Abstract
Although the cyclin G1 gene is known to be regulated at the transcriptional level by p53, less is understood about the turnover of its protein product. We found that ectopically and endogenously expressed cyclin G1 protein is highly unstable and is degraded by a proteasome-mediated pathway. The N-terminal 137 amino acids of cyclin G1 (cyclin G1-137) are necessary and sufficient for both cyclin G1 ubiquitination and turnover. Interestingly, a mutant cyclin G1 (8KR) in which all lysine residues in this region have been replaced with arginine can be both ubiquitinated in cells and stabilized by a proteasome inhibitor to a similar extent as wild-type cyclin G1-137. Furthermore, the presence of a six-Myc tag at the N terminus of cyclin G1-137 significantly inhibits the protein's turnover, suggesting a role for the extreme N terminus of the protein in ubiquitin-mediated proteolysis. Although we and others previously showed that cyclin G1 protein can bind to MDM2, which functions as an E3 ubiquitin ligase to p53 and itself, cyclin G1 protein can be degraded in cells without MDM2 and p53. Interestingly, the B′α1 subunit of the serine/threonine protein phosphatase 2A, which binds to cyclin G1, can stabilize cyclin G1 under unstressed conditions and upon DNA damage, as well as inhibit the ability of cyclin G1 to be ubiquitinated. Our results thus indicate that proteasomal turnover of cyclin G1 is regulated by noncanonical processes.
Cyclin G1 was one of the earliest p53 transcriptional targets to be identified (41), although to this day, its roles in cells are not fully understood. In some experimental settings, cyclin G1 may promote cell cycle arrest and apoptosis (23, 27, 42, 48, 50, 62), while in others it can increase cell proliferation (18, 26, 27, 51, 63) and even tumorigenesis (7, 22). Consistent with its possible pro-oncogenic roles, our group (43) and others (28, 31, 39, 40) have found evidence that cyclin G1 can serve as a negative regulator of p53, although it has also been reported that Mdm2 may downregulate growth arrest by p53 via cyclin G (62).
We previously showed that cyclin G1 binds to protein phosphatase 2A (PP2A) through its ability to interact with members of the B′α regulatory subunits (42). This interaction results in dephosphorylation of MDM2 at a minimum of two sites, T216 in mouse MDM2 and Ser166 in human MDM2 (HDM2) (43). We also showed that MDM2 phosphorylated at T216 binds less well to p53 (60). Correspondingly, MDM2 protein in cyclin G1 knockout mice is hyperphosphorylated at T216, and the levels of p53 are significantly higher than those in wild-type mouse embryonic fibroblasts (43).
Because of the complexity of its possible roles, we have investigated how the cyclin G1 protein itself is regulated. Cyclin G1 was so named because it has a typical cyclin box, although it lacks the PEST sequence that is required for degradation of the canonical cyclins. Thus, it is not clear how cyclin G1 protein turnover is controlled. Most proteins that are degraded by the proteasome need to first be conjugated to ubiquitin, a 9-kDa protein that can form polyubiquitination chains at an internal lysine residue(s) (19, 44). There are exceptions to this, and a small group of proteins are known to undergo lysineless ubiquitination and instead to have ubiquitin conjugated at the N-terminal methionine (11). Such proteins include p21 (5), the p14/19 ARF protein (29, 30), Id1 (52), Id2 (14), ERK3 (12), MyoD (6), LMP1 (1), LMP2A (21), the human papillomavirus 16 (HPV-16) oncoprotein E7 (3, 47), and HPV-58 E7 (1). Of these proteins, ARF and HPV-58 E7 are the most physiologically relevant examples because they are naturally occurring lysineless proteins that are degraded by the proteasome (29, 30), whereas for the other proteins, the mutated lysineless versions were identified as substrates for N-terminal ubiquitination.
Here we have examined the turnover of cyclin G1 and found that its degradation proceeds by lysineless ubiquitination and a proteasome-mediated degradation pathway that most likely involves its N terminus. Furthermore, the B′ subunit of PP2A stabilizes cyclin G1 by inhibiting its proteasome-mediated degradation.
MATERIALS AND METHODS
Expression plasmids.
Constructs containing full-length cyclin G1 cDNA (pCMV cyclin G1) and Flag-tagged human B′α1, B′α2, and Bα1 cDNAs (pCMV-F-B′α1, pCMV-F-B′α2, and pCMV-F-Bα1, respectively) have been described (42, 54). A hemagglutinin (HA)-tagged cyclin G1 construct was previously described (43). Flag-tagged human cyclin G1 was kindly provided by Karen Vousden, and the V5-tagged cyclin G2 cDNA expression vector was a generous gift from Mary Horn. The PCMV-HA-ubiquitin expression construct was obtained from D. Bohmann. BamHI-NotI sites were used to clone the human cyclin G1 cDNA into a pTRE2hyg vector (Clontech). HA-tagged N-terminal (residues 1 to 137) and C-terminal (residues 136 to 294) cyclin G1 portions were generated by PCR, using full-length cyclin G1 cDNA as the template. Multiple lysine mutations were generated by subsequent site-directed mutagenesis, using full-length cyclin G1 or the N-terminal portion (residues 1 to 137) as the template (Invitrogen). All mutant constructs were confirmed by sequencing.
Cells and culture conditions.
NIH 3T3 cells, p53- and MDM2-null mouse 2KO cells, HeLa Tet-on cells (Clontech), and H1299 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37°C in 5% CO2. Clone 6 rat cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 38°C in 5% CO2. MG132 (Sigma), E64 (Calbiochem), and cycloheximide (Sigma) were dissolved in dimethyl sulfoxide. LLnL (Sigma) was dissolved in 100% ethanol. Doxycycline (Sigma) and neocarzinostatin (NCS) (500 ng/ml; Kayaku Co., Tokyo, Japan) were dissolved in H2O. Transfections were performed using Lipofectamine (Invitrogen). In each experiment, 2 μg of DNA and 10 μl of Lipofectamine were introduced into 5 × 105 cells. To generate human cell lines with inducible cyclin G1, the pTRE2-hyg human Flag-tagged cyclin G1 plasmid was transfected into HeLa Tet-on cells (Clontech), and hygromycin B (400 μg/ml; Invitrogen)-resistant clones were isolated. Doxycycline-inducible Flag-tagged human cyclin G1 clones were selected by immunoblotting with M2 antibody (Sigma). One of the clones that expressed human Flag-cyclin G1 tightly controlled by doxycycline (low-level leakage) was selected for the experiments (clone 2), and additional clones were also characterized.
Antibodies.
Polyclonal rabbit anti-cyclin G1 antibody 178 was previously described (43). The following antibodies were obtained commercially: anti-HA (H11) antibody (Covance), anti-HA (3F10) antibody (Roche), M2 monoclonal antibody (Sigma), anti-green fluorescent protein (anti-GFP) antibody (Clontech), polyclonal rabbit antibody H-46 (Santa Cruz), and anti-V5 antibody (Invitrogen). The anti-mouse secondary antibody True Blot was obtained from Bioscience, and Alexa Fluor 680 or 800-conjugated goat anti-rabbit and -mouse immunoglobulin G secondary antibodies were purchased from Molecular Probes.
Immunoprecipitation and immunoblot analysis.
Cells were lysed in LB buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, aprotinin, phenylmethylsulfonyl fluoride, 1 mM EDTA, and 1 mM MgCl2. For immunoprecipitation, cells were lysed in TEGN buffer, containing 50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 10% glycerol, and 0.5% NP-40. Washing buffer contained an additional 200 mM NaCl. An Odyssey scanning system was used for quantitation of protein levels in immunoblots. Alexa Fluor 680 or 800-conjugated goat anti-rabbit or -mouse immunoglobulin G secondary antibodies were used, followed by Odyssey scanning and analysis by Odyssey software, version 2.0.
