Abstract
The translational GTPases promote initiation, elongation, and termination of protein synthesis by interacting with the ribosome. Mutations that impair GTP hydrolysis by eukaryotic translation initiation factor 5B/initiation factor 2 (eIF5B/IF2) impair yeast cell growth due to failure to dissociate from the ribosome following subunit joining. A mutation in helix h5 of the 18S rRNA in the 40S ribosomal subunit and intragenic mutations in domain II of eIF5B suppress the toxic effects associated with expression of the eIF5B-H480I GTPase-deficient mutant in yeast by lowering the ribosome binding affinity of eIF5B. Hydroxyl radical mapping experiments reveal that the domain II suppressors interface with the body of the 40S subunit in the vicinity of helix h5. As the helix h5 mutation also impairs elongation factor function, the rRNA and eIF5B suppressor mutations provide in vivo evidence supporting a functionally important docking of domain II of the translational GTPases on the body of the small ribosomal subunit.
Four universally conserved GTPases interact with the ribosome to coordinate the initiation, elongation, and termination of protein synthesis. Initiation factor 2/eukaryotic translation initiation factor 5B (IF2/eIF5B) initially binds to the small ribosomal subunit and promotes subunit joining during translation initiation (5, 30, 36). The factor EF-Tu in bacteria or eEF1A in eukaryotes delivers aminoacyl-tRNAs to the A site of the ribosome, while the bacterial factor EF-G, or eEF2 in eukaryotes, promotes translocation of peptidyl-tRNA from the A to the P site during translation elongation (reviewed in references 1, 37, and 38). Finally, the factor RF3 (or eRF3 in eukaryotes) functions with the stop codon recognition factors to promote translation termination (25). As expected, the conserved GTP binding (G) domains of these factors contain the sequence motifs that are hallmarks of all G proteins. In addition to the Walker A-box GXXXGK(T/S) motif (G-1), which interacts with α and β phosphates of GTP, and the G-4 motif NKXD, which specifies guanine nucleotide recognition, the G-3 motif DXXG within the mobile switch II element is also conserved (48). Finally, X-ray structure analyses revealed that in addition to the G domain, the four translation GTPases share a conserved β-barrel domain II (28, 40). Binding of GTP versus GDP to EF-Tu altered the position of the switch II element and induced a significant rearrangement of domain II relative to the G domain (9, 26). Likewise, GTP binding to eIF5B induced movement of switch II and a rotation-like movement of domain II (39). As these gross structural rearrangements alter the aminoacyl-tRNA and/or ribosome binding affinities of the factors, it can be proposed that GTP switches govern the interaction of the translational GTPases with the ribosome.
Cryo-electron microscopy (cryo-EM) studies have revealed the binding sites for EF-Tu, EF-G, IF2, and RF3 on the ribosome (2, 3, 20, 27, 33, 47, 49, 55). All four factors were found to bind in the ribosomal entry site, the cleft on the A site side of the interface between the two subunits. This position is consistent with EF-Tu and RF3 interacting with tRNA or release factors, respectively, in the A site and with EF-G translocating peptidyl-tRNA from the A to the P site. Binding of IF2 to the ribosomal entry site enables the factor to contact both Met-tRNA in the P site and the factor IF1 in the A site (3). Within the ribosomal entry site, the G domain of the translational GTPases was found in contact with the large ribosomal subunit in the vicinity of the sarcin-ricin loop and the ribosomal stalk (2, 13, 15, 47, 55). In contrast, domain II contacted the body of the small ribosomal subunit. In vitro chemical probing studies using tethered nucleic acid cleavage agents and footprinting techniques to introduce cleavages in the rRNA supported the findings of the cryo-EM studies and revealed IF2 and eIF5B G domain contacts with the large ribosomal subunit and eIF5B domain II contacts with the small subunit (29, 31, 54). However, it is noteworthy that no genetic data supporting the in vivo relevance of these statically determined binding sites have been reported.
In previous work, we demonstrated that a GTP/GDP switch governs the ribosome binding affinity of the eukaryotic IF2 ortholog eIF5B (44). The X-ray structure of the archaeal Methanobacterium thermoautotrophicum aIF5B resembled a chalice, with the G domain, domain II, and domain III forming the cup; helix H12 forming the stem; and domain IV forming the base. Comparison of the structures of aIF5B in the presence of GTP and GDP revealed a lever-type domain movement in which small perturbations in the position of the G domain and domains II and III triggered a swing of the helix H12 and larger movement of domain IV (39). Via an interaction between the C termini of eIF5B and eIF1A, the latter likely bound to the ribosomal A site, eIF5B is recruited to the small 40S ribosomal subunit and promotes large 60S subunit joining (19). Formation of the 80S ribosome triggers GTP hydrolysis by eIF5B and release of the factor enabling eIF1A to dissociate as well (19, 44). A point mutation (T439A) in the eIF5B G domain that impairs GTP hydrolysis does not impair subunit-joining activity in vitro; however, yeast cells expressing the eIF5B-T439A mutant exhibit a severe slow-growth phenotype due to drastically impaired protein synthesis. A second-site mutation in the eIF5B G domain suppressed the slow-growth phenotype associated with the eIF5B-T439A mutation but did not restore the factor's GTPase activity. Rather, the suppressor mutation lowered eIF5B ribosome binding affinity and enabled the factor to dissociate from the 80S ribosome in the absence of GTP hydrolysis (44). Interestingly, the suppressor mutation mapped to a region of the G domain that in the cryo-EM structures of EF-Tu and EF-G contacts the large ribosomal subunit.
Having identified an intragenic suppressor of eIF5B-T439A that restores cell growth by lowering the ribosome binding affinity of eIF5B, we reasoned that it should be possible to obtain mutations in the ribosome that likewise reduce eIF5B binding and suppress the toxic affects associated with expression of GTPase-defective mutants of eIF5B. Here we describe the isolation of a mutation in helix h5 of the 18S rRNA component of the small ribosomal subunit that restores the growth of yeast expressing the GTPase-deficient eIF5B-H480I mutant. Intragenic suppressors of the eIF5B-H480I mutant mapped to domain II, which in turn was found to bind the ribosome in the vicinity of helix h5 of the small subunit. Moreover, the helix 5 mutation in 18S rRNA impaired the binding of both eIF5B and elongation factor eEF2 to 80S ribosomes in vitro. Thus, we provide in vivo evidence supporting a binding interface between helix h5 of the 40S subunit and domain II of eIF5B, and based on the conservation of domain II among IF2/eIF5B, EF-Tu/eEF1A, EF-G/eEF2, RF3/eRF3, we propose that this interface is shared among all of the translational GTPases.
MATERIALS AND METHODS
rRNA suppressor analysis.
In strain NOY891 (MATa ade2-1 ura3-1 leu2-3 leu2-112 his3-11 can1-100 rdnΔΔ::HIS3), obtained from M. Nomura, the chromosomal rDNA repeats are completely deleted (rdnΔΔ), and the high-copy-number plasmid pNOY353 (PGAL7-35S rDNA, 5S rDNA, TRP1) expresses the 35S rRNA precursor under the control of galactose-inducible GAL7 promoter (57). The chromosomal FUN12 promoter in strain NOY891 was replaced with the GAL1 promoter by transformation with a KanMX6-PGAL1 PCR fragment generated using a primer complementary to the sequence from position −52 to −120 upstream of the FUN12 AUG start codon and a second primer corresponding the to ΔN-FUN12 open reading frame (residues 396 to 417). The presence of the KanMX6-PGAL1-ΔN-FUN12 allele in the resulting strain J215 was confirmed by colony PCR and DNA sequencing of the PCR fragments. Next, a derivative of the low-copy-number URA3 vector YCplac33 containing the ΔN-FUN12-H480I allele (encoding eIF5B397-1002-H480I) was introduced into strain J215. Random mutagenesis of the high-copy-number LEU2 plasmid pNOY373 (57), carrying a single rDNA repeat expressing both the 35S and 5S rRNAs from the native PolI and PolIII promoters, respectively, was performed using the bacterial mutator strain XL1-Red (Stratagene). The mutated plasmids were introduced into the derivative of strain J215 containing the ΔN-FUN12-H480I plasmid, and fast-growing transformants were selected on glucose medium, where expression of both PGAL1-ΔN-FUN12 and PGAL7-35S rDNA was repressed. To preferentially rescue the mutated pNOY373 plasmid from the fast-growing candidates, we relied on complementation of the leuB auxotrophic marker in Escherichia coli strain MC1066 by the yeast LEU2 allele on pNOY373.
Measurement of GTP binding affinity.
GTP binding affinities of eIF5B were measured using fluorescent N-methylanthraniloyl (Mant)-labeled GDP, as described previously (42). Briefly, initial reaction mixtures contained 2 μM eIF5B and 500 nM Mant-GDP in binding buffer (20 mM HEPES-KOH [pH 7.5], 50 mM potassium acetate, 2 mM dithiothreitol [DTT], and 2.5 mM magnesium acetate). Samples were excited at 360 nm in a Floromax-3 steady-state fluorometer, and fluorescence at 445 nm was monitored as a function of competitor unlabeled GTP added. Reactions equilibrated within 1 minute after addition of competitor, and the change in intensity (Ibound − Ifree) was plotted as a function of GTP concentration. The data were fit with the following formula by nonlinear regression using Kaleidagraph: 1 − [GTP]/(Kd + [GTP]) (where Kd is the dissociation constant).
Uncoupled GTPase assay.
