Abstract
Steroidogenic factor 1 (SF-1) is an orphan nuclear receptor selectively expressed in the adrenal cortex and gonads, where it mediates the hormonal stimulation of multiple genes involved in steroid hormone biosynthesis. SF-1 is the target of both phosphorylation and SUMOylation, but how these modifications interact or contribute to SF-1 regulation of endogenous genes remains poorly defined. We found that SF-1 is selectively SUMOylated at K194 in Y1 adrenocarcinoma cells and that although SUMOylation does not alter the subcellular localization of SF-1, the modification inhibits the ability of SF-1 to activate target genes. Notably, whereas SF-1 SUMOylation is independent of S203 phosphorylation and is unaffected by adrenocorticotropin (ACTH) treatment, loss of SUMOylation leads to enhanced SF-1 phosphorylation at serine 203. Furthermore, preventing SF-1 SUMOylation increases the mRNA and protein levels of multiple steroidogenic enzyme genes. Analysis of the StAR promoter indicates that blockade of SF-1 SUMOylation leads to an increase in overall promoter occupancy but does not alter the oscillatory recruitment dynamics in response to ACTH. Notably, we find that CDK7 binds preferentially to the SUMOylation-deficient form of SF-1 and that CDK7 inhibition reduces phosphorylation of SF-1. Based on these observations, we propose a coordinated modification model in which inhibition of SF-1-mediated transcription by SUMOylation in adrenocortical cancer cells is mediated through reduced CDK7-induced phosphorylation of SF-1.
Steroidogenic factor 1 (SF-1) (also called NR5A1 or Ad4BP) is an orphan nuclear receptor that plays a crucial role in the regulation of steroid hormone biosynthesis, as well as in the endocrine development of both the adrenal gland and gonads (68). Several genes, including the CYP17, DAX-1, CYP19, CYP11A1, MIS, 3β-HSD, CYP21, StAR, and Mc2R genes, have been identified as SF-1 target genes (8, 9, 38, 39, 43, 45, 62, 69, 70, 73). Regulation of these genes involves the concerted action of SF-1 with multiple transcription factors with which it can synergize, such as Sox9 (18), Wt1 (31, 48), Gata4 (65), EGR1 (19, 25), PITX1 (64), multiprotein bridging factor 1 (36), and TReP-132 (22). A number of coregulators, such as steroid receptor coactivator 1 (SRC-1) (16, 33), cyclic AMP response element-binding protein (CREB)-binding protein/p300 (47), transcriptional intermediary factor 2 (6), nuclear receptor corepressor (15), and β-catenin (46), have been reported to interact with SF-1 and likely participate in SF-1 gene activation. On the other hand, factors such as Dax-1 (34) and DP103 (50) appear to play an inhibitory role by limiting SF-1 function. The transcriptional capacity of SF-1 is influenced by posttranslational modifications, with phosphorylation at S203 playing a key stimulatory role (26). S203 phosphorylation serves to enhance coactivator binding and the transactivation potential of this receptor. Recent data indicate that SF-1 can be phosphorylated on residue S203 by either ERK1/2 or CDK7 (44). Given that CDK7 is a unique CDK kinase that functions both to facilitate cell cycle progression and to regulate transcriptional activation, it has been proposed that CDK7 serves to activate specific transcriptional programs that are critical for proliferation in a given organ (10, 44).
Recently, a novel posttranslational modification involving the conjugation of small ubiquitin-like modifier (SUMO) proteins has been identified and shown to regulate diverse cellular processes, including nuclear protein targeting, regulation of transcription, DNA repair, formation of subnuclear structures, chromosome segregation, and protein stability (21, 27, 35). In mammals, four SUMO proteins (SUMO1 to -4) are encoded by distinct genes. SUMO1 has ∼48% identity to either the closely related SUMO2 or -3 (57, 60). A fourth isoform, very similar to SUMO2/3, has also been identified (5). In contrast to SUMO1, SUMO2 and -3 contain a clear consensus SUMOylation site in their N-terminal regions. Current data suggest some selectivity in the SUMO1 and SUMO2/3 modification of proteins (57). Although the consequences of selective conjugation of different SUMO family proteins remain poorly defined, the functional effects of modification by SUMO2/3 can be distinguished from that of SUMO1 in transcriptional regulation (30).