In vivo ubiquitinylation assays.
Cells were lysed in buffer, followed by sonication three times for 10 s each time. Cell lysates were either subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or immunoprecipitated with the indicated antibodies and protein G-Sepharose 4 Fast (Pharmacia).
RESULTS
Cyclin G1 is an unusually unstable protein.
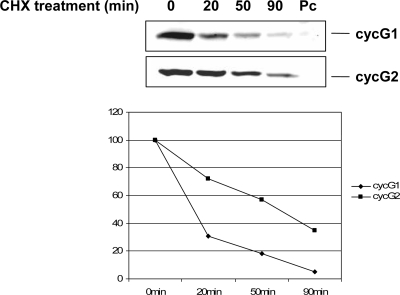
Endogenous cyclin G1 mRNA is induced rapidly upon p53 activation and is readily detectable (2). Yet we found that 30 h after transient transfection of a mouse cyclin G1 expression construct, cyclin G1 protein was virtually undetectable (data not shown). Expression of cyclin G1 from a recombinant retrovirus was also reported to be very poor (62). Nevertheless, when we looked at an earlier time point following introduction of a mouse cyclin G1 construct into NIH 3T3 cells (20 h after transfection), we were able to detect some cyclin G1 protein (Fig. 1). We used this condition to measure the half-life of cyclin G1 by treating cells with cycloheximide for various time periods (Fig. 1). This provided an approximate half-life for cyclin G1 of 20 min. In contrast, the half-life of the related protein cyclin G2, although this protein also is not very stable, is about 45 min (Fig. 1) and is closer to that of another well-studied p53 target protein, p21, which was reported to have a 50-min half-life (4, 29). Thus, compared with other p53-related proteins and other common cyclins (5, 29), cyclin G1 is exceptionally unstable.
FIG. 1.
Ectopically expressed murine cyclin G1 is short-lived. NIH 3T3 cells were transfected with constructs expressing mouse cyclin G1 (cycG1) tagged with an HA epitope at the C terminus, C-terminally V5-tagged cyclin G2 (cycG2), or empty pcDNA3 vector (Pc) for 20 h. Cells were then treated with 0.1 μg/ml cycloheximide (CHX) for the indicated times prior to lysis. Equivalent amounts of protein (30 μg) were separated by 10% SDS-PAGE and subjected to immunoblotting with polyclonal antibody 178 for cyclin G1 and anti-V5 monoclonal antibody for cyclin G2. Results were quantitated using an Odyssey scanning system.
Cyclin G1 protein can be stabilized by 26S proteasome inhibitors.
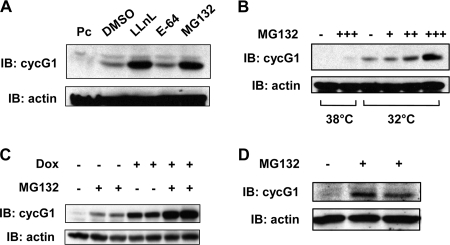
To further investigate the low levels of ectopically expressed cyclin G1, NIH 3T3 cells were transfected with a murine cyclin G1 expression plasmid and then treated with the 26S proteasome inhibitors LLnL and MG132 or the protease inhibitor E-64 (Fig. 2A). Cyclin G1 was stabilized by LLnL and MG132 but not by E-64. Clone 6 cells harbor a temperature-sensitive p53 mutant (A135V) in which p53 is in the mutant conformation at 38°C and is transcriptionally inactive (45). Under these conditions, cyclin G1 is not detectable. When these cells were treated with MG132 for 5 h, it became possible to detect a small amount of cyclin G1 protein (Fig. 2B) (43). When cells were shifted to 32°C, which results in production of transcriptionally active p53, and thereby induction of cyclin G1 transcription (41), protein levels of cyclin G1 were dramatically increased. Yet in this case, too, cyclin G1 protein levels were further increased upon treatment of cells with MG132. Thus, both ectopically and endogenously expressed rodent cyclin G1 protein turnover is regulated by proteasome-mediated degradation. To examine human cyclin G1 protein turnover, we generated clones of HeLa cell lines expressing doxycycline-inducible (Tet-on) Flag-tagged human cyclin G1, and one (clone 2) was selected for analysis (see Materials and Methods). In these cells, even uninduced human cyclin G1 protein was made detectable by adding MG132 (Fig. 2C). When the cells were treated with doxycycline to induce cyclin G1, there was a further dramatic increase of cyclin G protein upon treatment with the proteasome inhibitor. Thus, ectopically expressed human cyclin G1 protein is also degraded by the 26S proteasome pathway. Finally, we were also able to show that levels of endogenous human cyclin G1 protein in H1299 cells were increased approximately fivefold by MG132 treatment (Fig. 2D).
FIG. 2.
Exogenously and endogenously expressed cyclin G1 protein can be stabilized by inhibitors of the 26S proteasome. (A) NIH 3T3 cells were transfected with constructs expressing murine cyclin G1 or with empty pcDNA3 vector (Pc) for 20 h, after which cells were left untreated or treated with LLnL (25 μM), E-64 (50 μM), or MG132 (50 μM) for 5 h. Equivalent amounts of cell lysates (30 μg) were separated by 10% SDS-PAGE and immunoblotted with cyclin G1 (178) or actin antibodies, as indicated. DMSO, dimethyl sulfoxide. (B) Clone 6 rat cells expressing temperature-sensitive mutant p53 (A135V) were maintained at 38°C. Cells were shifted to 32°C for 5 h and then either left untreated or treated with 5 μM (+), 10 μM (++), or 50 μM (+++) MG132 for an additional 5 h. Cells were lysed and separated by 10% SDS-PAGE followed by immunoblotting with the indicated antibodies, as in panel A. Parallel cultures maintained at 38°C were left untreated or treated with 50 μM MG132 for 5 h and then treated as described above. (C) HeLa Tet-on cells (clone 2), engineered to express tetracycline/doxycycline-inducible human cyclin G1 as described in Materials and Methods, were plated for 24 h, followed by treatment with or without doxycycline (2 ng/ml) for 20 h. Cells were then either left untreated or treated with 25 μM MG132 for an additional 5 h prior to lysis. Equivalent amounts of cell extracts (30 μg) were separated by SDS-PAGE and immunoblotted with anti-human cyclin G1 polyclonal antibody H-46 or anti-actin antibody. (D) H1299 cells were left untreated or treated with 50 μM MG132 for 4 h prior to lysis and separation of cell extracts (50 μg) by 10% SDS-PAGE followed by immunoblotting with H-46 and antiactin antibodies.
Cyclin G1 protein is ubiquitinated in cells.
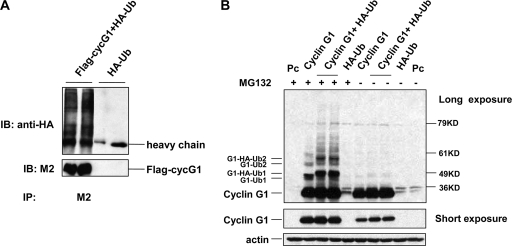
We further characterized cyclin G1 degradation by examining whether the protein can be ubiquitinated. When H1299 cells were cotransfected with Flag-tagged human cyclin G1 and HA-ubiquitin, significant amounts of HA-ubiquitin were present in anti-Flag immunoprecipitates (Fig. 3A). Further support that cyclin G1 is ubiquitinated was obtained by ectopic expression of murine cyclin G1 along with HA-ubiquitin in NIH 3T3 cells (Fig. 3B). In this case, direct immunoblotting with the anti-cyclin G1 antibody revealed multiple forms of cyclin G1 when the cells were treated with MG132. Even in the absence of ectopically expressed ubiquitin, cyclin G1 was detected as multiple species after MG132 treatment. This is likely due to the conjugation of endogenous ubiquitin to cyclin G1, since without MG132 treatment, no slower-migrating forms of cyclin G1 were detected.
