Abstract
Some human cancers maintain telomeres using alternative lengthening of telomeres (ALT), a process thought to be due to recombination. In Kluyveromyces lactis mutants lacking telomerase, recombinational telomere elongation (RTE) is induced at short telomeres but is suppressed once telomeres are moderately elongated by RTE. Recent work has shown that certain telomere capping defects can trigger a different type of RTE that results in much more extensive telomere elongation that is reminiscent of human ALT cells. In this study, we generated telomeres composed of either of two types of mutant telomeric repeats, Acc and SnaB, that each alter the binding site for the telomeric protein Rap1. We show here that arrays of both types of mutant repeats present basally on a telomere were defective in negatively regulating telomere length in the presence of telomerase. Similarly, when each type of mutant repeat was spread to all chromosome ends in cells lacking telomerase, they led to the formation of telomeres produced by RTE that were much longer than those seen in cells with only wild-type telomeric repeats. The Acc repeats produced the more severe defect in both types of telomere maintenance, consistent with their more severe Rap1 binding defect. Curiously, although telomerase deletion mutants with telomeres composed of Acc repeats invariably showed extreme telomere elongation, they often also initially showed persistent very short telomeres with few or no Acc repeats. We suggest that these result from futile cycles of recombinational elongation and truncation of the Acc repeats from the telomeres. The presence of extensive 3′ overhangs at mutant telomeres suggests that Rap1 may normally be involved in controlling 5′ end degradation.
Telomeres are the DNA-protein complexes present at the ends of linear chromosomes (11, 52, 83). Telomeric DNA is composed of short tandem repeats, commonly between 5 and 26 bp in size. Telomeres vary widely in size between organisms but are generally maintained at a relatively stable length within an organism. An essential role of telomeres is to protect chromosome ends from the homologous recombination (HR) and nonhomologous end joining, which normally act at broken double-stranded DNA ends (8, 21). However, the inability of replicative polymerases to fully replicate ends causes telomeres to shorten gradually over time, compromising their role in end protection. To prevent this problem, the great majority of eukaryotes use the ribonucleoprotein enzyme telomerase, which can add telomeric repeats to the telomere ends (1, 40). Telomerase is recruited to the telomere in large part via telomere binding proteins that bind to a short 3′ overhang in the DNA at the telomere end (71).
Telomeres are protected from degradation and repair by specific proteins. In humans, a complex of six proteins called shelterin caps and protects the telomere (15). In addition, a looped structure called a t-loop, thought to be a strand invasion of the 3′ DNA overhang into internal duplex telomeric DNA, seems to also facilitate end protection in many species (28). In yeasts such as Saccharomyces cerevisiae and Kluyveromyces lactis, the double-stranded telomeric DNA is bound by Rap1, while the single-stranded 3′ overhang is bound by the trimeric Cdc13/Stn1/Ten1 complex (4, 32). Rap1 participates in the negative regulation of telomere length, and its loss from the telomere results in telomere end-end fusions (37, 44, 51, 63). Proteins associated with Rap1 at the telomere include Rif1 and Rif2, which participate in telomere length regulation, and Sir2, -3, and -4, which participate in telomeric silencing (71). The Cdc13/Stn1/Ten1 complex is required to recruit telomerase and lagging-strand replication proteins to the telomere. It also protects the DNA end against recombination events (12, 19, 24-27, 64).
In humans, telomerase is present only at low levels in most somatic tissues, and telomeres become progressively shorter with each cell division (69, 70). Once telomeres reach a critically short length, they trigger a permanent growth arrest called replicative senescence. Because of this, immortalized cells, including the great majority of cancers, have a telomere maintenance method, most commonly telomerase (34). However, a significant minority of cancers use alternative lengthening of telomeres (ALT), to maintain telomeres (55). The telomeres in typical cells displaying ALT are highly heterogeneous in length, and while many are much longer than normal human telomeres, others are abnormally short (6, 38, 56, 65). The telomeres of ALT cancers can copy DNA from telomere to telomere and therefore are thought to maintain their lengths by recombination (17). Recombination proteins such as RAD51 and the MRN complex as well as telomere binding proteins are present in ALT cells in subnuclear bodies called ALT-associated PML bodies (89-91). Extrachromosomal telomeric DNA of both linear and circular form is also found abundantly in ALT cells (9, 62, 85).
Yeast mutants lacking telomerase have been important model systems for understanding how recombination can maintain telomeres. Upon deletion of telomerase in both Saccharomyces cerevisiae and Kluyveromyces lactis, cells display telomere shortening and growth senescence that is followed by the occasional production of better-growing postsenescence survivors that arise from recombinational telomere elongation (RTE) (42, 47, 49). RTE in yeast telomerase deletion mutants appears to be triggered by the telomeres becoming too short (50). Once below ∼100 bp in length, telomeres from both S. cerevisiae and K. lactis become able to initiate recombination (73, 77). In S. cerevisiae, two distinct types of survivors that differ both in their telomeric structure and in the genes required for their formation have been observed. Type I survivors display amplified subtelomeric Y′ elements and have short terminal tracts of telomeric repeats (42, 75). Their formation requires Rad52 and the canonical HR repair proteins Rad51, Rad55, and Rad57 (39). Type II survivors, in contrast, lack subtelomeric amplification and instead have elongated tracts of telomeric repeats. Their formation requires Rad52 and depends also on Rad50, Rad59, and Sgs1 instead of the Rad51 group of proteins (13, 74). In K. lactis, only type II survivors normally occur (47). A variety of experimental evidence with both K. lactis and S. cerevisiae suggests that type II postsenescence survivors arise through a “roll-and-spread” mechanism whereby an elongated telomere is first formed by a rolling-circle copying of a very small telomeric circle (t-circle) (29, 41, 58, 59). This is followed by additional break-induced replication events that copy the elongated sequence onto other telomeric ends (77).
More recently, it has become clear that RTE in yeast can become induced by certain perturbations in telomeric capping proteins even when telomeres are not abnormally short. For instance, in S. cerevisiae, a cdc13-1 yku70 mutation at the semipermissive temperature caused type II survivors to form after a period of senescence-like growth without appreciably shortened telomeres (23). In K. lactis, a mutation in the telomere-associated protein Stn1 (stn1-M1) led to RTE that produced highly elongated and unstable telomeres and other features that distinguished it from the RTE of telomerase deletion survivors (33). The stn1-M1 cells had a chronic moderate growth defect but failed to display the large changes in growth rate that characterize senescence and survivor formation. This unusual RTE, termed type IIR (“runaway”), is thought to be due to a telomere capping defect that renders telomeres prone to initiate HR in a manner largely or entirely independent of their size. Interestingly, in a recent finding, the stn1-281t allele of S. cerevisiae similarly led to long heterogeneous telomeres as well as inviability in the absence of RAD52 (66). The close similarity of type IIR RTE to the ALT phenotypes of certain human cancers and cell lines makes it an especially important phenomenon to understand.
It was recently shown that the sequences from a single telomere engineered to contain only mutationally tagged telomeric repeats could sometimes be spread to all other telomeres in the cell during the formation of postsenescence survivors in a K. lactis mutant lacking a functional TER1 gene encoding the telomerase RNA (77). Here, we have taken advantage of this technique to test whether either of two telomeric sequence mutations perturbs the manner in which RTE occurs in telomerase-negative cells. These mutations make base changes within each telomeric repeat that fall within the binding site of double-strand telomere binding protein Rap1. We demonstrate that both of these mutations can in fact lead to recombinational telomere maintenance with characteristics similar to those of type IIR RTE.
MATERIALS AND METHODS
Strains and culturing conditions.
All strains used in this study, with the exception of those used for Fig. 6C and 6D, are derivatives of 7B520 (ura3 his2-2 trp1) (88). The wild-type strain CBS 2359 was used for Fig. 6C and D, and the ku80Δ mutant used as a control was in a CBS 2359 background (36). The ter1-19A(Acc) and ter1-24T(SnaB) single mutants and heteroalleles were made by a plasmid loop-in replacement process using pTER-BX-UA, which was described previously (48, 50). All of the transformations of the single mutant telomere were made in 7B520 with a ter1 deletion mutation (68). TER1 was reintroduced into the cells with the plasmid pJR31, a derivative of pKL316, which contains a HIS3 gene (68). The SnaB and Acc mutant telomeres were constructed by performing oligonucleotide-directed mutagenesis on a plasmid (pAK25ΔB) that contained a cloned wild-type K. lactis telomere as described previously (77, 81). The pAK25ΔB plasmid was derived from pAK25 by filling in the overhangs of the unique BglII site next to the URA3 gene inserted into the subtelomeric sequence (50).
FIG. 6.
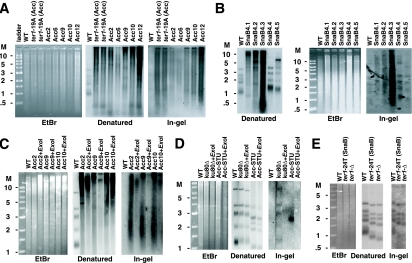
Telomeres in Acc and SnaB survivors have substantial amounts of single-stranded DNA. (A) Ethidium bromide-stained gel (EtBr), Southern blot, and in-gel hybridization of DNA from Acc survivors. The first lane in each is a wild-type (WT) control. The second and third lanes are two independent samples of ter1-19A(Acc) cells. The remaining lanes are the same survivors as shown in Fig. 4. (B) Ethidium bromide-stained gel, Southern blot, and in-gel hybridization of EcoRI-digested DNA from each of five streaks of SnaB survivor 4. The streak numbers are noted after the survivor number above the gel. Also shown is the wild-type strain 7B520. The Southern blot and in-gel hybridization only in panel B ran differently and therefore have different size markers. (C) Ethidium bromide-stained gel, Southern blot, and in-gel hybridization of EcoRI-digested and EcoRI- plus Exo I-digested DNAs of Acc survivors 2, 9, 10, and 12 along with those for wild-type strain CBS 2359. Both the Southern blot and in-gel hybridization were probed with a C-stranded telomeric oligonucleotide. (D) Ethidium bromide-stained gel, Southern blot, and in-gel hybridization of EcoRI-digested cells of the wild-type strain CBS 2359 and the EcoRI- and EcoRI- plus Exo I-digested DNAs of ku80Δ cells and senescent ter1-Δ cells containing an Acc-STU telomere that has not yet spread to other telomeres. (E) Ethidium bromide-stained gel, Southern blot, and in-gel hybridization of EcoRI-digested DNAs of the wild-type strain 7B520, a ter1-24T(SnaB) strain, and a ter1-Δ strain. Molecular weight markers (M) are shown in kilobases.