Despite limited sequence identity, SUMO proteins share with ubiquitin a common structural fold and use a parallel enzymological pathway of conjugation. Newly translated SUMO proteins are processed by specific SUMO proteases (SENPs) to remove C-terminal residues in SUMO and to expose a conserved diglycine motif (35). Notably, SUMO4 harbors a proline residue at position 90, which prevents initial processing by known SUMO protease enzymes and subsequent conjugation (51). Whether this member functions solely through noncovalent interactions remains to be determined. After this initial cleavage, SUMO is then activated in an ATP-dependent manner by the heterodimeric E1-activating enzyme SAE1/SAE2. The thioester-linked SUMO is then transferred to the SUMO-specific E2-conjugating enzyme Ubc9, which in turn recognizes specific substrates and catalyzes the formation of an isopeptide bond between SUMO and the target lysine. This step is facilitated by SUMO E3 ligases, such as RanBP2 and members of the protein inhibitor of activated STAT (signal transducers and activators of transcription) family (12, 52, 56). Covalent modification of proteins by SUMO is reversible through the action of members of the SENP family of proteases. SUMO modification of numerous transcription factors, including certain nuclear receptors, is associated with inhibition of transcription (4, 23, 49, 55). Notably, the consensus sequence for SUMOylation is found to be included in the definition of synergy control (SC) motifs, which are conserved regulatory features in multiple transcription factors (32). Recent data indicate that SC motifs function in a context-dependent manner to inhibit transcription by serving as sites for SUMO conjugation (13, 30, 61).
We previously determined that phosphorylation of SF-1 at S203 regulates SF-1-dependent recruitment of coregulatory proteins that ultimately engage the transcriptional machinery (70). Recent reports have demonstrated that SF-1 harbors functional SC motifs and that SUMOylation of SF-1 attenuates its transcriptional activity in a promoter context-dependent manner (11, 40, 42). However, the physiological function of this posttranslational modification and how it might functionally interact with other modifications, such as phosphorylation, remain to be elucidated. Thus, we have undertaken this study (i) to determine the role of SUMOylation in the cyclic recruitment, promoter occupancy, and clearance of SF-1 from endogenous target genes and (ii) to investigate the interplay between phosphorylation and SUMOylation of SF-1 in SF-1 activity. Our findings indicate that SUMOylation of SF-1 represses the transcription of target genes but does not alter nuclear localization. The SF-1 SUMOylation level is not altered by adrenocorticotropin (ACTH) induction or SF-1 phosphorylation. In contrast, we demonstrate that preventing SUMOylation of SF-1 enhances the level of SF-1 phosphorylation. Furthermore, we find that CDK7 kinase binds more avidly to a non-SUMOylatable form of SF-1 than to the wild-type (WT) protein. Thus, it appears that the inhibition of SF-1-mediated transcription by SUMOylation in adrenocortical cancer cells is mediated through inhibition of CDK7-induced phosphorylation of SF-1. Taken together, these results support a novel mechanism whereby SUMOylation serves to regulate the phosphorylation-dependent activation of this nuclear receptor.
MATERIALS AND METHODS
Reagents.
