Abstract
A significant fraction of human cancer cells and immortalized cells maintain telomeres in a telomerase-independent manner called alternative lengthening of telomeres (ALT). It has been suggested that ALT involves homologous recombination that is expected to generate unique intermediate DNAs. However, the precise molecular mechanism of ALT is not known. To gain insight into how telomeric DNAs (T-DNAs) are maintained in ALT, we examined the physical structures of T-DNAs in ALT cells. We found abundant single-stranded regions in both G and C strands of T-DNAs. Moreover, two-dimensional gel electrophoreses and native in-gel hybridization analyses revealed novel ALT-specific single-stranded T-DNAs, in addition to previously reported t-circles. These newly identified ALT-specific T-DNAs include (i) the t-complex, which consists of highly branched T-DNAs with large numbers of internal single-stranded portions; (ii) ss-G, which consists of mostly linear single-G-strand T-DNAs; and (iii) ss-C, which consists of most likely circular single-C-strand T-DNAs. Cellular-DNA fractionation by the Hirt protocol revealed that t-circles and ss-G exist in ALT cells as extrachromosomal and chromatin-associated DNAs. We propose that such ALT-specific T-DNAs are produced by telomere metabolism specific to ALT, namely, homologous recombination and the rolling-circle replication mechanism.
Telomeric DNA (T-DNA) in vertebrates consists of direct repeats of 5′-TTAGGG-3′/5′-CCCTAA-3′ in which the guanine-rich strand (the G strand) forms a single-stranded (ss) DNA overhang at the very ends (the G tail). The G tail is thought to invade telomeric duplex DNA to form a D-loop structure called the t-loop (8), thereby protecting natural DNA ends from unfavorable processing and preventing them from being recognized as a double-stranded (ds) DNA break (DSB). T-DNA length is decreased as cells divide due to the end replication problem. On the other hand, telomere lengths are maintained in approximately 90% of human cancer cells and immortalized cells by activating telomerase (12). A significant fraction of those cells, however, maintain telomeres in a telomerase-independent manner called alternative lengthening of telomeres (ALT) (10).
ALT cells are characterized by a set of unique features. T-DNAs in ALT cells are heterogeneous in length and change dynamically with time. Frequent reciprocal sister chromatid exchanges at telomeres (1, 14) and copying of a tag sequence in a telomere to other telomeres (5) are observed in ALT cells, suggesting the involvement of homologous recombination (HR) in ALT. In approximately 5 to 30% of interphase nuclei of ALT cells, a fraction of T-DNAs colocalize with a specific type of nuclear promyelocytic leukemia body called an ALT-associated promyelocytic leukemia body (APB) (32). Various recombination, repair, replication, and damage response proteins, as well as telomeric proteins, are present in APBs (20). These observations suggest that HR occurs at APBs to elongate T-DNAs in ALT cells.
Cells of the budding yeast Saccharomyces cerevisiae inactivated for telomerase survive by maintaining telomeres via telomerase-independent and HR-dependent pathways (15). Two genetically and mechanistically distinguishable survivors, type I and type II, have been reported (13, 27). Type I and type II cells maintain telomeres by amplifying subtelomeric Y′ elements or telomeric repeats, respectively. Type I depends on Rad51 and Rad52 but not Rad50, while type II requires Rad50 and Rad52 but not Rad51. Yeast Rad50 is a component of the Mre11-Rad50-Xrs2 complex, which is the counterpart of the metazoan Mre11-Rad50-Nbs1 complex. It is known that Rad50 and Nbs1 play important roles in ALT cells (11, 25). These observations suggest that human ALT cells are analogous to yeast type II survivors. Phosphatidylinositol 3-kinase-related protein kinases (PIKKs) of budding yeast, Mec1 and Tel1, are required by type II cells (29). DNA synthesis at APBs in ALT cells is inhibited by caffeine, an inhibitor of PIKKs (19), further emphasizing the similarity between yeast type II survivors and human ALT cells.
ALT cells possess extrachromosomal telomeric repeat (ECTR) DNAs, which may reflect the unique metabolism of T-DNAs. Both circular and linear ds ECTRs have been reported in ALT cells (2, 22, 28, 30). Circular ds ECTRs, referred to as t-circles, are detected not only in ALT cells, but also in telomerase-positive cells expressing a dominant-negative TRF2 mutant (30). In contrast, it was recently reported that APBs contain linear ds ECTRs (6). In yeast type I and type II survivors, circular telomeric repeats are utilized as a template for HR-dependent “roll-and-spread” telomere elongation (16).
In this study, to further understand the molecular mechanism of ALT, we analyzed the structures of T-DNAs in ALT cells in detail. Specifically, we exploited two-dimensional (2D) gel electrophoresis, as well as in-gel hybridization techniques. We demonstrate that ALT cells contain unusual T-DNAs that have not been reported before and discuss the possible mechanisms of de novo T-DNA synthesis in ALT cells implied by such novel T-DNAs.
MATERIALS AND METHODS
Cell culture.
GM847, WI38 VA13.2RA (VA13), GM639, and BFT-3B cells were obtained from Roger Reddel. All cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. U2-OS cells harboring the extrachromosomal plasmid pEBV-C (Invitrogen) were prepared by transfection and selection in the presence of 200 μg/ml G418.
Nucleases.
HinfI was used to liberate T-DNA and to measure its length in 1D gel electrophoresis in this study. Digestion with a mixture of AluI and MboI was performed for samples in 2D gel electrophoresis, which was used in most of the similar experiments appearing in the literature. BAL-31 and Escherichia coli exonuclease III (Exo III) were purchased from Takara, and E. coli exonuclease I (Exo I), RecJf (RecJ), and T7 endonuclease I (Endo I) were from New England Biolabs. Ten micrograms of cellular DNA containing 60 ng of 7.25-kb linear ss DNA was treated with 80 units of Exo I or 120 units of RecJ. Limited digestion of 10 μg of U2-OS cellular DNA with Exo III was performed by incubating DNA samples containing internal-control DNAs, 0.2 μg of 3.5-kb linear ds DNA, and 65 ng of 7.25-kb circular ss DNA with 170 units of Exo III at 37°C for 0 to 45 min. The mixture of 10 μg of U2-OS DNA and 1 μg of 7.3-kb linear ds DNA was treated with 2 units of BAL-31 at 30°C for 0 to 60 min. Ten micrograms of U2-OS DNA was incubated with 0 to 40 units of T7 Endo I at 37°C for 60 min. Marker and standard DNAs were prepared according to the method described in the supplemental material. The Hirt protocol was described previously (2).
Gel electrophoresis.