In general, cells were grown on yeast extract-peptone-dextrose (YPD) rich medium. The selective plates used were SD minimal medium plates lacking either histidine, uracil, or both. Transformations were plated on selective medium supplemented with 1 M sorbitol. YPD liquid medium was used for growing cells for genomic DNA preparations.
To generate cells with a single mutant telomere, DNA fragments containing a mutant telomere with a subtelomeric URA3 gene were transformed into K. lactis cells as described previously (50). pAK25 derivatives containing only Acc or SnaB repeats were cleaved with EcoRI and SacII to release the URA3-tagged telomeric fragment. This fragment was then transformed into a ter1-Δ mutant containing pJR31 and plated on medium lacking histidine and uracil and supplemented with 1 M sorbitol. Fragments used were ones that contained 15 or >28 SnaB repeats and one with 14 Acc telomeric repeats. Transformants were restreaked onto plates lacking uracil and histidine in order to eliminate any untransformed cells. Genomic DNA preparations were conducted to confirm that a single telomere had been replaced. In individual transformants, the numbers of mutant repeats retained on the mutant telomere after wild-type repeats were added to the end were commonly somewhat smaller than the number present on the transforming fragment.
Transformants confirmed to have a single mutant telomere were streaked on YPD medium and patched onto a plate lacking uracil and another lacking histidine. The YPD streaks were observed for the formation of senescing cells, as indicated by partially rough colonies. The rough edges were restreaked onto new YPD medium and also patched to the above-mentioned selective plates. The appearance of rough colonies corresponded with loss of the His+ phenotype as expected for loss of the TER1-containing pJR31 plasmid. The screening on medium lacking uracil confirmed that the single long telomere was still present. Senescing cells were then serially restreaked on YPD every 3 to 4 days, with each streak representing up to 20 to 25 cell divisions. The growth on these plates was then scored from 0 to 4 after loss of pJR31. A score of 0 represented no growth, and a score of 4 represented wild-type growth. Scores in between were based on the size and the degree of roughness of the colonies. After two or three streaks of senescence, when growth scores first leveled or began to improve, scoops of cells from the plates were taken for genomic DNA preparations. At this point, cells were considered to be “survivors” and were found to have telomeres that were lengthened relative to those of cells in a highly senescent state. Typically, multiple serial streaks were done on survivor cell lineages once they were formed.
Gel electrophoresis and Southern blotting.
Restriction digests of yeast genomic DNAs were carried out in the presence of RNase and were run on 0.8% agarose gels in Tris-borate buffer unless otherwise specified. They were visualized by ethidium bromide treatment prior to blotting. Gels were blotted onto a Hybond N+ (Amersham Biosciences, Piscataway, NJ) membrane in 0.4 M NaOH. They were allowed to transfer for 1 day and cross-linked using UV light from an electronic cross-linker.
Hybridizations were conducted in 7% sodium dodecyl sulfate, 0.5 M EDTA, and 0.5 M Na2HPO4 as described previously (14). For telomeric probes, the G-stranded Klac1-25 oligonucleotide (ACGGATTTGATTAGGTATGTGGTGT) was end labeled with [γ-32P]ATP and allowed to hybridize with the membrane for at least 4 h at 48°C. The membrane was then washed in 100 mM Na2HPO4 and 2% sodium dodecyl sulfate three times at 48°C and visualized using a Molecular Dynamics (Sunnyvale, CA) Storm phosphorimager. For subtelomeric probes, a 615-bp doubly cleaved HindIII subtelomeric fragment from plasmid pMya was gel purified using a QIAquick gel extraction kit (Qiagen, Valencia, CA). This fragment contains the K. lactis sequence between the EcoRI site and the URA3 gene (see Fig. 2A). pMya is a derivative of pAK25ΔB made by deleting the telomeric repeats after SacI digestion and religation. The subtelomeric fragment was labeled with [α-32P]dATP using the large Klenow fragment and a Stratagene (La Jolla, CA) Prime-It II random primer labeling kit.
FIG. 2.
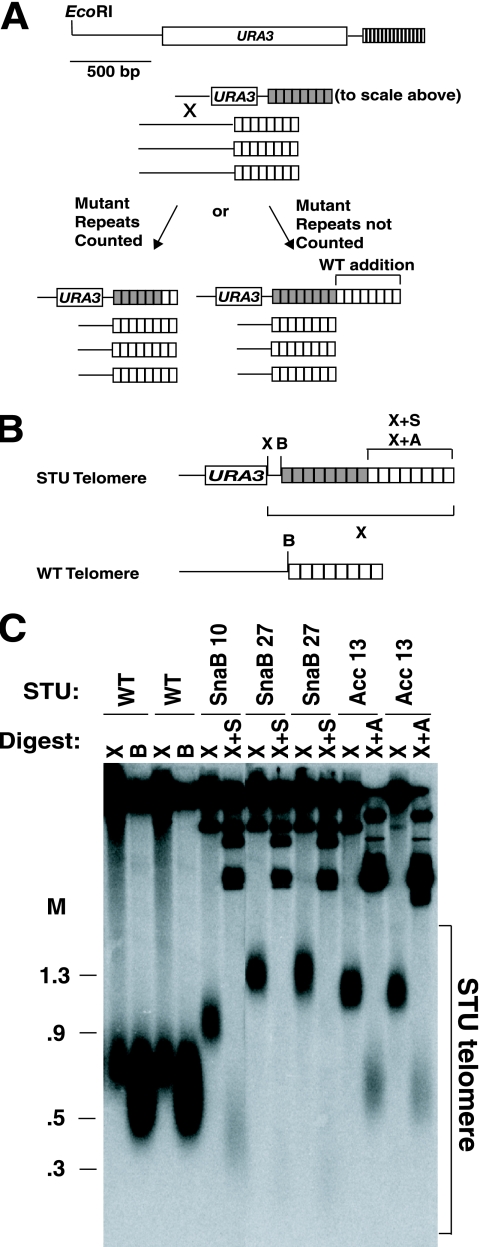
SnaB and Acc telomeric repeats are defective in regulating telomerase addition to their ends. (A) Diagram of the experimental method for replacing a native telomere with a mutant telomere (gray boxes represent mutant repeats, and white boxes represent wild-type repeats). Restriction fragments containing mutant telomeres were transformed into wild-type cells, where they each replaced a single native telomere by recombination between common subtelomeric sequences. Upon integration, the mutated telomeres acquire some number of terminal wild-type repeats (white boxes) from the resident wild-type telomerase. See text for details. A scale diagram of a STU telomeric fragment is shown at the top. (B) The STU telomeres have a unique XhoI (X) site at the end of the URA3 fragment. A BsrBI (B) site is located 3 bp upstream of the telomeric repeats that is present at 10 of 12 telomeres. The tagged repeats each have an Acc or SnaB (A or S) restriction site so that the wild-type (WT) addition onto them can be measured. The brackets represent fragments generated by particular digests. (C) Southern blot of SnaB and Acc telomeres. The leftmost four lanes show two independent wild-type STU telomere transformants cut by XhoI (X) or BsrBI (B). The band between 0.5 and 0.9 kb is the STU telomere in XhoI digests. The slightly smaller band in the BsrBI digests represents 10 of the 12 telomeres. The central six lanes show transformants that received a SnaB STU telomere with ∼10 SnaB repeats (SnaB 10) or ∼27 SnaB repeats (SnaB 27) cut by XhoI or by a double digest with XhoI and SnaBI (X+S). The rightmost lanes show XhoI and XhoI-AccI (X+A) digests of two transformants that received an Acc STU telomere with 13 Acc repeats. The wild-type addition in the different double-digest lanes can be seen as a light smear near the bottom of the gel. The bracket marks the range of positions of the STU telomere fragments. Markers (M) are shown in kilobases.
In-gel hybridization.
Agarose (0.7% or 0.8%) gels were run using the same protocol as that for the gels used for blotting. The EcoRI-digested DNA used for these gels was split in half, with half being used in each of two agarose gels, one that was subject to Southern blotting (denatured gels) and the other that was used as follows for in-gel hybridization. As described previously (16), gels were soaked in 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate, pH 7.0) for 30 min. They were then blotted to near dryness using Whatman 3-mm chromatography paper for approximately 30 to 90 min. The flattened gels were then hybridized in 10× SSC overnight with a C-strand-specific oligonucleotide (ACACCACATACCTAATCAAATCCGT) in order to visualize single-stranded telomeric G-strand DNA. The denatured gels were hybridized to either a C-strand-specific probe or a G-strand-specific probe. After hybridization, the gel was washed three or four times in 0.25× SSC for 1.5 h per wash, followed by visualization on a phosphorimager.
Exo I digestion.
