Abstract
Interactions between Z-disc proteins regulate muscle functions and disruption of these interactions results in muscle disorders. Mutations in Z-disc components myotilin, ZASP/Cypher, and FATZ-2 (calsarcin-1/myozenin-2) are associated with myopathies. We report here that the myotilin and the FATZ (calsarcin/myozenin) families share high homology at their final C-terminal five amino acids. This C-terminal E[ST][DE][DE]L motif is present almost exclusively in these families and is evolutionary conserved. We show by in vitro and in vivo studies that proteins from the myotilin and FATZ (calsarcin/myozenin) families interact via this novel type of class III PDZ binding motif with the PDZ domains of ZASP/Cypher and other Enigma family members: ALP, CLP-36, and RIL. We show that the interactions can be modulated by phosphorylation. Calmodulin-dependent kinase II phosphorylates the C terminus of FATZ-3 (calsarcin-3/myozenin-3) and myotilin, whereas PKA phosphorylates that of FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-1). This is the first report of a binding motif common to both the myotilin and the FATZ (calsarcin/myozenin) families that is specific for interactions with Enigma family members.
The sarcomere of striated muscle consists of strictly organized subunits, myosin-containing thick filaments and actin-containing thin filaments. The thin filaments are aligned and cross-linked at the Z-discs by a molecular complex in which α-actinin is one of the core structures. Since the contractile force is transduced via the Z-discs, this structure has special requirements. It must provide extensive stability and yet undergo modulation in response to external signals. The Z-discs also serve as important sensors of extracellular cues and mediators of cellular signals that result in various adaptive responses (37). Muscle cells are able to sense changes in their workload and adapt accordingly via complex signaling pathways, some involving calcium, since its level in muscle cells alters in response to nerve pulses and muscle contraction. Of special importance is calcineurin, a sarcomeric calcium/calmodulin-dependent phosphatase that can act as a sensor of change. It is involved in the regulation of genes affecting muscle differentiation and fiber-type specification (12, 13).
The special role of the Z-discs is indicated by the fact that mutations in several Z-disc proteins can result in neuromuscular disorders and cardiomyopathies. For instance, myofibrillar myopathy (desmin-related myopathy), a disease characterized by sarcomere disintegration and accumulation of thin filament material, is caused by dominantly inherited missense mutations in Z-disc proteins: myotilin, filamin-C, and Z-band alternatively spliced PDZ motif-containing protein (ZASP, also named LIM domain-binding factor 3, Cypher, or Oracle) (42, 43, 52). Missense mutations in myotilin can also result in limb-girdle muscular dystrophy 1A and spheroid body myositis (10, 18), while mutations in ZASP/Cypher (8, 57), myopalladin or FATZ-2 (calsarcin-1/myozenin-2) have been found to be associated with dominant familial dilated (7, 50) or hypertrophic cardiomyopathy (33). ZASP/Cypher knockout mice display a severe form of congenital myopathy and die postnatally (58), whereas myotilin knockout mice are virtually normal (31), suggesting redundancy between the myotilin family members and indicating that dysfunctional myotilin is more harmful to muscle cells than loss of the protein.
Myotilin (40), palladin (32, 34), and myopalladin (3) are homologous Z-disc proteins that form a novel family of immunoglobulin-domain-containing actin-binding proteins. Biochemical studies on the best-characterized family member, myotilin, have demonstrated an association with important components of the sarcomere: α-actinin (40), which is a core structural component of the Z-disc; filamins (15, 49); the proteins of the FATZ (calsarcin/myozenin) family (15); and actin (51). Myotilin is linked to signaling networks by binding to the ubiquitin ligases Murf-1 and Murf-2 (54) and indirectly via FATZ (calsarcin/myozenin). Experiments using myotilin fragments with dominant-negative effect have shown its critical involvement in sarcomere organization. Myotilin bundles and stabilizes actin effectively, which suggests a role for myotilin in the organization and maintenance of Z-disc integrity.
The FATZ (calsarcin/myozenin) proteins form another Z-disc family with structural and signaling functions. The three homologous members—FATZ-1 (calsarcin-2/myozenin-1), FATZ-2 (calsarcin-1/myozenin-2), and FATZ-3 (calsarcin-3/myozenin-3)—are localized in the Z-disc binding not only to myotilin but also to filamins A, B, and C (15), telethonin (T-cap), α-actinin, ZASP/Cypher, and calcineurin (9, 11, 12, 47). The three FATZ (calsarcin/myozenin) proteins share high homology at their N and the C terminals and, in fact, the binding sites for a variety of proteins occur in these regions. It has been suggested that the FATZ (calsarcin/myozenin) family may play a role in contributing to the formation and maintenance of the Z-disc, as well as in cell signaling, since its members bind calcineurin. FATZ-1 (calsarcin-2/myozenin-1) and FATZ-3 (calsarcin-3/myozenin-3) are highly expressed in skeletal muscle fast-twitch fibers, whereas FATZ-2 (calsarcin-1/myozenin-2) is highly expressed in cardiac muscle slow-twitch fibers. Mice lacking FATZ-2 (calsarcin-1/myozenin-2) showed an increase in the level of calcineurin, as well as a concurrent increase in the percentage of slow-twitch fibers (13). A recent report shows that FATZ-1 (calsarcin-2/myozenin-1) knockout mice have reduced body weight and fast-twitch muscle mass without exhibiting muscle atrophy (14). It is noteworthy that they also have the ability to run longer distances than control mice, thus exhibiting endurance to exercise. In fact, thus far only actinin-3 knockout mice have displayed this phenotype of endurance to exercise. FATZ-1 (calsarcin-2/myozenin-1)-deficient mice show an increase in oxidative muscle fibers and a switch from fast-twitch to slow-twitch fibers due to an increase in NFAT activity, as well as the regulator of calcineurin 1-4 (RCAN1-4), resulting in the concomitant increase in calcineurin signaling. Both FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2) can regulate calcineurin/NFAT activity, thus influencing the fiber type composition of skeletal muscle (14).
ZASP/Cypher (Oracle) (8, 35, 57) belongs to the Enigma family of proteins, the members of which all have a N-terminal PDZ domain and one or more LIM domains at the C-terminal (48). To date, there are six isoforms of ZASP/Cypher, all of which contain an N-terminal PDZ domain and none or three C-terminal LIM domains. ZASP/Cypher also contains a third domain known as the ZM motif which can also be found in ALP and CLP-36 (23, 24). It interacts with two different regions of α-actinin-2; its PDZ domain binds to the C-terminal EH-hand region of α-actinin-2, whereas its ZM motif binds to the rod region of α-actinin-2 (2, 23). ZASP/Cypher colocalizes with α-actinin-2 in the Z-disc, whereas the LIM domains interact with and are phosphorylated by all six isoforms of protein kinase C (PKC α, β1, γ, ζ, δ, and ɛ). ZASP/Cypher is important for the stability of the Z-disc; in fact, ZASP/Cypher knockout mice die in the first 24 h after birth as a result of functional failure of striated muscles caused by disruption of the Z-disc during muscle contraction (58). The PDZ of ZASP/Cypher is a classical type I PDZ domain that binds to the C-terminal of α-actinin-2.
To better understand the biology of the Z-disc and pathogenesis of muscle disorders, it is important to unravel the dynamic interplay of Z-disc components. In the present study, we demonstrate a novel PDZ domain-binding motif common to the myotilin and FATZ (calsarcin/myozenin) protein families. This domain mediates interaction with ZASP/Cypher in a phosphorylation-dependent manner and is also involved in targeting ZASP/Cypher.
MATERIALS AND METHODS
Plasmids.
Full-length myotilin (residues 1 to 498) and its variants encoding amino acids (aa) 1 to 270, 347 to 498, 441 to 498, and 214 to 250 or palladin (aa 715 to 772) were PCR amplified from human myotilin or palladin cDNA with the appropriate primer and subcloned into pAHP (38, 39, 41) for the expression of hemagglutinin (HA)-tagged proteins in eukaryotic cells or in vitro translation, into pEGFP-C2 (Clontech) for the expression of green fluorescent protein (GFP)-tagged proteins in eukaryotic cells, and into pGEX-4T1 (Pharmacia) glutathione S-transferase (GST) fusion vector for protein production in bacterial cells. Myotilin encoding aa 1 to 498 was cloned into the eukaryotic GST expression vector pDEST 27 vector (Gateway System; Invitrogen). The PDZ domain regions of human ZASP (1 to 255 bp), ALP (1 to 246 bp), and CLP-36 (1 to 255 bp) were amplified by PCR from their respective full-length cDNAs and subcloned into pQE30 (Qiagen) and a modified version of pGEX-6-P3 (GE Healthcare) for the expression of His and GST-tagged proteins, respectively. The full-length proteins of FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2) were cloned as previously described (9). FATZ-3 (calsarcin-3/myozenin-3) (1 to 756 bp), the C-terminal regions of FATZ-1 (calsarcin-2/myozenin-1) (730 to 897 bp), FATZ-2 (calsarcin-1/myozenin-2) (625 to 792 bp), and FATZ-3 (calsarcin-3/myozenin-3) (586 to 753 bp) were amplified by reverse transcription-PCR from human muscle mRNA (Clontech) and then cloned into pQE30 and the modified pGEX-6-P3. The site-directed mutations were generated by using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The authenticity of the constructs was verified by sequencing.
Yeast two-hybrid analysis.
Yeast transformation, mating, and detection of protein interactions by determining the β-galactosidase activity were as previously described (16, 39). The bait, myotilin cDNA encoding aa 102 to 498 in pGBKT7 vector, was used in screening a human skeletal muscle library in pACT2 (Clontech). Positive clones were sequenced.
Protein purification and in vitro binding assay.