The ribosome-dependent uncoupled GTPase activity of eIF5B was analyzed as described previously (43). Briefly, reaction mixtures contained 1 μM eIF5B and 0.4 μM 40S and 60S (or 80S) ribosomes in 1× reaction buffer (30 mM HEPES-KOH [pH 7.4], 50 mM potassium acetate, 2.5 mM magnesium acetate, and 2 mM DTT). Reactions were started by adding 50 nM [γ-33P]GTP and then quenched at various times by mixing with a 3× volume of stop solution (50 mM EDTA in 90% formamide). Progress of GTP hydrolysis was analyzed by polyethyleneimine cellulose thin-layer chromatography (TLC) (Selecto Scientific, GA) using a buffer containing 0.8 M lithium chloride and 0.8 M acetic acid. The fraction of GTP hydrolyzed was quantified by phosphorimager analysis, and the rate constant for GTP hydrolysis was determined by plotting the fraction of Pi released above background {[Pi]/([Pi] + [GTP])} as a function of the quench time. Data were fit using KaleidaGraph (Synergy Software) to the single exponential equation A[1 − exp(−kt)], in which A is the amplitude and k is the rate constant.
Polysome analysis.
Polysomes were analyzed by velocity sedimentation in 7 to 47% sucrose gradients as described previously (43). Briefly, cells were grown in synthetic complete medium to an optical density at 600 nm (OD600) of 1.0, and then, if required, cycloheximide (CHX) was added to a final concentration of 50 μg per ml of culture volume. Cells were collected by pouring the cultures into 500-ml centrifuge bottles that were two-thirds full of crushed ice and then pelleting by centrifugation at 5,000 rpm for 5 min at 4°C. Cells were suspended in breaking buffer [20 mM Tris-Cl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 2 mM DTT, 1× Complete Protease Inhibitor cocktail (EDTA free; Roche), 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), 5 μg/ml pepstatin, and 50 μg/ml CHX] and then broken by adding glass beads and mixing vigorously at 4°C using a vortex for five cycles of 1 min mixing followed by 1 min on ice. The clarified supernatants were layered on 8-ml 7 to 47% sucrose gradients and then subjected to centrifugation at 39,000 rpm for 2.5 h at 4°C in a Beckman SW41 rotor. Gradients were fractionated while monitoring absorbance at 254 nm. Polysome-to-monosome (P/M) ratios were calculated by measuring the area in the combined polysome fractions and 80S peak using ImageJ 1.40e software (NIH).
Derivatization of Fe(II)-BABE-eIF5B.
Derivatization of single cysteine mutants of eIF5B and cleavage reactions were performed essentially as described by Marzi et al. (31). The extent of Fe(II)-1-(p-bromoacetamidobenzyl)-EDTA [Fe(II)-BABE] modification for each mutant was examined using the thiol-specific fluorescent reagent 7-diethylamino-3-(4′-(iodoacetyl)amino)phenyl)-4-methylcoumarin (DCIA) (Molecular Probes, Invitrogen). Briefly, 25-μl reaction mixtures containing 500 pmol eIF5B and 4 nmol DCIA were incubated at 30°C in buffer containing 80 mM HEPES (pH 7.6), 1 M KCl, and 0.01% Nikkol. After 15 min, reactions were quenched by adding 7 μl of stop solution (25 mM Tris-HCl [pH 6.8], 6 M urea, 1% sodium dodecyl sulfate [SDS], 10% glycerol, 0.001% bromophenol blue, and 1% β-mercaptoethanol). Samples were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the extent of DCIA modification of eIF5B was detected by UV illumination and quantified by phosphorimager analysis. The gel was then stained using Coomassie brilliant blue (Sigma), and protein levels were quantified using ImageJ software. The relative extent of DCIA modification was determined after first subtracting the background obtained with the eIF5B-7CΔ mutant.
Directed hydroxyl radical mapping.
For hydroxyl radical cleavage of 18S rRNA, 2 μM Fe(II)-BABE-eIF5B was mixed with 1 μM reassociated 80S ribosomes in the presence of 1 mM GDPNP (a nonhydrolyzable analog of GTP) and incubated at 30°C for 10 min and then on ice for 10 min. Cleavage reactions were performed in the presence of H2O2 and ascorbic acid (31), and cleavage sites on the 18S rRNA were detected by primer extension using reverse transcriptase (50).
Purification of His6-tagged eEF2.
His6-tagged eEF2 was purified from yeast as described previously (35) with the following modifications. Following elution of His6-eEF2 from a HisTrap column (Amersham), the eEF2 fractions were applied to a HiTrap Q column (Amersham) and eluted using a 40 to 500 mM KCl gradient. The eEF2 fractions were dialyzed against 20 mM Tris-Cl (pH 7.5), 100 mM KCl, 1 mM DTT, and 10% glycerol and stored at −80°C.
Ribosome binding assay.
Purified eIF5B (0.5 μM) or eEF2 (0.7 μM) was incubated with purified yeast 80S ribosomes (0.5 μM), and ribosome binding activity was assessed by pelleting through a sucrose cushion (42, 43). Similar buffers were used for the eIF5B (30 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 2.5 mM magnesium acetate, 2 mM DTT, and 2 mM guanine nucleotide) and eEF2 (25 mM Tris-Cl [pH 7.5], 125 mM potassium acetate, 2.5 mM magnesium acetate and 1 mM DTT) binding reactions.
Measurement of ribosome transit time.
For measurement of ribosome transit time, exponentially growing cells (150 ml) were pulse-labeled with [35S]Met (Perkin-Elmer) at a concentration of 0.25 μCi per ml, and 10-ml samples were taken at 30-s intervals. Radioactivity incorporated into total or complete protein was measured using liquid scintillation counting as described previously (34, 41) with minor modifications. Cells were pelleted, suspended in 200 μl of lysis buffer (20 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 2 mM magnesium acetate, 1 mM DTT, and 0.5 mM Pefabloc SC [Roche]), and then broken following addition of a 60% volume of glass beads by vigorous mixing on a vortex for 15 min at 4°C. Following removal of the whole-cell extract, the glass beads were washed two times with 500 μl lysis buffer. The washes and whole-cell extract were combined and clarified by centrifugation at 4,000 rpm for 5 minutes in a microcentrifuge. [35S]Met incorporation into total protein was assessed using 400 μl of the clarified extract. To analyze completed protein production, 400 μl of the extract was layered on 320 μl of 60% sucrose solution (prepared in 1× binding buffer). Following centrifugation at 55,000 rpm for 30 min in a Beckman TLA120.2 rotor, polysomes and nascent proteins were pelleted, and completed proteins remained in the supernatant. The incorporation of [35S]Met was measured by trichloroacetic acid precipitation and liquid scintillation counting as described previously (34). Linear regression of the data was performed using KaleidaGraph.
RESULTS
Mutation of the conserved His residue in the D-X-X-G-H motif of translational GTPases impairs eIF5B function and yeast cell growth.
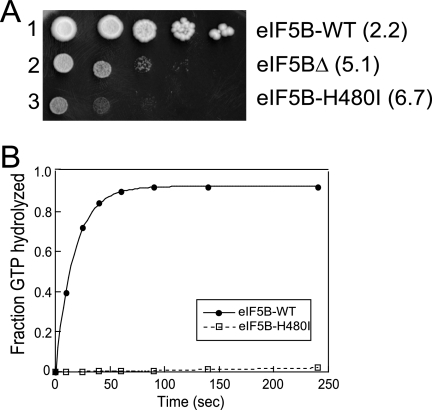
Previous studies revealed that removal of the weakly conserved N terminus of yeast eIF5B (residues 1 to 396), including all sequences preceding the GTP binding, domain has no effect on cell growth (12, 44). In this study all experiments employed an N-terminally truncated form of the factor (residues 397 to 1002). Comparison of the primary sequences of diverse GTPases revealed that a His residue in the four translational GTPases replaces the catalytically essential Gln residue that immediately follows the G-3 motif DXXG in the small GTPases of the Ras family (10). Plasmid-borne wild-type (WT) eIF5B and mutant versions of eIF5B, in which the corresponding His480 residue was mutated to Ile, were expressed under the control of the native FUN12 (eIF5B) promoter in the ΔeIF5B (fun12Δ) strain J111. As shown in Fig. 1A, expression of WT eIF5B complemented (doubling time = 2.2 h) the slow-growth phenotype of the ΔeIF5B strain (doubling time = 5.1 h), while expression of the eIF5B-H480I mutant resulted in a more severe slow-growth phenotype (doubling time = 6.7 h).
FIG. 1.
eIF5B-H480I mutation in the G-3 motif of switch II impairs yeast cell growth and ribosome-dependent GTPase Activity. (A) Growth rate analysis of yeast expressing the eIF5B-H480I mutant. The ΔeIF5B strain J111 (MATα ura3-52 leu2-3 leu2-112 fun12Δ) was transformed with empty vector YCplac33 (eIF5BΔ) or the same vector containing the indicated WT eIF5B, or eIF5B-H480I mutant allele. Strains were grown to saturation, and 5 μl of serial dilutions (OD600 = 1.0, 0.1, 0.01, 0.001, and 0.0001) was spotted on minimal medium supplemented with essential nutrients (SD). The plate was incubated for 4 days at 30°C. Doubling times during exponential growth in liquid SD medium are shown in parentheses. (B) Ribosome-dependent GTPase assays. Purified, recombinant eIF5B-WT or eIF5B-H480I (1 μM) was incubated with 0.4 μM 40S and 60S subunits and 50 nM [γ-33P]GTP. Aliquots from the reactions were quenched and analyzed at various time points by TLC, and the amount of phosphate released was quantified. The values were corrected by subtracting the GTPase activities observed in the absence of ribosomes or factors. The data presented are representative of three independent experiments.
Using unlabeled GTP to compete with fluorescent Mant-GDP revealed similar Kd values for GTP binding to WT eIF5B (10 μM) and eIF5B-H480I (20 μM) (data not shown). As the cellular concentration of GTP is estimated at 600 to 1,500 μM (17), the slightly elevated Kd value for eIF5B-H480I is likely not significant. In contrast, whereas WT eIF5B hydrolyzed GTP with a rate constant of 0.06 per s in the presence of purified yeast 80S ribosomes, the ribosome-dependent GTPase activity of the eIF5B-H480I mutant was at or below background levels (Fig. 1B). Thus, the His480 residue, which is conserved in all translational GTPases, plays an important role in GTP hydrolysis, perhaps reflecting a function in common with the corresponding Gln61 residue in Ras, which is known to stabilize the transition state of GTP hydrolysis (48).