A total of 13.3% of the total DNA from a genomic preparation from a 1.5-ml overnight culture was incubated with 20 units of EcoRI enzyme prepared by New England Biosciences (Ipswitch, MA) in NEBuffer EcoRI (50 mM NaCl, 100 mM Tris-HCl, 10 mM MgCl2, 0.025% Triton X-100 [pH 7.5 at 25°C]) for 3 hours prior to digestion with exonuclease I (Exo I). Twenty units of Exo I enzyme prepared by New England Biosciences was added after a buffer change from NEBuffer EcoRI to NEBuffer Exo I (67 mM glycine-KOH [pH 9.5], 6.7 mM MgCl2, 10 mM 2-mercaptoethanol) using Quantum Prep PCR Kleen spin columns (Bio-Rad Laboratories, Hercules, CA). Reaction mixtures were incubated for 3 hours at 37°C and run on agarose gels prepared for in-gel hybridization.
RESULTS
SnaB mutant repeats are infrequently incorporated at telomeres when synthesized by telomerase.
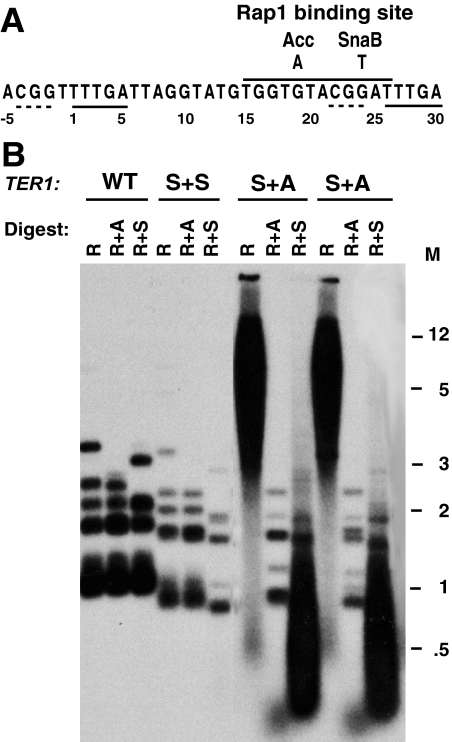
The sequence of telomeric repeats in an organism is specified by the template region of the RNA subunit of its telomerase. Mutational analysis of the template region of the K. lactis telomerase RNA (TER1) revealed that mutations within the left side of the Rap1 binding site, including ter1-19A(Acc) (Acc in Fig. 1A), lead to rapid and often severe telomere elongation that appears to be due to a disruption in Rap1 binding to the affected telomeric repeats (37, 48, 80). However, mutations in the right side of the Rap1 binding site, including ter1-24T(SnaB) (SnaB in Fig. 1A), lead to telomeres that stabilize at shorter-than-normal lengths (50, 80) (Fig. 1A and B). It was suggested that this region of the template encodes not only the Rap1 binding site but also another function that is required for the ability of telomerase to efficiently add sequence onto a telomere. As a test of this, ter1-24T(SnaB) cells were transformed with an integrative plasmid (pTER-BX-UA) containing the ter1-19A(Acc) gene. Transformants were found, as expected for homologous integration, to typically contain both the ter1-24T(SnaB) and the ter1-19A(Acc) alleles separated by vector sequences. Several such heteroallelic transformants were then examined for their telomere lengths. Results from this analysis (Fig. 1B) showed that telomeres in these strains invariably displayed a highly elongated smear of telomeric EcoRI fragments ranging from ∼3 kb to >12 kb (Fig. 1B, S+A samples). This contrasts with other experiments that showed that introducing ter1-19A(Acc) into cells with a wild-type TER1 produced a comparatively slight telomere elongation phenotype (53). These data indicated that the presence of the ter1-24T(SnaB) allele, unlike that of a wild-type TER1 allele, did not substantially interfere with the telomere elongation caused by the ter1-19A(Acc) allele. Further passaging of the ter1-24T(SnaB)/ter1-19A(Acc) heteroallelic strains for five serial restreaks (∼100 to 125 cell divisions) showed both continued telomere elongation and the appearance of some sharp telomere-hybridizing bands, which, based on previous work with ter1-19A(Acc) and other ter1 template mutations producing rapid telomere elongation, are likely to be telomere-telomere fusions (reference 51 and data not shown). Control transformants with an identical structure except for both ter1 alleles being ter1-24T(SnaB) exhibited a short-telomere phenotype similar to that of the original ter1-24T(SnaB) mutant (Fig. 1B, S+S sample). This indicated that the number of copies of ter1, by itself, was not leading to telomere elongation.
FIG. 1.
The long-telomere phenotype of the ter1-19A(Acc) mutation is dominant to the ter1-24T(SnaB) mutation in trans. (A) Diagram of the K. lactis telomerase RNA template region. The strand shown is the complement of that present in the RNA. The numbers shown signify the coordinates used for base positions in and around the template. Rap1 binds at the overlined sequence. Base substitutions making AccI and SnaBI restriction sites are indicated. The underlined sequences are involved in accurate alignment of the template with the telomeric DNA during telomerase translocation. (B) Southern blot of telomeric hybridization to DNA from ter1-24T(SnaB) and ter1-19A(Acc) cells created by integration of a ter1-19A(Acc)-containing plasmid into haploid ter1-24T(SnaB) cells. Two independent heteroallelic strains are shown (S+A). DNAs from a wild-type (WT) strain and a matching control containing two ter1-24T(SnaB) alleles are also shown. Each DNA was digested with EcoRI (R), EcoRI and AccI (R+A), and EcoRI and SnaBI (R+S). Markers (M) are shown in kilobases.
Digestion of the elongated telomeric fragments from ter1-24T(SnaB)/ter1-19A(Acc) heteroallelic strains with restriction enzymes (SnaBI and AccI) that specifically cleave each type of mutant repeat but not the wild-type repeats produced very different results. Digestion with AccI led to the disappearance of the great majority of the telomeric signal (not counting the residual wild-type telomeric repeats remaining at basal positions of the fragments) from each of several heteroallelic transformants that were examined (Fig. 1B and data not shown). In contrast, digestion with SnaBI produced a large smear of telomeric signal from ∼100 bp to >1 kb in size with a signal intensity roughly similar to that of the EcoRI-digested control with uncleaved telomeric repeats. These results are consistent with the great majority of the long telomeres in ter1-24T(SnaB)/ter1-19A(Acc) heteroallelic strains being composed of Acc repeats, with SnaB repeats only occasionally becoming incorporated. From these experiments, we conclude that either the SnaB telomerase is defective in synthesizing telomeric repeats or SnaB telomeric repeats are defective in being extended by telomerase.
Both SnaB and Acc repeats are defective in negatively regulating telomere length in the presence of telomerase.
Rap1 binding to telomeric repeats negatively regulates the ability of telomerase to extend yeast chromosome ends (44). Mutant telomeric repeats defective in binding Rap1 should therefore be poorly able to negatively regulate telomere length in the presence of telomerase. To test the Acc and SnaB repeats, cloned K. lactis telomeres were first constructed to be composed entirely of either Acc repeats or SnaB repeats. These telomeres, containing an S. cerevisiae URA3 gene inserted into adjacent subtelomeric sequence and referred to as STU (subtelomeric URA3) telomeres, were then transformed into K. lactis cells. This led to the transforming fragment integrating via subtelomeric homology and replacing one native telomere in each transformant (50) (diagrammed in Fig. 2A). The length of a STU telomere could be readily assessed by digestion with XhoI, which cleaves next to the URA3 gene and allows separation of the STU telomere from all other telomeres in the cell (Fig. 2B and C). When the STU telomere was composed of either wild-type repeats or the phenotypically silent Bcl mutant repeats, it was found, as expected, to be of wild-type length (53, 81) (Fig. 2C). In contrast, both Acc and SnaB repeats showed defects in telomere length regulation, with STU telomeres composed of them maintaining lengths substantially longer than wild-type telomeres (Fig. 2B and C). Acc repeats appeared to be completely “uncounted,” as the array of wild-type repeats present at the end of a STU telomere with 13 Acc repeats was the same size as wild-type telomeres (compare XhoI-plus-AccI digests of Acc13 clones in Fig. 2C with BsrBI digests of wild-type STU clones). This is consistent with the past observation that Acc repeats have a severe defect in binding Rap1 (37). The length regulation defect of SnaB repeats was less severe. A SnaB telomere estimated to have ∼10.2 SnaB repeats had a terminal wild-type repeat tract (X+S lane of SnaB 10) that was shorter than wild-type telomeres, and a telomere having ∼27 SnaB repeats had a terminal wild-type repeat tract that was shorter still (faint signal at ∼0.3 kb in X+S lanes of SnaB 27 samples). We conclude that the SnaB mutant repeats are partially defective in regulating telomere length. Using the amount of wild-type addition to the end of the mutant repeats, we established that SnaB repeats in the constructs tested retain roughly 40% of their ability to regulate telomere length. For example, a SnaB clone with ∼10.2 SnaB repeats was estimated to have ∼12.3 terminal wild-type repeats. Because our control wild-type telomeres averaged 15.9 repeats, the 10.2 SnaB repeats had the same length regulation ability as 3.6 wild-type repeats (15.9 minus 12.3).
Constructing telomerase deletion mutants with telomeres composed of Acc or SnaB mutant repeats.
We next examined the effect of the mutant telomeric repeats on recombinational telomere maintenance. We hypothesized that telomeres composed of mutant telomeric repeats might result in chronic telomere capping defects that in turn promoted the formation of very long telomeres by recombination (“runaway” type IIR RTE; outcomes 4 to 6 in Fig. 3A). To test this, we took advantage of a technique that we had previously shown to result in the spread of sequence from a single telomere to the 11 other telomeres in a K. lactis cell (77). Cells with a genomic ter1-Δ allele but having a wild-type copy of TER1 on a plasmid were first constructed to contain a single STU telomere containing SnaB or Acc repeats (Fig. 3A). As described above, these mutant STU telomeres were extended by the resident wild-type telomerase and had a total size (mutant plus wild-type repeats) that was longer than that of other telomeres in the cell. These transformants were then streaked on rich medium, and clones that had lost the TER1 plasmid were identified (by their rough colony phenotype and loss of the HIS3 marker).