FIG. 3.
Ubiquitination of cyclin G1. (A) H1299 cells were cotransfected with Flag-tagged human cyclin G1 and HA-tagged ubiquitin expression plasmids for 20 h, followed by treatment with 50 μM MG132 for 4 h. Cell lysates were immunoprecipitated with anti-Flag antibody (M2) and subjected to SDS-PAGE and immunoblotting with M2 or HA antibodies. (B) NIH 3T3 cells were cotransfected with untagged mouse cyclin G1 and HA-tagged ubiquitin expression plasmids or with empty pcDNA3 vector (Pc) for 20 h. Cells were left untreated or treated with 50 μM MG132 for 4 h. Cell lysates were separated by 8% SDS-PAGE and immunoblotted with cyclin G1 (178) or actin antibodies. The middle panel shows a shorter exposure of the same blot, showing cyclin G1 protein levels. G1-Ub1 and G1-HA-Ub1 indicate a single endogenous and ectopic HA-tagged ubiquitin molecule conjugated to ectopic cyclin G1, respectively. G1-Ub2 and G1-HA-Ub2 indicate two endogenous and ectopic HA-ubiquitin molecules conjugated to ectopic cyclin G1 protein, respectively.
The N terminus of cyclin G1 is required for proteasomal degradation.
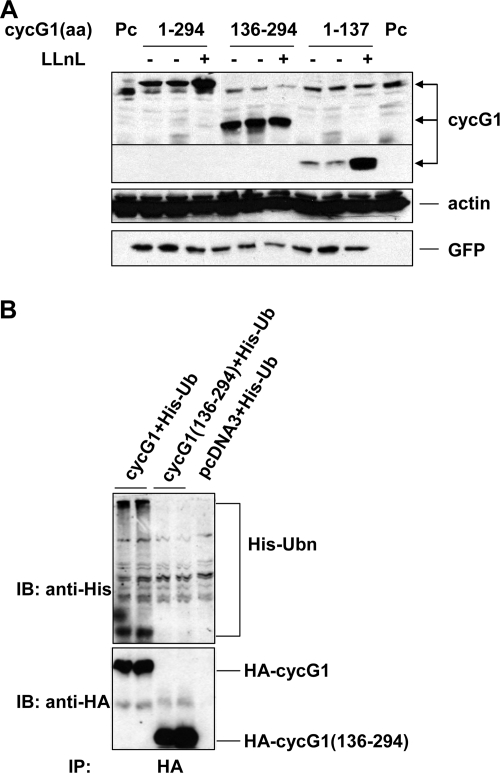
Since cyclin G1 protein is ubiquitinated and undergoes active proteasome-mediated degradation, we sought to identify which lysine residue(s) is ubiquitin conjugated. Murine, rat, and human cyclin G1 share 24 conserved lysine residues. To narrow down the critical lysine(s), we constructed two deletion mutants comprising either the N-terminal (residues 1 to 137; cyclin G11-137) or C-terminal (residues 136 to 294; cyclin G1136-294) portion of cyclin G1. Upon transfection of these constructs into NIH 3T3 cells followed by treatment with LLnL, it was clear that the N-terminal portion of cyclin G1 could be stabilized markedly by LLnL compared with its basal level (Fig. 4A). In contrast, there was a much greater initial quantity of the C-terminal portion of cyclin G1 than either the full-length protein or the N-terminal portion, and LLnL treatment had no effect on its levels. We concluded that the N terminus of cyclin G1 is the region of the protein that is targeted by the proteasome for degradation. These data are consistent with an experiment showing that in H1299 cells treated with MG132, it is possible to detect His-tagged ubiquitin conjugated to full-length cyclin G1 but not to the C-terminal portion (Fig. 4B). While we cannot formally rule out that the lysines within the C-terminal portion of cyclin G can be ubiquitinated in the context of the full-length protein, our results strongly indicate that this is not the case, but rather that cyclin G1 is ubiquitinated at a site(s) within its N terminus.
FIG. 4.
The N terminus of cyclin G1 is required for its ubiquitination and degradation. (A) NIH 3T3 cells were transfected with constructs expressing HA-tagged full-length (residues 1 to 294), C-terminal (residues 136 to 294), or N-terminal (residues 1 to 137) murine cyclin G1 or empty pcDNA3 vector (Pc) for 28 h and then were left untreated or treated with LLnL (25 μM) for 12 h. Equivalent amounts of cell lysates (30 μg) were separated by 12% SDS-PAGE and immunoblotted with anti-HA, antiactin, or anti-GFP antibodies (as a transfection control). (B) NIH 3T3 cells were transfected with constructs expressing HA-tagged full-length (residues 1 to 294) and C-terminal (residues 136 to 294) murine cyclin G1 with His-tagged ubiquitin for 24 h, followed by treatment with 25 μM MG132 for 5 h. Cell lysates were analyzed by immunoprecipitation with anti-HA antibody 3F10 and then by immunoblotting with anti-His antibody or anti-HA antibody.
Ubiquitin-mediated degradation of cyclin G1 can proceed efficiently in the absence of internal lysine residues.
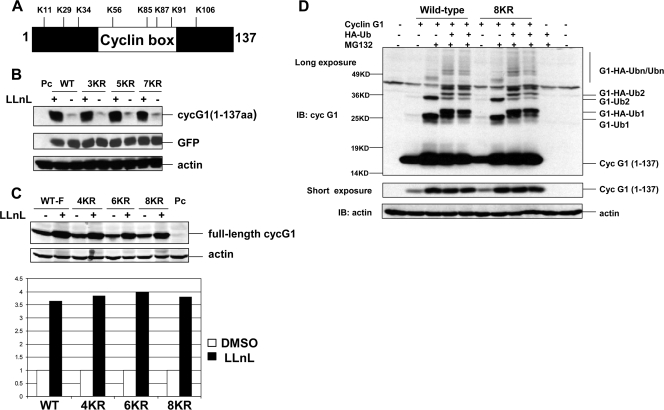
There are eight lysine residues within amino acids 1 to 137 of cyclin G1 (Fig. 5A), and these are highly conserved among the mouse, rat, and human homologues. To determine which lysine residue is critical for cyclin G1 ubiquitination and degradation, a series of lysine-to-arginine substitutions were generated in both the full-length and N-terminal versions of cyclin G1. We first made mutants with three, five, or seven lysines replaced with arginine within the N-terminal construct and unexpectedly found that each of these was equivalently stabilized by LLnL (Fig. 5B). We then constructed a series of K-to-R mutants in the context of full-length cyclin G1 in which four, six, or all eight of the lysines within the N-terminal 137 amino acids were mutated and again, surprisingly, found that LLnL treatment stabilized each, including the 8KR substitution mutant, to a similar extent to that with wild-type cyclin G1 (Fig. 5C). Furthermore, the truncated cyclin G1 construct [cyclin G11-137 (8KR)] in which all lysines were mutated showed similar stabilization and ubiquitination to those of the wild-type version of cyclin G11-137 (Fig. 5D). In fact, the cyclin G11-137 (8KR) mutant was conjugated to both endogenous ubiquitin and ectopic HA-ubiquitin to similar levels to those with wild-type cyclin G1 (Fig. 5D). Thus, although the N-terminal portion of cyclin G1 can be targeted efficiently by the proteasome for degradation, none of its eight lysine residues are apparently required for the ubiquitination and degradation of cyclin G1.
FIG. 5.