The pGEX expression plasmids containing cDNAs for α-actinin-2, myotilin, palladin, FATZ-1 (calsarcin-2/myozenin-1), FATZ-2 (calsarcin-1/myozenin-2), and FATZ-3 (calsarcin-3/myozenin-3) with or without the last 15 bp were transformed in Escherichia coli BL21(pLysS) cells. The pQE expression plasmids containing the cDNAs for the PDZ region of ZASP/Cypher, ALP, and CLP-36 were transformed in E. coli M15 cells. Protein expression was induced with 0.3 to 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 to 4 h at room temperature, and GST- and His-tagged proteins were purified according to the manufacturer's protocol for native proteins by using glutathione-Sepharose beads (GE Healthcare) and Ni-NTA resin (Qiagen), respectively. For the in vitro binding assay, the pCMV-myc-FATZ-3 (calsarcin-3/myozenin-3) or pAHP-myotilin or palladin plasmids were used as templates in a T3- or a T7-coupled rabbit reticulocyte transcription-translation system (Promega). Approximately 4 μg of GST fusion proteins on glutathione beads was incubated with 15 μl of in vitro-translated (IVT) 35S-labeled protein in 10 mM Tris-HCl (pH 7.5), 5 mM EDTA, 130 mM KCl, and 0.05% Tween 20. After the washes, the bound material was eluted by boiling in Laemmli buffer, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detected by autoradiography.
Cell culture, transfection, antibodies, and coprecipitation.
Rat cardiomyocytes were isolated by using the neonatal cardiomyocyte isolation system (Worthington Biochemical Corp., Lakewood, NJ) according to the manufacturer's instructions, except that the trituration step was done twice. Cardiomyocytes were transfected with GST-myotilin cDNA in pDEST 27 or with the pDEST 27 vector encoding mammalian GST alone, using Fugene HD (10 μg of DNA/10-cm plate, two plates for one sample). After 2 days, the cells were lysed into ELB (150 mM NaCl, 50 mM HEPES, 5 mM EDTA [pH 7.4])-0.5% NP-40 buffer supplemented with protease and phosphatase inhibitors. The lysates were briefly centrifuged (4,000 × g, 5 min), and glutathione beads were added to the supernatant, followed by incubation with mixing overnight at 4°C. The beads were then washed three times with lysis buffer, and the bound proteins were separated by SDS-PAGE, blotted, and detected with polyclonal anti-ZASP antibody.
Rat cardiomyocytes and COS7 (ATCC) cells were cultured, transfected, and fixed with 3.5% (vol/vol) paraformaldehyde as described previously (38, 41). Mouse muscle cryosections were prepared as described previously (31). Primary cardiomyocytes and cryosections were stained with a polyclonal antibody against myotilin residues 1 to 151 (29) and with a polyclonal rat antibody against ZASP (G. Faulkner, unpublished results). Transfected cells were stained with mouse anti-HA antibody recognizing the HA tag (Roche) and α-actinin-2 antibody (Sigma) to visualize the Z-discs. Alexa 488-, 568-, and 594-conjugated goat anti-mouse and goat anti-rabbit antibodies (Invitrogen; Molecular Probes) were used as secondary antibodies. Coverslips were mounted in DABCO (Sigma) and Mowiol (Calbiochem) and examined by using immunofluorescence (Zeiss Axiophot equipped with AxioCam cooled charge-coupled device camera).
Bioinformatics.
The program to extract from any database proteins with the last 5 aa having the motif E[ST][DE][DE]L was written by one of us (G. Valle). The last 8 aa were considered but only the terminal 5 aa were given the following weightings: position 0, L = 2; position 1, E or D = 1; position 2, E or D = 1; position 3, S or T = 1; and position 4, E = 1. A score of 6 was assigned when all of the criteria were met. This program was used to check the UniProt Knowledgebase release 11.3 (UniProtKB/Swiss-Prot release 53.3 and UniProtKB/TrEMBL release 36.3).
Peptides.
The peptides used for the present study were synthesized by the ICGEB peptide synthesis service using a Gilson AspecXL SPE robot. The linker made up of two gamma aminobutyric acid units was 12.3 Å in length. The following peptides were used: biotin-GABA-GABA-EpSEEE, -ESEEE, -EpSEEL, -ESEEL, -EpTEEL, -ETEEL, -EpSEDL,-ESEDL, -EpSDEL, and -ESDEL. For competition experiments the same peptides without biotin were used.
AlphaScreen.
AlphaScreen (Perkin-Elmer) is an amplified luminescent proximity homogeneous assay. Donor beads contain a photosensitizer, phthalocyanine, which converts ambient to singlet oxygen (an excited form) when illuminated at 680 nm. The singlet oxygen has a limited half-life (4 μs) prior to returning to ground state and can diffuse approximately 200 nm in that time. If an acceptor bead is within that distance, energy is transferred from the singlet oxygen to thioxene derivatives on the acceptor beads, initiating a cascade of events that results in the production of light at 520 to 620 nm. Both the GST and the His detection kits for AlphaScreen were used according to the manufacturer's protocol. The acceptor and donor beads were used at a concentration of 6.5 pM. The protein to be tested was added to the wells and then the acceptor beads. The following steps were done in the dark. The plate was incubated for 30 min at room temperature before the donor beads were added and then for a further 3 h, after which it was kept for 15 min at 28°C to equilibrate the temperature. The signal was read at 28°C using a Fusion AlphaMultilabel reader (Perkin-Elmer) at 300-ms excitation and 700-ms emission. When testing a protein for binding, it is necessary to titrate it against its partner to establish the concentration of both proteins, resulting in a significant value for the signal-to-noise ratio (S/N). The S/N normally used is in the range of 8 to 50. In every experiment, negative controls without one or both proteins were used to give the noise (background) level, and biotinylated GST (0.5 nM) or biotinylated His (1 nM) were used to as internal controls to normalize the signal. The experiments were repeated at least three times. In the competition experiments, the binding proteins were first added to the wells at a fixed concentration that would result in binding in the absence of a competitor, and then the protein used as competitor was added. The results were plotted as the ratio of the signal in the presence of the competitor divided by that of the signal in the absence of the competitor. These experiments were repeated at least three times, and the means and standard deviations from the mean were plotted. The ratio was calculated as the mean of the normalized signal divided by the mean of the normalized noise: R = Sm/Nm.
The confidence of the ratio was calculated from the standard deviation of the signal and the noise:
 |
where Sm is the normalized signal mean; δSm is the mean standard deviation of the normalized signal, Nm is the normalized noise mean, and δNm is the mean standard deviation of the normalized noise.
TranSignal PDZ array domains.
The PDZ array membranes (Panomics) were used according to the protocols in the manufacturer's handbook. The biotinylated peptides or His-tagged purified protein were used as ligands at 0.3 and 15 μg/ml, respectively. Detection was done by using chemiluminescence.
Phosphorylation experiments.
A total of 4 μg of GST fusion protein was used for each reaction. For phosphorylation studies, rat skeletal muscle extract was prepared from soleus and gastrocnemius muscle. The rat muscle was cut into 1- to 2-mm pieces, frozen in liquid nitrogen for 2 h, and maintained at −80°C until needed. calmodulin-dependent kinase II (CaMKII [Upstate Biotechnology], catalog no. 14-217, active, purified from rat forebrain) was used in phosphorylation assays as described by the manufacturer; the only modification was that analysis was performed by SDS-PAGE instead of using a scintillation counter. PKA phosphorylation was performed as described by Alfthan et al. (1). The C-terminal of GST-FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2), either wild-type or mutated S267A in FATZ-1 (calsarcin-2/myozenin-1), and S231A in FATZ-2 (calsarcin-1/myozenin-2) were expressed in E. coli and purified on glutathione beads, and equal amounts of each GST fusion protein were phosphorylated with PKA (1). Samples were analyzed by using SDS-PAGE, autoradiography, and ImageQuant software. The point mutations were made by substituting serine with alanine at sites based on the phosphorylation predictions for PKA. The mutations were generated by using a QuikChange site-directed mutagenesis kit according to the manufacturer's instructions. All constructs were verified by sequencing.
RESULTS
Identification of ZASP/Cypher as a new binding partner for myotilin.
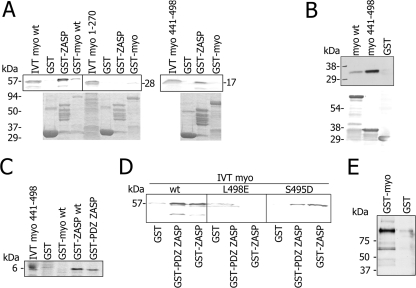
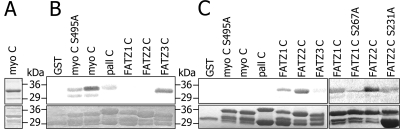
To search for novel binding partners to myotilin, we screened a human striated muscle library with the yeast two-hybrid method using a fragment of myotilin (aa 102 to 498) as bait. One of the positive clones encoded the N terminus of ZASP/Cypher (aa 1 to 169). A glutathione-bead pull-down assay with recombinant proteins was performed to verify the yeast two-hybrid result. Recombinant GST, GST-tagged ZASP-1, or GST-tagged myotilin were bound to beads and allowed to interact with 35S-labeled in-vitro translated myotilin. The bound proteins were detected by autoradiography (Fig. 1A). Wild-type myotilin bound GST-ZASP and GST-myotilin; the latter finding is in agreement with our previous results (41). The C terminus of myotilin (aa 441 to 498) was able to bind ZASP/Cypher, whereas the N terminus of myotilin (aa 1 to 270) could not bind either myotilin or ZASP/Cypher.
FIG. 1.
Myotilin binds the ZASP PDZ domain via its C terminus. (A to D) Affinity precipitation assays. In panel A, labeled IVT myotilin or its N-terminal (aa 1 to 270) or C-terminal (aa 441 to 498) fragments were allowed to bind to glutathione beads coated with recombinant GST-ZASP or GST. The top panels show autoradiographs of bound proteins (on the left is shown input IVT product), and the bottom panel shows Coomassie blue staining of the GST proteins. Full-length (aa 1 to 498) and C-terminal (aa 441 to 498) but not N-terminal (aa 1 to 270) myotilin binds to GST-ZASP (aa 1 to 283). As a positive control, the dimerization ability of wild-type myotilin is shown. In panel B, rat cardiomyocyte lysate was allowed to bind GST-myotilin, GST-C-terminal myotilin (aa 441 to 498), and GST. Bound ZASP/Cypher was detected with antibody. Endogenous ZASP/Cypher from the lysate bound to the GST-myotilin and GST-C-terminal myotilin (aa 441 to 498) but not GST. ZASP/Cypher binds to myotilin full-length (aa 1 to 498) as well as to its C-terminal (aa 441 to 498). Bottom panel as in panel A. In panel C, labeled IVT C-terminal myotilin (aa 441 to 498) was allowed to bind beads coated with full-length ZASP or the PDZ domain (aa 1 to 85) of ZASP. In panel D, IVT produced wild-type myotilin or myotilin with myotilin L498E or S495D substitutions was allowed to bind ZASP or ZASP PDZ domain. L498 is crucial for the interaction, since the mutation L498E eliminates binding. S495D modification does not affect binding. (E) In vivo interaction between myotilin and ZASP. Rat cardiomyocytes were transfected with GST or GST-myotilin in eukaryotic expression vectors Glutathione resin was used to pull down proteins from cardiomyocyte lysates. Bead-bound proteins were separated by SDS-PAGE, blotted, and probed using anti-ZASP polyclonal antibody. Cardiomyocyte ZASP precipitates with GST-myotilin but does not bind GST. wt, wild type.
