicate that the most potent PPMO with peptide motif 1 was (RFF)3RXB, which was about 2.5 times more potent than ampicillin on a molar basis. Furthermore, increasing the number of RFF repeat units from one to three reduced the MIC >32-fold from >80 to 2.5 μM. The replacement of arginine with lysine or ornithine increased the MIC by 4-fold and >32-fold, respectively. The substitution of valine, β-alanine, or 6-aminohexanoic acid for phenylalanine increased the MICs at least eightfold (compare the MICs of PPMOs 06-0079, 06-0064, and 23-248 with the MIC of PPMO 05-0200 in Table 1). Interestingly, the PPMO with the d-isomeric form of (RFF)3RXB (PPMO 05-0653) had an MIC that was only two times higher than that of the equivalent l-isomeric form (PPMO 05-0200).
The MIC of each PPMO with motif 2 (C-N) or motif 3 (C-N-C) was measured. PPMOs with (RX)6B, (RXR)4XB, or (RFR)4XB were more potent than PPMOs with (RXX)3B or (RFF)3RXB. The substitution of either lysine or ornithine for arginine in PPMOs with motif 2 or 3 reduced the potency. Two or four tandem copies of RX were not as potent as six tandem copies of RX.
Mouse peritonitis.
The (RX)6B-AcpP PMO, (RXR)4XB-AcpP PMO, and (RFR)4XB-AcpP PMO were tested in mice infected with E. coli. Groups of mice were treated with various doses of PPMOs ranging from 10 to 300 μg at 15 min and 12 h postinfection.
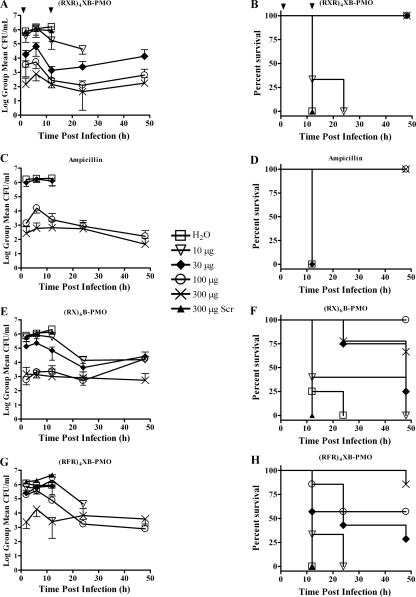
The results show that the (RXR)4XB-AcpP PMO was the most potent of the three PPMOs tested with mice (compare the effects of the 30-μg dose of each PPMO shown in Fig. 1A, B, E, F, G, and H). One 30-μg dose (1.5 mg/kg of body weight) of the (RXR)4XB-AcpP PMO significantly (P < 0.001) reduced the levels of bacteria in the blood by 98% within 2 h compared to the levels in the water- or scrambled-sequence-PPMO-treated controls. Higher doses (100 μg and 300 μg) of the (RXR)4XB-AcpP PMO further reduced CFU/ml in the blood proportionally by 2.5 to over 4 orders of magnitude over the first 12 h posttreatment. All mice treated with the 30-μg, 100-μg, or 300-μg dose survived the infection. There was no significant difference (P > 0.05) in the rates of survival between the group treated with water and that treated with 10 μg.
FIG. 1.
Bacteria in blood and survival of mice. Groups of mice (n = 3 to 7) were infected with E. coli and treated at 15 min and 12 h postinfection (arrows) with various PPMOs, a scrambled-base-sequence (Scr) (RXR)4XB PPMO, or ampicillin. Blood was collected at the indicated times and spread on LB plates to determine the numbers of CFU. (A and B) (RXR)4XB-PMO; (C and D) ampicillin; (E and F) (RX)6B-PMO; (G and H) (RFR)4XB-PMO. Error bars indicate standard deviations.
Groups of mice were treated with a 30-μg, 100-μg, or 300-μg dose of ampicillin for comparison of the results to those obtained for mice treated with a PPMO (Fig. 1C and D). Thirty-microgram doses of ampicillin were ineffective at reducing CFU/ml in the blood or increasing the rate of survival. Both of the higher doses of ampicillin significantly (P < 0.001) reduced CFU/ml in the blood by about 3 to 3.5 orders of magnitude and increased the rate of survival to 100% after 48 h.