Neutral and alkaline gel electrophoreses were performed as described in the supplemental material. 2D gel electrophoresis was performed according to the method of Cohen and Lavi (3) with minor modifications. In brief, DNA was resolved in the first dimension on a 0.4% agarose gel in 0.5× TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA) at 4°C and 1 V/cm. Gel slabs excised from appropriate lanes were immersed in a gel (1% agarose gel in 0.5× TBE, 0.3 μg/ml ethidium bromide), and electrophoresis in the second dimension was executed at 4°C and 5 V/cm. DNA was visualized by UV light imaging, a Typhoon 9400 imager, and ImageQuant software (Amersham/GE Healthcare).
Gel hybridization.
Southern hybridization to detect T-DNA was performed in Church's buffer as described in the supplemental material. For native in-gel hybridization analysis (4, 17), gels were dried and subjected to hybridization in Church's buffer. After analysis, the gels were soaked in solution containing 0.5 M NaOH and 0.15 M NaCl to denature the DNA in situ. The denatured gels were neutralized in 25 mM sodium phosphate (pH 6.5) and hybridized with probes. Hybridization signals were analyzed with a Typhoon 9400 imager and ImageQuant software.
RESULTS
Size distribution of G and C strands of T-DNA in ALT cells.
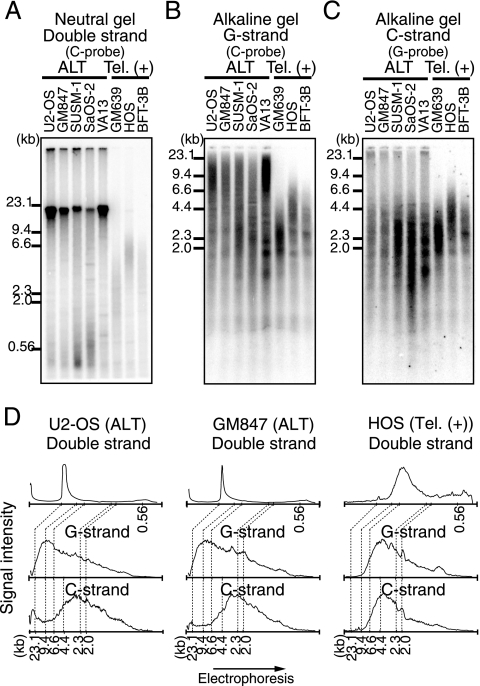
HinfI digests of DNAs prepared from five ALT (U2-OS, GM847, SUSM-1, SaOS-2, and VA13) and three telomerase-positive (GM639, HOS, and BFT-3B) cell lines were analyzed for T-DNA (Fig. 1). First, DNA digests separated on a neutral gel were analyzed by Southern hybridization using the (CCCTAA)4 oligonucleotide probe (C probe) (Fig. 1A). Strong DNA signals of approximately 20 kb were observed in all ALT cell lines. SUSM-1 and SaOS-2 cells showed relatively heterogeneous T-DNA lengths ranging from less than 0.5 to 20 kb. In contrast, T-DNA lengths in the telomerase-positive cell lines were distributed in narrower ranges of approximately 1 to 4 kb (GM639) and 2 to 10 kb (HOS). Next, DNA digests were denatured prior to sample loading, fractionated on alkaline gels, and analyzed by Southern hybridization using a C probe or a (TTAGGG)4 oligonucleotide probe (G probe) (Fig. 1B and C). The signal intensities of DNAs from U2-OS, GM847, and HOS cells were scanned along the direction of electrophoresis, and the quantified results were aligned to facilitate comparison (Fig. 1D). It was found that the size distribution of nondenatured T-DNAs correlated well with that of the G and C strands of denatured T-DNAs in the three telomerase-positive cell lines. These results indicate that the G and C strands of ds T-DNAs in telomerase-positive cells do not contain significant numbers of gaps or nicks. In contrast, the G and C strands of denatured T-DNAs migrated faster than those of nondenatured T-DNAs in the five ALT cell lines. Whereas most nondenatured T-DNA signals were observed in the range between 23.1- and 9.4-kb size markers, significant G- and C-strand signals were found in the range migrating faster than the 9.4-kb size marker. This was particularly prominent for the C strand: signal peaks were observed in the range smaller than the 4.4-kb size marker (Fig. 1D). These results suggest that both G and C strands of digested T-DNAs in ALT cells contain large numbers of internal gaps and/or nicks. Such gaps and nicks occur in greater frequency in the C strand than in the G strand and appear to be distributed throughout the lengths of ds T-DNAs. One potential explanation for the presence of gaps and nicks in the C strand is the ligation failure of Okazaki fragments during the lagging-strand synthesis of the C strand. However, the C and G strands of U2-OS cells did not change significantly during S-phase progression (see Fig. S1 in the supplemental material).
FIG. 1.
Size distribution of ds and ss telomere restriction fragments. (A to C) DNAs from five ALT (U2-OS, GM847, SUSM-1, SaOS-2, and VA13) and three telomerase-positive [Tel. (+)] (GM639, HOS, and BFT-3B) cell lines were subjected to neutral-gel (A) or alkaline-gel (B and C) electrophoresis and Southern hybridization using the indicated DNA probes. (D) Quantitation of Southern hybridization signals. Signals obtained by neutral-gel Southern hybridization (A) for two ALT cell lines (U2-OS and GM847) and one telomerase-positive cell line (HOS) are shown. The relative positions of size markers are also shown.
Gaps in T-DNA derived from ALT cells.
The native in-gel hybridization technique has been utilized to detect ss T-DNA. The G tail, the 3′-terminal overhang of the G strand, is characterized by its sensitivity to Exo I, which degrades ss DNAs from the 3′ termini. RecJ, in contrast, digests ss DNAs from the opposite direction, i.e., the 5′ termini.