In order to investigate whether binding between myotilin and ZASP/Cypher could occur in cardiomyocytes, an affinity precipitation assay was performed using a rat cardiomyocyte lysate. GST-myotilin, GST-C-terminal myotilin (aa 441 to 498), and GST were adsorbed to glutathione-Sepharose beads. Endogenous ZASP/Cypher from the lysate bound to the GST-myotilin and GST-C-terminal myotilin (aa 441 to 498) and was detected by SDS-PAGE and immunoblotting with a polyclonal rat antibody against ZASP/Cypher (Fig. 1B). ZASP/Cypher was able to bind both full-length and C-terminal myotilin but not GST.
We further mapped by affinity precipitation the regions mediating this interaction by using shorter protein fragments and residue substitutions (Fig. 1C). In these assays, the PDZ domain of ZASP/Cypher (aa 1 to 85) was shown to be sufficient for binding the C terminus of myotilin (aa 441 to 498). PDZ domains typically bind to motifs located at the extreme C terminus (44) and containing a crucial C-terminal leucine. The C-terminal residue of myotilin is a leucine and thus part of a potential PDZ binding motif. When the leucine in myotilin was replaced by glutamic acid, binding was lost both to the full-length ZASP/Cypher and the ZASP/Cypher PDZ domain (Fig. 1D). Instead, S495D substitution did not prevent binding. Hence, L498 in myotilin is needed for in vitro binding to both the full-length and the PDZ-domain of ZASP/Cypher.
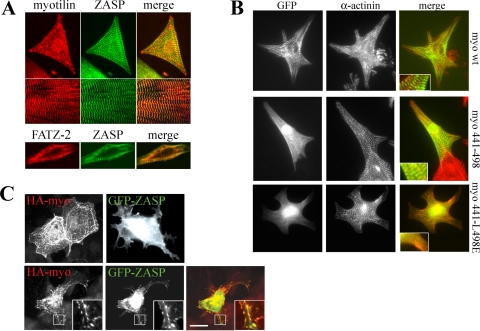
In order to show that the interaction between ZASP/Cypher and myotilin also occurs in cells, rat cardiomyocytes were transfected with pDEST27 eukaryotic vectors expressing GST-myotilin (aa 1 to 498) and GST alone. As can be seen in Fig. 1E, endogenous ZASP/Cypher was able to bind GST-myotilin in vivo, whereas it did not bind GST. To compare the distribution of myotilin and ZASP/Cypher in relevant cell types, we stained sections of mouse striated muscle and primary rat cardiomyocytes (Fig. 2A). The staining showed specific colocalization at the Z-disc of the endogenous myotilin and ZASP/Cypher in isolated cardiomyocytes and at the Z-disc of skeletal muscle tissue (Fig. 2A). In further experiments, primary rat cardiomyocytes and COS-7 cells were transfected with plasmids expressing myotilin and ZASP/Cypher fragments. In cardiomyocytes, both myotilin and its C-terminal (aa 441 to 498) localized exclusively to the Z-discs, as seen by colocalization with the staining pattern of the α-actinin antibody. However, a L498E substitution markedly reduced the Z-disc targeting of the C-terminal of myotilin (Fig. 2B). The result further indicates that the C-terminal sequence of myotilin is important for binding to ZASP/Cypher or some other Z-disc proteins.
FIG. 2.
Myotilin and ZASP/Cypher colocalize in muscle cells, and myotilin is involved in targeting ZASP/Cypher. (A) Rat neonatal cardiomyocytes (upper panel) and sections of skeletal muscle (middle panel) were stained myotilin (left, red) and ZASP/Cypher (middle, green). The bottom panel shows staining of cardiomyocytes for FATZ-2 (left, red) and ZASP/Cypher (middle, green). On the right, merged images are shown. (B) GFP-myotilin, GFP-myotilin 441-498, and GFP-myotilin 441-L498E were transfected to rat neonatal cardiomyocytes and stained for α-actinin as a Z-disc marker. Wild-type and C-terminal myotilin is targeted to the Z-discs in the sarcomere, whereas the C terminus with L498E substitution is not. The insets show larger magnifications of the merged images. (C) HA-myotilin and GFP-ZASP were transfected to COS-7 cells either (upper panel) or together (lower panel). Myotilin was detected with anti-HA antibody and Alexa-594-coupled secondary antibody (red). Single transfected myotilin induces thick filaments, whereas ZASP/Cypher is uniformly distributed in the cytoplasm. In double-transfected cells, ZASP/Cypher is targeted to the filaments, where it colocalizes with myotilin. wt, wild type. Scale bar, 5 μm.
In transfected COS-7 cells, myotilin induced formation of thick actin filament bundles as previously reported (41) (Fig. 2C). Instead, transfected ZASP/Cypher localized more diffusely in the cytoplasm. However, when the two proteins were transfected together, ZASP relocalized to the thick actin bundles and showed colocalization with myotilin (Fig. 2C). Taken together, the results indicate that the C terminus of myotilin binds the PDZ-domain of ZASP/Cypher via a targeting sequence and that myotilin plays a role in the subcellular distribution of ZASP/Cypher.
The conserved E[ST][DE][DE]L motif is shared by the FATZ (calsarcin/myozenin) and myotilin families.
We noted that the C-terminal 5 aa of FATZ-1 (calsarcin-2/myozenin-1; ETEEL), FATZ-2 (calsarcin-1/myozenin-2; ESEDL), FATZ-3 (calsarcin-3/myozenin-3; ESEEL), myotilin (ESEEL), myopalladin (ESDEL), and palladin (ESEDL) share high similarity (Fig. 3A). Notably those of FATZ-3 (calsarcin-3/myozenin-3) and myotilin are identical (ESEEL) as are those of FATZ-2 (calsarcin-1/myozenin-2) and palladin (ESEDL). This high similarity raised the question of whether these proteins could interact via their C termini with the PDZ domain of ZASP/Cypher or the PDZ domains of other proteins. In line with this hypothesis, FATZ-2 and ZASP/Cypher demonstrated in cardiomyotes an identical colocalization pattern, as seen with myotilin and ZASP/Cypher (Fig. 2A).
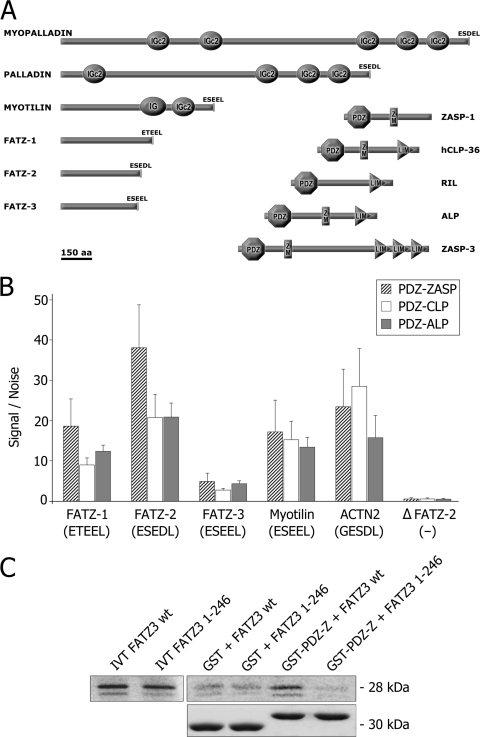
FIG. 3.
The FATZ (calsarcin/myozenin) and myotilin families interact with muscle-specific PDZ-domains. (A) A schematic diagram showing myotilin, FATZ, and Enigma family proteins drawn to scale. ZASP-1 (aa 1 to 283), hCLP-36 (aa 1 to 329), RIL (aa 1 to 330), ALP (aa 1 to 364), and ZASP-3 (aa 1 to 617) all have N-terminal PDZ domains (approximately aa 11 to 84) and C-terminal LIM domains, with the exception of the ZASP-1 isoform, which has no LIM domains. RIL has no ZASP-like motif (ZM). The myotilin family is composed of myotilin (aa 1 to 498), myopalladin (aa 1 to 1,320), and palladin (aa 1 to 1,001), and the FATZ family is composed of FATZ-1 (aa 1 to 299), FATZ-2 (aa 1 to 264), and FATZ-3 (aa 1 to 251). The FATZ family has no specific domains, whereas the myotilin family has immunoglobulinlike domains, IG and IGc2. (B) Interactions between His-PDZ domain proteins (ZASP/Cypher, CLP-36, and ALP) and GST-tagged proteins (FATZ-1, FATZ-2, FATZ-3, myotilin, and actinin-2) were measured by using the AlphaScreen technique. PDZ proteins and their ligands were used at 50 nM. The results shown are the means of nine different experiments, and the standard deviations of the means are plotted at the top of each column. (C) A GST pull-down experiment was performed as in Fig. 1. IVT 35S-labeled FATZ-3 (calsarcin-3/myozenin-3) protein binds to the PDZ domain of ZASP (GST-PDZ-Z), whereas the C-terminal truncated FATZ-3 (calsarcin-3/myozenin-3) protein (1 to 246 aa) lacking 5 aa does not. PDZ-ZASP constructs (PDZ-Z) contains the 85 N-terminal residues. Neither FATZ construct bound strongly to GST alone. The first two lanes show the input (50%) of the IVT proteins. The bottom row presents lanes from an SDS-PAGE gel stained with Coomassie blue showing amount of GST proteins used in experiments. wt, wild type.