Isolation of an rRNA suppressor of the eIF5B-H480I mutant.
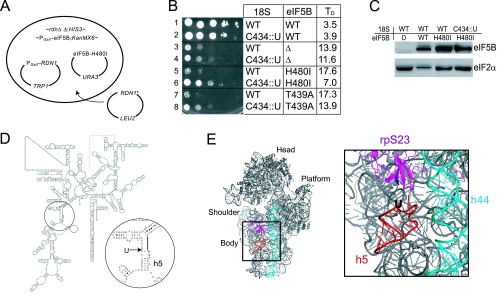
Based on the notion that the GTPase-deficient eIF5B-H480I mutant failed to release from the ribosome following subunit joining, we proposed that mutations in the eIF5B-ribosome interface that weakened eIF5B binding would suppress the slow-growth phenotype in the eIF5B-H480I mutant strain. Yeast ribosomes, like those found in higher eukaryotes, are composed of four rRNA molecules (18S in the small subunit and 5S, 5.8S, and 25S in the large subunit) and 78 different ribosomal proteins (52). Structural studies of prokaryotic ribosomes revealed that the ribosomal surface, including the so-called factor binding site, is composed of both protein and rRNA elements (7, 11, 13, 33, 37, 47). Thus, mutations in either ribosomal proteins or rRNA may suppress the eIF5B-H480I mutant. Because the 18S, 5.8S, and 25S rRNAs are cotranscribed as a single precursor 35S rRNA whereas a significant fraction of the ribosomal proteins are expressed from duplicated genes, we chose to screen for rRNA mutations that suppress the slow-growth phenotype associated with the eIF5B-H480I mutant.
In yeast, ∼150 copies of the RDN genes encoding the 35S rRNA precursor are tandemly repeated on chromosome XII, with the gene for the 5S rRNA residing in the intergenic regions. Wai et al. generated yeast strains (rdnΔΔ) in which all of the RDN genes were deleted from chromosome XII and the essential rRNAs were expressed from a plasmid-borne RDN1 allele (57). In the yeast strain NOY891, a TRP1-marked plasmid expresses 5S rRNA from the native PolIII promoter and expresses the 35S rRNA precursor from the galactose-inducible GAL7 promoter. While we were unable to delete the chromosomal FUN12 gene encoding eIF5B in this strain, perhaps indicating a synthetic growth defect, we were able to replace the chromosomal FUN12 promoter with the promoter from the galactose-inducible yeast GAL1 gene to make yeast strain J215 (Fig. 2A). We next introduced into strain J215 a URA3-marked plasmid that expresses either WT eIF5B or eIF5B-H480I under the control of the native FUN12 promoter and a LEU2 plasmid that expresses the 35S rRNA precursor under the control of the native PolI promoter (Fig. 2A). The transformants grew well on galactose medium due to the expression of both the chromosomal PGal1-eIF5B and the PGAL7-RDN1 genes (data not shown). However, on glucose medium, expression of both of these genes is repressed and cell growth is dependent on the eIF5B protein expressed from the URA3 plasmid and the RDN1 gene on the LEU2 plasmid. Transformants expressing WT eIF5B grew much better than transformants carrying an empty URA3 plasmid and expressing no eIF5B (Fig. 2B, compare rows 1 and 3). In addition, as observed above, yeast cells expressing GTPase-deficient eIF5B-H480I or the previous eIF5B-T439A mutant (44) grew even slower than cells not expressing eIF5B (Fig. 2B, compare rows 3, 5, and 7). Thus, the slow-growth phenotype associated with the eIF5B-H480I mutation is maintained in the rdnΔΔ strain.
FIG. 2.
rRNA Suppressor of the eIF5B-H480I mutant. (A) Screening for rRNA suppressors of the eIF5B-H480I mutant. In the yeast strain NOY891, the chromosomal rDNA repeats are deleted (rdnΔΔ::HIS3) and the 35S rRNA precursor is expressed under the control of the GAL7 promoter (PGAL7-RDN1) on the TRP1 plasmid pNOY353 (57). The chromosomal eIF5B (FUN12) promoter was replaced by the GAL1 promoter (PGAL1-eIF5B), and eIF5B-H480I was introduced on a URA3 plasmid (see text for details). The high-copy-number LEU2 plasmid pNOY373 carrying a single RDN1 repeat was randomly mutated and introduced into the yeast strain. Transformants that grew well on glucose medium, where PGAL1-eIF5B and PGAL7-RDN1 expression is repressed, were selected for further study (see text). (B) Growth rate analysis of yeast expressing various forms of eIF5B and 18S rRNA. Derivatives of yeast strain J215 carrying plasmids expressing WT eIF5B (eIF5B-WT), eIF5B-H480I, eIF5B-T439A, or the empty vector (eIF5BΔ) and plasmids expressing WT 18S rRNA or the 18S-C434::U insertion mutant, as indicated, were grown to saturation in glucose medium, and then 5 μl of serial dilutions (OD600 = 1.0, 0.1, 0.01, 0.001 and 0.0001) was spotted on SD medium. The plate was incubated for 4 days at 30°C. TD, doubling time during exponential growth in liquid SD medium. (C) Western blot analysis of eIF5B expression. Whole-cell extracts prepared from the transformants described in panel A were subjected to immunoblot analysis using anti-eIF5B or anti-eIF2α antiserum, as described previously (12). Immune complexes were visualized using enhanced chemiluminescence. (D) Secondary structure of yeast 18S rRNA (http://www.rna.icmb.utexas.edu), with the site of the suppressor mutation, a U insertion after C434 in helix h5, circled and shown in detail. (E) Ribbon diagram of the yeast 40S ribosomal subunit derived from the cryo-EM structure of the yeast eEF2-80S ribosome complex (47). Positions of helix h44 (blue), rpS23 (magenta), and the C434::U insertion in helix h5 (red) are highlighted and shown in detail.
To screen for rRNA mutations that suppress the slow-growth phenotype due to the eIF5B-H480I mutation, we randomly mutated the RDN1 gene on the LEU2 plasmid using the bacterial mutator strain XL1-Red and then introduced a library of mutant plasmids into a derivative of the yeast strain J215 expressing eIF5B-H480I. Transformants were screened on glucose medium to identify cells that grew faster than controls expressing WT rRNA. From a screen of ∼105 transformants, two fast-growing cells were identified and found to carry the same rRNA mutation, a U insertion following C434 in the 18S rRNA of the 40S ribosomal subunit (C434::U) (Fig. 2D). As shown in Fig. 2B, the rRNA suppressor mutation greatly improved the growth of yeast expressing the eIF5B-H480I mutant (compare rows 5 and 6) without significantly affecting eIF5B expression (Fig. 2C). Interestingly, the C434::U mutation in 18S rRNA also partially suppressed the slow-growth phenotype in cells expressing eIF5B-T439A (Fig. 2B, row 7 versus 8) and modestly enhanced the growth of cells lacking eIF5B (Fig. 2B, row 3 versus 4). In contrast, the C434::U mutation slightly impaired the growth of yeast expressing WT eIF5B (Fig. 2B, row 1 versus 2). Thus, the U insertion following C434 in 18S rRNA does not simply enhance the growth rate of yeast cells. Instead, the C434::U insertion suppresses the toxic effects of GTPase-deficient forms of eIF5B, converting these dominant-negative forms of the factor into stimulators of translation initiation.
The suppressor mutation was located on the secondary structure of yeast 18S rRNA at the top of helix h5 (Fig. 2D). To map the position of the suppressor mutation on the tertiary structure of the 40S subunit, the molecular model of the yeast eEF2-80S complex (47) was used as a guide. In this model, Spahn et al. (47) docked the X-ray crystal structure of eEF2-sordarin (23) and the molecular model of the yeast 80S ribosome (8, 46) into the cryo-EM map of their eEF2-80S structure. As shown in Fig. 2E (only the 40S portion of the eEF2-80S model is shown), the suppressor mutation is located on the body of the 40S subunit under ribosomal protein rpS23 (corresponding to S12 in the bacterial 30S subunit). Interestingly, the mutation is on the surface of the subunit that contacts domain II of eEF2 in the cryo-EM structure.
Isolation of intragenic suppressors of the eIF5B-H480I mutation.
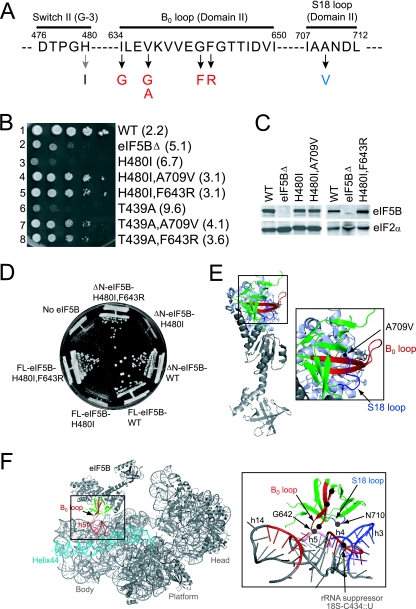
To complement the identification of the ribosomal suppressor of the eIF5B mutant, we screened for intragenic mutations in eIF5B-H480I that suppressed the slow-growth phenotype associated with this mutation. A plasmid-borne eIF5B-H480I allele was randomly mutated using the bacterial mutator strain XL1-Red and introduced into the ΔeIF5B strain J111. From a screen of ∼104 transformants, a single A709V mutation was identified as a strong suppressor of the eIF5B-H480I mutation (Fig. 3A and B, compare rows 3 and 4). As was observed with the rRNA suppressor mutation, the A709V mutation did not simply eliminate the toxicity associated with expression of the eIF5B-H480I mutant in yeast; instead, yeast expressing the eIF5B-H480I,A709V double mutant grew faster than cells expressing no eIF5B (Fig. 3B, compare rows 4 and 2). Thus, the suppressor mutation converted eIF5B-H480I from an inhibitor to an activator of translation. Western analyses revealed that the H480I and suppressor mutations did not alter the levels of eIF5B protein in yeast, indicating that the mutations altered eIF5B activity (Fig. 3C).