The (RX)6B-AcpP PMO significantly reduced CFU/ml in blood (Fig. 1E and F), although it was not as potent as the (RXR)4XB-AcpP PMO (compare Fig. 1E and F with Fig. 1A and B). The 30-μg dose of the (RX)6B-AcpP PMO significantly (P < 0.001) reduced CFU/ml blood by 88% within 2 h postinfection. The 100-μg and 300-μg doses each significantly (P < 0.001) reduced CFU/ml in blood equally by about 3 orders of magnitude at all times postinfection. All mice treated with 100 μg/dose (RX)6B-AcpP PMO survived, although significant numbers of mice did not survive treatment with 10 μg, 30 μg, or 300 μg per dose.
The (RFR)4XB-AcpP PMO also significantly reduced the numbers of bacteria in the blood and promoted survival (Fig. 1G and H). Doses of 10 μg or 30 μg did not significantly (P > 0.05) reduce CFU/ml in the blood at any time. Doses of 100 μg or 300 μg significantly (P < 0.01 and < 0.001, respectively) reduced CFU/ml in the blood within 2 h posttreatment. The CFU/ml of blood was reduced proportionally to the dose, and the 300-μg dose showed the most reduction (2 to 3 orders of magnitude) at 2, 6, and 12 h postinfection. The number of surviving mice was proportional to the dose.
DISCUSSION
The purpose of our experiments was to increase the potency of antisense PPMOs targeted to the essential gene acpP, which was previously shown to inhibit the growth of E. coli both in pure culture and in infected mice (22). We hypothesized that the efficacies of PPMOs might increase by varying the order of cationic and nonpolar amino acid residues within the repeating motif or by replacing arginine with other cationic amino acids, phenylalanine with other nonpolar amino acids, or naturally occurring amino acids with nonprotein amino acids. The rationale for including nonprotein amino acids in the peptide moiety was that proteolytic degradation might be reduced.
Previously, Vaara and Porro (25) suggested the possibility that the repeating amino acid sequence motifs (cationic-hydrophobic-hydrophobic) and (cationic-hydrophobic) might influence the membrane-penetrating characteristics of peptides. Their results suggest that the former motif is more effective than the latter motif in disorganizing the outer membrane of gram-negative bacteria. Although our experiments used a completely different end point than that used in the experiments of Vaara and Porro (25), our results show that the spacing of cationic and nonpolar residues does have an effect but support the suggestion that the motif with cationic-nonpolar-cationic residues is slightly more effective than the other two motifs tested.
The mechanisms by which amphipathic peptides penetrate biological membranes are controversial and depend on the structures and compositions of the peptides and target membranes (12, 29). Although our experiments do not directly address the mechanism of penetration, our results are consistent with those presented in other reports that demonstrate the importance of the number of repeating sequence motifs and the choice of cationic and nonpolar amino acids. In a recent study, the number of RW sequence repeats was varied from one to five (15). The optimal number was found to be three. Our results also show that one or two RFF repeats were ineffective but that three RFF repeats had significantly increased efficacy. However, two or four RX repeats were ineffective, but six RX repeats were highly effective. A similar effect has been shown for the uptake of PPMOs into HeLa cells (27). This suggests that it may be the length or the overall charge of the peptide that is more important than the number of repeats. Alternatively, the effect may be influenced by the bulkiness or hydrophobicity of the nonpolar side chains. Because 6-aminohexanoic acid has no side chain, the presumed interaction with the hydrophobic core of the bilayer may be quite different from that of phenylalanine. Previous results have concluded that for short membrane-penetrating peptides (such as the ones that we used), the composition of the amino acids is more important than the order of the amino acids (19, 20).