Our findings raised the hypothesis that the C-terminal final 5 aa in myotilin/FATZ families would represent a new binding motif involved in protein interactions. To investigate, whether this putative binding motif was shared by other proteins, a program was written to extract from databases proteins with the final 5 aa of their C-terminal having the motif E[ST][DE][DE]L. This program was used to check the UniProt Knowledgebase release 11.3. Although this E[ST][DE][DE]L motif could be found in some bacteria, in vertebrates it was restricted to the proteins of the FATZ (calsarcin/myozenin) and myotilin families, with the exception of histidine ammonia lyase in which the final C-terminal amino acids (ESEDL) are identical to those of FATZ-2 (calsarcin-1/myozenin-2) and palladin. The data reported in Table 1 show only the proteins with an exact match at their C termini for the motif E[ST][DE][DE]L. Synonymous proteins were removed for clarity. It is notable that there are very few proteins that have exact matches at their C termini for the motif: these are FATZ-1 (myozenin-1/calsarcin-2), FATZ-2 (myozenin-2/calsarcin-1), FATZ-3 (myozenin-3/calsarcin-3), myotilin, myopalladin, palladin, and histidine ammonia lyase. All of these proteins, except histidine ammonia lyase, are localized in the Z-disc of striated muscle and have a role in muscle or cytoskeletal structure and/or function.
TABLE 1.
The E[ST][DE][DE]L motif in the FATZ (calsarcin/myozenin) and myotilin families is conserved in vertebrates
| C-terminal end | Accession no. | Matching proteina |
|---|---|---|
| LDGETEEL | Q9NP98 | MYOZ1-HUMAN, FATZ-1 (calsarcin-2/myozenin-1) |
| LDGETEEL | Q1AG03 | Q1AG03-CANFA, calsarcin-2, Canis familiaris |
| LDGETEEL | Q8SQ24 | MYOZ1-BOVIN, myozenin-1, Bos taurus |
| LDGETEEL | Q4PS85 | MYOZ1-PIG, myozenin-1, Sus scrofa |
| LDGETEEL | Q1AG02 | Q1AG02-RABIT, calsarcin-2, Oryctolagus cuniculus |
| LDGETEEL | Q9JK37 | MYOZ1-MOUSE, myozenin-1, Mus musculus |
| VDGETEEL | Q6DIU0 | Q6DIU0-XENTR, myozenin-1, Xenopus tropicalis |
| MDGETEEL | Q7SYY0 | Q7SYY0-XENLA, Myoz1-prov, Xenopus laevis |
| FDGETDDL | Q6DHF0 | Q6DHF0-DANRE, Zgc:92347, Danio rerio |
| SSEETDDL | Q1JQ62 | Q1JQ62-DANRE, Si-Danio rerio (H-L-H protein) |
| FDGETDEL | Q4SQM4 | Q4SQM4-TETNG, chromosome 17 Calsarcin rel. |
| TVPESEDL | Q9NPC6 | MYOZ2-HUMAN, FATZ-2 (calsarcin-1/myozenin-2) |
| TVPESEDL | Q5R6I2 | MYOZ2-PONPY, myozenin-2, Pongo pygmaeus |
| TIPESEDL | Q5E9V3 | MYOZ2-BOVIN, myozenin-2, Bos taurus |
| TIPESDDL | Q1AG08 | Q1AG08-PIG, calsarcin-1, Sus scrofa |
| TVPESDDL | Q9JJW5 | MYOZ2-MOUSE, myozenin-2, Mus musculus |
| EIPESDDL | Q8AVF9 | Q8AVF9-XENLA, Myoz2-prov protein, Xenopus laevis |
| EIPESDDL | Q5I0T4 | Q5I0T4-XENTR, myozenin-2, Xenopus tropicalis |
| FIPESDDL | Q6P2T2 | Q6P2T2-DANRE, myozenin-2, Danio rerio |
| ALVESEDL | Q7L3E0 | Q7L3E0-HUMAN, palladin protein, Homo sapiens |
| ALVESEDL | Q4R5Y9 | Q4R5Y9-MACFA, testis cDNA, clone |
| GLVESEDL | Q3MHW8 | Q3MHW8-BOVIN, Palld protein, Bos taurus |
| GLVESEDL | Q6DFX7 | Q6DFX7-MOUSE, Palld protein, Mus musculus |
| GLVESDDL | Q4RKT9 | Q4RKT9-TETNG, chromosome 1, SCAF15025 |
| KIPESEDL | Q4VB93 | Q4VB93-HUMAN, histidine ammonia-lyase, Homo sapiens |
| TIPESDDL | P35492 | HUTH-MOUSE, histidine ammonia-lyase, Mus musculus |
| TIPESDDL | P21213 | HUTH-RAT, histidine ammonia-lyase, Rattus norvegicus |
| TIPESDDL | Q76N86 | Q76N86-RAT, histidase, Rattus norvegicus |
| SVVESDEL | Q86TC9 | MYPN-HUMAN, myopalladin, Homo sapiens |
| SVVESDEL | Q5DTJ9 | MYPN-MOUSE, myopalladin, Mus musculus |
| SVVESDEL | Q86TC92 | MYPN-HUMAN, isoform 2 of Q86TC9, Homo sapiens |
| NLPESEEL | Q8TDC0 | MYOZ3-HUMAN, FATZ-3 (calsarcin-3/myozenin-3) |
| NLPESEEL | Q8TDC02 | MYOZ3-HUMAN, isoform 2 of Q8TDC0, Homo sapiens |
| NLPESEEL | Q08DI7 | Q08DI7-BOVIN, myozenin-3, Bos taurus |
| NLPESEEL | Q1AG05 | Q1AG05-PIG, calsarcin-3, Sus scrofa |
| KLPESEEL | Q8R4E4 | MYOZ3-MOUSE, myozenin-3, Mus musculus |
| KLPESEEL | Q9Z3272 | SYNPO-RAT, isoform 2 of Q9Z327, Rattus norvegicus |
| GLYESEEL | Q9UBF9 | MYOTI-HUMAN, myotilin, Homo sapiens |
| GLYESEEL | Q0VCX9 | Q0VCX9-BOVIN, similar to titin immunoglobulin domain |
| GLYESEEL | Q9JIF9 | MYOTI-MOUSE, myotilin, Mus musculus |
That is, vertebrate proteins with an exact match for the C-terminal motif E[ST][DE][DE]L.
The FATZ (calsarcin/myozenin) family of proteins is known to bind ZASP/Cypher, but these interactions have not been mapped either on ZASP/Cypher or on the FATZ (calsarcin/myozenin) family members (12). Since here we show that myotilin binds to ZASP/Cypher through its C terminus, we speculated that the final 5 aa of the C-terminal of all myotilin and FATZ (calsarcin/myozenin) family members could have a role in the interaction with ZASP/Cypher. This hypothesis was further strengthened by information obtained by means of the ELM program (36), a resource for predicting functional sites in eukaryotic proteins, which predicted that the terminal 4 aa of myotilin, myopalladin, palladin, and the FATZ (calsarcin/myozenin) family constituted a binding motif for class III PDZ domain proteins. This binding motif is X[DE]X[IVL], where X is any amino acid.
Characterization of the interaction between the FATZ (calsarcin/myozenin) and myotilin families and muscle-specific PDZ domains.
In addition to ZASP/Cypher, Z-disc proteins ALP and CLP-36 are also members of the Enigma family of PDZ proteins. Enigma family members ZASP-1 (aa 1 to 283), hCLP-36 (aa 1 to 329), RIL (aa 1 to 330), ALP (aa 1 to 364), and ZASP-3 (aa 1 to 617) all have N-terminal PDZ domains (approximately aa 11 to 84) and C-terminal LIM domains, with the exception of the ZASP-1 isoform, which has no LIM domains (Fig. 3A). RIL has no ZASP-like motif (i.e., ZM).
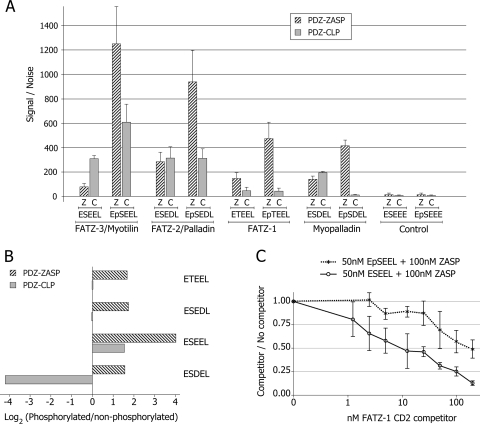
We decided to determine whether the interactions detected with ZASP/Cypher could apply in a more general way to other Enigma family members; we therefore used the AlphaScreen method (as described in Materials and Methods) as a quantitative method for measuring binding between the PDZ domains of members of the Enigma family and proteins of the FATZ (calsarcin/myozenin) family and myotilin. The results of these experiments (Fig. 3B) showed that the FATZ (calsarcin/myozenin) family, myotilin, and α-actinin-2 behave as ligands for the PDZ domains of Enigma family members ZASP/Cypher, CLP-36, and ALP. The AlphaScreen binding experiments were done using both full-length and truncated (lacking the C-terminal 5 aa's) protein; the His-PDZ domain proteins were bound to the donor beads, and the GST-protein ligands were bound to the acceptor beads. Since the native proteins used in these experiments were expressed in bacteria, they were nonphosphorylated. The PDZ domain of ZASP/Cypher showed much stronger binding to the ligand proteins than the PDZ domains of ALP or CLP-36, with the exception of α-actinin2, which shoed stronger binding to the PDZ domain of CLP-36. The best ligand protein for overall binding was FATZ-2 (calsarcin-1/myozenin-2), whereas FATZ-3 (calsarcin-3/myozenin-3) bound poorly to the three PDZ domains studied. The C-terminal truncated forms of each of the proteins were produced and tested, but only the result for the truncated FATZ-2 (calsarcin-1/myozenin-2) protein is shown in the interests of clarity (Fig. 3B). The AlphaScreen result that the FATZ-3 (calsarcin-3/myozenin-3) truncated protein was unable to bind the PDZ domain of ZASP was confirmed by a GST pull-down experiment (Fig. 3C) in which IVT 35S-labeled FATZ-3 (calsarcin-3/myozenin-3) full-length (aa 1 to 251) and truncated (aa 1 to 246) proteins were used. As expected, the full-length FATZ-3 (calsarcin/myozenin-3) but not the truncated FATZ-3 (calsarcin-3/myozenin-3) pulled down the GST PDZ-ZASP (Fig. 3C). Neither protein bound strongly to GST alone. The lower panel shows the amounts of GST proteins used in the experiment.