All lysines in the N terminus of cyclin G1 are dispensable for its degradation and ubiquitination. (A) Positions of the eight lysine residues in the N-terminal 137 amino acids of the murine cyclin G1 protein. (B) NIH 3T3 cells were transfected with constructs expressing an N-terminal version (residues 1 to 137) of wild-type or lysine substitution (3KR, 5KR, and 7KR) mutant cyclin G1 or with empty pcDNA3 vector (Pc) for 28 h and then were left untreated (−) or treated (+) with LLnL (+) (25 μM) for 12 h. Cell extracts (30 μg) were separated by 12% SDS-PAGE and immunoblotted with anti-cyclin G1 (178), anti-GFP, or antiactin antibodies. For N-terminal (residues 1 to 137) cyclin G1 substitution mutants, 3KR represents K11R, K29R, and K34R mutations; 5KR represents K11R, K29R, K34R, K56R, and K85R mutations; and 7KR represents K11R, K29R, K34R, K56R, K85R, K87R, and K91R mutations. (C) NIH 3T3 cells were transfected with constructs expressing full-length versions of wild-type or N-terminal lysine substitution (4KR, 6KR, or 8KR) mutant cyclin G1 or with empty pcDNA3 vector (Pc) for 28 h and then were left untreated (−) or treated with LLnL (+) (25 uM) for 12 h. Cell extracts (30 μg) were separated by 10% SDS-PAGE and immunoblotted with anti-cyclin G1 (178) or antiactin antibodies. The graph shows the increase (fold change) in wild-type and mutant cyclin G1 proteins compared with untreated levels, as analyzed by an Odyssey scanning system. For full-length cyclin G1 substitution mutants, 4KR represents K11R, K29R, K34R, and K56R mutations; 6KR represents K11R, K29R, K34R, K56R, K85R, and K87R mutations; and 8KR represents K11R, K29R, K34R, K56R, K85R, K87R, K91R, and K106R mutations. (D) NIH 3T3 cells were transfected with wild-type or mutant (8KR) N-terminal (residues 1 to 137) cyclin G1 as in panel C, with or without HA-ubiquitin, for 24 h, followed by treatment with MG132 (25 μM) for 5 h. Cell lysates were separated by 12% SDS-PAGE and immunoblotted with anti-cyclin G1 (178) or antiactin antibodies. The middle panel shows a shorter exposure of the cyclin G1 portion of the blot. G1-Ub1, G1-Ub2, G1-HA-Ub, and G1-HA-Ub2 are described in the legend to Fig. 3B.
Adding an extra sequence at the N terminus of cyclin G1 inhibits its degradation.
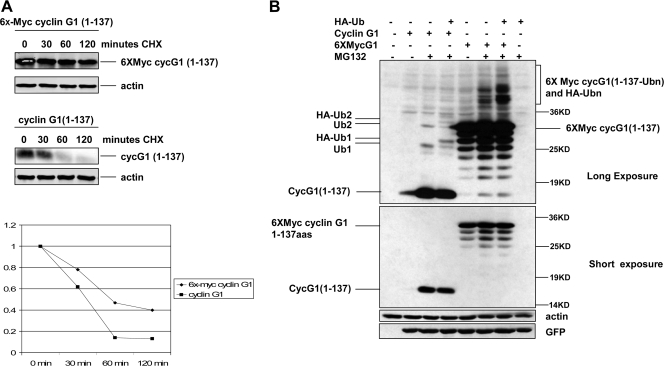
As mentioned earlier, p21 and ARF have been shown to undergo lysineless proteasomal degradation and can be conjugated to ubiquitin at their N termini (5, 29, 30). One line of evidence to support this is that both proteins can be stabilized by the addition of six copies of the Myc tag (Met-Glu-Gln-Lys-Leu-Ile-Ser-Glu-Glu-Asp-Leu-Asn-Glu) fused to the N-terminal methionine (5). Presumably, the six-Myc-tagged protein blocks the recognition site for ubiquitin or ubiquitin ligase. To test whether the addition of an N-terminal tag similarly stabilizes cyclin G1, we generated a construct expressing amino acids 1 to 137 of cyclin G1 with a six-Myc tag fused to the N-terminal methionine. Indeed, as shown in Fig. 6A, we found that six-Myc-tagged cyclin G1-137 has a longer half-life (60 min) in transfected NIH 3T3 cells than that of untagged cyclin G11-137 (30 min). Furthermore, six-Myc-tagged cyclin G11-137 was stabilized to a lesser extent by MG132 treatment (about 1.5-fold) (Fig. 6B) than was nontagged cyclin G11-137 (about 10- to 25-fold) (Fig. 6B). This indicates that ubiquitin conjugation to cyclin G1 is inhibited by the extra sequence at the N terminus, even though endogenous ubiquitin and ectopic HA-ubiquitin are conjugated to the modified cyclin G11-137 in the presence of MG132 (Fig. 6B). This could be related to the fact that there is one lysine residue in each Myc tag. Nevertheless, our data suggest that when it is unmodified, the N terminus of cyclin G1 is the ubiquitin conjugating site for cyclin G1 degradation.
FIG. 6.
A six-Myc epitope tag at the N terminus stabilizes cyclin G11-137. (A) NIH 3T3 cells were transfected with six-Myc-tagged cyclin G11-137 or untagged cyclin G11-137 for 24 h, followed by treatment with cycloheximide (0.1 μg/ml). Cells were lysed at the indicated times posttreatment, and lysates were separated by 12% SDS-PAGE followed by immunoblotting with cyclin G1 and actin antibodies as described above. Cyclin G1 protein was quantitated using an Odyssey system. (B) NIH 3T3 cells were transfected with constructs expressing six-Myc-tagged and untagged cyclin G11-137, with or without HA-ubiquitin, for 24 h and then either left untreated or treated with 25 μM MG132 in dimethyl sulfoxide for 5 h. Cell lysates were separated by 12% SDS-PAGE and immunoblotted with anti-cyclin G1, antiactin, or anti-GFP antibodies, as indicated on the right. On the left are indicated one or two molecules of untagged endogenous ubiquitin (Ub1 and Ub2) or ectopic HA-tagged ubiquitin (HA-Ub1 and HA-Ub2) conjugated to cyclin G1. Long and short exposures of the immunoblot of cyclin G1 are indicated.
Proteasome-mediated degradation of cyclin G1 is p53 and MDM2 independent.
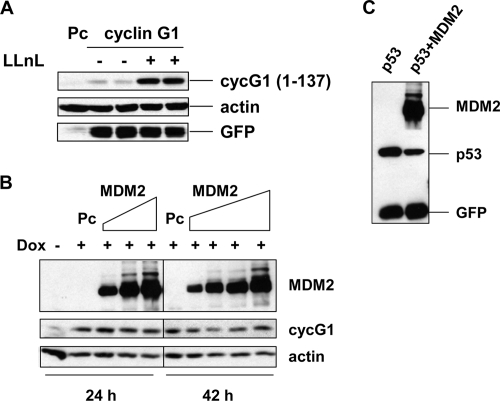
MDM2, the main negative regulator of p53, is an E3 ubiquitin ligase (15, 20, 46, 55). Under normal growth conditions, p53 protein levels are kept low by MDM2 targeting p53 for proteasome degradation. MDM2 can also function as a ubiquitin E3 ligase for itself. Previously, we showed that MDM2 and cyclin G1 can bind each other in vitro and in vivo and that the acidic region and the RING domain of MDM2 are involved in the interaction (43). To investigate whether MDM2 plays a role in cyclin G1 turnover, we first used a mouse cell line (2KO) in which both p53 and MDM2 are homozygously deleted (35, 38, 53). In these cells, the N-terminal cyclin G11-137 was stabilized by LLnL (Fig. 7A). To test whether human MDM2 can target human cyclin G1 for degradation, cyclin G1-inducible HeLa cells were transfected with increasing amounts of MDM2 and cells were collected at 24 and 42 h posttransfection. Doxycycline-induced cyclin G1 protein levels were not affected by Mdm2 at the 24-h time point (Fig. 7B) and were only marginally impacted after 42 h. As expected, Mdm2 targeted coexpressed p53 for degradation in the same cells (Fig. 7C). Based on this observation, it is possible that under some conditions Mdm2 can target cyclin G1 for degradation, but the ratio of cyclin G1 and Mdm2 may be very critical for this to occur. This is not likely to be a key activity of MDM2, since when we introduced small interfering RNA against Mdm2 into p53-null H1299 cells, there was a significant increase in p21, previously shown to be an Mdm2 target for degradation (24, 53), but there was no detectable increase in cyclin G1 protein (data not shown).