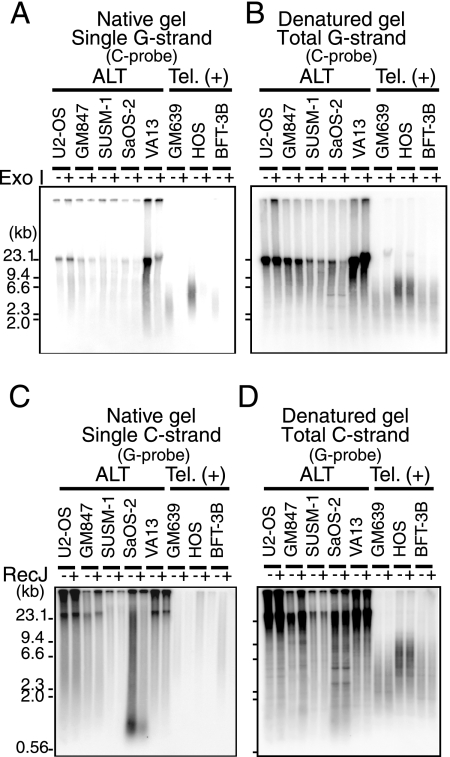
HinfI digests of DNA prepared from the five ALT cell lines and the three telomerase-positive cell lines were resolved by neutral-gel electrophoresis. The gels were in-gel hybridized with the G or C probe under native conditions to detect ss T-DNA (Fig. 2A and C; see Fig. S2A and C in the supplemental material). Then, the same gels were denatured and hybridized with the same probe to detect the total amounts of ss and ds T-DNAs (Fig. 2B and D; see Fig. S2B and D in the supplemental material). In the three telomerase-positive cell lines subjected to native in-gel hybridization, significant strand-specific signals were observed for the G strand, but not for the C strand. The G-strand signals were sensitive to Exo I treatment prior to restriction digestion, but not to RecJ, indicating that they corresponded to the G tail (Fig. 2A; see Fig. S2C in the supplemental material). In contrast, significant signals of both single G and C strands were detected in the native in-gel hybridization of DNAs derived from the five ALT cell lines, although the intensities varied among the cell lines. These signals were largely insensitive to excess amounts of Exo I or RecJ (Fig. 2; see Fig. S2 in the supplemental material). The activities and specificities of Exo I and RecJ were demonstrated using an internal control DNA contained in the sample (see Fig. S5A and B in the supplemental material). These results suggest that most of these signals were derived from internal gapped T-DNA instead of 5′ or 3′ overhangs, such as the G tail.
FIG. 2.
In-gel hybridization analyses of gapped T-DNA in ALT cells. DNAs from various ALT and telomerase-positive [Tel. (+)] cell lines were subjected to gel electrophoresis and in-gel hybridization. Samples were treated (+) or not treated (−) with Exo I or RecJ. (A) The single G strand and its sensitivity to Exo I were analyzed in a native gel with the C probe. (B) Total G strand was detected using a denatured gel from panel A. (C) The single C strand and its sensitivity to RecJ were analyzed in a native gel with the G probe. (D) Total C strand in the denatured gel from panel C. The positions of size markers are shown on the left.
Unusual strand-specific T-DNA in ALT cells.
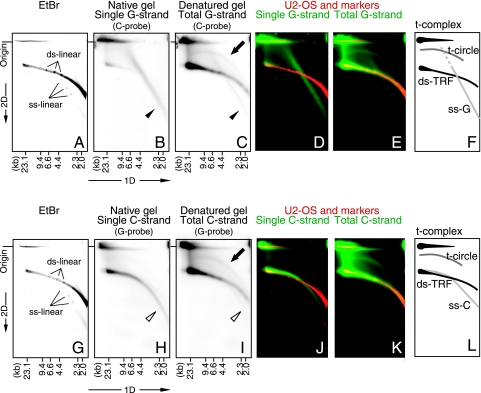
To further analyze T-DNA structures specifically found in ALT cells, DNA prepared from U2-OS cells was digested with AluI and MboI and then resolved by 2D gel electrophoresis. Electrophoresis in the first and second dimensions was performed in the absence and presence of ethidium bromide (EtBr) to estimate DNA size and conformation, respectively. EtBr detected a single intense arc signal that overlapped with linear marker ds DNAs (linearized plasmid DNAs) included in the sample (Fig. 3A and G), indicating that bulk linear restricted ds DNAs comprised the EtBr-stained arc signal. The gels were then subjected to in-gel hybridization with the G or C probe under native and denaturing conditions. In the denatured gels (Fig. 3C and I), both probes detected an intense arc signal overlapping with the EtBr-stained arc signal (Fig. 3E and K), indicating that the intense arc signal represented abundant linear restricted ds T-DNAs (ds telomeric restriction fragments [ds-TRFs]). At the origins of the gels, signals migrating in the first dimension but not in the second (hereafter called the t-complex) were observed. The t-complex and ds-TRFs hybridized with G and C probes under native conditions (Fig. 3B and H), suggesting that they contain significant numbers of ss portions in both the G and C strands. The relative signal intensities in native and denatured gels were higher for the t-complex than for ds-TRFs, suggesting that ds portions are more limited in the t-complex than in ds-TRFs. It is known that DNAs with complex structures, such as branched ds DNAs, migrate slowly in the presence of EtBr, as in the case of the second-dimension gel electrophoresis (7). We therefore believe that the t-complex that migrated little in the second dimension represents complex DNA structures containing significant amounts of ss T-DNAs.
FIG. 3.
2D gel electrophoresis and in-gel hybridization analysis of T-DNA of U2-OS cells. Linear marker ds DNA (ds-linear) (4.14, 5.49, and 7.30 kb) and marker ssDNA (ss-linear) (4.14, 5.49, and 7.30 kb) were simultaneously subjected to electrophoresis with AluI- and MboI-digested U2-OS DNA. (A to E) Results of G-strand analysis using the C probe. (G to K) Results of C-strand analysis using the G probe. Signals of U2-OS and marker DNA by EtBr staining (A and G), single G or C strand observed in a native gel (B and H), and total G or C strand observed in a denatured gel (C and I) are shown. The arrows, filled arrowheads, and open arrowheads indicate signals of the t-circle, ss-G, and ss-C structures, respectively. Panels A to C and G to I were each processed by pseudocolor imaging and merged. (D) Bulk and marker DNAs (A) (red) and single G strand (B) (green). (E) Bulk and marker DNAs (A) (red) and total G strand (C) (green). (J) Bulk and marker DNAs (G) (red) and single C strand (H) (green). (K) Bulk and marker DNAs (G) (red) and total C strand (I) (green). (F and L) Schematic representations of the results for G and C strands. Origin, the position of the origin of the second-dimension gel electrophoresis. The positions of the size markers for the first-dimension gel electrophoresis are shown at the bottom.
Open circular ds DNAs of various sizes were observed as an arc signal in 2D gel electrophoresis (3). We detected arc T-DNA signals that migrated in a manner similar to that of open circular marker DNAs and called them t-circles (Fig. 3C and I; see Fig. S3B and E in the supplemental material). t-circles were detected by G and C probes in U2-OS and the other ALT cell lines, but not in the telomerase-positive cell lines (see Fig. S4 in the supplemental material). These results indicate that t-circles are specific to ALT cells, as reported previously (2, 30). Interestingly, the mobilities of t-circles were slightly different from those of open circular marker ds DNAs (see Fig. S3A to F in the supplemental material). We therefore suggest that t-circles in ALT cells are not genuine open circular DNAs but some specific structures closely akin to open circles. We also observed faint T-DNA arc signals that overlapped with covalently closed circular marker DNAs in U2-OS (see Fig. S3 in the supplemental material) and SUSM-1 (see Fig. S4B in the supplemental material) cells. These results suggest that small amounts of closed circular T-DNAs are present in ALT cells.