α-Amanitin was purchased from Sigma (St. Louis, MO). All cell culture reagents, protein A-agarose, and TRIzol were purchased from Invitrogen (Carlsbad, CA). Human ACTH (amino acids 1 to 24) was from Calbiochem (La Jolla, CA). The antibodies used were SF-1 and RNA polymerase II, CTD4H8 (Upstate Biochemistry Inc., Charlottsville, VA); phospho-ERK1/2 (Thr202/Tyr204) (Cell Signaling Technology Inc., Beverly, MA); FLAG and β-actin (Sigma); and CDK7, SRC-1, SUMO2, SUMO3, lamin, and tubulin (Santa Cruz Biotechnology). Anti-StAR antibody was kindly provided by D. M. Stocco (Texas Tech University). Anti-phospho-SF-1 antibody was kindly provided by H. A. Ingraham (University of California, San Francisco). Luciferase activity was measured using the Dual Luciferase Assay System (Promega, Madison, WI).
DNA constructs.
Mouse SF-1 cDNA was PCR amplified using the forward primer 5′-TCGTGGATCCATGGACTACTCGTACGACGAG-3′ and the reverse primer 5′-ACGAAAGCTTTCAAGTCTGCTTGGCCTGCAG-3′. N-terminally hemagglutinin (HA)-tagged SUMO1-SF-1 expression plasmids were generated by ligating the BamHI/HindIII fragment from SF-1 cDNA into the same sites of pcDNA3 HA-SUMO1(-Gly) Gal4 (14, 30). N-terminally HA-tagged SUMO2-SF-1 expression plasmid was generated the same way using the corresponding pcDNA3 HA-SUMO(-Gly) Gal4 vector (14, 30).
Oligonucleotides.
5′-TCGTGGTACCATGCATCACCACCATCATCATGATTACAAGGATGACGACGATAAGGGATCCATGGACTACTCG-3′ and 5′-CGAGTAGTCCATGGATCCCTTATCGTCGTCATCCTTGTAATCATGAT GATGGTGGTGATGCATGGTACCACGA-3′, which contain a starting ATG codon followed by hexahistidine (HIS) and FLAG tags, were used as adapter primers. For N-terminal HIS-FLAG-tagged mouse SF-1 expression plasmids, adapter primers were annealed and digested with KpnI/BamHI and then ligated into the same sites of the pcDNA3-HA-SUMO2-SF-1 plasmid to create the pcDNA3-HIS-FLAG-SF-1 expression plasmid. The pcDNA3-HIS-FLAG-SF-1 K119R, K194R, 2KR, S203A, S203D, K194RS203A, and K194RS203D; pcDNA3-HA-SUMO1-SF-1 K194R; and pcDNA3-HA-SUMO2-SF-1 K194R plasmids were derived from the WT HIS-FLAG-SF-1, HA-SUMO1-SF-1, and HA-SUMO2-SF-1 vectors by the QuikChange site-directed mutagenesis approach (Strategene). The pcDNA3-HA-SUMO3 vector used for in vivo SUMOylation assays was described previously (61). All vectors were verified by nucleotide sequencing.
The mouse Mc2R-luciferse reporter plasmid containing 1 kb of the mouse Mc2R promoter was a kind gift of F. Beuschlein (University of Freiburg, Freiburg, Germany). The mouse StAR luciferase reporter gene was kindly provided by Kenneth Escudero (Texas A&M University, Kingsville).
Cell culture and transfections.
Cos-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) in the presence of 10% fetal bovine serum and antibiotics (Gibco) in humidified air containing 5% CO2 at 37°C. Mouse Y1 adrenocortical carcinoma cells and stable Y1 cell lines were maintained in DMEM supplemented with 7.5% horse serum, 2.5% fetal bovine serum, and antibiotics in humidified air containing 5% CO2 at 37°C. After incubation, the cells were transfected using Fugene HD transfection reagent (Roche). Approximately 40 to 48 h after transfection, the cells were harvested. Luciferase activity was measured and normalized with Renilla activity. All experiments were performed three times in triplicate.
In vivo SUMOylation assay.