The results of in vitro testing showed that among the PPMOs with an amino acid sequence repeat similar to that of (RFF)3RXB-AcpP (repeat motif 1; Table 1), arginine is superior to lysine or ornithine and phenylalanine is superior to valine, β-alanine, or 6-aminohexanoic acid. Among the PPMOs with repeat motifs 2 and 3, the trends were similar to those for motif 1, although valine and β-alanine were not evaluated as substitutes for nonpolar residues. Unfortunately, attempts to synthesize the (RF)6XB-AcpP PMO were unsuccessful because it was insoluble in aqueous solution. These effects have also been observed in antimicrobial peptides (18, 26). Because the role of cationic side chains is to make initial electrostatic contact with the anionic moieties of lipids (3, 29), our results are consistent with the suggestion that the more dispersed positive charge of the guanidinium group of arginine may electrostatically interact with the anionic moieties of the lipids in a more productive manner than either lysine or ornithine (18, 26). The superior effect of phenylalanine is thought to be attributable to its bulkiness (19, 20).
Another interesting result was that d-isomers in place of l-isomers of arginine and phenylalanine increased the MIC by only a factor of 2. This suggests that the mechanism of entry of PPMOs across the outer membrane of E. coli is not receptor mediated. Our result is consistent with previous results and models that have predicted that cationic and amphipathic peptides penetrate directly through membrane lipids (6, 16, 29, 30).
The (RX)6B-, (RXR)4XB-, and (RFR)4XB-AcpP PMOs each significantly reduced the infections in mice. The (RXR)4XB-AcpP PMO reduced the levels of bacteremia in a dose-dependent manner, whereas the (RX)6B-AcpP PMO reduced CFU/ml in blood to similar extents at both 100 μg and 300 μg per dose. The response to treatment with the (RFR)4XB-AcpP PMO was proportional to the dose at early times after treatment, and this was reflected in the rate of survival. The (RXR)4XB-AcpP PMO was the most potent of the three PPMOs tested, and 100% of the mice survived treatment with a dose as low as 30 μg (1.5 mg/kg).
The (RXR)4XB-AcpP PMO was more potent than ampicillin. In vitro MIC analysis showed that on a molar basis, the PPMO was about five times more potent. Because the molecular weight of a (RXR)4XB-AcpP PMO is about 16 times greater than that of ampicillin, on the basis of weight, the (RXR)4XB-AcpP PMO is about three times less potent in vitro. However, in the mouse model of infection used for our experiments, the (RXR)4XB-AcpP PMO was more potent, even on a weight basis. Clearly, 30 μg of the (RXR)4XB-AcpP PMO was more effective in lowering CFU/ml in the blood and promoting survival than 30 μg of ampicillin (compare Fig. 1A and B with Fig. 1C and D). We conclude that the (RXR)4XB-AcpP PMO is more potent than ampicillin for reducing CFU/ml in blood and increasing the rate of survival, at least in the model of infection that was used for testing.
Although the (RFR)4XB-AcpP PMO had the lowest MIC, it was not the most effective PPMO in vivo. The reason for this is unclear, but one possibility is that the composition and repeat pattern of all natural amino acids made it more susceptible to proteolysis in animals. Alternatively, it may have less favorable pharmacokinetics or bioavailability than the other PPMOs.
Thirty percent of the mice treated with 300 μg/dose (RX)6B-AcpP PMO died, despite having levels of bacteria in the blood comparable to those in the blood of mice treated with 100 μg/dose, all of which survived. These results suggest that the (RX)6B-AcpP PMO may be toxic to mice at the 300-μg/dose (∼15 mg/kg), which may account for its lack of an ability to further reduce CFU/ml in the blood compared to the numbers in the group treated with 100 μg/dose. Similar PPMOs with the RX repeating motif were not toxic in tissue culture (27). The apparent toxicity in mice is likely caused by the peptide moiety or the conjugate, because the PMO without an attached peptide is nontoxic at these doses (8).
If the (RX)6B-AcpP PMO was toxic at 15 mg/kg, the relatively short time to death (12 h) is a bit unusual and suggests a rapid event such as a lipopolysaccharide-induced cytokine cascade. Alternatively, it is plausible that the arginines of (RX)6 could be converted to NO by nitric oxide synthase. This would lead to critical events in the lung, heart, or brain, all of which could lead to death in 12 h.