FIG. 3.
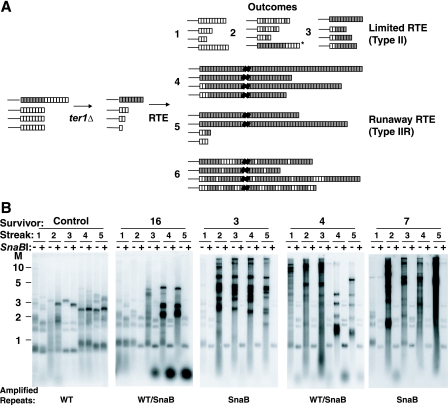
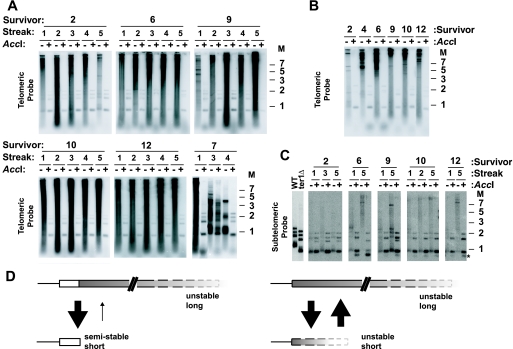
SnaB telomeric repeats can promote the formation of long, unstable telomeres through RTE. (A) Scheme for generating ter1-Δ cells containing mutant repeats. Cells containing a single STU telomere with mutant telomeric repeats (left drawing) were deleted for telomerase and allowed to senesce (middle drawing). The long size of the mutant telomere greatly enhances the likelihood that the mutant repeats will spread to all other chromosome ends during RTE. The drawings at the right show several possible outcomes for the telomere structures. Outcomes 1 to 3 depict typical moderate telomere lengthening seen in type II survivors with only wild-type repeats (outcome 1), interspersed wild-type and mutant repeats (outcome 2), or only amplification of mutant repeats (outcome 3). The asterisk depicts a different possibility that defective mutant repeats could be effectively “capped” by wild-type repeats. Outcomes 4 to 6 depict potential results with type IIR RTE generating very long telomeres. Outcome 4, all telomeres mutant and long. Outcome 5, mix of mutant long telomeres and shortened telomeres. Outcome 6, long mutant telomeres with some interspersed wild-type repeats. Gray and white boxes are mutant repeats and wild-type repeats, respectively. (B) Southern blot hybridized to a telomeric probe of ter1-Δ survivors with telomeres containing SnaB repeats. Each gel shows a separate SnaB survivor followed for five serial restreaks after senescence. The first gel is a control survivor that retained only wild-type (WT) repeats, while the other gels show examples of some of the spreading patterns. DNA from each sample was digested with EcoRI (−) and with EcoRI plus SnaBI (+). Underneath the gels is indicated the type of repeat primarily amplified. Markers (M) are shown in kilobases.
In the absence of telomerase, growth senescence ensued, and after two or three serial streaks it was possible to identify postsenescence survivors that showed improved growth and contained telomeres that had been elongated by recombination. As observed previously when a single abnormally long, but functionally wild-type, telomere was present, the presence of the Acc and SnaB STU telomere led to a detectable partial suppression in the senescence of a ter1-Δ mutant (reference 77 and data not shown). Topcu et al. also showed that the sequence of the single abnormally long telomere is spread to all 11 other K. lactis telomeres during postsenescence survivor formation at a frequency exceeding 90% (77). We therefore expected that the mutant telomeric repeats from the abnormally long Acc and SnaB STU telomeres would commonly be spread to all telomeres in the ensuing survivors. The efficiency of spreading could be judged by the frequency with which telomeric restriction fragments could be shortened by the restriction enzyme able to specifically cleave the Acc or SnaB telomeric repeats.
Figure 3A diagrams several conceptual outcomes of the fate of the telomeres after the mutant repeats have spread to all of the telomeres. Among “limited RTE” scenarios, telomeres never become more elongated than has been observed in type II RTE in K. lactis regardless of whether the mutant repeats constitute none, some, or all (outcomes 1 to 3) of the newly acquired telomeric sequences. Alternatively, type IIR “runaway” RTE might occur as a result of the presence of mutant repeats. This might involve the extreme elongation of all telomeres with uniform tracts of the mutant telomeric repeat (outcome 4). Two alternative possibilities are that some telomeres remain short (perhaps because of sufficient numbers of residual wild-type repeats) (outcome 5) or that elongated telomeres are not all homogeneous in sequence, with some or all containing interspersed wild-type repeats (outcome 6).
We observed that in two separate experiments, 30% (7/23) of survivors and 58% (7/12) of survivors derived from cells with an Acc telomere had spread mutant repeats to all telomeres (Fig. 3 and data not shown; see also Fig. 4). Among survivors derived from cells with a SnaB telomere, our results varied depending upon the length of the mutant telomere. Only 8% (1/12) of survivors derived from a cell starting with a STU telomere estimated to have 10 SnaB repeats were found to have spread SnaB repeats. However, 62% (8/13) of survivors derived from a cell starting with a STU telomere estimated to have ∼27 SnaB repeats were found to have spread SnaB repeats. The low frequency of spreading in survivors derived from the shorter SnaB telomere is similar to that previously observed with a STU telomere carrying a wild-type length of phenotypically normal Bcl repeats (77). The survivors derived from cells with Acc or the longer SnaB STU telomeres displayed a spreading frequency somewhat lower than the 94% observed with an elongated Bcl telomere. This could reflect an increased instability of the mutant SnaB and Acc telomeres once gradual sequence loss had eliminated the terminal wild-type repeats.
FIG. 4.
Acc survivors exhibit type IIR RTE after spreading but can have persistent short telomeres. (A) Southern blots of six independent Acc survivors. Each gel shows DNA from an independent survivor that was serially restreaked five times after survivor formation. Samples were digested with EcoRI (−) and EcoRI plus AccI (+). A telomeric probe was used for hybridization. (B) Southern blot of telomeres of Acc survivors after 10 streaks. The digests and probe are the same as for panel A. (C) Persistent short telomeres in Acc survivors. Southern blots of DNA from wild-type (WT) cells, ter1-Δ cells, and Acc survivors were hybridized to a subtelomeric sequence common to 11 of 12 telomeres. The dot indicates the position of a group of telomeric fragments when they contain only a small number of telomeric repeats. The signal in this band in the EcoRI-AccI digests represents the total amount of this group of telomeres and the signal in the EcoRI digests represents the fraction of the telomeres that are very short even without cleavage of the Acc repeats. The asterisk marks the position of subtelomeric fragments in EcoRI-AccI digests that have lost all detectable wild-type repeats and that consequently are not detectable with the telomeric probe. Molecular weight markers (M) are shown in kilobases. (D) Model for persistent short telomeres in early Acc survivors. In the case on the left, a telomere is shown with a basal region of wild-type repeats (white box) and a long terminal region of Acc repeats (gray region) that is unstable and, as a consequence, highly heterogeneous in length in a population of cells. Truncated forms of the telomere that retain only the basal wild-type repeats may confer a semistable state that is relatively resistant to being reelongated by recombination. However, in the situation on the right, an unstable long telomere with no basal wild-type repeats could be subject to a similarly high rate of truncation events but be unable to stabilize any particular short telomere.
SnaB repeats can promote the formation of longer telomeres through RTE than can wild-type repeats.
Nine ter1-Δ survivor clones that had acquired SnaB repeats at most or all telomeres (henceforth called SnaB survivors) were followed by serial restreaking on YPD plates. The telomeres from each streak were then examined by Southern blotting. Figure 3B shows genomic DNA from four representative SnaB survivors and a control survivor that retained only wild-type telomeric repeats, each digested with EcoRI alone and also with EcoRI plus SnaBI. The double digestion cleaved all SnaB mutant telomeric repeats and typically left behind much shorter fragments containing subtelomeric sequence and a small number of wild-type repeats. Results from this analysis (Fig. 3B) showed that SnaB survivors showed a more variable range of telomere lengths than has been seen in ordinary ter1-Δ survivors containing only wild-type telomeric repeats.
Three of the nine SnaB survivors, including survivor 16 (Fig. 3B), displayed telomeres that never became more than moderately elongated (with estimated sizes of telomeric repeat arrays remaining typically not more than several hundred base pairs). This result is similar to the limited type II RTE (outcomes 1 to 3 in Fig. 3A) that occurs in survivors of telomerase deletion with wild-type repeats (47) and the control survivor in Fig. 3B. These three SnaB survivors all displayed quite similar outcomes. In streaks 1 and 2, telomeres remained very short but typically were cut slightly shorter still upon digestion with SnaBI (see streak 2 for survivor 16 in Fig. 3B), consistent with many or all telomeres having acquired one or more SnaB repeats. In subsequent streaks, however, moderate telomere elongation (telomeric EcoRI fragments all <∼5 kb) was present and was invariably accompanied by a fragment less than ∼0.2 kb in size in EcoRI-SnaBI double digests that hybridized intensely to a telomeric probe (see streaks 3 to 5 for survivor 16 in Fig. 3B). This fragment almost certainly represents short blocks of wild-type repeats that are interspersed among SnaB repeats in many or all of the telomeres of these survivors (type II RTE outcome 2 in Fig. 3A). Similar interspersion of wild-type repeats was observed in many survivors derived from ter1-Δ cells with basal wild-type repeats and terminal Bcl repeats, where it was thought to be a consequence of roll-and-spread amplification (59). Telomeric circles with both wild-type and SnaB repeats most likely arise from a single telomere containing both repeat types and would be predicted by the roll-and-spread model to produce repeating patterns of the two repeat types if copied by a rolling-circle gene conversion. We suggest that sufficient concentrations of interspersed wild-type repeats in SnaB survivors render those telomeres relatively resistant to further telomeric recombination.