The importance of the final 5 aa of the C-terminal of the ligand proteins was shown both by the AlphaScreen method (Fig. 3B) and by the GST pull-down experiments (Fig. 3C) since all of the proteins studied except the C-terminal truncated proteins bound to the three PDZ domains (ZASP/Cypher, ALP, and CLP-36).
PDZ binding specificity of the E[ST][DE][DE]L motif.
The results of the database search shown in Table 1 suggested that myopalladin and palladin also share the motif E[ST][DE][DE]L and therefore would be able to interact with PDZ domains. To further study the specificity of the motif, peptides were made that corresponded to the C-terminal final 5 aa of FATZ-1 (calsarcin-2/myozenin-1; ETEEL), FATZ-2 (calsarcin-1/myozenin-2)/palladin (ESEDL), FATZ-3 (calsarcin-3/myozenin-3)/myotilin (ESEEL), myopalladin (ESDEL), and also a mutated peptide ESEEE that had the final amino acid changed from an L to an E. These peptides, as well as native His-tagged C-terminal FATZ-3 (calsarcin-3/myozenin-3) (aa 81 to 251) protein, were used to probe commercial PDZ arrays; each array contained 28 different GST-tagged PDZ domain proteins spotted in duplicate, as well as positive and negative controls (Fig. 4). The PDZ array contained the CLP-36 (hCLIM1) protein but not ALP or ZASP/Cypher. However, the array contained another member the Enigma family of PDZ proteins, RIL (21).
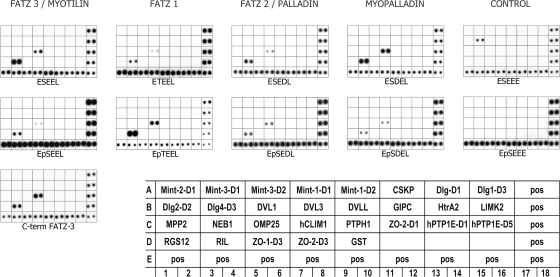
FIG. 4.
PDZ binding specificity of the E[ST][DE][DE]L motif. A commercial PDZ domain array I (TranSignal, Panomics) was probed using His-tagged C-terminal FATZ-3 (calsarcin-3/myozenin-3) or peptide ligands (either unphosphorylated or phosphorylated as indicated by “p”). The positive controls were His-tagged protein spots (100 ng/spots) in the bottom row and on the right column. The GST fusion proteins were spotted in duplicate, and spots with stronger intensities indicate a higher binding affinity of the ligand of interest to PDZ domain(s). Specific binding was seen with the PDZ domains of RIL (reversion-induced LIM protein) and hCLIM1 (human 36-kDa carboxyl-terminal LIM domain protein, CLP-36). The mutated peptide ESEEE bound only to Dlg4-D3 (human postsynaptic density-95, PDZ domain 3). A diagram on the right-hand side of the figure shows the proteins present in PDZ domain array I.
All of the peptides with the exception of the mutated peptide bound to the PDZ domain proteins on the array, CLP-36 (hCLIM1) and RIL, whereas the mutated peptide interacted only with the PDZ domain of Dlg4-D3 protein (Fig. 4, top panel). Both the FATZ-3 (calsarcin-3/myozenin)/myotilin peptide (ESEEL) and the C-terminal FATZ-3 (calsarcin-3/myozenin-3) protein behaved similarly, binding only the CLP-36 (hCLIM1) and RIL proteins on the array (Fig. 4). No interaction was detected between the peptides corresponding to the FATZ (calsarcin/myozenin) and myotilin families and any of the other PDZ domain proteins of the array. These results clearly demonstrate the specificity of the E[ST][DE][DE]L motif for binding PDZ domain proteins of the Enigma family, as well as the importance of the last amino acid of the motif.
Phosphorylation modulates binding of peptide ligands of the FATZ (calsarcin/myozenin)-myotilin families to the PDZ domains of Enigma family members.
The interaction between the PDZ domain and its ligand is often regulated by phosphorylation of the ligand sequence (22). The sequences of the C termini of the myotilin and FATZ (calsarcin/myozenin) protein families contain a potential phosphorylation site (S/T). We synthesized the corresponding phosphorylated peptides and probed the PDZ arrays with them (Fig. 4, middle panel). To test the specificity of binding, we also designed a phosphopeptide in which the last amino acid was changed from leucine to glutamic acid (EpSEEE).
Phosphorylation modulated the ability of the peptides to bind the PDZ domains (Fig. 4, middle panel). As can be seen in Fig. 4, phosphorylation affected the binding of the peptide ligands to the PDZ domains of ZASP/Cypher and CLP-36 both negatively and positively for the myopalladin and FATZ-1 (calsarcin-2/myozenin-1) peptides, respectively. When phosphorylated, the FATZ-1 (calsarcin-2/myozenin-1) peptide (EpTEEL) bound better to both RIL and CLP-36, whereas the interaction of the phosphorylated FATZ-3 (calsarcin-3/myozenin-3)/myotilin (EpSEEL) and myopalladin (EpSDEL) peptides was markedly reduced to both PDZ domains. The interaction of the FATZ-2 (calsarcin-1/myozenin-2)/palladin peptide (EpSEDL) was not affected by phosphorylation. Thus, although phosphorylation modulated strength of binding, it did not induce new interactions within the tested PDZ domains.
To further study the role of phosphorylation in interactions between the PDZ domains and their phosphorylated and nonphosphorylated peptide ligands, we used the AlphaScreen method, which gives a more quantitative measure of the bindings. In the AlphaScreen experiments the biotinylated peptides and the His-tagged PDZ proteins were bound to the donor and acceptor beads, respectively. The PDZ domain proteins were used at a concentration of 50 nM, whereas the peptides were used at a concentration of 10 nM, except for ESEEE and ESDEL, which were used at concentrations of 50 and 25 nM, respectively. As controls, we used both the phosphorylated and the nonphosphorylated mutated peptides (ESEEE and EpSEEE), neither of which bound to the PDZ domains.
It is striking that although both the phosphorylated and the nonphosphorylated ligands bound to the ZASP/Cypher PDZ domain, the phosphorylated ligand bound much better (Fig. 5A); however, this is not the case for the ligand with the PDZ domain of CLP-36. For the PDZ domain of CLP-36, the strength of binding varied depending on the peptide ligand used; both the FATZ-1 (calsarcin-2/myozenin-1) and the FATZ-2 (calsarcin-1/myozenin-2)/palladin peptide ligands bound equally well whether phosphorylated or not. However, the FATZ-3 (calsarcin-3/myozenin-3)/myotilin ligand bound much better when phosphorylated, whereas the myopalladin ligand was the opposite: it bound better when nonphosphorylated. In the AlphaScreen experiments all of the nonphosphorylated peptides bound poorly to PDZ domain of ALP and, although the phosphorylated peptide ligands showed better binding, this did not reach the values seen for the other PDZ domains, and therefore these data are not shown.
FIG. 5.
Phosphorylation modulates the interaction between members of the Enigma family and peptides with the E[ST][DE][DE]L motif. (A) Interactions between the His-PDZ domain proteins (50 nM) of ZASP/Cypher (Z) and CLP-36 (C) and nonphosphorylated and phosphorylated peptide ligands (10 nM) were measured by using the AlphaScreen technique. The myopalladin and mutated peptides (ESEEE and EpSEEE) were used at 25 and 50 nM, respectively. The peptide ligands corresponded to the final C-terminal 5 aa of FATZ-1 (ETEEL, EpTEEL), FATZ-2/palladin (ESEDL, EpSEDL), and FATZ-3/myotilin (ESEEL, EpSEEL) and myopalladin (ESDEL, EpSDEL). The results shown are the mean of at least three different experiments. (B) An overview of the changes seen in the strength of binding due to phosphorylation of the ligand. The results are given as the log2 of the ratio between phosphorylated and nonphosphorylated peptides. (C) C-terminal FATZ-1 (CD2) protein competes with the interaction between ZASP/Cypher (100 nM) and the nonphosphorylated and phosphorylated FATZ-3/myotilin peptide ligands (100 nM, ESEEL, EpSEEL). The S/N is the signal obtained in the presence of the competitor divided by that of the signal obtained without the competitor. The results shown are the mean of at least three different experiments, and the standard deviations of the mean are given for each point.
An overview of the effect of phosphorylation on the binding of the peptide ligands to the PDZ domains of ZASP/Cypher and CLP-36 is shown in Fig. 5B. The results are given as the log2 of the ratio between phosphorylated and nonphosphorylated peptides. In practical terms, the log2 values indicate how many times you have to double (or half if negative) the nonphosphorylated value to equal that of the phosphorylated. In the AlphaScreen experiments (Fig. 5A), all of the tested phosphopeptides showed increased binding to the ZASP/Cypher PDZ domain. The highest increase being that of the binding of the phosphorylated FATZ-3 (calsarcin-3/myozenin-3)/myotilin peptide ligand to PDZ-ZASP; it increased 16-fold over that of the nonphosphorylated ligand. The binding of FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2)/palladin peptides to the CLP-36 PDZ-domain was not affected by phosphorylation, whereas the binding of the myopalladin nonphosphorylated peptide ligand was 18-fold greater than that of the phosphorylated ligand.
The C-terminal of FATZ-1 (calsarcin-2/myozenin-1) competes with the interaction between ZASP-1 and FATZ-3 (calsarcin-3/myozenin-3)/myotilin peptide ligand.