FIG. 3.
Intragenic suppressors of the eIF5B-H480I mutant. (A) Amino acid sequence of the switch II (G-3 sequence motif), the B0 loop in domain II, and the S18 loop in domain II of yeast eIF5B. The H480I mutation (gray arrow) and suppressor mutations (black arrows) are shown, and the position numbers in yeast eIF5B are indicated. (B) Growth rate analysis of yeast expressing WT and mutant forms of eIF5B. The ΔeIF5B strain J111 was transformed with the empty vector YCplac33 (eIF5BΔ) or the same plasmid containing the indicated WT or mutant eIF5B genes. Transformants were grown, spotted on SD medium, and incubated at 30°C for 4 days as described for Fig. 1. Doubling times during exponential growth in liquid SD medium are shown in parentheses. (C) Western blot analysis of eIF5B expression. Whole-cell extracts prepared from the transformants described for panel B were subjected to immunoblot analysis using anti-eIF5B or anti-eIF2α antiserum, as described previously (12). Immune complexes were visualized using enhanced chemiluminescence. (D) Analysis of intragenic suppressors in full-length eIF5B-H480I. The ΔeIF5B strain J111 was transformed with the empty vector YCplac33 (No eIF5B) or the same plasmid containing the indicated full-length (FL, residues 1 to 1002) or N-terminally truncated (ΔN, residues 397 to 1002) versions of WT or mutant eIF5B genes. Transformants were streaked on SD medium and incubated at 30°C for 4 days. (E) Ribbon diagram of archaeal aIF5B. Domain II is depicted in green, and the locations of the B0 loop (red), the S18 loop (blue), and the A709V intragenic suppressor mutation (black dot) are indicated and shown in detail. (F) Model of domain II of eIF5B binding near helix h5 of the 40S subunit. Left, the eIF5B-40S complex was modeled based on the cryo-EM structure of the eEF2-80S complex (PDB code 1S1H) (47) by aligning the G domain and domain II of aIF5B (PDB code 1G7T) (39) with the corresponding domains of eEF2 using the align program in PyMol software (14). Domain II of eIF5B is colored green, with the B0 loop in red. The head, platform, and body regions of the 40S subunit are indicated, and helix h5 of the 16S rRNA is shown in red, with helix h44 in cyan. Right, blow-up view of the boxed region, showing the locations of hydroxyl radical cleavage sites in 18S rRNA. The locations of intragenic suppressor mutations in the B0 (red) and S18 (blue) loops of eIF5B-H480I and the location of the C434::U 18S rRNA suppressor mutation are shown as black dots. The positions of the G642C and N710C mutations in eIF5B, where Fe(II)-BABE was attached, are labeled and shown as gray dots. For clarity, rRNA is colored gray and ribosomal proteins are not shown. The regions of 18S rRNA cleaved by Fe(II)-BABE tethered to residues 642 and 710 in eIF5B are colored red and blue, respectively. The region around 18S rRNA helix h5 was cleaved by Fe(II)-BABE tethered at either position 642 or 710 in eIF5B and is shown in pink.
The location of the suppressor mutation was mapped on the structure of archaeal M. thermoautotrophicum aIF5B. As shown in Fig. 3E, the A709 residue is located in a loop following β-strand S18 in domain II of aIF5B. Interestingly, the corresponding region of domain II of eEF2 is near the contact site for the 40S subunit in the eEF2-80S cryo-EM structure (47). Thus, we propose that the A709V mutation, and the h5 mutation in 18S rRNA, may weaken the binding of eIF5B to the ribosome. Based on the structure of the eEF2-80S complex, we docked domain II of aIF5B on the 40S subunit (Fig. 3F). This model predicted that a β-hairpin loop connecting β-strands S10 and S11 in domain II of eIF5B would interact with the 40S subunit in the vicinity of helix h5 (Fig. 3F). Interestingly, this β-hairpin loop (which we will refer to as the B0 loop [Fig. 3A and E]) is conserved in all translational GTPases (see Fig. 6A), including the elongation factor EF-Tu (4), and mutation of Gly222 in the B0 loop of EF-Tu (referred to as the B0 mutation) inactivated the factor apparently by weakening the interaction of EF-Tu with the ribosome (51). We hypothesized that mutations in the B0 loop of eIF5B would, like the helix h5 mutation, suppress the toxic phenotype associated with the eIF5B-H480I mutation. To test this proposal, the B0 loop of eIF5B-H480I was randomly mutated and the mutants were screened in the ΔeIF5B strain J111. Five independent suppressor mutations were identified (Fig. 3A and B): I634G, V637A, V637G, G642F, and F643R. All of the suppressors conferred better growth than an eIF5BΔ strain, and the proteins were expressed at levels equivalent to those of WT eIF5B and eIF5B-H480I (in Fig. 3B and C only the F643R suppressor is shown; similar results were obtained for the other four suppressors [data not shown]). Interestingly, the V637G and F643R mutations were also able to suppress the slow-growth phenotype associated with the eIF5B-T439A mutation (Fig. 3B, row 8 versus 6, and data not shown), and the F643R mutation was also able to suppress the slow-growth phenotype associated with the H480I mutation in full-length eIF5B (Fig. 3D).
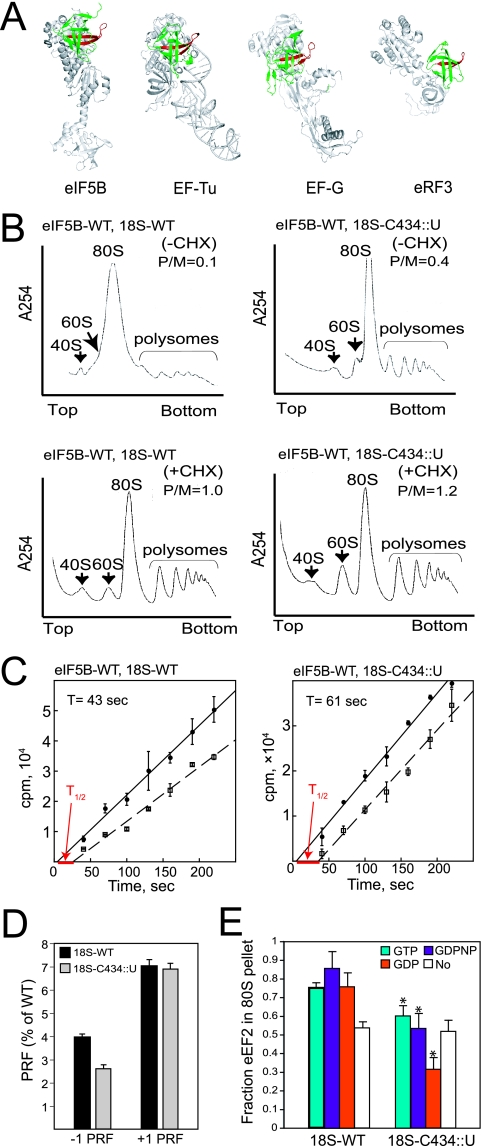
FIG. 6.
The C434::U Insertion in 18S rRNA impairs translation elongation. (A) Conservation of the B0 loop structure in translational GTPases. Ribbon diagrams of the structures of aIF5B (PDB code 1G7T), EF-Tu (1TTT), EF-G (2EFG), and eRF3 (1R5B) were aligned such that the protruding B0 loop (red) in domain II (green) of the GTPases face the same direction. The small black dot in the B0 loop of EF-Tu indicates the site of the original B0 (G222D) mutation. (B) Impaired translation elongation in yeast expressing 18S rRNA bearing the C434::U mutation. Polysome profiles of derivatives of yeast strain J215 expressing WT eIF5B and either WT rRNA (left panels, 18S-WT) or the C434::U rRNA suppressor allele (right panels, 18S-C434::U) are shown. Cells were grown exponentially in SD medium and either treated (lower panels) or not treated (upper panels) with 50 μg/ml CHX for 5 min prior to harvesting. Whole-cell extracts from the strains were resolved by velocity sedimentation in 7 to 47% sucrose gradients as previously described (6). Gradients were fractionated while scanning at 254 nm, and the positions of the 40S, 60S, 80S, and polysomes are indicated. P/M ratios were calculated by measuring the area in the combined polysome fractions and in the 80S peak. (C) Ribosomal transit time analysis. Exponentially growing cells expressing either WT 18S (left panel) or the 18S-C434::U mutant (right panel) were pulse-labeled with [35S]Met, and incorporation of radioactivity into the total (•) and completed protein (□) was plotted as a function of the time. The data presented are the averages and standard errors from at least three independent experiments. The fitting lines obtained by linear regression were extrapolated to the x axis to determine the half-transit time (T1/2) and to calculate the transit time (T). (D) Analysis of PRF using dual-luciferase reporter assays. Cell extracts were obtained from the WT or 18S-C434::U mutant strains bearing the dual-luciferase reporter plasmids designed to analyze the −1 (L-A) or +1 (Ty1) PRF, as described previously (21). The efficiency of PRF was determined as the percentage of a 0-frame control in which the two luciferase reporters are fused in frame. The results shown are the averages from three independent experiments with standard deviations. (E) Ribosome binding assay. Purified eEF2 was mixed, in the presence of GTP, GDPNP, or GDP as indicated, with yeast 80S ribosomes prepared from strains expressing either WT or C434::U mutant 18S rRNA. Binding reaction mixtures were loaded on a 10% sucrose cushion, and following centrifugation the amounts of eEF2 recovered in the pellet and supernatant fractions were determined by SDS-PAGE and quantitative densitometry. The data presented represent the fraction of eEF2 present in the ribosomal pellet and are the averages and standard deviations from three independent experiments. *, P < 0.05 by t test (obtained by comparison with 18S-WT).