In native 2D gel hybridization, the C probe detected another type of T-DNA signal not observed in the telomerase-positive cell lines (Fig. 3B; see Fig. S4 in the supplemental material). The signal migrated diagonally in the gel as a straight line and overlapped with the signals of linear marker ss DNAs (Fig. 3D). These results suggest that the T-DNAs are mostly or exclusively composed of single G-strand DNAs (ss-G). The G probe also detected a similar ALT-specific T-DNA signal (Fig. 3H; see Fig. S4 in the supplemental material). This arc-shaped signal partly overlapped with ds-TRF signals in the low-mobility range of the second dimension of gel electrophoresis. In the high-mobility range, however, the signal gradually branched from the ds-TRF arc to form an independent arc that was positioned in the region between the ds-TRF arc and the linear marker ss DNAs (Fig. 3G to K). The smooth nature of the arc suggests that the T-DNAs consist of a spectrum of DNAs having similar conformations. The close apposition of T-DNAs with ds-TRFs for the slowly migrating ones suggests that they are linear ds T-DNAs with single-C-strand T-DNAs. The relative straight character of the arc signal for the fast-migrating ones suggests that they are similar to genuine ss DNAs. Taken together, the evidence suggests that these T-DNAs are a population of C-strand DNAs that increase their fraction of ds portions as their sizes increase, and they will be referred to hereafter as ss-C. The signal of ss-G or ss-C did not overlap with the signals of circular marker ss DNAs (see Fig. S3G to L in the supplemental material). ss-G and ss-C were observed in all five ALT cell lines but not in the telomerase-positive cell lines we examined (see Fig. S4 in the supplemental material). Taken together, ALT cell lines contain (i) gapped ds-TRFs, (ii) complex-structured DNAs having significant amounts of ss portions in both G and C strands (t-complex), (iii) t-circles, (iv) mostly or exclusively single G strands (ss-G), and (v) single C strands with increasing fractions of ds portions as the size increases (ss-C). Importantly, these features are characteristic of ALT cells and are not observed in telomerase-positive cells.
Terminal structures of ALT-specific T-DNA.
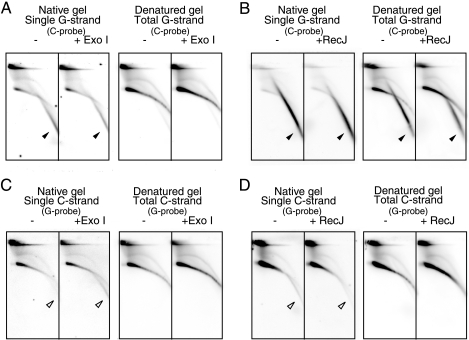
To analyze the terminal structures of unusual T-DNAs in ALT cells, we examined their sensitivities to the ss DNA-specific exonucleases Exo I and RecJ. Cellular DNAs from U2-OS cells containing internal-control DNAs were digested with AluI and MboI and resolved by 2D gel electrophoresis and in-gel hybridization analyses (Fig. 4). The signals of ss-G (Fig. 4A and B) and ss-C (Fig. 4C and D) were insensitive to Exo I or RecJ, indicating that these DNAs do not contain significant numbers of ssDNA termini. The activities and specificities of Exo I and RecJ were confirmed by monitoring internal control DNAs included in the sample (see Fig. S5A and B in the supplemental material). Given that ss-G mostly consists of linear ss DNAs, it is suggested that ss-G contains short ds DNA patches and/or forms complex structures, such as the G quartet, that protect the end from ss DNA-specific exonucleases (see Fig. 9). On the other hand, ss-C was resistant to Exo I and RecJ and was less mobile than linear ss DNAs, suggesting that ss-C consists of circular ss DNAs. However, the arc signal of ss-C was less mobile than the signal of ss circular markers in the second-dimension gel electrophoresis (see Fig. S3J to L in the supplemental material). Finally, ds-TRFs and the t-complex were resistant to Exo I and RecJ, although they contained significant numbers of ss regions. Therefore, ss DNAs mostly exist as internal gaps.
FIG. 4.
Sensitivity of U2-OS T-DNA to ss DNA-specific exonucleases. U2-OS DNAs were treated with (+) Exo I (A and C) or RecJ (B and D) or mock treated (−) prior to AluI and MboI digestion and 2D gel electrophoresis, as indicated. Single- and total-G-strand (A and B) and C-strand (C and D) T-DNA signals were analyzed by in-gel hybridization using the indicated probes. The filled and open arrowheads indicate specific signals of ss-G and ss-C, respectively.
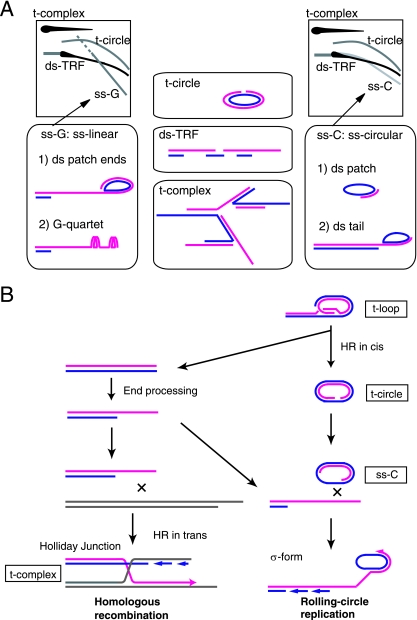
FIG. 9.
Model of telomere maintenance in ALT cells. (A) Models of structures of ALT-specific T-DNAs. The red and blue lines indicate G and C strands, respectively. (B) Model of ALT-specific T-DNAs in telomere elongation. T-DNA is synthesized by HR between distinct telomeres (HR in trans) or the rolling-circle mechanism using the t-circle as a template.