The most common outcome for SnaB survivors, including survivors 3, 4, and 7 in Fig. 3B, was that by streak 2, they displayed telomeres of unusually large sizes, which frequently migrated at positions above 5 kb in the gel. Characteristically, the telomeric hybridization signal in these survivors extended to limit mobility (>20 kb) in gels and also appeared in the wells of the gel. These features have not been seen in K. lactis telomerase deletion survivors with wild-type repeats, and they argue that SnaB repeats can lead to the formation of much longer telomeres from RTE than can wild-type repeats (47). We conclude that these SnaB survivors display a telomere length phenotype that is intermediate between those for limited type II RTE and runaway type IIR (outcomes 1 to 3 and 4 to 6 in Fig. 3A, respectively).
The elongated telomeres in SnaB survivors appear to have variable degrees of stability. In some cases, such as survivor 3 in Fig. 3B, telomeres appeared relatively stable over many cell divisions, with changes in the telomeric fragment pattern largely limited to the gradual shortening expected for cells lacking telomerase. Many SnaB survivors with long telomeres showed little if any sign of wild-type repeats except those few that are adjacent to subtelomeric sequences. This may imply that telomeres composed solely of SnaB repeats can often resist uncapping and engaging in recombination for many consecutive cell divisions. In other cases, however, long telomeres in SnaB survivors can be highly unstable. A particularly dramatic example of this can be observed between the third and fourth streaks for survivor 4 in Fig. 3B. Here, a very long and heterogeneous telomeric signal, commonly reaching to limit mobility, changed abruptly into a pattern of much shorter telomeric fragments, migrating at positions below 4 kb in the gel. This indicates that telomeres composed of SnaB repeats can be subject to high rates of becoming truncated. Interestingly, the shortened telomeres of survivor 4 at streaks 4 and 5 appeared to contain interspersed wild-type repeats as indicated by the short fragment hybridizing to a telomeric probe in the EcoRI-SnaBI digest (Fig. 3B). This adds further support to the idea that interspersed blocks of wild-type repeats can prevent further elongation by recombinational processes of telomeres containing SnaB repeats.
ter1-Δ SnaB survivors with long telomeres displayed growth characteristics that were different than those of telomerase deletion survivors with wild-type repeats. Whereas ter1-Δ survivors with wild-type repeats vary widely from streak to streak, from having highly senescent very slow growth to growth indistinguishable from that of wild-type cells (47), SnaB survivors with long telomeres showed more constant growth characteristics, from slightly senescent to normal (data not shown). SnaB survivors with short telomeres and interspersed wild-type repeats instead appeared to more closely resemble telomerase deletion survivors with wild-type repeats in having more variable growth characteristics.
Acc repeats promote type IIR RTE.
Seven independent ter1-Δ survivor clones that had acquired Acc repeats at all telomeres (henceforth called Acc survivors) were followed by serial restreaking and Southern blotting. The results from this analysis showed that Acc survivors showed a highly elongated and heterogeneous pattern beginning from the first streak examined when hybridized with a telomeric probe (Fig. 4A). Cleavage of DNA from these survivors with AccI eliminated almost all telomeric signal, consistent with the long telomeric sequences being composed almost entirely of Acc repeats. The bands remaining after AccI cleavage correspond to shortened telomeric fragments that retained small numbers of wild-type telomeric repeats adjacent to subtelomeric sequence. The smear of telomeric signal in the EcoRI digests visible below ∼0.7 kb in Acc survivors is too small to have intact subtelomeric sequences and has been shown to be at least largely circular in nature (L. Bechard, E. Basenko, and M. McEachern, unpublished data). Past work has shown that the small extrachromosomal telomeric sequence present in long telomere mutants is primarily double- and single-stranded circles (29). In six out of the seven Acc survivors, while the telomeric signal sometimes varied in intensity between streaks, the general pattern of long and heterogeneous telomeric signal did not vary greatly either between survivors or between streaks 1 to 5 of the same survivor. After 10 streaks, however, most Acc survivors showed a reduced amount of low-molecular-weight telomeric signal, and the high-molecular-weight telomeric signal was more frequently in sharp bands (Fig. 4B). These results suggest that the telomere phenotype of the Acc survivors might gradually change over continued passaging, most likely toward a state favoring a more stable telomere function. The colony phenotypes of Acc survivors always showed a slight to moderate senescence phenotype (rough colonies and slightly slower growth). This supports the idea that their telomeres were never completely wild type in their function.
In one Acc survivor (survivor 7 in Fig. 4A), substantial telomere shortening was seen between the first and third streaks. The reason for this change is not clear. One possibility is that, as appears to occur in some SnaB survivors, the mutant phenotype is suppressed by the presence of a sufficient number of wild-type repeats interspersed with the mutant repeats.
DNA samples from the Acc survivors were next probed with a subtelomeric probe. Unlike the telomeric probe, this probe does not exaggerate the abundance of long telomeric fragments, nor was it expected to detect extrachromosomal telomeric DNA. The results from this (Fig. 4C) were striking. As expected, smears of signal extending to high molecular weights were observed, consistent with the presence of long telomeric fragments. However, at most time points for most Acc survivors examined, a substantial amount of signal in EcoRI digests was seen to run at short sizes that were nearly identical to those of the bands that had been digested with EcoRI plus AccI. This indicated that many of the telomeres in Acc survivors were often very short and contained few if any Acc repeats. Those Acc survivor samples that showed few or no short EcoRI fragments (most notably Acc survivor 6, streak 5, and Acc survivor 12, streak 5, in Fig. 4C) instead showed another, still-shorter fragment in EcoRI-AccI digests (Fig. 4C). This additional band was never present in digests with EcoRI alone and was not detectable with a telomeric probe. It therefore likely represents chromosome ends that contain no basal wild-type repeats. Our results suggest either that telomeres in Acc survivors tend to exist in a heterogeneously long state or, if some wild-type repeats remain basally, they remain very short and essentially without Acc repeats. The results also suggest that the presence of even small numbers of basal wild-type repeats can stabilize telomeres to persist at short lengths for long enough periods of time to permit their detection as a significant fraction of the total telomere population in Acc survivors. SnaB survivors were also probed with a subtelomeric probe and were not seen to contain telomeres that were very short among the long telomeres, which is consistent with the lesser recombination defect in SnaB survivors (data not shown).
Wild-type repeats are variably present within the long tracts of Acc repeats in Acc survivors.
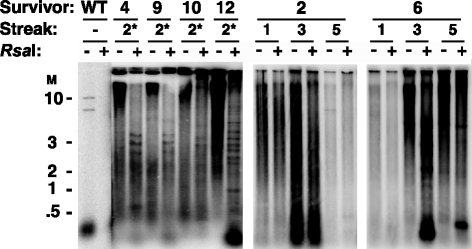
To further examine the structure of telomeres in Acc survivors, RsaI digests were performed. This restriction enzyme cleaves wild-type K. lactis telomeric repeats but not Acc repeats. Figure 5 shows BsrBI digests of DNA from a number of Acc survivors alone or in combination with RsaI digests. As expected, the telomeric signal from a wild-type control was completely eliminated by RsaI digestion (Fig. 5, leftmost lanes). Also as expected, telomeric signal from each Acc survivor was found to be substantially resistant to RsaI digestion. With Acc survivor 2, for example, there was little or no sign of RsaI cleavage of telomeric arrays at any of three streaks examined, consistent with the long telomeres containing Acc repeat tracts uninterrupted by wild-type repeats. Acc survivor 10 similarly shows little evidence of telomere cleavage by RsaI. However, with the other four Acc survivors examined (survivors 4, 9, 12, and 6), there was a pronounced shift of high-molecular-weight signal to smaller sizes after cleavage with RsaI, which was often accompanied by the appearance of some sharp bands below 5 kb. These results indicate that at least small numbers of wild-type repeats can often be interspersed within the long Acc telomeres. Clearly the wild-type repeats in these clones are unable to provide proper telomere function. Presumably they are not present in sufficient concentrations or at the correct positions (the ends) to be able to correct the defects caused by the more abundant Acc repeats. The more severe defect of Acc repeats may also act to render interspersed wild-type repeats less able to provide telomere function than is the case with SnaB survivors.
FIG. 5.
Acc survivors can contain wild-type repeats within their long tracts of Acc repeats. Southern blots show Acc survivors digested with BsrBI (−) or BsrBI plus RsaI (+) and hybridized to a telomeric probe. All survivor numbers correspond to those in Fig. 4. At left is a wild-type (WT) control where the telomeres are completely cut away by RsaI. The central and right gels show results from serial restreaks of Acc survivors 2 and 6. Asterisks with the survivor numbers indicate that the DNAs shown are from different subclones of the Acc survivors than are shown in Fig. 4. Markers (M) are shown in kilobases.
Very small bands produced by RsaI digestion (most prominent in Acc survivor 12 and streaks 3 and 5 of Acc survivor 6) conceivably could represent small tandemly repeating units containing both wild-type and Acc repeats, similar to the repeating arrays observed in some telomerase deletion survivors with wild-type repeats generated from cells with two types of telomeric repeats (59). The changes in the RsaI digestion profile of Acc survivor 6 over a five-streak growth course showed dramatic changes in the telomeric signal at <0.5 kb. These data indicate that there can be rapid and substantial turnover of telomeric sequences in Acc survivors.
Abundant single-stranded DNA is seen at telomeres in Acc and SnaB survivors.