The intragenic and rRNA suppressor mutations restore general translation by lowering the ribosomal binding affinity of eIF5B.
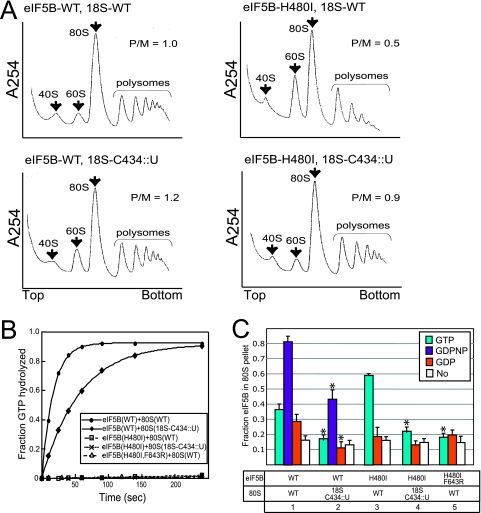
To address the mechanism of the growth suppression by the eIF5B intragenic and rRNA suppressor mutations, we first examined cellular protein synthesis using polysome profile analysis. Cells were treated with CHX to prevent elongation, and then cell extracts were fractionated on sucrose density gradients. Mutations that cause a rate-limiting impairment in translation initiation will result in a reduction of polysomes and an increase in 80S levels, whereas mutations that primarily impair translation elongation or termination will increase the P/M ratio. The eIF5B-H480I mutation resulted in a reduction in polysomes and a corresponding increase in the amount of 80S complexes not engaged in protein synthesis (P/M ratio of 1.0 in WT cells, compared to a ratio of 0.5 in the eIF5B-H480I mutant [Fig. 4A, top panels]). As the polysome profiles monitor all mRNAs in the cell, the drop in P/M ratio indicates a defect in general translation initiation in the eIF5B-H480I mutant. Consistent with the improved growth rate of the eIF5B-H480I strain expressing the C434::U mutant form of 18S rRNA, the P/M ratio was increased from 0.5 to 0.9 in this strain (Fig. 4A, right panels), indicating restored translation of a majority of the mRNAs in the cell. Similarly, the P/M ratio increased ∼4-fold in strains carrying the F643R intragenic suppressor mutation in eIF5B (0.2 for the eIF5B-H480I strain versus 0.8 for the eIF5B-H480I,F643R strain in the J111 strain background [data not shown]). The increased P/M ratio in the 18S-C434::U mutant compared to the WT strain (Fig. 4A, left panels) likely reflects a defect in translation elongation (see below).
FIG. 4.
Analysis of rRNA and intragenic suppressors of the eIF5B-H480I mutant. (A) Restoration of translational activity by the 18S-C434::U rRNA suppressor mutation. Polysome profiles of derivatives of yeast strain J215 expressing WT rRNA (top profiles, 18S-WT) or the C434::U rRNA suppressor allele (bottom profiles, 18S-C434::U) and carrying plasmids expressing WT eIF5B (left, eIF5B-WT) or eIF5B-H480I (right), as indicated, are shown. Whole-cell extracts from the strains were resolved by velocity sedimentation in 7 to 47% sucrose gradients (6). Gradients were fractionated while scanning at 254 nm, and the positions of the 40S, 60S, 80S ribosomes and polysomes are indicated. P/M ratios were calculated by measuring the area in the combined polysome fractions and in the 80S peak. (B) Ribosome-dependent GTPase assays. Purified, recombinant eIF5B-WT, eIF5B-H480I, or eIF5B-H480I,F643R was incubated with [γ-33P]GTP and reconstituted 80S ribosomes (43) prepared from strains expressing either WT or C434::U mutant 18S rRNA. Assays were conducted as described for Fig. 1B. The data presented are representative of three independent experiments. (C) Ribosome binding assay. Purified eIF5B-WT, eIF5B-H480I, or eIF5B-H480I,F643R was mixed, in the presence of GTP, GDPNP, GDP, or no nucleotide, as indicated, with yeast 80S ribosomes prepared from strains expressing either WT or C434::U mutant 18S rRNA. Binding reactions were loaded on a 10% sucrose cushion, and following centrifugation the amount of eIF5B recovered in the pellet and supernatant fractions was determined by SDS-PAGE and quantitative densitometry. The data presented represent the fraction of eIF5B present in the ribosomal pellet and are the averages and standard deviations from three independent experiments. *, P < 0.05 by t test (lanes 2 and 4 were compared with lanes 1 and 3, respectively; lane 5 was compared with lane 3).
To further assess the mechanism of suppression, we examined the impact of the rRNA and intragenic suppressor mutations on eIF5B GTPase activity. WT eIF5B showed good GTPase activity with WT ribosomes (0.06 s−1) and slightly lower activity with the C434::U 18S rRNA mutant ribosomes (0.02 s−1) (Fig. 4B). Neither the mutant ribosomes nor the intragenic F643R mutation restored the GTPase activity of the eIF5B-H480I mutant to detectable levels (Fig. 4B). Thus, both the rRNA and the intragenic suppressor mutations restore eIF5B-H480I function in protein synthesis without restoring the GTPase activity of the factor.
Overexpression of eIF5B-H480I in yeast expressing WT eIF5B causes a slow-growth phenotype indicating a dominant-negative phenotype. As reported previously for the GTPase-deficient eIF5B-H480E and eIF5B-T439A mutants (44), we found here that eIF5B-H480I substituted for WT eIF5B and stimulated ribosomal subunit joining in a reconstituted yeast in vitro translation system (data not shown). To test whether the rRNA and the intragenic mutations altered the ribosomal binding affinity of the factor, we used a sucrose cushion assay to monitor eIF5B binding to purified yeast 80S ribosomes. WT and mutant forms of eIF5B were mixed with WT or mutant 80S ribosomes in the presence of GTP, nonhydrolyzable GDPNP, GDP, or no nucleotide, and the complexes were isolated following sedimentation through a 10% sucrose cushion. WT eIF5B showed good binding to ribosomes in the presence of GDPNP and less binding in the presence of GTP, GDP, or no nucleotide (Fig. 4C). As WT eIF5B readily hydrolyzes GTP in the presence of 80S ribosomes (Fig. 4B), the low binding observed in the presence of GTP is likely due to GTP hydrolysis. The expected GTP switch behavior of WT eIF5B binding to the ribosome is observed by comparing the binding in the presence of GDPNP and GDP. For the GTPase-defective eIF5B-H480I mutant, good binding was observed in the presence of GTP and much less binding in the presence of GDP or no nucleotide (Fig. 4C). The binding of both WT eIF5B and eIF5B-H480I was significantly reduced with the mutant ribosomes containing the C434::U insertion in the 18S rRNA. Likewise, the ribosome binding affinity of the eIF5B-H480I,F643R mutant (selected among the domain II intragenic suppressors for biochemical analysis) in the presence of GTP was much lower than that of the eIF5B-H480I mutant (Fig. 4C). Notably, the binding of eIF5B-H480I,F643R to WT ribosomes and the binding of eIF5B-H480I to mutant ribosomes in the presence of GTP was comparable to the low-level binding of WT eIF5B or eIF5B-H480I to WT ribosomes in the presence of GDP (Fig. 4C). These results indicate that the rRNA suppressor mutation, as well as the intragenic domain II F643R mutation, lowered the ribosomal binding affinity of eIF5B, and they are consistent with the notion that the suppressor mutations enable the release of eIF5B from the ribosome following subunit joining in the absence of GTP hydrolysis.
Hydroxyl radical probing reveals binding of the B0 loop in domain II of eIF5B to helix h5 of the 18S rRNA in the 40S subunit.
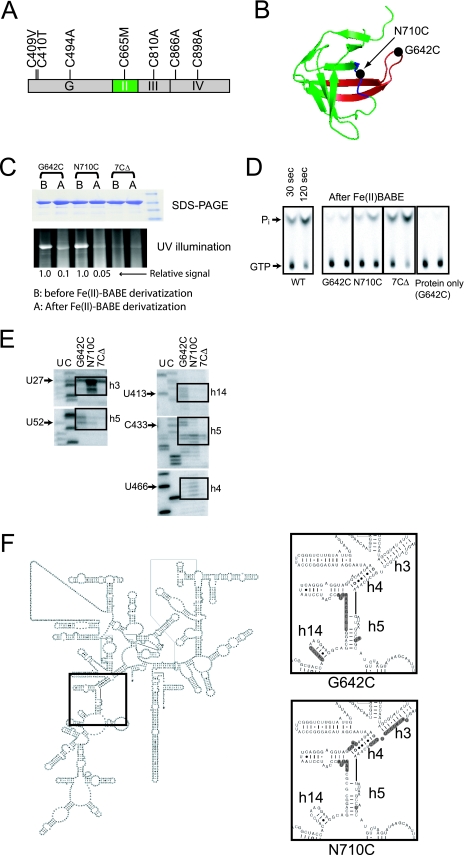
Based on the cryo-EM image of eEF2 bound to yeast 80S ribosomes (47) as well as the images of IF2, EF-G, and EF-Tu bound to 70S ribosomes (3, 13, 55), domain II of these related translational GTPases is thought to contact the body of the small ribosomal subunit in the vicinity of helix h5. Thus, we considered the possibility that the intragenic suppressor mutations in domain II of eIF5B and the rRNA mutations define a surface for interaction between eIF5B and the 40S subunit. To define the binding site of domain II of eIF5B on the ribosome, we used directed hydroxyl radical probing (22). First, a cysteine-less version of yeast eIF5B397-1002 was generated by mutating the seven cysteine residues and incorporating residues found in eIF5B from other organisms as follows: C409V, C410T, C494A, C665M, C810A, C866A, and C898A (Fig. 5A). Expression of cysteine-less eIF5B (eIF5B-7CΔ) fully complemented the slow-growth phenotype of an eIF5BΔ strain (data not shown), and the ribosome-dependent GTPase activity of purified eIF5B-7CΔ was comparable to that of WT eIF5B (Fig. 5D, WT versus 7CΔ). Using the eIF5B-7CΔ mutant as a backbone, two mutants containing a single exposed cysteine residue in domain II were generated. First, Gly642 at the tip of the B0 loop (Fig. 5B) was mutated to Cys. This residue, which corresponds to the Gly222-to-Asp mutation site in the original B0 mutant of EF-Tu (18), is immediately adjacent to the F643R suppressor site. Second, Asn710 was mutated to cysteine. While mutation of Ala709 suppressed the eIF5B-H480I mutant, the Ala709 side chain is directed toward the interior of the β-barrel fold of domain II. In contrast, the side chain of Asn710, immediately adjacent to Ala709, is exposed on the surface of eIF5B (Fig. 5B). The single N710C and G642C mutant forms of eIF5B-7CΔ either fully or partially, respectively, complemented the growth defect in strains lacking eIF5B (data not shown).