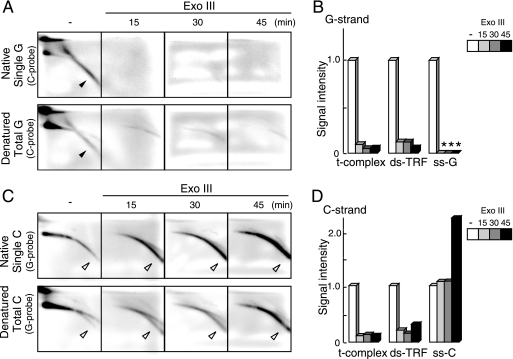
Exo III degrades one strand of ds DNA processively in the 3′-to-5′ direction. Cellular DNAs from U2-OS cells containing internal-control DNAs were incubated with Exo III for various times, digested with AluI and MboI, and resolved by 2D gel electrophoresis and in-gel hybridization analyses (Fig. 5A and C). It was shown, by monitoring internal control DNAs, that Exo III reacted specifically as expected (see Fig. S5C in the supplemental material). As the four images for a particular set of hybridization conditions at different time points were derived from a single gel, we were able to directly compare their signal intensities. Signals of ds-TRF in the denatured gels and those of ss-G and ss-C in the native gels were scanned and quantitated (Fig. 5B and D). The signal intensities of the t-complex and ds-TRFs decreased rapidly during Exo III treatment for both G- and C-strand signals. These results are consistent with the notion that the t-complex and ds-TRF consist of ds T-DNAs containing significant numbers of nicks and/or gaps. The Exo III treatment also reduced the signal intensities of ss-G to an undetectable level at 15 min. The kinetics of the ss-G degradation was similar to that of the internal control of linear ds DNA (see Fig. S5C in the supplemental material). The ss-G sensitivity to Exo III is explained in Discussion below. In contrast, the signal intensities of ss-C did not change at 15 and 30 min and increased at 45 min, consistent with the notion that ss-C consists of circular C-strand ss DNAs. However, given that the internal control of circular ss DNA was slowly degraded by Exo III (see Fig. S5C in the supplemental material), we suggest that the amount of ss-C increased during the treatment. Presumably, ds-TRFs and the t-complex may have been converted into ss-C via degradation of the nicked/gapped strands.
FIG. 5.
Sensitivity of U2-OS T-DNA to Exo III. U2-OS DNAs were treated with Exo III for 0 (−), 15, 30, and 45 min prior to AluI and MboI digestion and 2D gel electrophoresis. (A and C) Single (upper) and total (lower) G-strand (A) and C-strand (C) T-DNA signals were analyzed by in-gel hybridization using the indicated probes. The filled and open arrowheads indicate specific signals of ss-G and ss-C, respectively. (B and D) Quantification of the signal intensities of various types of G-strand (B) and C-strand (D) T-DNAs. The asterisks indicate that the signal was not detected. The signal intensities were normalized to those without Exo III treatment.
Gapped and unpaired T-DNAs in ALT cells.
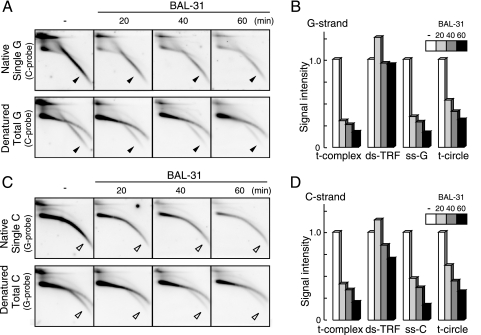
BAL-31 exhibits processive ds exonucleolytic and ss endonucleolytic activities. Cellular DNAs from U2-OS cells containing internal control DNAs were subjected to limited digestion with BAL-31. The DNA was processively digested at ∼15 bp/min during incubation, as evidenced by the gradual shortening of the internal control DNA (see Fig. S5D in the supplemental material). The sample was further digested with AluI and MboI and resolved by 2D gel electrophoresis and in-gel hybridization analyses (Fig. 6). Signals of the t-complex, ds-TRF, and t-circle in the denatured gels and ss-G and ss-C in the native gels were scanned and quantitated (Fig. 6B and D). While the signal intensity of ds-TRF did not markedly change, those of ss-G, ss-C, and the t-complex were significantly reduced with increasing BAL-31 incubation time. It is possible that the t-complex is converted into ds-TRFs by BAL-31 digestion of ss portions. The signals of the t-circle were also reduced with incubation time, suggesting that the ss portion of the t-circle was cleaved under these conditions.
FIG. 6.
Sensitivity of U2-OS T-DNA to BAL-31. U2-OS DNAs were treated with BAL-31 for 0 (−), 20, 40, and 60 min prior to AluI and MboI digestion and 2D gel electrophoresis. (A and C) Single (upper) and total (lower) G-strand (A) and C-strand (C) T-DNA signals were analyzed by in-gel hybridization using the indicated probes. The filled and open arrowheads indicate specific signals of ss-G and ss-C, respectively. (B and D) Quantification of signal intensities of various types of G-strand (B) and C-strand (D) T-DNAs. The signal intensities were normalized to that without BAL-31 treatment.
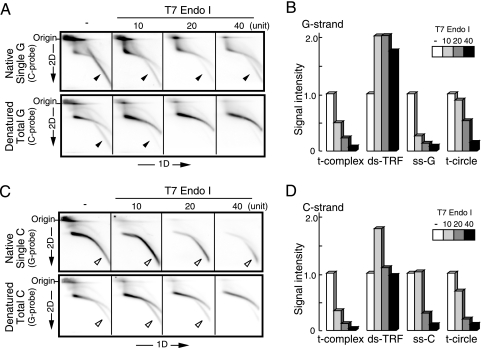
T7 Endo I cleaves preferentially unpaired ds DNAs, such as mismatches, heteroduplexes, and cruciform DNAs. It also digests nicked DNAs, albeit slowly (24). DNAs prepared from U2-OS cells were subjected to digestion with T7 Endo I at various concentrations, treated with AluI and MboI, and analyzed by 2D gel electrophoresis and in-gel hybridization (Fig. 7A and C). The signals of the t-complex, ds-TRF, and t-circle in the denatured gels and ss-G and ss-C in the native gels were scanned and quantitated (Fig. 7B and D). The t-complex, ss-G, and ss-C were sensitive to T7 Endo I, suggesting the presence of unpaired ds DNA regions in these T-DNAs. In contrast, T7 Endo I did not significantly digest ds-TRFs, suggesting a lack of unpaired ds DNAs in ds-TRFs. Interestingly, the ds-TRF signal intensity increased proportionally to the decrease of t-complex signal intensity during the T7 Endo I reaction. It is possible that the t-complex was converted into linear ds T-DNAs by the endonucleolytic digestion. Based on these observations, we propose that the t-complex consists of groups of ds T-DNAs connected by unpaired DNAs, such as branches (see Fig. 9). It was found that slowly migrating ss-C molecules whose signals overlapped with the ds-TRF arc were preferentially digested by T7 Endo I rather than fast-migrating ss-C molecules, suggesting that unpaired regions are frequently contained in or close to the ds regions of ss-C molecules. We demonstrated that most ss-G DNAs are linear single-G-strand T-DNAs. The sensitivity of ss-G to T7 Endo I suggests that the target of the enzyme is formed by single G strands, such as the G quartet (see Fig. 9).