FIG. 7.
Cyclin G1 degradation by the 26S proteasome is p53 and MDM2 independent. (A) MDM2−/− p53−/− (2KO) cells were transfected with the N-terminal cyclin G11-137 construct or empty pcDNA3 vector (Pc) for 28 h and either left untreated or treated with LLnL (25 μM) for 12 h. Cell extracts (30 μg) were separated by 12% SDS-PAGE and analyzed by immunoblotting. (B) HeLa Tet-on cells expressing human cyclin G1 (clone 2) were left untreated or treated with doxycycline (2 ng/ml) and, at the same time, were transfected with increasing amounts of Flag-HDM2 for 24 h (0.5 μg, 1.0 μg, and 2.0 μg) or 42 h (0.2 μg, 0.5 μg, 1.0 μg, and 2.0 μg) or with empty pcDNA3 vector (Pc). Cell lysates were separated by 10% SDS-PAGE and analyzed by immunoblotting with anti-Flag (M2), anti-human cyclin G1 (H-46), or antiactin antibodies. (C) HeLa Tet-on cells (clone 2) without doxycycline were transfected with 0.3 μg of p53-expressing plasmid, with or without 1.5 μg of HDM2-expressing plasmid, for 24 h. Cell lysates were separated by 10% SDS-PAGE and analyzed by immunoblotting with anti-Flag (M2), anti-p53 (DO-1), or anti-GFP antibodies.
Cyclin G1 protein turnover is negatively regulated by the B′α1 subunit of PP2A.
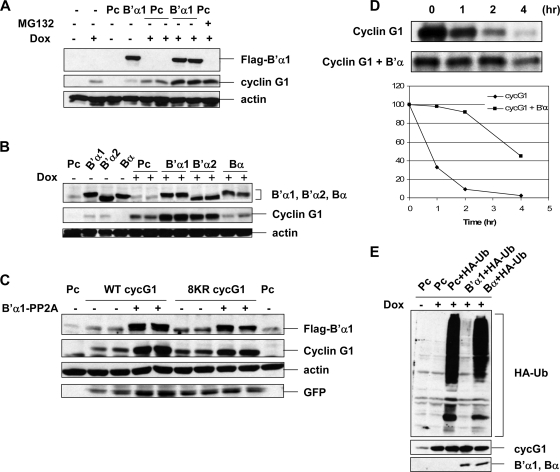
While free p21 and ARF can be targeted by the 26S proteasome, binding of these proteins to cdk2, cdk4 (p21), and B23 (ARF) stabilizes them (5, 29). As mentioned above, members of the B′α subfamily of PP2A phosphatase can interact with cyclin G1. The interaction between cyclin G1 and B′α1 is reasonably strong, as the cyclin G-B′α1 complexes are stable even in 2% NP-40 and 400 mM NaCl (42), and the majority of cyclin G1 protein is associated with high-molecular-weight complexes containing PP2A (42, 43). We therefore considered that members of the B′α family may regulate cyclin G1 turnover. To test this hypothesis, ectopic Flag-tagged B′α proteins were introduced into HeLa cells expressing inducible human cyclin G, and their impact on cyclin G1 protein levels was assessed. Indeed, cyclin G1 levels were markedly increased by Flag-B′α1 under both noninduced and doxcycline-induced conditions (Fig. 8A). Augmentation of cyclin G1 by B′α1 was comparable to the effect of treating cells with MG132. Furthermore, we found that B′α2 expression increased cyclin G1 levels to the same extent as B′α1 (Fig. 8B), whereas another version of the PP2A B subunit (Bα) that cannot bind cyclin G1 had no such stabilizing effect. The effect of the B′α subunit was p53 and MDM2 independent, since the same results were obtained with 2KO cells (Fig. 8C). Notably, the 8KR mutant form of full-length cyclin G1, described in Fig. 5, was stabilized to the same extent as wild-type cyclin G1. Most importantly, evidence that cyclin G1 proteasomal turnover was regulated by B′α was derived from experiments showing that the half-life of cyclin G1 was greatly increased in the presence of B′α (Fig. 8D) and that association of ectopically expressed HA-ubiquitin with cyclin G1 was markedly reduced in the presence of B′α1 but not Bα (Fig. 8E). Interestingly, B′α1 did not stabilize either the N-terminal (residues 1 to 137) or C-terminal (residues 136 to 294) portion of cyclin G1 (data not shown), suggesting that it requires interaction with both parts of the cyclin G1 protein. This is consistent with the observation that unlike full-length cyclin G1, neither cyclin G11-137 nor cyclin G1136-294 can form stable interactions with B′α1 (data not shown). Taken together, these data demonstrate that the B′α1 and B′α2 regulatory subunits of PP2A stabilize cyclin G1 by inhibiting its ubiquitination and degradation by the 26S proteasome.
FIG. 8.
B′α family subunits of PP2A stabilize human cyclin G1 protein. (A) HeLa Tet-on cells (clone 2) that were untreated or treated with doxycycline (Dox; 2 ng/ml) were transfected with empty pcDNA3 vector (Pc) or a vector expressing the Flag-tagged B′α1 subunit of PP2A (B′α1) for 42 h. Equivalent amounts of cell lysates (30 μg) were separated by 10% SDS-PAGE and immunoblotted (IB) with anti-Flag (M2), anti-human cyclin G1 (H-46), or antiactin antibodies. The final lane contained an extract of cells transfected with pcDNA3 and treated with doxycycline as described above, but MG132 (25 μM) was added for another 4 h. (B) HeLa Tet-on cells (clone 2) were left untreated or treated with 2 ng/ml doxycycline and, at the same time, were transfected with pcDNA3 (Pc), Flag-B′α1, Flag-B′α2, or Flag-Bα for 42 h. Equivalent amounts (30 μg) of cell lysates were separated by 10% SDS-PAGE and immunoblotted with anti-Flag (M2), anti-human cyclin G1 (H-46), or antiactin antibodies. (C) 2KO cells were transfected with empty pcDNA3 vector (Pc) or with constructs expressing wild-type (WT) or 8KR mutant full-length mouse cyclin G1, with or without the Flag-B′α1 subunit of PP2A, for 24 h. Cell lysates were separated by 10% SDS-PAGE and immunoblotted with anti-Flag (M2), anti-cyclin G1 (178), antiactin, or anti-GFP antibodies. (D) (Top) COS-1 cells were transfected with constructs expressing wild-type full-length mouse cyclin G1, with or without Flag-B′α1. Forty-eight hours after transfection, the transfected cells were labeled with [35S]methionine for 3 h and further incubated with unlabeled medium for the indicated chase periods. Subsequently, lysates prepared from the labeled cells were used for immunoprecipitation with anti-cyclin G1 antibody (178). The immunoprecipitated cyclin G1 was then separated by 10% SDS-PAGE and processed for fluorography. (Bottom) The intensity of the labeled cyclin G1 after each chase time was quantified by densitometry, and the quantified results of the pulse-chase analyses are presented on a logarithmic scale. The presented values were calculated as follows: level of labeled cyclin G1 at the indicated time for the chase period/level of cyclin G at the start of the chase period. (E) HeLa Tet-on cells (clone 2) were left untreated (−) or treated (+) with doxycycline (Dox; 2 ng/ml) as indicated and, at the same time, were transfected with constructs expressing either empty pcDNA3 vector (Pc) or HA-ubiquitin (HA-Ub), with or without either Flag-B′α1 or Flag-Bα, for 24 h as indicated, followed by treatment with MG132 (25 nM) for 4 h. Equivalent amounts (1 mg) of cell lysates were immunoprecipitated with anti-cyclin G1 antibody H-46. The immunoprecipitates were separated by 10% SDS-PAGE and immunoblotted with anti-HA and anti-human cyclin G1 (H-46) antibodies. Flag-B′α1 and Flag-Bα were immunoblotted with anti-Flag (M2) antibody from 30 μg of cell lysates.