FIG. 5.
Directed hydroxyl radical probing of eIF5B domain II interaction with the 40S ribosomal subunit. (A) Construction of cysteine-less eIF5B (eIF5B-7CΔ). A schematic of ΔN-eIF5B is shown; the G domain and domains II, III, and IV are labeled. The seven cysteine residues in ΔN-eIF5B were mutated as indicated. (B) Ribbon model of domain II of eIF5B. The B0 loop is colored red, and the black circles indicate the sites of intragenic suppressor mutations and where single cysteine mutations were introduced for attaching Fe(II)-BABE. (C) Extent of Fe(II)-BABE derivatization examined by reactivity with the thiol-specific fluorescent reagent DCIA. Following BABE derivatization, samples were treated with DCIA and then subjected to SDS-PAGE. The extent of DCIA modification was detected by UV illumination (bottom panel) and quantified by phosphorimager analysis. The gel was then stained with Coomassie brilliant blue (upper panel). The relative extent of DCIA modification was determined after subtracting background obtained with the eIF5B-7CΔ mutant. (D) Analysis of GTPase activity of Fe(II)-BABE-derivatized eIF5B. Ribosome-dependent GTPase activities of unmodified WT eIF5B and Fe(II)-BABE modified eIF5B-7CΔ, eIF5B-7CΔ-G624C, and eIF5B-7CΔ-N710C were measured in the presence or absence (protein only) of 80S ribosomes, as indicated. Reactions were quenched at 30 or 120 s, and the degree of GTP hydrolysis was analyzed by TLC as described for Fig. 1B. The positions of GTP and Pi are indicated. (E) Directed hydroxyl radical probing of 18S rRNA obtained from Fe(II)-BABE-eIF5B-40S ribosome complexes. Primer extension analysis of hydroxyl radical cleavage in helices h3, h4, h5, and h14 of 18S rRNA in eIF5B-80S complexes using Fe(II)-BABE-derivatized eIF5B-7CΔ-G642C, eIF5B-7CΔ-N710C, or mock-derivatized eIF5B-7CΔ (7CΔ) is shown. Lanes U and C represent 18S rRNA sequence generated using the same primers, and the boxes highlight the positions of cleaved nucleotides. (F) Secondary structure of yeast 18S rRNA. Shaded nucleotides in the insets show the sites of hydroxyl radical cleavage in helices h3, h4, h5, and h14 by Fe(II)-BABE-derivatized eIF5B-7CΔ-G642C and eIF5B-7CΔ-N710C, as indicated.
To perform hydroxyl radical mapping, Fe(II) was linked to the unique cysteine residues in the eIF5B-7CΔ-G642C and eIF5B-7CΔ-N710C mutants using the linker BABE. The eIF5B-7CΔ was used as a negative control in these experiments. The extent of Fe(II)-BABE derivatization was estimated at 90% based on the reactivity of the labeled proteins with the thiol-specific fluorescent molecule DCIA (Fig. 5C). The ribosome-dependent GTPase activities of Fe(II)-BABE-derivatized eIF5B-7CΔ-N710C and eIF5B-7CΔ-G642C were reduced compared to those of WT eIF5B and eIF5B-7CΔ (Fig. 5D), consistent with the finding that mutations at the adjacent residues impaired eIF5B binding to 80S ribosomes (Fig. 5D). Fe(II)-BABE-derivatized eIF5B-7CΔ-G642C and eIF5B-7CΔ-N710C, as well as the control protein eIF5B-7CΔ, were incubated with yeast 80S ribosomes in the presence of GDPNP to form eIF5B-80S complexes (and mimic the nonhydrolyzed state of the eIF5B-H480I mutant). The rRNA in the complexes was cleaved by hydroxyl radicals formed in the vicinity of the tethered Fe(II), and the sites of cleavages in the 18S rRNA were mapped using primer extension analysis. As shown in Fig. 5E, in the presence of eIF5B-7CΔ, a few minor cleavage sites were mapped in helices h3 and h5. With eIF5B-7CΔ-G642C, rRNA cleavages were mapped to helix h5 near residues U52 and C433 and to helix h14 near residue U413 (Fig. 5E). Finally, eIF5B-7CΔ-N710C yielded prominent rRNA cleavages in helix h3 near residues U27 and weaker cleavages in helix h5 and in helix h4 near residue U466 (Fig. 5E). Three independent repetitions of this experiment yielded similar results, and no specific cleavages were observed in other regions of the 18S rRNA (cleavages in the 25S rRNA of the large ribosomal subunit were not analyzed).
A summary of the rRNA cleavages on the secondary structure of yeast 18S rRNA is presented in Fig. 5F. Interestingly, helices h3, h4, h5, and h14 cluster in the 5′ domain of the 18S rRNA (Fig. 5F) and come together to form a surface on the body of the 40S subunit (Fig. 3F). Attaching Fe(II)-BABE at residue 642 in the B0 loop generated cleavages in helices h5 and h14, while Fe(II)-BABE linked to residue 710 generated cleavages in helices h3 and h4. The specificity of these cleavages fit well with the model docking eIF5B on the 80S ribosome (Fig. 3F). Taken together, our hydroxyl radical mapping results support the results of the modeling and suppressor analyses and indicate that domain II of eIF5B, and in particular the B0 loop, contacts the body of the 40S subunit in the vicinity of helix h5 (Fig. 3F).
The C434::U mutation in the small ribosomal subunit impairs translation elongation factor function.
The position we mapped for eIF5B binding to the 40S subunit using hydroxyl radical mapping resembles the position where domain II of eEF2 contacts the 40S subunit in the cryo-EM structure of eEF2 bound to the 80S ribosome (47, 53). Interestingly, the B0 loop structure, though not sequence, is conserved in all translational GTPases (4) and projects from the surface of domain II as shown in Fig. 6A. Based on these similarities, we proposed that the conserved domain II of all the translational GTPases (including EF-Tu [eEF1A], EF-G [eEF2], IF2 [eIF5B], and RF3 [eRF3]) binds to the same region on the body of the small subunit in the vicinity of helix h5, and we reasoned that the C434::U mutation would affect the function of the translation elongation factors. As shown in Fig. 4A (and repeated in the lower panels of Fig. 6B), in cells treated with CHX to block translation elongation, similar amounts of polysomes were observed in sucrose gradient analyses of WT and C434:U mutant cells (note that the P/M ratio was slightly increased in the 18S-C434::U mutant cells). However, when the experiment was performed in the absence of CHX, ribosomes continued to elongate during extract preparation from WT cells (Fig. 6B, upper left panel), resulting in polysome runoff and accumulation of an 80S peak consisting primarily of inactive 80S couples (associated 40S and 60S subunits not engaged with mRNA). In contrast, a small but significant amount of polysomes was maintained in the extracts from the C434::U mutant cells even in the absence of CHX (Fig. 6B, upper right panel). These results indicate that there is a defect in translation elongation in the cells expressing the C434:U mutant form of 18S rRNA.
To further assess the impact of the 18S-C434::U mutation on translation elongation, we measured the time required for a ribosome following initiation to release a completed protein. The ribosomal transit time was determined by comparing the rate of [35S]Met incorporation into total protein (nascent plus completed protein) versus completed protein (not associated with ribosomes) (see Materials and Methods) (34). As shown in Fig. 6C, we reproducibly observed an increased average transit time in the 18S-C434::U mutant cells (61 s) compared to the WT cells (43 s). This increased ribosomal transit time is consistent with both the modest growth defect in the FUN12+ C434::U mutant strain (Fig. 2B) and the idea that the18S-C434::U mutation impaired translation elongation.
Mutations in elongation factors eEF1A and eEF2 have been reported to affect the fidelity of translation elongation (16, 35), resulting in either increased or decreased rates of programmed ribosomal frameshifting (PRF) on the −1 PRF site from the yeast L-A virus and the +1 PRF site from the yeast Ty1 retrotransposon. Using a dual-luciferase reporter assay in which the Renilla and firefly luciferase reporters were separated by the +1 PRF site (21), no differences in frameshifting between the WT and C434::U mutant cells were detected (Fig. 6D). In contrast, the C434::U mutation decreased −1 PRF by 37.5% compared to the WT control (Fig. 6D). A similar impairment in −1 PRF was previously reported for mutations in ribosome-associated chaperones (32). It was proposed that lack of the chaperones impaired nascent peptide progression through the exit tunnel, leading to altered P-site tRNA positioning and impaired accommodation of the A-site tRNA. As it is thought that −1 PRF occurs after aminoacyl-tRNA accommodation in the A site and prior to eEF2-mediated translocation, perhaps the rRNA mutation alters eEF1A interactions with the ribosome in a manner that impairs tRNA accommodation in the A site. Taken together, the results in Fig. 6B to D indicate that the C434::U mutation in the 18S rRNA affects both the rate and fidelity of translation elongation, consistent with the notion that the mutation alters the interaction of elongation factors with the ribosome.