One of the advantages of the AlphaScreen technique is that it can be used for competition studies to examine the strength of binding. We used the C-terminal domain of the GST-tagged FATZ-1 (calsarcin-2/myozenin-1) protein (CD2) as a competitor of the interaction between the His-tagged ZASP-1 protein and the phosphorylated and nonphosphorylated peptides of FATZ-3 (calsarcin-3/myozenin-3)/myotilin (Fig. 5C). This CD2 region of FATZ-1 (calsarcin-2/myozenin-1) is the C-terminal part of the protein (aa 171 to 299), and therefore it has the ETEEL motif, which is capable of binding the PDZ of ZASP/Cypher. In fact, the FATZ-1 (calsarcin-2/myozenin-1)-CD2 fragment was able to compete with the interactions between ZASP-1 and both the nonphosphorylated (ESEEL) and phosphorylated (EpSEEL) peptide ligands. However, 10 times less (10 nM versus 100 nM) FATZ-1(calsarcin-2/myozenin-1)-CD2 protein was required to markedly reduce the binding of ZASP/Cypher to the nonphosphorylated ligand compared to that of the phosphorylated ligand. These data demonstrate that the nonphosphorylated ESEEL peptide binds less avidly to ZASP than the phosphorylated peptide, findings correlating well with the observation that the ZASP PDZ domain bound better to the phosphorylated peptide (Fig. 5A).
CaMKII and PKA induce phosphorylation of the PDZ-binding motif.
It was of interest to test whether muscle cells contain kinase activity toward the ligand sequences since we found that phosphorylation of the C termini of the myotilin and FATZ (calsarcin/myozenin) family proteins is a mechanism that regulates their interaction with the Enigma family PDZ domains. We first tested whether muscle lysate contains kinase activity that can phosphorylate myotilin. The C-terminal myotilin fragment (aa 441 to 498) was expressed as a GST fusion protein and subjected to in vitro phosphorylation reaction using [α-33P]ATP and rat striated muscle lysate. Autoradiography of the SDS-PAGE gels demonstrated that muscle lysate contains kinase activity that can phosphorylate C-terminal myotilin (Fig. 6A).
FIG. 6.
CaMKII phosphorylates the C-terminal end of myotilin and FATZ-3 (calsarcin-3/myozenin-3), whereas PKA phosphorylates the C-terminal ends of FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2). (A) Myotilin 441-498 was produced as a GST fusion protein and allowed to react with muscle lysate in the presence of [γ-33P]ATP. The top panel shows an autoradiograph, and the bottom panel shows Coomassie blue staining of the same sample. (B and C) The C-terminal fragments (final 56 aa's) of myotilin and FATZ (calsarcin/myozenin) family members were produced as GST fusion proteins and allowed to react with CaMKII (B) or PKA (C). The serine of the C-terminal CaMKII site in myotilin was mutated S495A, as were the serines at the PKA sites in FATZ-1 (calsarcin-2/myozenin-1) and in FATZ-2(calsarcin-1/myozenin-2), respectively: S267A and S231A. The top and bottom panels are as described in panel A. The myotilin and FATZ family proteins are differentially phosphorylated by CaMKII and PKA. Serine mutations result in significant reductions in the phosphorylation of each protein.
We then tested whether kinases associated with muscle pathophysiology can phosphorylate the myotilin C terminus and other C termini binding to Enigma family PDZ domains. Recombinant active CaMKII (Fig. 6B) and PKA (Fig. 6C), respectively, were allowed to phosphorylate the C termini of myotilin (aa 441 to 498), palladin (aa 715 to 772), FATZ-1 (calsarcin-2/myozenin-1; aa 244 to 299), FATZ-2 (calsarcin-1/myozenin-2; aa 209 to 264), and FATZ-3 (calsarcin-3/myozenin-3; aa 196 to 251) fused to GST. CaMKII readily phosphorylated in vitro the C terminus of myotilin, as well as that of FATZ-3 (calsarcin-3/myozenin-3), which shares the last 5 aa of the C-terminal sequence with myotilin. The C terminus of palladin was weakly phosphorylated, whereas no phosphorylation of the C termini of FATZ-1 (calsarcin-2/myozenin-1), FATZ-2 (calsarcin-1/myozenin-2), or control GST was seen. The C-terminal myotilin sequence contains six potential phosphorylation sites (serines or threonines), including serine 495 at the PDZ-binding domain. The residues were mutated individually to alanine, and the fragments were expressed as GST fusion proteins. Mutation of S495 to alanine greatly reduced the CaMKII catalyzed phosphorylation of the fragment, suggesting that S495 is the target residue (Fig. 6B), whereas mutation of other residues did not significantly alter phosphorylation (data not shown). The C-terminal regions of FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2) (final 56 aa of C termini) had only one potential PKA phosphorylation site at S267 and S231, respectively. Mutating S267A FATZ-1 (calsarcin-2/myozenin-1) and S231A FATZ-2 (calsarcin-1/myozenin-2) greatly reduced PKA phosphorylation of these C termini (Fig. 6C). The substrate specificity of PKA differs from that of CaMKII. Interestingly, PKA phosphorylated the C termini of FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2), which did not serve as substrates for CaMKII. On the other hand, the CaMKII substrates myotilin and FATZ-3 (calsarcin-3/myozenin-3) were not phosphorylated by PKA.
DISCUSSION
A striking feature of the Z-disc is the multiplicity of protein-protein interactions that form part of a complex three-dimensional network. The nature of the Z-disc allows it to serve both as a structural unit and as a coordinator of intracellular signaling. The Z-disc can respond to external stimuli by dissolution and reorganization, which requires dynamic and coordinated dissociation and association of molecular interactions. Minor modifications caused by missense mutations in several Z-disc proteins can lead to autosomal-dominant fully penetrant diseases of the skeletal muscle and/or the heart. An insufficient knowledge of the Z-disc interactome and its regulation has thus far hampered understanding of the events that allow Z-disc modulation in response to physiological stimuli, as well as in the pathogenesis of muscle disorders. In the present study, we have identified a unique binding motif common to the C terminus of myotilin and FATZ (calsarcin/myozenin) family proteins that binds specifically to the PDZ domain of the Enigma family members ZASP/Cypher, CLP-36, ALP, and RIL.
Proteins of the Enigma family possess both an N-terminal PDZ domain and one or three C-terminal LIM domains that are modular protein interaction motifs present in proteins with a wide range of functions (Fig. 3A). The Enigma family is composed of two subfamilies based on the number of LIM domains. The ALP subfamily with one LIM domain includes ALP (56), RIL (21), CLP-36 (hCLIM1, Elfin) (53), and Mystique (27), whereas the Enigma subfamily with three LIM domains has three members: Enigma (55), Enigma homologue (26), and ZASP/Cypher (Oracle) (8, 35, 57). We did not test whether Mystique or Enigma would bind myotilin or FATZ (calsarcin/myozenin) family members, since these Enigma proteins are not expressed in the striated muscle. Such an interaction, however, might be relevant for the function of palladin, which is widely expressed in different cell types.
PDZ domains are structurally conserved 80- to 100-aa modules present in a wide range of proteins either singly or tandemly. In most cases, the PDZ domains recognize very short C-terminal sequence motifs (3 to 7 aa) via specific side chain interactions between the amino acids of the ligand and those of the hydrophobic pocket formed between a beta B strand and an alpha B helix at the surface of the PDZ domain (6, 17). For instance, in the Dlg PDZ domains the four residues that reside in the carboxylate-binding loop are GLGF (5), whereas in Enigma family PDZ domains they are [P/S]WGF (2). We postulate that these four residues form the basis of why the Enigma family members interact in a specific way with the proteins of the myotilin and FATZ (calsarcin/myozenin) families.
The C terminus PDZ-binding motifs have been classified into three groups with the consensus sequence: S/T-X-hydrophobic-COOH for type I; hydrophobic-X-hydrophobic-COOH for type II and D/E-X-hydrophobic for type III (20, 46). Also, residues other than the extreme C-terminal amino acids may affect specificity (4, 45). Based on its interaction with the C terminus of α-actinin (classified as a type I ligand), the PDZ domain of ZASP is considered a type I PDZ domain. However, our ligand that binds ZASP is classified as a type III motif; therefore, ZASP is both a type I and a type III PDZ domain. This dual specificity for PDZ domains has also been reported for the syntenin protein.
The myotilin and FATZ (calsarcin/myozenin) proteins share the C-terminal motif E[ST][DE][D/E]L, which has the characteristics of a class III type PDZ domain binding sequence. The canonical PDZ ligand motif is usually 4 aa; for example, that of α-actinin is ESDL (by convention L = 0, D = 1, S = 2, and E = 3). However, our putative motif, E[ST][DE][DE]L, is composed of 5 aa (L = 0, D/E = 1, D/E = 2, S/T = 3, and E = 4). We consider the E at position 4 of our motif important since it is conserved from humans to zebrafish in both the myotilin and FATZ families. We saw from a database scan that this E[ST][DE][DE]L C-terminal ligand motif in vertebrates is restricted to the FATZ (calsarcin/myozenin) and myotilin families with the exception of histidine ammonia lyase that has its final C-terminal amino acids (ESEDL) identical to that of FATZ-2 (calsarcin-1/myozenin-2)/palladin (Table 1). l-Histidine ammonia-lyase is a cytosolic enzyme catalyzing the first reaction in histidine catabolism; the nonoxidative deamination of l-histidine to trans-urocanic acid. It is probable that histidine lyase can bind the same PDZ proteins as the FATZ (calsarcin/myozenin) and myotilin families, but whether it does so in reality would depend on its location. Although there is a report of histidine lyase in the sarcoplasm of the muscle (25), it is not normally found in muscle, whereas proteins of both the FATZ (calsarcin/myozenin) and the myotilin families are located in the Z-disc.