DNA damage-induced degradation of cyclin G1 is attenuated by the B′α subunit of PP2A.
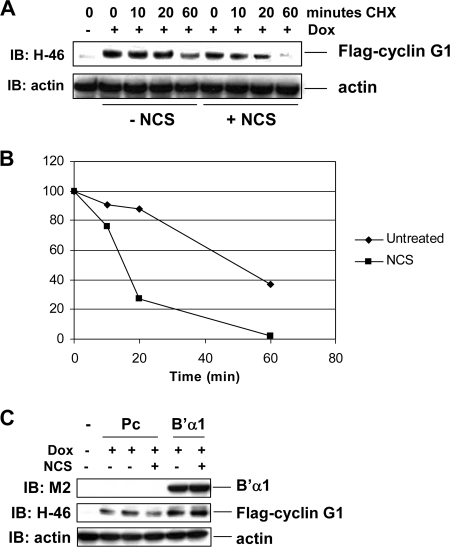
Finally, we wanted to determine whether DNA damage affects the protein stability of cyclin G1. For this purpose, we used our HeLa cells expressing doxycycline-inducible Flag-tagged human cyclin G1. These cells were induced with doxycycline for 48 h and mock treated or treated with the radiomimetic agent NCS for 5 h, and cyclin G1 protein stability was analyzed by a cycloheximide chase. We found that the half-life of N-terminally Flag-tagged human cyclin G1 in untreated cells was approximately 50 min (Fig. 9A and B), similar to the half-life of six-Myc-tagged cyclin G11-137 (Fig. 6A). However, upon NCS treatment, the half-life of Flag-tagged human cyclin G1 was reduced to 15 min (Fig. 9A and B), showing that DNA damage destabilizes cyclin G1 protein. In addition, NCS destabilized ectopic mouse cyclin G1 in NIH 3T3 cells (data not shown). More interestingly, we found that in the presence of ectopic B′α1, NCS could not destabilize cyclin G1 compared with pcDNA3 vector (Fig. 9C). These data indicate that the B′α1 regulatory subunit of PP2A also plays a role in stabilizing cyclin G1 in the DNA damage environment.
FIG. 9.
The B′α1 subunit of PP2A stabilizes human cyclin G1 protein under DNA damaging conditions. (A) HeLa Tet-on cells (clone 2) expressing Flag-tagged human cyclin G1 were plated with or without 2 ng/ml of doxycycline (Dox) for 48 h. The cells were either left untreated or treated with 330 ng/ml of NCS for 5 h and then were treated with cycloheximide (CHX) (0.1 μg/ml) and harvested at the indicated time points. Cell lysates (20 μg) were separated by 10% SDS-PAGE and immunoblotted with anti-cyclin G1 polyclonal (H-46) or antiactin antibodies. (B) Cyclin G1 protein was quantitated using NIH ImageJ software. (C) HeLa Tet-on cells expressing Flag-tagged human cyclin G1 were plated with or without 2 ng/ml of doxycycline (Dox) for 16 h, followed by transfection with vector pcDNA3 (Pc) or B′α1 for 24 h. NCS (330 ng/ml) was added 5 h prior to lysis. Equivalent amounts of cell lysates (30 μg) were separated by 10% SDS-PAGE and immunoblotted with the indicated antibodies.
DISCUSSION
Most proteins whose turnover is mediated by the 26S proteasome are ubiquitinated before being degraded by the proteasome (19). In agreement with this, we found that cyclin G1 is ubiquitinated in cells, as determined by immunoprecipitation experiments in which we showed that HA-ubiquitin could be conjugated to Flag-cyclin G1 and by MG132 treatment of cells, which led to the accumulation of multiple forms of cyclin G1. A closer examination of the cyclin G1 protein revealed that the N-terminal portion, but not the C-terminal portion, of cyclin G1 could be stabilized by the proteasome inhibitor LLnL, and correspondingly, endogenous ubiquitin could only be conjugated to the full-length and the N-terminal portion of cyclin G1.
Our further investigations into the mechanisms involved in cyclin G1 degradation revealed that cyclin G1 is not degraded through the internal lysine-dependent ubiquitination pathway. Strikingly, mutation of all eight internal lysine residues in the context of the full-length or N-terminal portion of cyclin G1 did not affect the extent to which these proteins were stabilized by LLnL or ubiquitinated, indicating that the ubiquitination and degradation of cyclin G1 occur in a lysine-independent manner. As mentioned earlier, lysineless ubiquitination has been reported for other proteins. Instead, these proteins are ubiquitinated at their N termini and the addition of an N-terminal tag stabilizes them, most probably by blocking access of ubiquitin to the N-terminal residue (11). Here we show that placing an extra sequence at the N terminus of cyclin G1 stabilizes the protein and reduces its degradation by the proteasome, suggesting that cyclin G1 is also ubiquitinated at its N terminus. The physiological relevance of N-terminal ubiquitination remains to be determined. However, Sun et al. recently showed that glucocorticoids differentially regulate MyoD and Id1 degradation by N-terminal ubiquitination in myotubes, demonstrating a key role for the N terminus-dependent ubiquitination pathway in the physiology of muscle protein degradation (52). In addition to MyoD, which can be degraded by both the lysine-dependent and N-terminal ubiquitination pathways, multiple mechanisms of degradation have been reported for p21 (9, 10, 32, 49). Deciphering the functional significance of these alternate degradation pathways for cyclin G1 and other proteins remains a future challenge.
The MDM2 protein, a p53 target and E3 ubiquitin ligase, is the main negative regulator of p53 (15, 16, 37). In addition to p53, MDM2 functions as a ubiquitin ligase for pRB (56, 57), Numb (59), histone 2B (36), MDMX (13), and MDM2 itself (15, 20). It was reported that p21 physically binds to MDM2, and although p21 degradation does not require MDM2 ubiquitin ligase activity, MDM2 promotes p21 degradation by facilitating binding of p21 with the proteasome C8 subunit (61). ARF also binds to MDM2, although its turnover does not require MDM2 (29). We found that cyclin G1 binds to MDM2 at both its C-terminal RING and central acidic regions (43), although whether MDM2 could target cyclin G1 for degradation was not examined. We showed here that intermediate levels of overexpressed MDM2 had a slight impact on cyclin G1 levels. Similarly, Zhao et al. showed that cyclin G1 expression was progressively reduced as Mdm2 expression was increased (62). Taken together, these data suggest that perhaps MDM2 can directly (or indirectly) impact cyclin G1 degradation under certain conditions. However, data obtained using the 2KO cell line, which lacks both the MDM2 and p53 genes, and a small interfering RNA against MDM2 indicate that MDM2 is unlikely to be the primary E3 ligase for cyclin G1. It remains to be determined whether MDMX has a role to play in the ubiquitination and degradation of cyclin G1.