FIG. 7.
Sensitivity of U2-OS T-DNA to T7 Endo I. U2-OS DNAs were treated with various amounts of T7 Endo I prior to AluI and MboI digestion and 2D gel electrophoresis (−, untreated). (A and C) Single (upper) and total (lower) G-strand (A) and C-strand (C) T-DNA signals were analyzed by in-gel hybridization using the indicated probes. The filled and open arrowheads indicate specific signals of ss-G and ss-C, respectively. (B and D) Quantification of signal intensities of various types of G-strand (B) and C-strand (D) T-DNAs observed.
Chromosomal or extrachromosomal origin of T-DNAs.
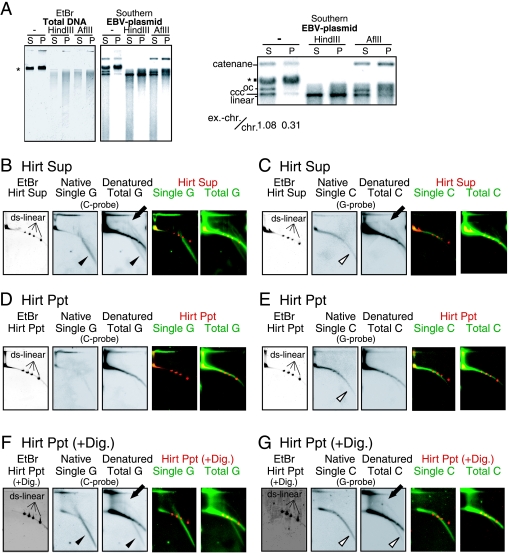
To examine whether T-DNAs were extrachromosomal in origin, we prepared DNAs from U2-OS cells according to the Hirt protocol (treatment of cell lysate with high concentrations of sodium dodecyl sulfate and NaCl at 4°C) and examined T-DNAs in the supernatant fraction (Sup) and the precipitate fraction (Ppt) (Fig. 8). It is known that low-molecular-weight DNAs, including extrachromosomal DNAs, are enriched in the Sup whereas undegraded chromosomal DNAs are concentrated in the Ppt. Precipitated DNA obtained by the Hirt protocol can be dissolved and analyzed further. We used U2-OS cells containing an Epstein-Barr virus (EBV)-derived vector that served as an extrachromosomal marker DNA. We found that total DNA was fractionated to the Sup and Ppt at a ratio of 1 to 2.2, suggesting that a significant amount of chromosomal DNA is present in the Hirt Sup, as reported previously (2). Equal amounts of Sup and Ppt DNAs (0.1 μg each) were examined by gel electrophoresis and Southern hybridization analysis of the EBV vector. When the DNAs were not treated, extrachromosomal marker DNA was detected at the positions expected for open circular, covalently closed circular, and, presumably, catenated DNAs (Fig. 8A). In addition, we observed a signal that comigrated with bulk U2-OS genomic DNA signal. This signal was not detected when the fractionated DNAs were treated with AflII, which does not digest the EBV vector DNA, suggesting that it was derived from the marker DNA closely associated or concatenated, for example, with the bulk cellular chromatin DNA. Because the intensities of signals corresponding to catenanes were increased in the AflII-digested Ppt, it is likely that a fraction of chromatin-associated marker DNAs were present as catenanes. When the fractionated DNAs were digested with HindIII, which cleaves the EBV vector DNA at a unique site, most signals were represented as a single band corresponding to the linear marker DNA, as expected. Together, all behaviors of the marker DNAs after restriction digestion were compatible with the assignment of signals to the DNA configurations indicated in Fig. 8A. The relative ratios of the signal intensities of extrachromosomal marker DNA (i.e., catenanes, open circular, and covalently closed circular) to the chromatin-bound marker DNA were 1.08 and 0.31 in untreated Sup and Ppt, respectively. Therefore, the Hirt protocol enriched extrachromosomal DNAs by approximately 3.5-fold.
FIG. 8.
Hirt fractionation of U2-OS T-DNA. (A) Fractionation of EBV-based vector DNAs contained in U2-OS. Hirt Sup (S) and Ppt (P) DNAs (0.1 μg of each) and their HindIII or AflII digests were analyzed by gel electrophoresis. Images of the EtBr-stained gel (EtBr) and hybridization with the EBV vector probe (Southern) are shown. Part of the hybridization signals was enlarged and is shown on the right. The positions of open circular (oc), closed circular (ccc), ds linear (linear), and catenated (catenane) EBV DNAs are indicated. The asterisk indicates the signals of bulk chromosomal DNAs. The numbers indicate the signal intensities of extrachromosomal EBV DNAs (ex.-chr.; oc, ccc, and catenane) relative to those of chromatin-associated EBV DNAs (chr.). (B to E) Ten micrograms of undigested DNA derived from the Hirt Sup and 10 μg of undigested DNA from the Hirt Ppt were analyzed by 2D gel electrophoresis and in-gel hybridization using the indicated probes. Linear size marker ds DNAs (ds-linear) (2.69, 4.14, 5.49, and 7.30 kb) were simultaneously loaded on a gel. (F and G) Ten micrograms of Ppt DNA was digested with AluI and MboI prior to 2D gel electrophoresis. G-strand signals (B, D, and F) and C-strand signals (C, E, and G) were analyzed for the Sup (B and C) and Ppt (D to G). Merged signals of EtBr (red) and hybridization (green) images are also shown. The solid arrows, filled arrowheads, and open arrowheads indicate t-circle, ss-G, and ss-C signals, respectively.
We next analyzed 10 μg each of Sup and Ppt DNAs using 2D gel electrophoresis and in-gel hybridization (Fig. 8B to G). Comparable amounts of ss-C were observed in both the Sup and Ppt (Fig. 8C and E), suggesting that not all ss-C existed as extrachromosomal DNAs. In contrast, the t-circle and ss-G were largely fractionated to the Sup and scarcely to the Ppt (Fig. 8B and C), suggesting that they exist as extrachromosomal DNAs and that t-circles correspond to previously reported ECTRs. When Ppt DNAs were treated with AluI and MboI, t-circle and ss-G signals were detected in digested Ppt DNAs (Fig. 8F and G). This result suggested that some fractions of ss-G and t-circles are closely associated with chromosomal DNAs.