There was no apparent short-term, gross toxicity with the (RXR)4XB-AcpP PMO, even at the highest dose tested (300 μg [15 mg/kg]), as determined by the 100% survival rate of mice in this treatment group. Previous results have shown that a (RXR)4XB PMO causes significant toxicity at 150 mg/kg in rats and has a 50% lethal dose of about 210 mg/kg (2). If these safety values are converted to those for mice by the use of conventional surface area calculations (24), the toxicity in mice would translate to about 300 mg/kg. Thus, the estimated therapeutic index (50% lethal dose/50% effective dose) of the (RXR)4XB-AcpP PMO may be greater than 200 (300 mg/kg/1.5 mg/kg).
Recently, (RXR)4 has been found to efficiently deliver PMOs through eukaryotic cell plasma membrane and into HeLa or CHO cells (1). Although the mechanism of entry across the eukaryotic cell plasma membrane is apparently mediated by binding to proteoglycan, its mechanism of subsequent transit across internal membranes and into the nucleus is unknown. The ability of (RXR)4XB to mediate the entry of PMOs into both eukaryotic and prokaryotic cells suggests that antibacterial PPMOs might be designed for use against intracellular bacteria. Indeed, preliminary data indicate that (RXR)4XB PMOs targeted to specific essential genes significantly reduce intracellular Salmonella, Chlamydia, and Mycobacterium levels (G. Mitev and B. L. Geller, unpublished data).
Acknowledgments
We thank the entire chemistry department at AVI BioPharma for making and purifying the PPMOs. We thank Alix Gitelman for advice on statistical analysis.
This work was funded by AVI BioPharma.
Bruce Geller was employed at both AVI BioPharma and Oregon State University.
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Abes, S., H. M. Moulton, P. Clair, P. Prevot, D. S. Youngblood, R. P. Wu, P. L. Iversen, and B. Lebleu. 2006. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Control. Release 116:304-313. [DOI] [PubMed] [Google Scholar]
- 2.Amantana, A., H. M. Moulton, M. L. Cate M. T. Reddy, T. Whitehead, J. N. Hassinger, D. S. Youngblood, and P. L. Iversen. 2007. Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate. Bioconjug. Chem. 18:1325-1331. [DOI] [PubMed] [Google Scholar]
- 3.Brown, K. L., and R. E. W. Hancock. 2006. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 18:24-30. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed., approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Deere, J., P. Iversen, and B. L. Geller. 2005. Antisense phosphorodiamidate morpholino oligomer length and target position effects on gene-specific inhibition in Escherichia coli. Antimicrob. Agents Chemother. 49:249-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennison, S. R., J. Wallace, F. Harris, and D. A. Phoenix. 2005. Amphiphilic α-helical antimicrobial peptides and their structure/function relationships. Protein Pept. Lett. 12:31-39. [DOI] [PubMed] [Google Scholar]
- 7.Geller, B. L., J. D. Deere, D. A. Stein, D. Kroeker, H. M. Moulton, and P. L. Iversen. 2003. Inhibition of gene expression in Escherichia coli by antisense phosphorodiamidate morpholino oligomers. Antimicrob. Agents Chemother. 47:3233-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geller, B. L., J. Deere, L. Tilley, and P. L. Iversen. 2005. Antisense phosphorodiamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J. Antimicrob. Chemother. 55:983-988. [DOI] [PubMed] [Google Scholar]
- 9.Good, L., and P. E. Nielsen. 1998. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. USA 95:2073-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Good, L., S. K. Awasthi, R. Dryselius, O. Larsson, and P. E. Nielsen. 2001. Bacterial antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19:360-364. [DOI] [PubMed] [Google Scholar]
- 11.Gruegelsiepe, H., O. Brandt, and R. K. Hartmann. 2006. Antisense inhibition of RNase P: mechanistic aspects and application to live bacteria. J. Biol. Chem. 281:30613-30620. [DOI] [PubMed] [Google Scholar]