Previously, it was shown that ter1 long-telomere mutants of two distinct classes {immediate [e.g., ter1-19A(Acc)] and delayed elongation} have an abundance of single-stranded telomeric DNA, specifically of the G-rich strand, at their telomeres (80). To test for the presence of single-stranded DNA in Acc and SnaB survivors, in-gel hybridizations were performed using a telomeric oligonucleotide probe complementary to the sequence of the G-rich telomeric strand. The results obtained from Acc survivors and ter1-19A(Acc) mutants are shown in Fig. 6A. The ethidium bromide gel picture is provided as a loading control. EcoRI digestions were prepared, and half of each was run on a gel for standard Southern blotting (denatured gel) and the other half was run on a gel for the in-gel hybridization. The wild-type control showed telomeric signal in the denatured gel but little or no detectable telomeric signal in the in-gel hybridization. However, the in-gel hybridization showed that appreciable amounts of single-stranded telomeric DNA were present both in the ter1-19A(Acc) strains and in the Acc survivors. Some variability in the extent of the single-stranded telomeric DNA was evident and likely reflects variations in the length or other features of the telomeres in these cells. Figure 6B illustrates the presence of abundant single-stranded DNA in a SnaB survivor (survivor 4 in Fig. 3B) followed for five streaks. As can be seen, telomeres in these SnaB survivors display abundant single-stranded telomeric DNA, particularly when the telomeres are very long. Our results demonstrate that elevated levels of single-stranded telomeric DNA are present in Acc and SnaB survivors and that it forms in a telomerase-independent manner.
We next tested whether the single-stranded telomeric DNA that we detected in Acc survivors existed as 3′ overhangs by digesting with Exo I. Figure 6C shows digestion of Acc2, Acc9, and Acc10 with EcoRI and with EcoRI plus Exo I. The denatured and in-gel hybridizations to a C-strand telomeric probe as well as the ethidium bromide-stained control are shown. Digestion with Exo I produced a 36 to 55% decrease in the signal present in the in-gel hybridizations. These results are consistent with at least a sizable fraction of the single-stranded telomeric DNA in Acc survivors existing as long 3′ single-stranded overhangs. Partial resistance of telomeric single-stranded DNA to Exo I digestion was also observed in stn1-t mutants with elongated telomeres in S. cerevisiae (66). Conceivably, the resistant fraction in both cases might represent single-stranded gaps.
We also examined the senescing ter1-Δ precursors to Acc survivors that contained a single mutant telomere for the presence of long 3′ overhangs. We found that the telomere with the Acc repeats produced a prominent in-gel hybridization signal to a C-strand telomeric probe that was sensitive to Exo I digestion. This signal was not seen in the same cells at the earliest stages of senescence (when the telomere would still be capped with many wild-type repeats), nor was it seen in the short telomeres with only wild-type repeats that were present in the same cell (Fig. 6C and data not shown). Similar results were found in senescing cells with the single SnaB telomere (data not shown). On the other hand, neither ter1-24T(SnaB) cells, which have very few SnaB repeats at the ends of each of the 12 telomeres, nor ter1-Δ cells lacking any mutant telomere show an increased 3′ overhang signal relative to wild-type controls (Fig. 6E). We conclude that telomeres terminating in extended tandem arrays of Acc or SnaB repeats are not able to protect telomeres from extensive degradation of 5′ ends.
DISCUSSION
Length regulation defects of Acc and SnaB repeats in the presence of telomerase are consistent with Rap1 binding defects.
Our results here show that both SnaB and Acc mutant telomeric repeats are defective in regulating telomere length in cells expressing telomerase. While SnaB repeats retain a partial ability to negatively regulate telomere length when present basally at a telomere, Acc repeats appear to be completely defective in this function. Telomeres with an array of Acc repeats thus acquire a terminal array of wild-type telomeric repeats that is the full size of normal telomeres.
The defect of the SnaB and Acc repeats in regulating telomere length in the presence of telomerase is very likely the result of defects in their ability to bind the Rap1 protein. Rap1 is well known to be a key negative regulator of telomere length in S. cerevisiae and K. lactis through its ability to bind double-stranded telomeric repeats (37, 44). Also, the base changes of both telomeric mutations fall within the Rap1 binding site and disrupt Rap1 binding in vitro, with the extent of disruption of binding being greater for the Acc mutation than for the SnaB mutation (37; A. Krauskopf and E. H. Blackburn, personal communication). Although a basal array of Acc repeats is completely “uncounted” with respect to regulating telomere length, this may not mean that there is a complete absence of Rap1 bound to them in vivo. Not all sequences able to bind Rap1 appear able to regulate telomere length (30, 37, 80). In K. lactis, for example, a basal array of “Kpn” mutant repeats, each with two base changes near but not in the Rap1 binding site, are strongly defective at being “counted” yet bind Rap1 with at least normal affinity as individual repeats in vitro (30, 37, 80). This lack of counting could result from cooperative interactions between Rap1 molecules or from Rap1's known ability to bend DNA (22, 54, 84).
What is the defect of SnaB repeats that interferes with telomerase-mediated telomere maintenance?
A number of lines of evidence argue that SnaB mutant telomeric repeats have a second defect that inhibits telomerase's ability to add sequence onto the mutant telomeric ends. The ter1-24T(SnaB) mutant produces telomeres that are substantially shorter than wild type, a characteristic shared with several other ter1 template mutations that alter the right side of the Rap1 binding site (50, 80). In addition, combining the ter1-24T(SnaB) base change in cis with the ter1-19A(Acc) mutation almost completely blocks the extreme elongation normally caused by the latter allele (51). Also, our data here show that cells containing separate ter1-24T(SnaB) and ter1-19A(Acc) alleles exhibit extensive telomere elongation through incorporation of Acc repeats but little incorporation of SnaB repeats. The very poor accumulation of SnaB repeats in a ter1-24T(SnaB) mutant presumably accounts for why telomeres can remain short in this mutant despite the mutant repeats being defective in the negative regulation of both telomerase and RTE.
The defect, or defects, in SnaB repeats that block sequence addition by telomerase is not fully understood. One recently discovered defect is that the ter1-24T(SnaB) mutation interferes with the base pairing between the telomerase RNA and the 3′ end of the telomeric DNA. Recent evidence has indicated that telomeric DNA copied from positions 22 to 24 of the template base pairs with positions adjacent to the template and that this partially explains the short-telomere phenotypes of mutations at positions 22 to 24 (82; Z. Wang and M. McEachern, unpublished data) (dotted lines in Fig. 1A show alignment regions). An additional possibility is that the SnaB mutation also interferes with the binding of Cdc13 or Est1, proteins that interact with the 3′ single-stranded tail of telomeres and are required for recruitment or activation of telomerase (19, 20, 64, 72). The binding site of Cdc13 within S. cerevisiae telomeric sequences does appear to overlap the Rap1 binding site (18). Whether this is true in K. lactis has not been determined.
Both SnaB and Acc telomeres are defective at regulating RTE.
SnaB and Acc repeats can promote the formation of telomeres by RTE that are much longer than those seen in comparable telomerase deletion mutants containing only wild-type telomeric repeats. This elongation, particularly in the case of Acc survivors, appears to be similar to that of the type IIR RTE originally described for the stn1-M1 mutant of K. lactis (33). Type IIR RTE was defined as RTE that produces very long telomeric repeat tracts due to a capping defect that makes telomeres prone to inducing recombination in a manner independent of their length. This contrasts with “ordinary” type II RTE, where the telomeric recombination is initiated by a capping defect brought on by critically short telomeres and appears to be suppressed by even modestly elongated telomeres. It could be noted, though, that standard type II RTE could potentially produce a long telomere but only in a single step, such as perhaps copying a circular template or copying another telomere that was already long.
Other facts in addition to the abnormally long telomeres of SnaB and Acc survivors suggest that both types of mutant repeats produce type IIR RTE. Both SnaB and Acc survivors that have amplified mutant but not wild-type telomeric repeats normally lack the irregular cycles of growth senescence and recovery typical of telomerase deletion mutants with only wild-type repeats. Instead, they have no obvious growth defects or else modest chronic growth defects that do not appreciably change with passaging. This is similar to the case for the stn1-M1 mutant, which had continuous moderate growth and cellular defects presumably stemming from the continual presence of telomeric ends that triggered a DNA damage response.
Another key feature predicted for type IIR RTE is extreme and constitutive telomere instability. Even relatively long telomeres are expected to be highly unstable in length and not able to shorten gradually over multiple cell divisions as occurs in ordinary K. lactis type II survivors. This certainly appears to be the case with Acc survivors. Although the long smeared telomeric signal of Acc survivors in Southern blots remained relatively constant over time, we interpret this not as telomeric stability but rather as a steady state of high instability. The presence of telomeric DNA in highly smeared signal indicates that telomeres in Acc survivors (at least those composed of primarily Acc repeats) are typically too unstable to exist at or near a discrete size for the entire 20 to 25 cell divisions needed to generate enough cells for Southern analysis. However, with the extreme size and continuous presence of long telomeres in these cells, it is not currently possible to fully exclude the possibility that a minority of relatively stable telomeres could be present in Acc survivors. The abundance of extrachromosomal telomeric DNA in Acc survivors serves as further evidence of frequent telomeric recombination. SnaB survivors, though clearly less extreme than Acc survivors, also showed instances of sudden large changes in the sizes of telomeres that in survivors with wild-type repeats would have been long enough to be relatively resistant to such changes.
Our results here with Acc survivors raise the question of the relative contributions of telomerase and HR to the long- telomere phenotype of the ter1-19A(Acc) mutant. The “immediate-elongation” phenotype has been shown to arise independently of the critical recombination gene RAD52 (80). This implies that the extreme telomere elongation in the immediate-elongation mutants very likely can arise independently by two completely different mechanisms, type IIR RTE and unregulated telomerase addition. The abundant extrachromosomal telomeric circles in these mutants, in contrast, are present in RAD52 cells but absent in rad52 mutants. The high rate of telomeric recombination in ter1-19A(Acc) mutants suggests that it is highly likely that the type IIR RTE is actively occurring in the presence of telomerase, as is the case for the type IIR RTE in the stn1-M1 mutant (33).
The unusual persistence of short wild-type telomeres in early Acc survivors.