DISCUSSION
In this study, we analyzed the physical structures of T-DNAs in ALT cells. 2D gel electrophoreses and native in-gel hybridization analyses enabled us to describe ALT-specific T-DNAs in detail and to identify novel ALT-specific T-DNAs. In addition to the t-circle, which has been reported, four distinct types of T-DNAs were identified in ALT cells, namely, gapped ds-TRFs, t-complex, ss-G, and ss-C. Importantly, those T-DNAs were not detected in telomerase-positive cells. Although ds-TRFs were observed in both ALT and telomerase-positive cell lines, those in ALT cells are unique in that they contain internal DNA gaps on both strands. Such T-DNAs were not detected in cellular DNAs prepared from telomerase-positive cells when fivefold more DNAs were examined with longer exposure times (data not shown), highlighting the specificity to ALT cells. The features of these T-DNAs in ALT cells revealed in this study are summarized in Table 1. These T-DNAs in ALT cells are characterized by the presence of significant numbers of ss and/or unpaired regions. Below, we discuss models of the DNA structures of the T-DNA species (Fig. 9).
TABLE 1.
Sensitivities of unique T-DNAs
| Treatment | Specificitya | Sensitivityb
|
||||
|---|---|---|---|---|---|---|
| ss-G | ss-C | t-complex | t-circle | gapped ds-TRF | ||
| Exo I | 3′-to-5′ ss-Exo | − | − | − | − | − |
| RecJ | 3′-to-5′ ss-Exo | − | − | − | − | − |
| Exo III | ds-dependent 3′-to-5′ ss-Exo | + | − | + | ND | + |
| BAL-31 | Exo and ss-Endo | + | + | + | ± | − |
| T7 Endo I | Unpaired ds | + | ± | + | ± | − |
ds, ds DNA; ss, ss DNA.
Sensitivity is expressed as + (sensitive), ± (moderately sensitive), and − (insensitive). ND, not determined.
Gapped ds-TRFs.
ds-TRFs in ALT cells and telomerase-positive cells migrated similarly in 2D gel electrophoresis, indicating that ds-TRFs in ALT cells consist of linear ds T-DNAs. However, ds-TRFs in ALT cells hybridized with both probes (particularly with the G probe) in native gels, and the signals were resistant to Exo I and RecJ. Taking these facts together, we conclude that ds-TRFs in ALT cells are linear ds T-DNAs containing significant numbers of internal gaps, particularly in the C strand (gapped ds-TRFs). The presence of gaps is unique to telomeres, since we did not detect ss DNA at other repetitive DNA loci, alphoid repeats, and ribosomal DNA by alkaline gel electrophoresis (data not shown).
The t-complex.
The t-complex contains large amounts of ssT-DNAs. Indeed, the t-complex produced strong signals in the native hybridization of 2D gels. It is not sensitive to Exo I or RecJ, suggesting that the ss T-DNAs exist as internal gaps rather than ss termini. The fact that the t-complex is the most sensitive to T7 Endo I among the T-DNA species in ALT cells also suggests the presence of many target sites of the endonuclease, such as Holliday junctions. Taking these facts together, we propose that the t-complex is a mixture of highly branched T-DNAs with large numbers of internal ss portions. One potential source of the t-complex is multiple T-DNAs undergoing HR with each other (Fig. 9). HR is initiated by the formation of DSBs, and DSBs are potentially formed when the replication fork is stalled. We have previously shown that the replication fork frequently stalls at telomeric repeats in vitro (23). However, the amount of ss T-DNAs was not markedly changed during S phase, as shown by alkaline (see Fig. S1 in the supplemental material) and 2D (data not shown) gel analyses.
ss-G.
ss-G was insensitive to Exo I and RecJ and sensitive to Exo III, BAL-31, and T7 Endo I. The diagonal signal of ss-G in the 2D gel overlapped with linear ss DNA markers. These results suggest that ss-G consists of mostly linear single-G-strand T-DNAs with ds termini. The G-rich ss regions may form non-Watson-Crick base pairs, such as the G quartet (31), thereby providing the targets for T7 Endo I. It is not clear how ss-G is sensitive to Exo III, because Exo III, a ds DNA-specific exonuclease, does not digest ss DNAs. One possibility is that small ds DNA regions are distributed throughout ss-G. Another possibility, which we favor, is that ss-G was digested by Exo III when its ss DNA regions hybridized with ss-C. The Hirt experiment demonstrated that ss-G exists as both extrachromosomal DNA and DNA closely associated with bulk chromosomes. Strand-specific diagonal signals similar to ss-G in the 2D gel have been reported in mitochondrial DNA of the yeast Candida parapsilosis (21). It is possible that a common DNA replication mechanism operates in both telomerase-independent systems, ALT and mitochondrial DNAs.
ss-C.
ss-C was insensitive to Exo I, RecJ, and Exo III; partially sensitive to T7 Endo I; and sensitive to BAL-31. A structural model consistent with these observations is circular single-C-strand T-DNA. The arc signal of ss-C did not exactly overlap with linear and circular marker ss DNAs (Fig. 3J; see Fig. S3 in the supplemental material). Slowly migrating ss-C molecules were positioned close to ds-TRFs, suggesting that such ss-C molecules are mostly linear ds DNAs. Fast-migrating ss-C molecules showed a diagonal pattern in 2D gel electrophoresis, suggesting that ss portions are dominant. Since ss-C formed a smooth arc as a whole, we suggest that it is a spectrum of DNAs composed of mostly single C strands having large fractions of ds portions as the size increases. ss-C of relatively large size was sensitive to T7 Endo I, suggesting that it contains unpaired regions in the ds portion. ss-C was detected in both Hirt Sup and Hirt Ppt DNAs. Because the Hirt Sup contained significant amounts of bulk chromosomal DNAs, it is difficult to speculate on the origin of ss-C from these results. Recently, it was reported that the nematode Caenorhabditis elegans has RecJ-sensitive C-strand ss DNAs, called the C tail (26). As described above, ss-C in human ALT cells is distinct from the C tail of the nematode.
The t-circle.
t-circles were insensitive to Exo I and RecJ and moderately sensitive to BAL-31 and T7 Endo I. The t-circle in ALT cells has been described as open circular ds DNAs (2, 30). The sensitivity of t-circles to BAL-31 supports the existence of nicks and/or gaps in the t-circles. We found that t-circle arc signals were positioned close to, but did not exactly overlap with, open circular DNA markers in 2D gels (see Fig. S3 in the supplemental material), suggesting that the t-circle may not be a simple open circular DNA species but a more complex one. t-circles were found in Hirt Sup DNA, but not in the undigested Hirt Ppt DNA, consistent with its extrachromosomal origin. These molecules were also detected in digested Hirt Ppt DNA, suggesting that a fraction of t-circle molecules are closely associated with chromosomal DNA through catenation, for example.
Implications of ALT-specific T-DNAs.