Consistent with these in vivo defects in translation elongation, the C434::U mutation in the 18S rRNA impaired the binding of elongation factor eEF2 to purified yeast 80S ribosomes. Using the sucrose cushion assay we previously described for analyzing eIF5B binding to purified yeast 80S ribosomes, we monitored the binding of purified eEF2 to WT or mutant 80S ribosomes in the presence of GTP, nonhydrolyzable GDPNP, GDP, or no nucleotide. Ribosome binding was assessed after pelleting the eEF2-ribosome complexes through a 10% sucrose cushion. The eEF2 readily bound to WT 80S ribosomes in the presence of GTP, GDPNP, and GDP, and the binding was significantly lower in the absence of a nucleotide (Fig. 6E, left). This nucleotide (GTP or GDP) versus no-nucleotide switch behavior of eEF2 contrasts with the GTP-versus-GDP switch governing eIF5B binding to ribosomes (Fig. 4C). However, this alternate switch property of eEF2 is consistent with the finding that the kinetics of EF-G association with bacterial ribosomes is the same in the presence of GTP and GDP (24). The C434::U mutation lowered eEF2 binding in the presence of GTP or GDPNP down to the basal level observed with WT ribosomes in reactions lacking nucleotide (Fig. 6E) and, for unknown reasons, to even lower levels in reactions containing GDP. Thus, the helix h5 mutation in 18S rRNA weakened the ribosome binding of both eIF5B (Fig. 4C) and eEF2 (Fig. 6E).
DISCUSSION
While structural and biochemical studies of static translational GTPase-ribosome complexes have provided valuable insights into the function of the translation factors, in vivo evidence supporting the functional importance of these interactions has not been described. Our identification of an rRNA mutation in helix h5 and domain II mutations in eIF5B, combined with the hydroxyl radical mapping experiments placing eIF5B domain II in the vicinity of helix h5, provide strong genetic evidence supporting the functional importance of this interaction.
Suppressor mutation in the 18S rRNA bypasses the requirement for the GTP/GDP switch governing eIF5B ribosome binding affinity.
The eIF5B-H480I mutation blocks the ribosome-dependent GTPase activity of the factor (Fig. 1B), and, consistently, expression of the mutant factor inhibits yeast cell growth (Fig. 1A). Here we describe intragenic suppressor mutations in domain II of eIF5B as well as a U insertion mutation in helix h5 of the 18S rRNA that suppress the toxic phenotypes associated with the H480I mutation. The suppressor mutations do not restore eIF5B GTPase activity (Fig. 4B) but instead restore translation activity (Fig. 4A) by decreasing the ribosome binding affinity of eIF5B (Fig. 4C). We propose that the eIF5B and ribosomal suppressor mutations bypass the requirement for eIF5B GTPase activity by enabling the factor to dissociate from the ribosome in the absence of GTP hydrolysis. Thus, these results provide independent support for the notion that the eIF5B GTP switch governs the ribosome binding affinity of the factor (44).
A conserved element in domain II of the translational GTPases contacts the body of the small ribosomal subunit.
Our findings that eIF5B domain II and rRNA helix h5 mutations decrease the ribosomal binding affinity of eIF5B (Fig. 4C) and that the B0 loop of eIF5B is in close contact with the 40S subunit in the vicinity of helix h5 (Fig. 3F and 5) are supported by cryo-EM and mapping studies of the IF2/aIF5B/eIF5B-ribosome complexes. Cryo-EM studies of Thermus thermophilus IF2-mRNA-fMet-tRNAiMet-70S ribosome complexes revealed that domain II of IF2-GTP is close to helices h5 and h17 in the body of the 30S subunit (33), and similar complexes of IF2 with the 30S subunit placed domain II in the vicinity of helices h5 and h14 (45). Likewise, in E. coli initiation complexes consisting of IF2-GDPNP, mRNA, fMet-tRNAiMet, IF1, IF3, and 70S ribosomes, domain II of IF2 covered an area that included helix h5 (3). In addition, two studies chemically mapped the binding site of IF2/eIF5B on the ribosome. First, linking the chemical nuclease Cu(II)-phenanthraline to residue 451 in domain II of B. stearothermophilus IF2 resulted in hydroxyl radical-induced cleavages in helix h3 (31). Second, linking Fe(II)-BABE at multiple positions in domain II of human eIF5B resulted in rRNA cleavages in helices h3, h4, h5, h15, and h17 (54). Though none of the sites of derivatization in either of these studies mapped close to the G642 and N710 sites employed in our studies, our mutual identification of helices h3, h4, and h5 provides strong support for docking of eIF5B domain II on the body of the 40S subunit (Fig. 3F).
Previously we identified three intragenic mutations that suppressed the translation defect and slow-growth phenotype of the GTPase-deficient eIF5B-T439A switch I mutant (44). One suppressor, H505Y, mapped to the G domain, and biochemical analyses revealed that this mutation lowered the ribosomal binding of eIF5B. The other two suppressors, F643S and A709V, mapped to domain II; however, their mechanism of suppression was not explored. Interestingly, in this work we independently isolated the same A709V and the related F643R mutations as suppressors of the GTPase-deficient eIF5B-H480I switch II mutant. Thus, the domain II suppressor mutations are not specific to the eIF5B-H480I mutant but instead can apparently restore the function of any eIF5B GTPase-deficient mutant.
In addition to the F643R mutation, we also identified the G642F suppressor mutation, whose position corresponds to the site of the so-called B0 mutation (G222D) in the E. coli tufB gene encoding EF-Tu. Biochemical analyses revealed that the EF-Tu-G222D mutant requires a higher Mg2+ concentration than WT EF-Tu to catalyze polyphenylalanine synthesis and to hydrolyze GTP in the presence of ribosomes (51). In addition, pre-steady-state kinetic analyses revealed that the B0 mutation impaired the high-affinity interactions of EF-Tu with the ribosome and apparently altered the transmission of a signal coupling codon recognition to GTP hydrolysis and release of aminoacyl-tRNA from EF-Tu to the ribosomal A site (56). These results demonstrate that the B0 loop is an important determinant of EF-Tu function on the ribosome.
Interestingly, the B0 loop structure, but not sequence, is conserved in all four translational GTPases (4), and several structural studies have revealed contacts between domain II of the translational GTPases and the body of the small ribosomal subunit. In cryo-EM structures of RF3-GTP bound to a posttermination ribosome (20) and of EF-G-GTP bound to the 70S ribosome (13), the B0 loop was docked in the vicinity of helix h5 on the small ribosomal subunit. Moreover, in the structures of an EF-Tu-GTP-aminoacyl-tRNA complex bound to the E. coli 70S ribosome in the presence of kirromycin (55) and of eEF2 bound to the yeast 80S ribosome in the presence of the drug sordarin (47), domain II of the GTPases contacted the helix h5 region of the ribosomal subunit. Taken together, the cryo-EM studies, the nuclease mapping experiments, and our genetic suppressor analyses reveal a common and functionally important binding site for the translational GTPases on the ribosome.
Further characterization of the 18S-C434::U mutant provided the first evidence that the interaction between domain II of the translational GTPases and the body of the small subunit is also functionally important. The presence of stable polysomes in the absence of CHX in the C434::U mutant cells (Fig. 6B) indicates a defect in translation elongation. Likewise, the increased P/M ratio in the presence of CHX (Fig. 4A, left panels) and the increased ribosomal transit time (Fig. 6C) in the 18S-C434::U mutant cells are consistent with a defect in translation elongation or termination (as the mutant cells display a defect in release of the completed proteins). As both elongation (factors eEF1A and eEF2) and termination (factor eRF3) are dependent on GTPase function, the observed defect in ribosomal transit time supports the notion that the rRNA mutation alters the functional interaction of the translational GTPases with the ribosome. Consistently, the helix h5 mutation decreased the affinity of eEF2 binding to the ribosome (Fig. 6E). Thus, in addition to altering the interaction of eIF5B, the helix h5 mutation affects elongation factor function on the ribosome. Together these results demonstrate that the domain II contacts with the small subunit contribute significantly to the interaction of the GTPases with the ribosome. In the absence of this supporting contact, premature or unregulated release of the translational GTPases from the ribosome negatively affects the rate and alters the fidelity of protein synthesis.
Acknowledgments
We thank our colleagues in the Dever, Lorsch, and Hinnebusch laboratories for advice and helpful discussions; Jon Dinman for the PRF reporter plasmids; Preeti Saini for advice on the transit time measurements; and Alan Hinnebusch for comments on the manuscript.
This work was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Development, National Institutes of Health (T.E.D.), and by American Cancer Society grant RSG-03-156-01-GMC (J.R.L.).
Footnotes
Published ahead of print on 24 November 2008.
REFERENCES
- 1.Abel, K., and F. Jurnak. 1996. A complex profile of protein elongation: translating chemical energy into molecular movement. Structure 4229-238. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, R. K., P. Penczek, R. A. Grassucci, and J. Frank. 1998. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl. Acad. Sci. USA 956134-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, G. S., A. Zavialov, R. Gursky, M. Ehrenberg, and J. Frank. 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121703-712. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, G. R., and J. Nyborg. 2001. Structural studies of eukaryotic elongation factors. Cold Spring Harbor Symp. Quant. Biol. 66425-437. [DOI] [PubMed] [Google Scholar]
- 5.Antoun, A., M. Y. Pavlov, K. Andersson, T. Tenson, and M. Ehrenberg. 2003. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 225593-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asano, K., J. Clayton, A. Shalev, and A. G. Hinnebusch. 2000. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 142534-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289905-920. [DOI] [PubMed] [Google Scholar]
- 8.Beckmann, R., C. M. Spahn, N. Eswar, J. Helmers, P. A. Penczek, A. Sali, J. Frank, and G. Blobel. 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107361-372. [DOI] [PubMed] [Google Scholar]
- 9.Berchtold, H., L. Reshetnikova, C. O. Reiser, N. K. Schirmer, M. Sprinzl, and R. Hilgenfeld. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365126-132. [DOI] [PubMed] [Google Scholar]
- 10.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349117-127. [DOI] [PubMed] [Google Scholar]
- 11.Cate, J. H., M. M. Yusupov, G. Z. Yusupova, T. N. Earnest, and H. F. Noller. 1999. X-ray crystal structures of 70S ribosome functional complexes. Science 2852095-2104. [DOI] [PubMed] [Google Scholar]
- 12.Choi, S. K., D. S. Olsen, A. Roll-Mecak, A. Martung, K. L. Remo, S. K. Burley, A. G. Hinnebusch, and T. E. Dever. 2000. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 207183-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connell, S. R., C. Takemoto, D. N. Wilson, H. Wang, K. Murayama, T. Terada, M. Shirouzu, M. Rost, M. Schuler, J. Giesebrecht, M. Dabrowski, T. Mielke, P. Fucini, S. Yokoyama, and C. M. Spahn. 2007. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol. Cell 25751-764. [DOI] [PubMed] [Google Scholar]
- 14.DeLano, W. 2004. The PyMol user's manual. DeLano Scientific, San Carlos, CA.