Many Acc survivors exhibit persistent short telomeric bands in the first several streaks after they are generated. These short telomeres are composed largely or entirely of wild-type repeats, and their behavior is therefore not likely to be representative of telomeres containing appreciable numbers of Acc repeats. The persistence of these short wild-type telomeres remains difficult to fully explain. Almost certainly, the presence of a small number of wild-type repeats provides a degree of telomere function that allows the short telomere to be somewhat resistant to recombinational processes. Consistent with this, disappearance of the persistent short telomeres is correlated with loss of the wild-type repeats from those telomeres.
What is more perplexing is how the persistently short telomeric bands can remain a constant size over several streaks despite the absence of telomerase. We have previously seen an example of short telomeres in K. lactis persisting for a number of streaks after the introduction of a particular ter1 template mutation that produced mutant telomeric repeats and elongated telomeres (51). However, in this case, the persistent short telomeres clearly displayed gradual shortening prior to the point where they became elongated. This was interpreted as indicating that some telomeres did not acquire mutant telomeric repeats for many cell divisions (perhaps because of inefficiency of the mutant telomerase template) and instead underwent gradual replicative sequence loss until either telomerase or recombination finally made them much longer. This explanation cannot account for the persistent short telomeres in Acc survivors, because those telomeres do not undergo gradual shortening over the multiple streaks where they persist (Fig. 4C). This forces us to the conclusion that the persistent short telomeres are being actively elongated by recombination during the time period when they appear to be persisting at very short sizes.
One possible explanation is that the same set of short telomeres is repeatedly elongated by recombination to only very small extents so as to maintain their short sizes. This seems implausible given that telomerase is absent and other telomeres in the same cells undergo RTE that routinely adds kilobases to their lengths. An alternative possibility is that the persistently short telomeric bands represent a semistable intermediate state of telomeres that are otherwise regularly undergoing recombination events that may either greatly lengthen or greatly truncate them (Fig. 4D). The persistent short telomeric bands would therefore represent a percentage of all telomeres that retained a minimal number of basal wild-type repeats. Given the sizable percentage of total subtelomeric signal present in the persistently short telomeric bands (Fig. 4C), this model would seem to require that telomeric truncations could routinely remove essentially all but the basal-most wild-type repeats from a telomere with terminal Acc repeats. Supporting the possibility of such large deletions is the observation that “immediate-elongation” ter1 template mutations, including ter1-19A(Acc), undergo turnover of basal wild-type repeats in spite of having highly elongated telomeres (53). That wild-type repeats in Acc survivors might be particularly resistant to loss would not be entirely surprising given that protecting chromosome ends from degradation and recombination is their normal function. Precedents are known in both K. lactis and S. pombe where defects in telomere binding proteins can give rise to very rapid and dramatic shortening of all long telomeres in the cell (2, 3, 33, 57).
Disruption of Rap1 binding is the most likely cause of the type IIR RTE of SnaB and Acc survivors.
The simplest possible explanation for the enhanced tendency of the Acc and SnaB repeats to recombine is a defect in Rap1 binding. Both mutations fall within the Rap1 binding site, and both interfere with Rap1 binding in vitro. Moreover, the more modest RTE phenotype in SnaB survivors relative to Acc survivors correlates with the SnaB mutant having a lesser Rap1 binding defect as judged by both telomere length defects in the presence of telomerase and in vitro binding studies. In S. cerevisiae, the Rif1 and Rif2 proteins bind to the Rap1 C terminus and play crucial roles in mediating Rap1's role in the negative regulation of telomere length in the presence of telomerase (31, 74, 87). In the K. lactis genome, RIF1, but not RIF2, has been identified. We have found that deletion of RIF1 in ter1-Δ mutants does not produce an obvious type IIR RTE phenotype (O. Sprusansky and M. McEachern, unpublished data). Thus, we conclude that the type IIR RTE phenotypes of SnaB and Acc repeats act independently, or at least not primarily, through effects on Rif1 interactions at the telomere. It also seems unlikely that the additional defects of SnaB repeats that interfere with telomere elongation by telomerase in ter1-24T(SnaB) cells could be solely responsible for the weaker RTE phenotype of SnaB survivors relative to Acc survivors. If disrupted Cdc13 binding leads to both the short telomeres of ter1-24T(SnaB) cells and a type IIR phenotype, it would predict, contrary to our observations, that SnaB survivors would have a more extreme RTE phenotype than Acc survivors. We cannot rule out, however, that defects in Cdc13 binding make contributions to the RTE phenotypes of Acc or SnaB survivors. Finally, the anticipated base-pairing defect of SnaB repeats with the region next to the Ter1 template (Fig. 1A) would be expected to be specific to telomerase-mediated telomere elongation and not affect telomeric recombination.
How might Rap1 negatively regulate telomeric recombination?
An important conclusion from our results is that the SnaB and Acc telomeric repeat mutations disrupt the negative regulation of both telomerase- and recombination-mediated telomere elongation. This suggests that there is some overlap in the negative regulation of telomere elongation by telomerase and by recombination. Previously reported data would also seem to support this idea. The “Kpn” mutation of the K. lactis telomeric repeat (a double base change that does not affect Rap1 binding in vitro [37]) also disrupts both elongation processes (48, 77). In S. cerevisiae, the MRX complex and the Tel1 and Mec1 kinases contribute to both telomerase- and recombination-mediated telomere maintenance (5, 35, 43, 61, 67, 78, 79), while the Rif1 and Rif2 proteins act to inhibit both processes (31, 74, 87).
A known overlap between sequence addition by telomerase and recombination is that both require the formation of 3′ overhangs. Normal telomeres of S. cerevisiae are exonucleolytically processed to acquire 3′ overhangs of >25 nucleotides during S phase (10). Recent data have suggested that formation of these short overhangs may be a key regulated step in telomerase-mediated telomere elongation (60). These 3′ overhangs are thought to be good substrates for binding and sequence addition by telomerase but too short to be efficient substrates for recombination. In contrast, nontelomeric broken DNA ends in yeast (at least outside of G1 phase) are well known to be degraded at their 5′ ends to produce long 3′ overhangs that can serve as substrates for Rad51 binding and strand invasion (86). Thus, the size of the 3′ telomeric overhang is likely to be critical for determining whether it can be elongated by telomerase or recombination.
Our results support the possibility that Rap1 acts to prevent recombination from initiating at telomeres by helping block the action of one or more exonucleases that degrade 5′ strand ends. By interfering with Rap1 binding, SnaB and Acc mutant repeats present at telomeric ends would allow the formation of 3′ overhangs long enough to provoke the telomeres to initiate HR. In the case of at least the Acc repeats, the long overhangs may also stimulate sequence addition by telomerase. Precedent exists for a protein that binds the double-stranded part of telomeric DNA to protect against formation of 3′ overhangs. Absence of the Taz1 protein in the fission yeast S. pombe leads to longer 3′ overhangs than are present at telomeres of wild-type cells (76). One possibility for an exonuclease that might be blocked by Rap1 is Exo1. Exo1 is known to be an important contributor to the 3′ overhangs that are generated at broken ends and certain dysfunctional telomeres, though it seems not to be required for the short overhangs of normal yeast telomeres (45, 46, 76).
The different roles that Rap1 plays at yeast telomeres likely depend on different regions of the telomeric repeat tract. The negative regulation of telomere length in the presence of telomerase can clearly be carried out by Rap1 binding sites located within the most basal part of the telomere. In contrast, the blockage by Rap1 of telomere-telomere fusions from nonhomologous end joining appears to be carried out by only the most terminal telomeric repeats (7, 51, 63). It would seem highly likely that the proposed ability of Rap1 to regulate the nucleolytic degradation of the 5′ end of the telomere also is localized to the more terminal part of the telomere. This may explain why the ter1-19A(Acc) mutant displays massive telomere elongation in spite of having near-normal numbers of wild-type repeats basally at telomeres (48).
It remains to be determined whether type IIR RTE occurs through a roll-and-spread mechanism as has been suggested for type II RTE (59). Although telomeric circles may be abundant in K. lactis mutants undergoing type IIR RTE, the continuous presence of long telomeres in these cells would seem to provide suitable alternative templates for short telomeres to copy to become long. Perhaps an important place for rolling circle synthesis to contribute to type IIR RTE is at the very early stages after a mutant's creation during the formation of the first long telomere in the cell.
Human ALT cells utilize recombination to maintain their telomeres in the manner that is often highly reminiscent of type IIR RTE in yeast. The ALT phenotype does not occur in most human cells that undergo severe telomere shortening, suggesting that it may require one or more mutations in order to occur. Although ALT cells are unlikely to have mutant telomeric repeat sequences, our results here strengthen the idea that mutations that cause chronic telomere capping defects that promote telomeric recombination and are not suppressed by telomere elongation are likely required. Future studies with yeast type IIR may therefore be of considerable importance in gaining insight to telomere maintenance in human ALT cancers.
Acknowledgments
This work was supported by grants to M.J.M. from the National Institutes of Health (GM 61645) and the American Cancer Society (024255-01) and to Z.T. from The Scientific and Technological Research Council of Turkey (TUBITAK) (SBAG-2791).
We thank Elizabeth Blackburn, in whose lab M.J.M. constructed the ter1-24T(SnaB)/ter1-19A(Acc) heteroallelic strains. We also thank Evelina Basenko for helpful comments on the manuscript and Rolf Kooistra for providing us with the wild-type strain CBS 2359 and the ku80Δ strain.
Footnotes
Published ahead of print on 24 November 2008.