The in vivo SUMOylation assay was carried out as previously described (14, 30). Briefly, Cos-7 cells (2 × 106) were seeded in 10-cm plates and transfected 24 h later with 5 μg of the indicated receptor and HA-SUMO3 expression vectors. Y1 cells (2 × 106) were seeded in 10-cm plates and 24 h later were transfected with 3 μg of the indicated receptor and HA-SUMO3 expression vectors. After 48 h, cells were harvested in 700 μl lysis buffer (500 mM NaCl, 10 mM imidazole, 45 mM Na2HPO4, 5 mM Na2H2PO4, 8 M urea, pH 8) containing complete protease inhibitors without EDTA (1 tablet/10 ml; Roche) and sonicated. The lysates were cleared and incubated with 100 μl of 50% Ni2+-nitrilotriacetic acid agarose (Qiagen) at room temperature for 60 min on a rotator. The resin was washed three times in wash buffer 1 (400 mM NaCl, 10 mM imidazole, 17.6 mM Na2HPO4, 32.4 mM Na2H2PO4, 8 M urea, pH 6.75) and two times in wash buffer 2 (150 mM NaCl, 10 mM imidazole, 17.6 mM Na2HPO4, 32.4 mM Na2H2PO4, pH 6.75). Samples were resuspended in 3× EDTA sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Samples (15 μl) were resolved by 10% SDS-PAGE and processed for immunoblotting using monoclonal anti-FLAG immunoglobulin G (Sigma) or anti-HA-11 (Covance) primary antibodies and anti-goat peroxidase conjugate (Santa Cruz Biotechnology) and anti-mouse immunoglobulin G-peroxidase conjugate (Bio-Rad) secondary antibodies. Images were captured in a Kodak Image Station 440 CF using Super Signal West Femto substrates (Pierce). For ACTH treatment experiments, Y1 cells (2 × 106) were seeded in 10-cm plates and, 24 h later, serum deprived in DMEM supplemented with 0.05% bovine serum albumin, followed by transfection with 3 μg HA-SUMO3 expression vector and the indicated receptor expression vector. Twenty-four hours after transfection, the cells were treated with 2.5 μM α-amanitin for 2 h. The cells were washed twice with phosphate-buffered saline (PBS), and fresh serum-free medium was added 30 min prior to ACTH (10 nM) stimulation for the indicated times.
Immunoprecipitation assays.
Stable Y1 cells (2 × 106) were seeded onto 10-cm plates. After 24 h, cells were harvested and lysed in lysis buffer (40 mM HEPES, 120 mM sodium chloride, 10 mM sodium pyrophosphate, 10 mM sodium glycerophosphate, 1 mM EDTA, 50 mM sodium fluoride, 0.5 mM sodium orthovanadate, 1% Triton X-100) containing protease inhibitor cocktail (Sigma), followed by rotation for 1 h at 4°C to solubilize the proteins. Soluble protein was collected and immunoprecipitated with the indicated antibody overnight. Protein A-agarose beads were added to the protein lysates for 2 h in the cold room. The beads were centrifuged and washed at least three times with lysis buffer. The proteins were eluted by boiling them in 50 μl of 2× Laemmli sample buffer, resolved by 7.5% SDS-PAGE, and processed for immunoblotting as described below.
Immunoblotting.
Protein lysates were allowed to rotate at 4°C for 30 min, and the protein contents of the high-speed supernatant were determined using the Bradford protein assay (Bio-Rad Laboratories, Inc., Hercules, CA). Equivalent quantities of protein (20 to 45 μg) were resolved on polyacrylamide-SDS gels, transferred to nitrocellulose membranes (Bio-Rad), and immunoblotted with specific antibodies. The results were visualized using the Supersignal West Dura Extended Duration Substrate kit (Pierce Chemical Co., Rockford, IL).