Initially, we checked whether proteins of the FATZ (calsarcin/myozenin) family and myotilin could bind ZASP/Cypher and then extended this experiment to include two other Enigma family members, ALP and CLP-36. From our results, it can be seen that all of the ligand proteins were able to bind the three PDZ domain proteins, although the FATZ-3 (calsarcin-3/myozenin-3) protein did not bind as well as myotilin. However, it is important to note that both the PDZ proteins and the protein ligands are produced in bacteria and therefore nonphosphorylated. The interaction of the FATZ (calsarcin/myozenin) and myotilin families with the Enigma family members is highly specific, as verified by the restricted number of PDZ domain proteins with which their peptide ligands will bind when given the opportunity. In fact, only 2 of the 28 PDZ domain proteins on the PDZ array bound to the peptide ligands. Interestingly, these were the only two Enigma family members on the array: RIL and CLP-36 (hCLIM1). We confirmed by array experiments the importance for binding specificity of the extreme C-terminal amino acid, in this case L. The main contrast between the AlphaScreen results and the arrays was the binding with the FATZ-3 (calsarcin-3/myozenin-3)/myotilin peptide since in the arrays the phosphorylated peptide did not bind well. This discrepancy could be due to variability in the amount of protein spotted.
Myocytes respond to external stimuli by activating intracellular pathways, which modify cellular functions. For instance, stimulation of α-adrenergic receptors on myocytes activates protein kinase A (PKA), which phosphorylates several sarcomeric proteins and results in enhanced cardiac performance (30). Similarly, CaMKII, the predominant CaMK isoform in the heart, is an important modulator of both electrophysiological and contractile properties of the myocytes (28). Extended activation of both kinases is thought to be important in the events that lead to myocyte dysfunction and to heart failure. Except for the finding that Src phosphorylates palladin (39); no information on the interplay between kinases and the myotilin or FATZ (calsarcin/myozenin) families has been available. We identify here CaMKII and PKA as kinases that are able to phosphorylate the C termini of myotilin and FATZ (calsarcin/myozenin) family members. In myotilin, the residue phosphorylated by CaMKII is S495 within the ESEEL motif. CaMKII also phosphorylates FATZ-3 (calsarcin-3/myozenin-3), whose last five C-terminal residues are identical to those of myotilin but not to those of FATZ-1 (calsarcin-2/myozenin-1) or FATZ-2 (calsarcin-1/myozenin-2), whose C-terminal sequences are conserved but are slightly different. Interestingly, PKA phosphorylates FATZ-1 (calsarcin-2/myozenin-1) and FATZ-2 (calsarcin-1/myozenin-2) at residues within PDZ-binding motif, but not myotilin or FATZ-3 (calsarcin-3/myozenin-3), indicating that different cellular stimuli result in modification of different myotilin and FATZ (calsarcin/myozenin) family members.
PDZ-based interactions can be regulated by phosphorylation of the C-terminal PDZ-binding motifs (20). Typically, phosphorylation results in the disruption of binding to the PDZ domain (22). This is the case also in the interaction between CLP-36 and the C-terminal myopalladin peptide, which is lost upon phosphorylation. However, the effect of phosphorylation on binding varies depending on the PDZ domain and the ligand. In fact, phosphorylation significantly increases binding between of all FATZ (calsarcin/myozenin) and myotilin family peptides and the PDZ domain of ZASP/Cypher. Thus, not only the stimulus-kinase connection can regulate PDZ interactions within the Z disc, but various stimuli can result in a differential preference in the PDZ domain interactions. Such a preference may not be limited to the myotilin and FATZ (calsarcin/myozenin) family members, since these proteins apparently compete with α-actinin for binding to ZASP/Cypher.
The main finding of this research is the identification of a group of striated muscle proteins, consisting of the three FATZ (calsarcin/myozenin) family proteins and also myotilin, palladin, and myopalladin that selectively interact via a C-terminal motif with the PDZ domains of several Enigma proteins, ZASP/Cypher, CLP-36, ALP, and RIL. Based on the work presented here, we propose that the FATZ (calsarcin/myozenin) and myotilin families bind to members of the Enigma family of PDZ proteins via the E[ST][DE][DE]L C-terminal ligand motif that can be considered a novel class III PDZ domain binding motif. In muscle, PDZ proteins function as adaptors in translating mechanical stress signals from the Z-disc to the nucleus (19). Some members of the Enigma family of PDZ proteins, for example, ZASP/Cypher, are known to bind protein kinases via their C-terminal LIM domains. Therefore, it is possible that ZASP/Cypher and some members of the Enigma family of PDZ-proteins link the proteins of the myotilin and FATZ (calsarcin/myozenin) families to signaling events such as PKC phosphorylation. Since mutations in myotilin, myopalladin, ZASP/Cypher, and FATZ-2 (calsarcin-1/myozenin-2) can lead to myopathies, knowledge of these protein interactions is highly relevant to understanding muscle and cardiac disorders. We demonstrate here that several of the known disease-associated Z-disc proteins—myotilin, myopalladin, FATZ-2 (calsarcin-1/myozenin-2), and ZASP/Cypher—are part of the same structural complex, whose composition can be regulated by signaling molecules associated with pathophysiological stimuli.
Acknowledgments
The skillful technical assistance of H. Ahola is gratefully acknowledged.
This study was supported by grants from the Telethon Foundation of Italy (grant GGP04088 to G.F. and grant GSP042894B to G.V.) and by grants from the Academy of Finland, the Sigrid Juselius Foundation, the Finnish Heart Association, and the Association Française Contre les Myopathies to O.C.
Footnotes
Published ahead of print on 1 December 2008.
REFERENCES
- 1.Alfthan, K., L. Heiska, M. Grönholm, G. H. Renkema, and O. Carpén. 2004. Cyclic AMP-dependent protein kinase phosphorylates merlin at serine 518 independently of p21-activated kinase and promotes merlin-ezrin heterodimerization. J. Biol. Chem. 27918559-18566. [DOI] [PubMed] [Google Scholar]
- 2.Au, Y., R. A. Atkinson, R. Guerrini, G. Kelly, C. Joseph, S. R. Martin, F. W. Muskett, A. Pallavicini, G. Faulkner, and A. Pastore. 2004. Solution structure of ZASP PDZ domain; implications for sarcomere ultrastructure and enigma family redundancy. Structure 12611-622. [DOI] [PubMed] [Google Scholar]
- 3.Bang, M. L., R. E. Mudry, A. S. McElhinny, K. Trombitas, A. J. Geach, R. Yamasaki, H. Sorimachi, H. Granzier, C. C. Gregorio, and S. Labeit. 2001. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J. Cell Biol. 153413-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuming, T., L. Skrabanek, M. Y. Niv, P. Mukherjee, and H. Weinstein. 2005. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics 21827-828. [DOI] [PubMed] [Google Scholar]
- 5.Cho, K. O., C. A. Hunt, and M. B. Kennedy. 1992. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9:929-942. [DOI] [PubMed] [Google Scholar]
- 6.Doyle, D. A., A. Lee, J. Lewis, E. Kim, M. Sheng, and R. MacKinnon. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 851067-1076. [DOI] [PubMed] [Google Scholar]
- 7.Duboscq-Bidot, L., P. Xu, P. Charron, N. Neyroud, G. Dilanian, A. Millaire, V. Bors, M. Komajda, and E. Villard. 2008. Mutations in the Z-band protein myopalladin gene and idiopathic dilated cardiomyopathy. Cardiovasc. Res. 77118-125. [DOI] [PubMed] [Google Scholar]
- 8.Faulkner, G., A. Pallavicini, E. Formentin, A. Comelli, C. Ievolella, S. Trevisan, G. Bortoletto, P. Scannapieco, M. Salamon, V. Moulay, G. Valle, and G. Lanfranchi. 1999. ZASP: a new Z-band alternatively spiced PDZ-motif protein. J. Cell Biol. 146:465-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faulkner, G., A. Pallavicini, A. Comelli, M. Salamon, G. Bortoletto, C. Ievolella, S. Trevisan, S. Kojic, F. Dalla Vecchia, P. Laveder, G. Valle, and G. Lanfranchi. 2000. FATZ: a filamin, actinin, and telethonin binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 27541234-44142. [DOI] [PubMed] [Google Scholar]
- 10.Foroud, T., N. Pankratz, A. P. Batchman, M. W. Pauciulo, R. Vidal, L Miravalle, H. H. Goebel, L. J. Cushman, B. Azzarelli, H. Horak, M. Farlow, and W. C. Nichols. 2005. A mutation in myotilin causes spheroid body myopathy. Neurology 651936-1940. [DOI] [PubMed] [Google Scholar]
- 11.Frey, N., J. A. Richardson, and E. N. Olson. 2000. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc. Natl. Acad. Sci. USA 9714632-14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey, N., and E. N. Olson. 2002. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 27713998-14004. [DOI] [PubMed] [Google Scholar]
- 13.Frey, N., T. Barrientos, J. M. Shelton, D. Frank, H. Rütten, D. Gehring, C. Kuhn, M. Lutz, B. Rothermel, R. Bassel-Duby, J. A. Richardson, H. A. Katus, J. A. Hill, and E. N. Olson. 2004. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 10:1336-1343. [DOI] [PubMed] [Google Scholar]
- 14.Frey, N., D. Frank, S. Lippl, C. Kuhn, H. Kögler, T. Barrientos, C. Rohr, R. Will, O. J. Müller, H. Weiler, R. Bassel-Duby, H. A. Katus, and E. N. Olson. 9 October 2008. Calsarcin-2 deficiency increases exercise capacity in mice through calcineurin/NFAT activation. J. Clin. Investig. 118:3598-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gontier, Y., A. Taivainen, L. Fontao, A. Sonnenberg, A. van der Flier, O. Carpen, G. Faulkner, and L. Borradori. 2005. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J. Cell Sci. 1183739-3749. [DOI] [PubMed] [Google Scholar]