REFERENCES
- 1.Autexier, C., and N. F. Lue. 2006. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75493-517. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2921171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Beernink, H. T., K. Miller, A. Deshpande, P. Bucher, and J. P. Cooper. 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol. 13575-580. [DOI] [PubMed] [Google Scholar]
- 4.Bertuch, A. A., and V. Lundblad. 2006. The maintenance and masking of chromosome termini. Curr. Opin. Cell Biol. 18247-253. [DOI] [PubMed] [Google Scholar]
- 5.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 171819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan, T. M., A. Englezou, J. Gupta, S. Bacchetti, and R. R. Reddel. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 144240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, S. D., S. Iyer, J. Xu, M. J. McEachern, and S. U. Astrom. 2007. The role of nonhomologous end-joining components in telomere metabolism in Kluyveromyces lactis. Genetics 1751035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cervantes, R. B., and V. Lundblad. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14351-356. [DOI] [PubMed] [Google Scholar]
- 9.Cesare, A. J., and J. D. Griffith. 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 249948-9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakhparonian, M., and R. J. Wellinger. 2003. Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet. 19439-446. [DOI] [PubMed] [Google Scholar]
- 11.Chan, S. R., and E. H. Blackburn. 2004. Telomeres and telomerase. Philos. Trans. R. Soc. London B 359109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 211819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 811991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 192100-2110. [DOI] [PubMed] [Google Scholar]
- 16.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 9313902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26447-450. [DOI] [PubMed] [Google Scholar]
- 18.Eldridge, A. M., W. A. Halsey, and D. S. Wuttke. 2006. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry 45871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286117-120. [DOI] [PubMed] [Google Scholar]
- 20.Evans, S. K., and V. Lundblad. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 1621101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira, M. G., K. M. Miller, and J. P. Cooper. 2004. Indecent exposure: when telomeres become uncapped. Mol. Cell 137-18. [DOI] [PubMed] [Google Scholar]
- 22.Gilson, E., M. Roberge, R. Giraldo, D. Rhodes, and S. M. Gasser. 1993. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J. Mol. Biol. 231293-310. [DOI] [PubMed] [Google Scholar]
- 23.Grandin, N., and M. Charbonneau. 2003. The Rad51 pathway of telomerase-independent maintenance of telomeres can amplify TG1-3 sequences in yku and cdc13 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 233721-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandin, N., C. Damon, and M. Charbonneau. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 208397-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 206127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandin, N., C. Damon, and M. Charbonneau. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 201173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandin, N., S. I. Reed, and M. Charbonneau. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11512-527. [DOI] [PubMed] [Google Scholar]
- 28.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97503-514. [DOI] [PubMed] [Google Scholar]
- 29.Groff-Vindman, C., A. J. Cesare, S. Natarajan, J. D. Griffith, and M. J. McEachern. 2005. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol. Cell. Biol. 254406-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossi, S., A. Bianchi, P. Damay, and D. Shore. 2001. Telomere formation by Rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol. 218117-8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy, C. F., L. Sussel, and D. Shore. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6801-814. [DOI] [PubMed] [Google Scholar]
- 32.Hug, N., and J. Lingner. 2006. Telomere length homeostasis. Chromosoma 115413-425. [DOI] [PubMed] [Google Scholar]
- 33.Iyer, S., A. D. Chadha, and M. J. McEachern. 2005. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell. Biol. 258064-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 2662011-2015. [DOI] [PubMed] [Google Scholar]
- 35.Kironmai, K. M., and K. Muniyappa. 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2443-455. [DOI] [PubMed] [Google Scholar]
- 36.Kooistra, R., P. J. Hooykaas, and H. Y. Steensma. 2004. Efficient gene targeting in Kluyveromyces lactis. Yeast 21781-792. [DOI] [PubMed] [Google Scholar]
- 37.Krauskopf, A., and E. H. Blackburn. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383354-357. [DOI] [PubMed] [Google Scholar]
- 38.Lansdorp, P. M., S. Poon, E. Chavez, V. Dragowska, M. Zijlmans, T. Bryan, R. Reddel, M. Egholm, S. Bacchetti, and U. Martens. 1997. Telomeres in the haemopoietic system. Ciba Found. Symp. 211209-218. (Discussion, 211:219-222.) [DOI] [PubMed] [Google Scholar]
- 39.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Legassie, J. D., and M. B. Jarstfer. 2006. The unmasking of telomerase. Structure 141603-1609. [DOI] [PubMed] [Google Scholar]
- 41.Lin, C. Y., H. H. Chang, K. J. Wu, S. F. Tseng, C. C. Lin, C. P. Lin, and S. C. Teng. 2005. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot. Cell 4327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73347-360. [DOI] [PubMed] [Google Scholar]
- 43.Lustig, A. J., and T. D. Petes. 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 831398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275986-990. [DOI] [PubMed] [Google Scholar]
- 45.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 161919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maringele, L., and D. Lydall. 2004. EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics 1661641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 101822-1834. [DOI] [PubMed] [Google Scholar]
- 48.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376403-409. [DOI] [PubMed] [Google Scholar]
- 49.McEachern, M. J., and J. E. Haber. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75111-135. [DOI] [PubMed] [Google Scholar]
- 50.McEachern, M. J., and S. Iyer. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7695-704. [DOI] [PubMed] [Google Scholar]
- 51.McEachern, M. J., S. Iyer, T. B. Fulton, and E. H. Blackburn. 2000. Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc. Natl. Acad. Sci. USA 9711409-11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34331-358. [DOI] [PubMed] [Google Scholar]
- 53.McEachern, M. J., D. H. Underwood, and E. H. Blackburn. 2002. Dynamics of telomeric DNA turnover in yeast. Genetics 16063-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller, T., E. Gilson, R. Schmidt, R. Giraldo, J. Sogo, H. Gross, and S. M. Gasser. 1994. Imaging the asymmetrical DNA bend induced by repressor activator protein 1 with scanning tunneling microscopy. J. Struct. Biol. 1131-12. [DOI] [PubMed] [Google Scholar]
- 55.Muntoni, A., and R. R. Reddel. 2005. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14(Spec. No. 2)R191-R196. [DOI] [PubMed] [Google Scholar]
- 56.Murnane, J. P., L. Sabatier, B. A. Marder, and W. F. Morgan. 1994. Telomere dynamics in an immortal human cell line. EMBO J. 134953-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282493-496. [DOI] [PubMed] [Google Scholar]
- 58.Natarajan, S., C. Groff-Vindman, and M. J. McEachern. 2003. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot. Cell 21115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Natarajan, S., and M. J. McEachern. 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 224512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negrini, S., V. Ribaud, A. Bianchi, and D. Shore. 2007. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 21292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8657-660. [DOI] [PubMed] [Google Scholar]
- 62.Ogino, H., K. Nakabayashi, M. Suzuki, E. Takahashi, M. Fujii, T. Suzuki, and D. Ayusawa. 1998. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem. Biophys. Res. Commun. 248223-227. [DOI] [PubMed] [Google Scholar]
- 63.Pardo, B., and S. Marcand. 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 243117-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104387-396. [DOI] [PubMed] [Google Scholar]
- 65.Perrem, K., L. M. Colgin, A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2001. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 213862-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petreaca, R. C., H. C. Chiu, and C. I. Nugent. 2007. The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics 1771459-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritchie, K. B., and T. D. Petes. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy, J., T. B. Fulton, and E. H. Blackburn. 1998. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 123286-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shay, J. W., and W. E. Wright. 2005. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 26867-874. [DOI] [PubMed] [Google Scholar]
- 70.Shay, J. W., and W. E. Wright. 2001. Telomeres and telomerase: implications for cancer and aging. Radiat. Res. 155188-193. [DOI] [PubMed] [Google Scholar]
- 71.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73177-208. [DOI] [PubMed] [Google Scholar]
- 72.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 2971023-1026. [DOI] [PubMed] [Google Scholar]
- 73.Teixeira, M. T., M. Arneric, P. Sperisen, and J. Lingner. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117323-335. [DOI] [PubMed] [Google Scholar]
- 74.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6947-952. [DOI] [PubMed] [Google Scholar]
- 75.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 198083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomita, K., A. Matsuura, T. Caspari, A. M. Carr, Y. Akamatsu, H. Iwasaki, K. Mizuno, K. Ohta, M. Uritani, T. Ushimaru, K. Yoshinaga, and M. Ueno. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 235186-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Topcu, Z., K. Nickles, C. Davis, and M. J. McEachern. 2005. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc. Natl. Acad. Sci. USA 1023348-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsai, Y. L., S. F. Tseng, S. H. Chang, C. C. Lin, and S. C. Teng. 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 225679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsukamoto, Y., A. K. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 111328-1335. [DOI] [PubMed] [Google Scholar]
- 80.Underwood, D. H., C. Carroll, and M. J. McEachern. 2004. Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres. Eukaryot. Cell 3369-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Underwood, D. H., and M. J. McEachern. 2001. Totally mutant telomeres: single-step mutagenesis of tandem repeat DNA sequences. BioTechniques 30934-935, 938. [DOI] [PubMed] [Google Scholar]
- 82.Underwood, D. H., R. P. Zinzen, and M. J. McEachern. 2004. Template requirements for telomerase translocation in Kluyveromyces lactis. Mol. Cell. Biol. 24912-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Verdun, R. E., and J. Karlseder. 2007. Replication and protection of telomeres. Nature 447924-931. [DOI] [PubMed] [Google Scholar]
- 84.Vignais, M. L., and A. Sentenac. 1989. Asymmetric DNA bending induced by the yeast multifunctional factor TUF. J. Biol. Chem. 2648463-8466. [PubMed] [Google Scholar]
- 85.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119355-368. [DOI] [PubMed] [Google Scholar]
- 86.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11748-760. [DOI] [PubMed] [Google Scholar]
- 88.Wray, L. V., Jr., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 71111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu, G., W. H. Lee, and P. L. Chen. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 27530618-30622. [DOI] [PubMed] [Google Scholar]
- 90.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 594175-4179. [PubMed] [Google Scholar]
- 91.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. de Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25347-352. [DOI] [PubMed] [Google Scholar]