ChIP assays and re-ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described in our laboratory (70). Briefly, stable Y1 cells were synchronized by α-amanitin, fixed with formaldehyde, and harvested. For immunoprecipitation, anti-SF-1, anti-RNA polymerase II, and anti-SRC-1 antibodies were used. The extracted DNA fragments were quantified by real-time PCR using pairs of primers that covered the SF-1 response region within the mouse StAR proximal promoter (nucleotides −80 to −10). The primers used for PCR were 5′-AATGACTGATGACTTTTTTATCTCAAGTG-3′ (forward) and 5′-AAGTGCGCTGCCTTAAATGC-3′ (reverse).
Reverse transcription-PCR and real-time PCR.
Cells were washed once with PBS and then directly harvested in TRIzol reagent with vigorous pipetting to homogenize the cellular lysates. Total RNA was treated with DNase (Ambion) to remove any residual genomic DNA and quantified by UV spectrometry. One microgram of total RNA was used to synthesize cDNA using the iScript kit (Bio-Rad, Hercules, CA) according to the manufacturer's recommended protocol. The final cDNA product was purified and eluted in 50 μl of Tris-EDTA buffer using PCR purification columns (Qiagen, Hilden, Germany).
For quantitative real-time PCR analysis of mRNA transcript abundance, PCRs were made up using a 2× Sybr green PCR master mix (Applied Biosystems, Foster City, CA), along with gene-specific primers, and thermocycling was performed in the ABI 7300 thermocycler system (Applied Biosystems). All data were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as an internal standard. The results were expressed as the mean ± standard error of the mean of three independent experiments. The primer sequences for each gene were as follows: mouse StAR, 5′-GTGGTGTCATCAGAGCTGAACACGGCCCCAC-3′ (forward) and 5′-CTGCGATAGGACCTGGTTGATGATTGTC-3′ (reverse); mouse 3β-HSD, 5′-CTGGAAACTGTGAGCTTCCTCCTGAGTCCA-3′ (forward) and 5′-CTCCCAGCTGACAAGTGGCTCATAGCCCAG-3′ (reverse); mouse CYP21, 5′-CTGGAACCTGGGAAGAATCCCAGAACACCA-3′ (forward) and 5′-CAGGGTTCCATCTGGTGGAGGCAGCAGAGTG-3′ (reverse); and mouse CYP17, 5′-CGTGCATTGGAGAGGCTCTGGCCCG-3′ (forward) and 5′-GGGTCGATCAGAAAGACCACCTTGGGG-3′ (reverse).
RESULTS
SUMO modification of SF-1 in Y1 adrenocortical cells.
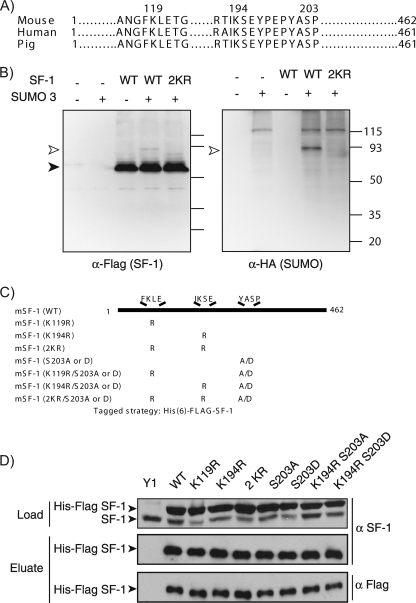
SF-1 harbors two evolutionarily conserved sequences that conform to the SUMOylation/SC motif consensus (Fig. 1A). To examine the SUMO modification of SF-1, we isolated His-FLAG-tagged SF-1 from Cos-7 cells coexpressing HA-SUMO3 by Ni2+ chelate chromatography under denaturing conditions. Western blot analysis of the preparations using anti-HA antibodies (Fig. 1B) revealed that a major HA-immunoreactive band was detected only in samples derived from cells coexpressing SUMO and SF-1. This band corresponded to covalently SUMO-modified SF-1, since it was also visible as a minor Flag-immunoreactive species (Fig. 1B). It is important to note that the migration of such branched, SUMO-modified proteins during SDS-PAGE is highly anomalous, and their molecular weights cannot be interpolated from linear polypeptide data. Thus, we have found that the behavior of such branched chains depends strongly and importantly, in a highly nonlinear manner, on the relative length of each of the arms (61). Given the central position of the SUMOylation sites in SF-1 (see below), the conjugation of a single SUMO moiety can give rise to a large reduction in mobility, as we have observed previously (61). Notably, and as is the case for most SUMOylated proteins, the extent of SF-1 modification appeared to be relatively low (<10%). Importantly, the SUMO modification was not observed in a mutant SF-1 in which the acceptor lysines within the two SUMOylation motifs were replaced with arginines. These results are consistent with observations made by other groups using similar heterologous systems (11, 40, 42). Whether this modification occurs in cellular contexts in which SF-1 is normally expressed, however, has not been examined. SF-1 is selectively expressed in the adrenal cortex, and in this regard, Y1 cells, derived from an adrenocortical carcinoma, are a useful model, since they retain expression of multiple adrenal-specific genes.