Our findings strongly implicate PP2A as a major regulator of cyclin G1 stability. First, increasing the levels of the regulatory B′α subunits of PP2A promotes the accumulation of cyclin G1 by interfering with its turnover. Moreover, association of cyclin G1 with the B′α subunit of PP2A inhibited its ubiquitination. In addition, B′α1 was found to stabilize cyclin G1 under DNA damaging conditions. How might PP2A regulate the turnover of cyclin G1? PP2A regulates the stability of other proteins, including c-myc, the Pim family of protein kinases, and securin (17, 34, 58). Each of these proteins interacts with the enzymatically active PP2A and is a substrate of PP2A phosphatase. The dephosphorylation of c-myc and the Pim kinases by PP2A leads to their proteolysis, whereas PP2A stabilizes securin by removing phosphate groups that target the protein for degradation via the SCF ubiquitin ligase. We previously showed that upon p53 induction, cyclin G1 forms a complex with enzymatically active PP2A, which dephosphorylates MDM2 and thereby negatively regulates p53 function (43). It remains to be determined whether cyclin G1 itself is a substrate of PP2A and if the stability of cyclin G1 is regulated by phosphorylation.
Given the complexity of proposed functions for cyclin G1, the physiological significance of cyclin G1 stability control is not immediately obvious. Since the discovery of cyclin G1 as a target of p53, it has been described as both a positive and negative regulator of cell growth (8). Most recently, Jones et al. showed that cyclin G1 drives apoptosis of neural crest cells, which can be prevented by p53 inhibition (25). Cyclin G1 has also been associated with genomic instability and is frequently overexpressed in human tumors, such as cervical carcinoma (33) and human hepatocellular carcinoma (18). In addition to cyclin G1, other negative regulators of p53, including Mdm2 and COP1, are also found overexpressed in tumors, and their increased levels have been shown to downregulate p53 function. Similar to cyclin G1, these proteins are also rapidly turned over, and this may be important to allow full activation of p53-dependent pathways, thereby preventing tumorigenesis. Cyclin G1 is also involved in various p53-independent activities, which may not be surprising given the complexity of its roles in cells and its many interacting partners. Unraveling the mechanisms involved in cyclin G1 stability control will likely provide further insight into the role of cyclin G1 during normal differentiation and malignant transformation.
Acknowledgments
Ella Freulich is thanked for expert technical assistance. Thanks are extended to Marshall Urist for helpful discussions and to Max Li for assistance with some experiments.
This work was supported by a C. J. Martin fellowship from the National Health and Medical Research Council of Australia to M.J.P. and by NIH grants CA58316 and CA87497.
Footnotes
Published ahead of print on 3 November 2008.
REFERENCES
- 1.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 27523491-23499. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S., S. Rowan, and K. H. Vousden. 1996. Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 131103-1109. [PubMed] [Google Scholar]
- 3.Ben-Saadon, R., I. Fajerman, T. Ziv, U. Hellman, A. L. Schwartz, and A. Ciechanover. 2004. The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J. Biol. Chem. 27941414-41421. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, C., T. Hay, D. W. Meek, and D. P. Lane. 2002. Hypophosphorylation of Mdm2 augments p53 stability. Mol. Cell. Biol. 226170-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom, J., V. Amador, F. Bartolini, G. DeMartino, and M. Pagano. 2003. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 11571-82. [DOI] [PubMed] [Google Scholar]
- 6.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 175964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, D. S., N. L. Zhu, G. Hung, M. J. Skotzko, D. R. Hinton, V. Tolo, F. L. Hall, W. F. Anderson, and E. M. Gordon. 1997. Retroviral vector-mediated transfer of an antisense cyclin G1 construct inhibits osteosarcoma tumor growth in nude mice. Hum. Gene Ther. 81667-1674. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X. 2002. Cyclin G: a regulator of the p53-Mdm2 network. Dev. Cell 2518-519. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., L. F. Barton, Y. Chi, B. E. Clurman, and J. M. Roberts. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell 26843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, X., Y. Chi, A. Bloecher, R. Aebersold, B. E. Clurman, and J. M. Roberts. 2004. N-Acetylation and ubiquitin-independent proteasomal degradation of p21(Cip1). Mol. Cell 16839-847. [DOI] [PubMed] [Google Scholar]
- 11.Ciechanover, A., and R. Ben-Saadon. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14103-106. [DOI] [PubMed] [Google Scholar]
- 12.Coulombe, P., G. Rodier, E. Bonneil, P. Thibault, and S. Meloche. 2004. N-terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol. Cell. Biol. 246140-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Graaf, P., N. A. Little, Y. F. Ramos, E. Meulmeester, S. J. Letteboer, and A. G. Jochemsen. 2003. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J. Biol. Chem. 27838315-38324. [DOI] [PubMed] [Google Scholar]
- 14.Fajerman, I., A. L. Schwartz, and A. Ciechanover. 2004. Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitination. Biochem. Biophys. Res. Commun. 314505-512. [DOI] [PubMed] [Google Scholar]
- 15.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2758945-8951. [DOI] [PubMed] [Google Scholar]
- 16.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2569-573. [DOI] [PubMed] [Google Scholar]
- 17.Gil-Bernabe, A. M., F. Romero, M. C. Limon-Mortes, and M. Tortolero. 2006. Protein phosphatase 2A stabilizes human securin, whose phosphorylated forms are degraded via the SCF ubiquitin ligase. Mol. Cell. Biol. 264017-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gramantieri, L., M. Ferracin, F. Fornari, A. Veronese, S. Sabbioni, C. G. Liu, G. A. Calin, C. Giovannini, E. Ferrazzi, G. L. Grazi, C. M. Croce, L. Bolondi, and M. Negrini. 2007. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 676092-6099. [DOI] [PubMed] [Google Scholar]
- 19.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 20.Honda, R., and H. Yasuda. 2000. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 191473-1476. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, M., A. Ikeda, and R. Longnecker. 2002. Lysine-independent ubiquitination of Epstein-Barr virus LMP2A. Virology 300153-159. [DOI] [PubMed] [Google Scholar]
- 22.Jensen, M. R., V. M. Factor, A. Fantozzi, K. Helin, C. G. Huh, and S. S. Thorgeirsson. 2003. Reduced hepatic tumor incidence in cyclin G1-deficient mice. Hepatology 37862-870. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, M. R., V. M. Factor, and S. S. Thorgeirsson. 1998. Regulation of cyclin G1 during murine hepatic regeneration following Dipin-induced DNA damage. Hepatology 28537-546. [DOI] [PubMed] [Google Scholar]
- 24.Jin, Y., H. Lee, S. X. Zeng, M. S. Dai, and H. Lu. 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 226365-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, N. C., M. L. Lynn, K. Gaudenz, D. Sakai, K. Aoto, J. P. Rey, E. F. Glynn, L. Ellington, C. Du, J. Dixon, M. J. Dixon, and P. A. Trainor. 2008. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 14125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kampmeier, J., A. Behrens, Y. Wang, A. Yee, W. F. Anderson, F. L. Hall, E. M. Gordon, and P. J. McDonnell. 2000. Inhibition of rabbit keratocyte and human fetal lens epithelial cell proliferation by retrovirus-mediated transfer of antisense cyclin G1 and antisense MAT1 constructs. Hum. Gene Ther. 111-8. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, S. H., M. Ikawa, A. Ito, M. Okabe, and H. Nojima. 2001. Cyclin G1 is involved in G2/M arrest in response to DNA damage and in growth control after damage recovery. Oncogene 203290-3300. [DOI] [PubMed] [Google Scholar]