It is well established that human ALT cells contain t-circles (2, 30). It has been proposed that t-circles are formed by telomere metabolism specific to ALT cells, such as XRCC3-dependent HR of the t-loop, and that they are involved in rapid telomere-shortening events (10, 18). Now that we have identified novel T-DNAs in ALT cells, we discuss whether they are implicated in any aspects of telomere metabolism in ALT cells.
ss T-DNA and the t-circle are present not only in ALT cells, but also in telomerase-independent yeast systems (9, 21). It has been proposed that ss T-DNA is a replicating intermediate that uses t-circles as template DNAs. In analogy, ss T-DNAs in ALT cells may represent intermediate DNAs synthesized by the rolling-circle replication model (Fig. 9B). When a t-loop undergoes HR in cis, it produces a t-circle and a newly formed T-DNA end. After the 5′ end of the C strand is resected, the resultant ss G-strand end may undergo strand invasion into two T-DNA duplex targets. In the first case, the G strand invades another telomere to perform HR between telomeres. Holliday junction T-DNAs are expected to be produced as an intermediate and may represent the t-complex. In the second case, the ss G-strand end invades a t-circle and accomplishes the rolling-circle replication. Initially, the G strand is synthesized by using the circular C strand as a template. Once the G-strand synthesis occurs for a single round of the circular DNA, the G strand will be continuously replicated while displacing the preexisting G strand to form a σ-form-like structure. Subsequently, C-strand synthesis occurs to form a σ-form-like structure. The series of DNAs replicated by the rolling-circle mechanism may represent the spectrum of DNAs consisting of ds-TRFs and ss-C. Electron microscopic studies have shown that ALT cells frequently contain looped and tailed DNA molecules (2). We propose that ss-C is an intermediate DNA in the rolling-circle replication and that the t-complex is the Holliday junction intermediate in telomeric HR in trans (Fig. 9B). It is possible that ss-G could be formed as an intermediate DNA of rolling-circle replication and HR in trans.
The ssT-DNAs reported here provide a clue to elucidating the molecular mechanism of ALT. Future study is necessary to demonstrate the model provided here.
Supplementary Material
Acknowledgments
We are grateful to R. Reddel for providing cell lines and to Ishikawa laboratory members for helpful discussion. The excellent secretarial work of A. Katayama, M. Sasaki, and F. Maekawa and the technical assistance of M. Tamura are also acknowledged.
This work was supported by a Center of Excellence grant and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to F.I. and A.N.).
Footnotes
Published ahead of print on 17 November 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bechter, O. E., Y. Zou, W. Walker, W. E. Wright, and J. W. Shay. 2004. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 643444-3451. [DOI] [PubMed] [Google Scholar]
- 2.Cesare, A. J., and J. D. Griffith. 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 249948-9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, S., and S. Lavi. 1996. Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol. Cell. Biol. 162002-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 9313902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26447-450. [DOI] [PubMed] [Google Scholar]
- 6.Fasching, C. L., A. A. Neumann, A. Muntoni, T. R. Yeager, and R. R. Reddel. 2007. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 677072-7077. [DOI] [PubMed] [Google Scholar]
- 7.Friedman, K. L., and B. J. Brewer. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262613-627. [DOI] [PubMed] [Google Scholar]
- 8.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97503-514. [DOI] [PubMed] [Google Scholar]
- 9.Groff-Vindman, C., A. J. Cesare, S. Natarajan, J. D. Griffith, and M. J. McEachern. 2005. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol. Cell. Biol. 254406-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21598-610. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, W. Q., Z. H. Zhong, J. D. Henson, A. A. Neumann, A. C. Chang, and R. R. Reddel. 2005. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 252708-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 2662011-2015. [DOI] [PubMed] [Google Scholar]
- 13.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Londoño-Vallejo, J. A., H. Der-Sarkissian, L. Cazes, S. Bacchetti, and R. R. Reddel. 2004. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 642324-2327. [DOI] [PubMed] [Google Scholar]
- 15.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73347-360. [DOI] [PubMed] [Google Scholar]
- 16.McEachern, M. J., and J. E. Haber. 2006. Telomerase-independent telomere maintenance in yeast, p. 199-224. In T. de Lange, V. Lundblad, and E. H. Blackburn (ed.), Telomeres, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 163705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murnane, J. P., L. Sabatier, B. A. Marder, and W. F. Morgan. 1994. Telomere dynamics in an immortal human cell line. EMBO J. 134953-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nabetani, A., O. Yokoyama, and F. Ishikawa. 2004. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J. Biol. Chem. 27925849-25857. [DOI] [PubMed] [Google Scholar]
- 20.Neuman, A. A., and R. R. Reddel. 2006. Telomerase-independent maintenance of mammalian telomeres, p. 163-198. In T. de Lange, V. Lundblad, and E. H. Blackburn (ed.), Telomeres, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Nosek, J., A. Rycovska, A. M. Makhov, J. D. Griffith, and L. Tomaska. 2005. Amplification of telomeric arrays via rolling-circle mechanism. J. Biol. Chem. 28010840-10845. [DOI] [PubMed] [Google Scholar]
- 22.Ogino, H., K. Nakabayashi, M. Suzuki, E. Takahashi, M. Fujii, T. Suzuki, and D. Ayusawa. 1998. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem. Biophys. Res. Commun. 248223-227. [DOI] [PubMed] [Google Scholar]
- 23.Ohki, R., and F. Ishikawa. 2004. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 321627-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkinson, M. J., and D. M. Lilley. 1997. The junction-resolving enzyme T7 endonuclease I: quaternary structure and interaction with DNA. J. Mol. Biol. 270169-178. [DOI] [PubMed] [Google Scholar]
- 25.Potts, P. R., and H. Yu. 2007. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14581-590. [DOI] [PubMed] [Google Scholar]
- 26.Raices, M., R. E. Verdun, S. A. Compton, C. I. Haggblom, J. D. Griffith, A. Dillin, and J. Karlseder. 2008. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132745-757. [DOI] [PubMed] [Google Scholar]
- 27.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 198083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokutake, Y., T. Matsumoto, T. Watanabe, S. Maeda, H. Tahara, S. Sakamoto, H. Niida, M. Sugimoto, T. Ide, and Y. Furuichi. 1998. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem. Biophys. Res. Commun. 247765-772. [DOI] [PubMed] [Google Scholar]
- 29.Tsai, Y. L., S. F. Tseng, S. H. Chang, C. C. Lin, and S. C. Teng. 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 225679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119355-368. [DOI] [PubMed] [Google Scholar]
- 31.Williamson, J. R., M. K. Raghuraman, and T. R. Cech. 1989. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59871-880. [DOI] [PubMed] [Google Scholar]
- 32.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 594175-4179. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.