- 16.Grönholm. M., M. Sainio, F. Zhao, L. Heiska, A. Vaheri, and O. Carpén. 1999. Homotypic and heterotypic interaction of the neurofibromatosis 2 tumor suppressor protein merlin and the ERM protein ezrin. J. Cell Sci. 112895-904. [DOI] [PubMed] [Google Scholar]
- 17.Harris, B. Z., and W. A. Lim. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 1143219-3231. [DOI] [PubMed] [Google Scholar]
- 18.Hauser, M. A., S. K. Horrigan, P. Salmikangas, U. M. Torian, K. D. Viles, R. Dancel, R. W. Tim, A. Taivainen, L. Bartoloni, J. M. Gilchrist, J. M. Stajich, P. C. Gaskell, J. R. Gilbert, J. M. Vance, M. A. Pericak-Vance, O. Carpen, C. A. Westbrook, and M. C. Speer. 2000. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum. Mol. Genet. 92141-2147. [DOI] [PubMed] [Google Scholar]
- 19.Hoshijima, M. 2006. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am. J. Physiol. Heart Circ. Physiol. 290H1313-H13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, A. Y., and M. Sheng. 2002. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2775699-5702. [DOI] [PubMed] [Google Scholar]
- 21.Kiess, M., B. Scharm, A. Aguzzi, A. Hajnal, R. Klemenz, I. Schwarte-Waldhoff, and R. Schäfer. 1995. Expression of ril, a novel LIM domain gene, is down-regulated in Hras-transformed cells and restored in phenotypic revertants. Oncogene 1061-68. [PubMed] [Google Scholar]
- 22.Kim, E., and M. Sheng. 2004. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5771-781. [DOI] [PubMed] [Google Scholar]
- 23.Klaavuniemi, T., A. Kelloniemi, and J. Ylänne. 2004. The ZASP-like motif in actinin-associated LIM protein is required for interaction with the alpha-actinin rod and for targeting to the muscle Z-line. J. Biol. Chem. 27926402-26410. [DOI] [PubMed] [Google Scholar]
- 24.Klaavuniemi, T., and J. Ylänne. 2006. Zasp/Cypher internal ZM-motif containing fragments are sufficient to colocalize with alpha-actinin-analysis of patient mutations. Exp. Cell Res. 3121299-1311. [DOI] [PubMed] [Google Scholar]
- 25.Krishnamoorthy, R. V. 1977. Increased histaminase activity in the atrophic muscle of denervated frog. Enzyme 2273-79. [DOI] [PubMed] [Google Scholar]
- 26.Kuroda, S., C. Tokunaga, Y. Kiyohara, O. Higuchi, H. Konishi, K. Mizuno, G. N. Gill, and U. Kikkawa. 1996. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J. Biol. Chem. 27131029-33132. [DOI] [PubMed] [Google Scholar]
- 27.Loughran, G., N. C. Healy., P. A. Kiely, M. Huigsloot, N. L. Kedersha, and R. O'Connor. 2005. Mystique is a new insulin-like growth factor-I-regulated PDZ-LIM domain protein that promotes cell attachment and migration and suppresses Anchorage-independent growth. Mol. Biol. Cell 16:1811-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maier, L. S., and D. M. Bers. 2007. Role of Ca2+/calmodulin-dependent protein kinase (CaMK) in excitation-contraction coupling in the heart. Cardiovasc. Res. 73631-640. [DOI] [PubMed] [Google Scholar]
- 29.Mologni, L., E. Sala, B. Riva, L. Cesaro, S. Cazzaniga, S. Redaelli, O. Marin, N. Pasquato, A. Donella-Deana, and C. Gambacorti-Passerini. 2005. Expression, purification, and inhibition of human RET tyrosine kinase. Protein Expr. Purif. 41177-185. [DOI] [PubMed] [Google Scholar]
- 30.Mongillo, M., and M. Zaccolo. 2006. A complex phosphodiesterase system controls beta-adrenoceptor signalling in cardiomyocytes. Biochem. Soc. Trans. 34510-511. [DOI] [PubMed] [Google Scholar]
- 31.Moza, M., L. Mologni, R. Trokovic, G. Faulkner, J. Partanen, and O. Carpen. 2007. Targeted deletion of the muscular dystrophy gene myotilin does not perturb muscle structure or function in mice. Mol. Cell. Biol. 27244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mykkänen, O.-M., M. Grönholm, M. Rönty, M. Lalowski, P. Salmikangas, H. Suila, and O. Carpén. 2001. Characterization of human palladin, a microfilament-associated protein. Mol. Biol. Cell 123060-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osio, A., L. Tan, S. N. Chen, R. Lombardi, S. F. Nagueh, S. Shete, R. Roberts, J. T. Willerson, and A. J. Marian. 2007. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ. Res. 100766-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parast, M. M., and C. A. Otey. 2000. Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J. Cell Biol. 150643-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passier, R., J. A. Richardson, and E. N. Olson. 2000. Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech. Dev. 92277-284. [DOI] [PubMed] [Google Scholar]
- 36.Puntervoll, P., R. Linding, C. Gemünd, S. Chabanis-Davidson, M. Mattingsdal, S. Cameron, D. M. Martin, G. Ausiello, B. Brannetti, A. Costantini, F. Ferrè, V. Maselli, A. Via, G. Cesareni, F. Diella, G. Superti-Furga, L. Wyrwicz, C. Ramu, C. McGuigan, R. Gudavalli, I. Letunic, P. Bork, L. Rychlewski, B. Küster, M. Helmer-Citterich, W. N. Hunter, R. Aasland, and T. J. Gibson. 2003. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 313625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyle, W. G., and R. J. Solaro. 2004. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 94296-305. [DOI] [PubMed] [Google Scholar]
- 38.Rönty, M., A. Taivainen, M. Moza, G. D. Kruh, E. Ehler, and O. Carpén. 2005. Involvement of palladin and apha-actinin in targeting of the Abl/Arg kinase adaptor ArgBP2 to the actin cytoskeleton. Exp. Cell Res. 31088-98. [DOI] [PubMed] [Google Scholar]
- 39.Rönty, M., A. Taivainen, L. Heiska, C. Otey, E. Ehler, W. K. Song, and O. Carpen. 2007. Palladin interacts with SH3 domains of SPIN90 and Src and is required for Src-induced cytoskeletal remodeling. Exp. Cell Res. 3132575-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmikangas, P., O.-M. Mykkänen, M. Grönholm, L. Heiska, J. Kere, and O. Carpen. 1999. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet. 81329-1336. [DOI] [PubMed] [Google Scholar]
- 41.Salmikangas, P., P. F. van der Ven, M. Lalowski, A. Taivainen, F. Zhao, H. Suila, R. Schroder, P. Lappalainen, D. O. Furst, and O. Carpen. 2003. Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum. Mol. Genet. 12189-203. [DOI] [PubMed] [Google Scholar]
- 42.Selcen, D., and A. G. Engel. 2004. Mutations in myotilin cause myofibrillar myopathy. Neurology 621363-1371. [DOI] [PubMed] [Google Scholar]
- 43.Selcen, D., and A. G. Engel. 2005. Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 57:269-276. [DOI] [PubMed] [Google Scholar]
- 44.Sheng, M., and C. Sala. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 241-29. [DOI] [PubMed] [Google Scholar]
- 45.Skelton, N. J., M. F. Koehler, K. Zobel, W. L. Wong, S. Yeh, M. T. Pisabarro, J. P. Yin, L. A. Lasky, and S. S. Sidhu. 2003. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J. Biol. Chem. 278:7645-7654. [DOI] [PubMed] [Google Scholar]
- 46.Songyang, Z., A. S. Fanning, C. Fu, J. Xu, S. M. Marfatia, A. H. Chishti, A. Crompton, A. C. Chan, J. M. Anderson, and L. C. Cantley. 1997. Recognition of unique C-terminal motifs by distinct PDZ domains. Science 27573-77. [DOI] [PubMed] [Google Scholar]
- 47.Takada, F., D. L. Vander Woude, H. Q. Tong, T. G. Thompson, S. C. Watkins, L. M. Kunkel, and A. H. Beggs. 2001. Myozenin: an alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc. Natl. Acad. Sci. USA 981595-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.te Velthuis, A. J., T. Isogai, L. Gerrits, and C. P. Bagowski. 2007. Insights into the molecular evolution of the PDZ/LIM family and identification of a novel conserved protein motif. PLoS ONE 2e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Ven, P. F., S. Wiesner, P. Salmikangas, D. Auerbach, M. Himmel, S. Kempa, K. Hayess, D. Pacholsky, A. Taivainen, R. Schroder, O. Carpen, and D. O. Furst. 2000. Indications for a novel muscular dystrophy pathway. gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell Biol. 151235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vatta. M., B. Mohapatra, S. Jimenez, X. Sanchez, G. Faulkner, Z. Perles, G. Sinagra, J. H. Lin, T. M. Vu, Q. Zhou, K. R. Bowles, A. Di Lenarda, L. Schimmenti, M. Fox, M. A. Chrisco, R. T. Murphy, W. McKenna, P. Elliott, N. E. Bowles, J. Chen, G. Valle, and J. A. Towbin. 2003. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 422014-2027. [DOI] [PubMed] [Google Scholar]
- 51.von Nandelstadh, P., M. Grönholm, M. Moza, A. Lamberg, H. Savilahti, and O. Carpen. 2005. Actin-organising properties of the muscular dystrophy protein myotilin. Exp. Cell Res. 310:131-139. [DOI] [PubMed] [Google Scholar]
- 52.Vorgerd, M., P. F. van der Ven, V. Bruchertseifer, T. Lowe, R. A. Kley, R. Schroder, H. Lochmuller, M. Himmel, K. Koehler, D. O. Furst, and A. Huebner. 2005. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am. J. Hum. Genet. 77297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, H., D. C. Harrison-Shostak, J. J. Lemasters, and B. Herman. 1995. Cloning of a rat cDNA encoding a novel LIM domain protein with high homology to rat RIL. Gene 165267-271. [DOI] [PubMed] [Google Scholar]
- 54.Witt, S. H., H. Granzier, C. C. Witt, and S. Labeit. 2005. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: toward understanding MURF-dependent muscle ubiquitination. J. Mol. Biol. 350713-722. [DOI] [PubMed] [Google Scholar]
- 55.Wu, R. Y., and G. N. Gill. 1994. LIM domain recognition of a tyrosine-containing tight turn. J. Biol. Chem. 26925085-25090. [PubMed] [Google Scholar]
- 56.Xia, H., S. T. Winokur, W. L. Kuo, M. R. Altherr, and D. S. Bredt. 1997. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol. 139:507-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, Q., P. Ruiz-Lozano, M. E. Martone, and J. Chen. 1999. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 27419807-19813. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, Q., P. H. Chu, C. Huang, C. F. Cheng, M. E. Martone, G. Knoll, G. D. Shelton, S. Evans, and J. Chen. 2001. Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell Biol. 155605-612. [DOI] [PMC free article] [PubMed] [Google Scholar]