- 28.Kimura, S. H., and H. Nojima. 2002. Cyclin G1 associates with MDM2 and regulates accumulation and degradation of p53 protein. Genes Cells 7869-880. [DOI] [PubMed] [Google Scholar]
- 29.Kuo, M. L., W. den Besten, D. Bertwistle, M. F. Roussel, and C. J. Sherr. 2004. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 181862-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, M. L., W. den Besten, and C. J. Sherr. 2004. N-terminal polyubiquitination of the ARF tumor suppressor, a natural lysine-less protein. Cell Cycle 31367-1379. [DOI] [PubMed] [Google Scholar]
- 31.Lafarga, V., A. Cuadrado, and A. R. Nebreda. 2007. p18(Hamlet) mediates different p53-dependent responses to DNA-damage inducing agents. Cell Cycle 62319-2322. [DOI] [PubMed] [Google Scholar]
- 32.Li, X., L. Amazit, W. Long, D. M. Lonard, J. J. Monaco, and B. W. O'Malley. 2007. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell 26831-842. [DOI] [PubMed] [Google Scholar]
- 33.Liang, J., M. L. Bian, Q. Y. Chen, X. Liu, H. Ou, M. Li, and J. Liu. 2006. Relationship between cyclin G1 and human papilloma virus infection in cervical intraepithelial neoplasia and cervical carcinoma. Chin. Med. Sci. J. 2181-85. [PubMed] [Google Scholar]
- 34.Losman, J. A., X. P. Chen, B. Q. Vuong, S. Fay, and P. B. Rothman. 2003. Protein phosphatase 2A regulates the stability of Pim protein kinases. J. Biol. Chem. 2784800-4805. [DOI] [PubMed] [Google Scholar]
- 35.McDonnell, T. J., R. Montes de Oca Luna, S. Cho, L. L. Amelse, A. Chavez-Reyes, and G. Lozano. 1999. Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. J. Pathol. 188322-328. [DOI] [PubMed] [Google Scholar]
- 36.Minsky, N., and M. Oren. 2004. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell 16631-639. [DOI] [PubMed] [Google Scholar]
- 37.Momand, J., H. H. Wu, and G. Dasgupta. 2000. MDM2—master regulator of the p53 tumor suppressor protein. Gene 24215-29. [DOI] [PubMed] [Google Scholar]
- 38.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378203-206. [DOI] [PubMed] [Google Scholar]
- 39.Ohtsuka, T., M. R. Jensen, H. G. Kim, K. T. Kim, and S. W. Lee. 2004. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene 235405-5408. [DOI] [PubMed] [Google Scholar]
- 40.Ohtsuka, T., H. Ryu, Y. A. Minamishima, A. Ryo, and S. W. Lee. 2003. Modulation of p53 and p73 levels by cyclin G: implication of a negative feedback regulation. Oncogene 221678-1687. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto, K., and D. Beach. 1994. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 134816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto, K., C. Kamibayashi, M. Serrano, C. Prives, M. C. Mumby, and D. Beach. 1996. p53-dependent association between cyclin G and the B′ subunit of protein phosphatase 2A. Mol. Cell. Biol. 166593-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto, K., H. Li, M. R. Jensen, T. Zhang, Y. Taya, S. S. Thorgeirsson, and C. Prives. 2002. Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol. Cell 9761-771. [DOI] [PubMed] [Google Scholar]
- 44.Pickart, C. M., and M. J. Eddins. 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 169555-72. [DOI] [PubMed] [Google Scholar]
- 45.Pinhasi-Kimhi, O., D. Michalovitz, A. Ben-Zeev, and M. Oren. 1986. Specific interaction between the p53 cellular tumour antigen and major heat shock proteins. Nature 320182-184. [DOI] [PubMed] [Google Scholar]
- 46.Poyurovsky, M. V., and C. Prives. 2006. Unleashing the power of p53: lessons from mice and men. Genes Dev. 20125-131. [DOI] [PubMed] [Google Scholar]
- 47.Reinstein, E., M. Scheffner, M. Oren, A. Ciechanover, and A. Schwartz. 2000. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 195944-5950. [DOI] [PubMed] [Google Scholar]
- 48.Seo, H. R., D. H. Lee, H. J. Lee, M. Baek, S. Bae, J. W. Soh, S. J. Lee, J. Kim, and Y. S. Lee. 2006. Cyclin G1 overcomes radiation-induced G2 arrest and increases cell death through transcriptional activation of cyclin B1. Cell Death Differ. 131475-1484. [DOI] [PubMed] [Google Scholar]
- 49.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5403-410. [DOI] [PubMed] [Google Scholar]
- 50.Shimizu, A., J. Nishida, Y. Ueoka, K. Kato, T. Hachiya, Y. Kuriaki, and N. Wake. 1998. CyclinG contributes to G2/M arrest of cells in response to DNA damage. Biochem. Biophys. Res. Commun. 242529-533. [DOI] [PubMed] [Google Scholar]
- 51.Smith, M. L., H. U. Kontny, R. Bortnick, and A. J. Fornace, Jr. 1997. The p53-regulated cyclin G gene promotes cell growth: p53 downstream effectors cyclin G and Gadd45 exert different effects on cisplatin chemosensitivity. Exp. Cell Res. 23061-68. [DOI] [PubMed] [Google Scholar]
- 52.Sun, L., J. S. Trausch-Azar, L. J. Muglia, and A. L. Schwartz. 2008. Glucocorticoids differentially regulate degradation of MyoD and Id1 by N-terminal ubiquitination to promote muscle protein catabolism. Proc. Natl. Acad. Sci. USA 1053339-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao, W., and A. J. Levine. 1999. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 966937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tehrani, M. A., M. C. Mumby, and C. Kamibayashi. 1996. Identification of a novel protein phosphatase 2A regulatory subunit highly expressed in muscle. J. Biol. Chem. 2715164-5170. [DOI] [PubMed] [Google Scholar]
- 55.Toledo, F., and G. M. Wahl. 2006. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6909-923. [DOI] [PubMed] [Google Scholar]
- 56.Uchida, C., S. Miwa, T. Isobe, K. Kitagawa, T. Hattori, T. Oda, H. Yasuda, and M. Kitagawa. 2006. Effects of MdmX on Mdm2-mediated downregulation of pRB. FEBS Lett. 5801753-1758. [DOI] [PubMed] [Google Scholar]
- 57.Uchida, C., S. Miwa, K. Kitagawa, T. Hattori, T. Isobe, S. Otani, T. Oda, H. Sugimura, T. Kamijo, K. Ookawa, H. Yasuda, and M. Kitagawa. 2005. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 24160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeh, E., M. Cunningham, H. Arnold, D. Chasse, T. Monteith, G. Ivaldi, W. C. Hahn, P. T. Stukenberg, S. Shenolikar, T. Uchida, C. M. Counter, J. R. Nevins, A. R. Means, and R. Sears. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6308-318. [DOI] [PubMed] [Google Scholar]
- 59.Yogosawa, S., Y. Miyauchi, R. Honda, H. Tanaka, and H. Yasuda. 2003. Mammalian Numb is a target protein of Mdm2, ubiquitin ligase. Biochem. Biophys. Res. Commun. 302869-872. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, T., and C. Prives. 2001. Cyclin a-CDK phosphorylation regulates MDM2 protein interactions. J. Biol. Chem. 27629702-29710. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Z., H. Wang, M. Li, S. Agrawal, X. Chen, and R. Zhang. 2004. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem. 27916000-16006. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, L., T. Samuels, S. Winckler, C. Korgaonkar, V. Tompkins, M. C. Horne, and D. E. Quelle. 2003. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol. Cancer Res. 1195-206. [PubMed] [Google Scholar]
- 63.Zhu, N. L., L. Wu, P. X. Liu, E. M. Gordon, W. F. Anderson, V. A. Starnes, and F. L. Hall. 1997. Downregulation of cyclin G1 expression by retrovirus-mediated antisense gene transfer inhibits vascular smooth muscle cell proliferation and neointima formation. Circulation 96628-635. [DOI] [PubMed] [Google Scholar]