- 15.Diaconu, M., U. Kothe, F. Schlunzen, N. Fischer, J. M. Harms, A. G. Tonevitsky, H. Stark, M. V. Rodnina, and M. C. Wahl. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121991-1004. [DOI] [PubMed] [Google Scholar]
- 16.Dinman, J. D., and T. G. Kinzy. 1997. Translational misreading: mutations in translation elongation factor 1alpha differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA 3870-881. [PMC free article] [PubMed] [Google Scholar]
- 17.Ditzelmuller, G., W. Wohrer, C. P. Kubicek, and M. Rohr. 1983. Nucleotide pools of growing, synchronized and stressed cultures of Saccharomyces cerevisiae. Arch. Microbiol. 13563-67. [DOI] [PubMed] [Google Scholar]
- 18.Duisterwinkel, F. J., B. Kraal, J. M. De Graaf, A. Talens, L. Bosch, G. W. Swart, A. Parmeggiani, T. F. La Cour, J. Nyborg, and B. F. Clark. 1984. Specific alterations of the EF-Tu polypeptide chain considered in the light of its three-dimensional structure. EMBO J. 3113-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fringer, J. M., M. G. Acker, C. A. Fekete, J. R. Lorsch, and T. E. Dever. 2007. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol. 272384-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, H., Z. Zhou, U. Rawat, C. Huang, L. Bouakaz, C. Wang, Z. Cheng, Y. Liu, A. Zavialov, R. Gursky, S. Sanyal, M. Ehrenberg, J. Frank, and H. Song. 2007. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129929-941. [DOI] [PubMed] [Google Scholar]
- 21.Harger, J. W., and J. D. Dinman. 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 91019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilek, G. M., R. Marusak, C. F. Meares, and H. F. Noller. 1995. Directed hydroxyl radical probing of 16S rRNA using Fe(II) tethered to ribosomal protein S4. Proc. Natl. Acad. Sci. USA 921113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen, R., P. A. Ortiz, A. Carr-Schmid, P. Nissen, T. G. Kinzy, and G. R. Andersen. 2003. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 10379-385. [DOI] [PubMed] [Google Scholar]
- 24.Katunin, V. I., A. Savelsbergh, M. V. Rodnina, and W. Wintermeyer. 2002. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 4112806-12812. [DOI] [PubMed] [Google Scholar]
- 25.Kisselev, L., M. Ehrenberg, and L. Frolova. 2003. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 22175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kjeldgaard, M., P. Nissen, S. Thirup, and J. Nyborg. 1993. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 135-50. [DOI] [PubMed] [Google Scholar]
- 27.Klaholz, B. P., A. G. Myasnikov, and M. Van Heel. 2004. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427862-865. [DOI] [PubMed] [Google Scholar]
- 28.Kong, C., K. Ito, M. A. Walsh, M. Wada, Y. Liu, S. Kumar, D. Barford, Y. Nakamura, and H. Song. 2004. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell 14233-245. [DOI] [PubMed] [Google Scholar]
- 29.La Teana, A., C. Gualerzi, and A. Dahlberg. 2001. Initiation factor IF2 binds the α-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 71173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, J. H., T. V. Pestova, B. S. Shin, C. Cao, S. K. Choi, and T. E. Dever. 2002. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. USA 9916689-16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marzi, S., W. Knight, L. Brandi, E. Caserta, N. Soboleva, W. E. Hill, C. O. Gualerzi, and J. S. Lodmell. 2003. Ribosomal localization of translation initiation factor IF2. RNA 9958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muldoon-Jacobs, K. L., and J. D. Dinman. 2006. Specific effects of ribosome-tethered molecular chaperones on programmed −1 ribosomal frameshifting. Eukaryot. Cell 5762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myasnikov, A. G., S. Marzi, A. Simonetti, A. M. Giuliodori, C. O. Gualerzi, G. Yusupova, M. Yusupov, and B. P. Klaholz. 2005. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol. 121145-1149. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, P. J., and E. H. McConkey. 1980. Evidence for control of protein synthesis in HeLa cells via the elongation rate. J. Cell Physiol. 104269-281. [DOI] [PubMed] [Google Scholar]
- 35.Ortiz, P. A., R. Ulloque, G. K. Kihara, H. Zheng, and T. G. Kinzy. 2006. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 28132639-32648. [DOI] [PubMed] [Google Scholar]
- 36.Pestova, T. V., I. B. Lomakin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403332-335. [DOI] [PubMed] [Google Scholar]
- 37.Ramakrishnan, V. 2002. Ribosome structure and the mechanism of translation. Cell 108557-572. [DOI] [PubMed] [Google Scholar]
- 38.Rodnina, M. V., and W. Wintermeyer. 2001. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 70415-435. [DOI] [PubMed] [Google Scholar]
- 39.Roll-Mecak, A., C. Cao, T. E. Dever, and S. K. Burley. 2000. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103781-792. [DOI] [PubMed] [Google Scholar]
- 40.Roll-Mecak, A., B. S. Shin, T. E. Dever, and S. K. Burley. 2001. Engaging the ribosome: universal IFs of translation. Trends Biochem. Sci. 26705-709. [DOI] [PubMed] [Google Scholar]
- 41.Shenton, D., J. B. Smirnova, J. N. Selley, K. Carroll, S. J. Hubbard, G. D. Pavitt, M. P. Ashe, and C. M. Grant. 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 28129011-29021. [DOI] [PubMed] [Google Scholar]
- 42.Shin, B. S., M. G. Acker, D. Maag, J. R. Kim, J. R. Lorsch, and T. E. Dever. 2007. Intragenic suppressor mutations restore GTPase and translation functions of a eukaryotic initiation factor 5B switch II mutant. Mol. Cell. Biol. 271677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin, B. S., and T. E. Dever. 2007. Molecular genetic structure-function analysis of translation initiation factor eIF5B. Methods Enzymol. 429185-201. [DOI] [PubMed] [Google Scholar]
- 44.Shin, B. S., D. Maag, A. Roll-Mecak, M. S. Arefin, S. K. Burley, J. R. Lorsch, and T. E. Dever. 2002. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 1111015-1025. [DOI] [PubMed] [Google Scholar]
- 45.Simonetti, A., S. Marzi, A. G. Myasnikov, A. Fabbretti, M. Yusupov, C. O. Gualerzi, and B. P. Klaholz. 2008. Structure of the 30S translation initiation complex. Nature 455416-420. [DOI] [PubMed] [Google Scholar]
- 46.Spahn, C. M., R. Beckmann, N. Eswar, P. A. Penczek, A. Sali, G. Blobel, and J. Frank. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107373-386. [DOI] [PubMed] [Google Scholar]
- 47.Spahn, C. M., M. G. Gomez-Lorenzo, R. A. Grassucci, R. Jorgensen, G. R. Andersen, R. Beckmann, P. A. Penczek, J. P. Ballesta, and J. Frank. 2004. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 231008-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66639-678. [DOI] [PubMed] [Google Scholar]
- 49.Stark, H., M. V. Rodnina, J. Rinke-Appel, R. Brimacombe, W. Wintermeyer, and M. van Heel. 1997. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389403-406. [DOI] [PubMed] [Google Scholar]
- 50.Stern, S., D. Moazed, and H. F. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164481-489. [DOI] [PubMed] [Google Scholar]
- 51.Swart, G. W., A. Parmeggiani, B. Kraal, and L. Bosch. 1987. Effects of the mutation glycine-222→aspartic acid on the functions of elongation factor Tu. Biochemistry 262047-2054. [DOI] [PubMed] [Google Scholar]
- 52.Taylor, D., J. Frank, and T. Kinzy. 2007. Structure and function of eukaryotic ribosome and elongation factors, p. 59-85. In M. Mathews, N. Sonenberg, and J. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 53.Taylor, D., J. Nilsson, A. Merrill, G. Andersen, P. Nissen, and J. Frank. 2007. Structures of modified eEF2 · 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 262421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unbehaun, A., A. Marintchev, I. B. Lomakin, T. Didenko, G. Wagner, C. U. Hellen, and T. V. Pestova. 2007. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 263109-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valle, M., A. Zavialov, W. Li, S. M. Stagg, J. Sengupta, R. C. Nielsen, P. Nissen, S. C. Harvey, M. Ehrenberg, and J. Frank. 2003. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 10899-906. [DOI] [PubMed] [Google Scholar]
- 56.Vorstenbosch, E., T. Pape, M. V. Rodnina, B. Kraal, and W. Wintermeyer. 1996. The G222D mutation in elongation factor Tu inhibits the codon-induced conformational changes leading to GTPase activation on the ribosome. EMBO J. 156766-6774. [PMC free article] [PubMed] [Google Scholar]
- 57.Wai, H. H., L. Vu, M. Oakes, and M. Nomura. 2000. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 283524-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]