- 12.Henriques, S. T., M. N. Melo, and M. A. R. B. Castanho. 2006. Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem. J. 399:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulyte, A., N. Nekhotiaeva, S. K. Awasthi, and L. Good. 2005. Inhibition of Mycobacterium smegmatis gene expression and growth using antisense peptide nucleic acids. J. Mol. Microbiol. Biotechnol. 9:101-109. [DOI] [PubMed] [Google Scholar]
- 14.Kurupati, P., K. S. W. Tan, G. Kumarasinghe, and C. L. Poh. 2007. Inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant β-lactamase-producing Klebsiella pneumonia strain. Antimicrob. Agents Chemother. 51:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, Z., A. Brady, A. Young, B. Rasimick, K. Chen, C. Zhou, and N. R. Kallenbach. 2007. Length effects in antimicrobial peptides of the (RW)n series. Antimicrob. Agents Chemother. 51:597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maloy, W. L., and U. P. Kari. 1995. Structure-activity studies on magainins and other host defense peptides. Biopolymers 37:105-122. [DOI] [PubMed] [Google Scholar]
- 17.Nekhotiaeva, N., S. K. Awasthi, P. E. Nielsen, and L. Good. 2004. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 10:252-259. [DOI] [PubMed] [Google Scholar]
- 18.Shafer, W. M., F. Hubalek, M. Huang, and J. Pohl. 1996. Bactericidal activity of a synthetic peptide (CG 117-136) of human lysosomal cathepsin G is dependent on arginine content. Infect. Immun. 64:4842-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strom, M. B., B. E. Haug, O. Rekdal, M. L. Skar, W. Stensen, and J. S. Svendsen. 2003. Important structural features of 15-residue lactoferricin derivatives and methods for improvement of antimicrobial activity. Biochem. Cell Biol. 80:65-74. [DOI] [PubMed] [Google Scholar]
- 20.Strom, M. B., O. Rekdal, and J. S. Svendsen. 2002. Antimicrobial activity of short arginine- and tryptophan-rich peptides. J. Pept. Sci. 8:431-437. [DOI] [PubMed] [Google Scholar]
- 21.Tan, X.-X., J. K. Actor, and Y. Chen. 2005. Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob. Agents Chemother. 49:3203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilley, L. D., B. L. Mellbye, S. E. Puckett, P. L. Iversen, and B. L. Geller. 2007. Antisense peptide-phosphorodiamidate morpholino oligomer conjugate: dose-response in mice infected with Escherichia coli. J. Antimicrob. Chemother. 59:66-73. [DOI] [PubMed] [Google Scholar]
- 23.Tilley, L. D., O. S. Hine, J. A. Kellogg, J. N. Hassinger, D. D. Weller, P. L. Iversen, and B. L. Geller. 2006. Gene-specific effects of antisense phosphorodiamidate morpholino oligomer-peptide conjugates on Escherichia coli and Salmonella enterica serovar Typhimurium in pure culture and in tissue culture. Antimicrob. Agents Chemother. 50:2789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.U.S. Food and Drug Administration, Center for Drug Evaluation and Research. 2008. Dose calculator to convert animal dose to human equivalent dose. Center for Drug Evaluation and Research, U.S. Food and Drug Administration, Rockville MD. http://www.fda.gov/cder/cancer/animalframe.htm. Date last accessed, 7 July 2008.
- 25.Vaara, M., and M. Porro. 1996. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 40:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel. H. J., D. J. Schibli, W. G. Jing, E. M. Lohmeier-Vogel, R. F. Epand, and R. M. Epand. 2002. Towards a structure-function analysis of bovine lactoferricin and related tryptophan- and arginine-containing peptides. Biochem. Cell Biol. 80:49-63. [DOI] [PubMed] [Google Scholar]
- 27.Wu, R. P., D. S. Youngblood, J. N. Hassinger, C. E. Lovejoy, M. H. Nelson, P. L. Iversen, and H. M. Moulton. 2007. Cell-penetrating peptides as transporters for morpholino oligomers: effect of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res. 35:5182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue-Wen, H., P. Jie, A. Xian-Yuan, and Z. Hong-Xiang. 2007. Inhibition of bacterial translation and growth by peptide nucleic acids targeted to domain II of 23S rRNA. J. Pept. Sci. 13:220-226. [DOI] [PubMed] [Google Scholar]
- 29.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]
- 30.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]