FIG. 1.
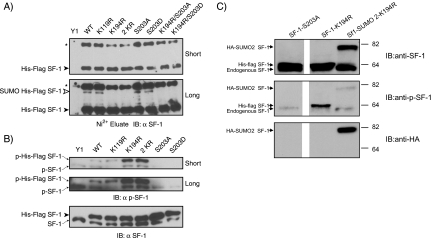
SF-1 SUMOylation. (A) Sequence alignment of the human, mouse, and pig SF-1 proteins showing the regions that contain the two potential SUMO sites (K119 and K194) and the phosphorylation site (S203). (B) COS-7 cells (2 × 106) were seeded in 10-cm plates and transfected 24 h later with 5 μg of SF-1 receptor and HA-SUMO3 expression vectors as indicated. After 48 h, cells were harvested and subjected to an in vivo SUMOylation assay as described in Materials and Methods. The cell lysates were subjected to Ni2+ bead pulldown, followed by anti-FLAG (left) or anti-HA (right) immunoblotting. The empty arrowheads indicate SUMOylated SF-1. The solid arrowhead indicates non-SUMOylated SF-1. (C) Schematic representation of the mouse SF-1 protein with the lysine-to-arginine and serine-to-alanine/aspartate SF-1 mutants generated in this study to determine potential SUMOylation and phosphorylation sites on SF-1. (D) Expression of HIS-FLAG-tagged SF-1 in stable Y1 cell lines. Lysates of Y1 cells (1 × 106) expressing HIS-FLAG-tagged WT SF-1 (WT), SUMO mutant SF-1 (K119R, K194R, and 2KR), phosphomutant SF-1 (S203A and S203D), or combined SUMO and phosphomutant SF-1 (K194RS203A and K194RS203D) were subjected to either anti-SF-1 immunoblotting (top) or Ni2+ bead pulldown, followed by anti-SF-1 immunoblotting (middle) or by anti-FLAG immunoblotting (bottom).
To facilitate the analysis of SF-1 SUMOylation, we created stable Y1 adrenocortical carcinoma cells expressing HIS-FLAG-tagged forms of SF-1. By expressing various forms of SF-1 (Fig. 1C), this approach permits the analysis of the interplay between SUMOylation and phosphorylation. Moreover, it allows an assessment of the functional consequences of these modifications, especially with respect to the dynamic SF-1 recruitment to, and regulation of, relevant endogenous genes. As can be seen in Fig. 1D, we generated lines expressing WT SF-1 or mutants bearing single or compound substitutions in SUMOylation sites alone or in combination with S203 mutations that prevent (S203A) or mimic (S203D) phosphorylation. Importantly, these forms are expressed at levels greater than those of the endogenous SF-1 and can be readily isolated and distinguished by virtue of the associated His and FLAG tags (Fig. 1D).
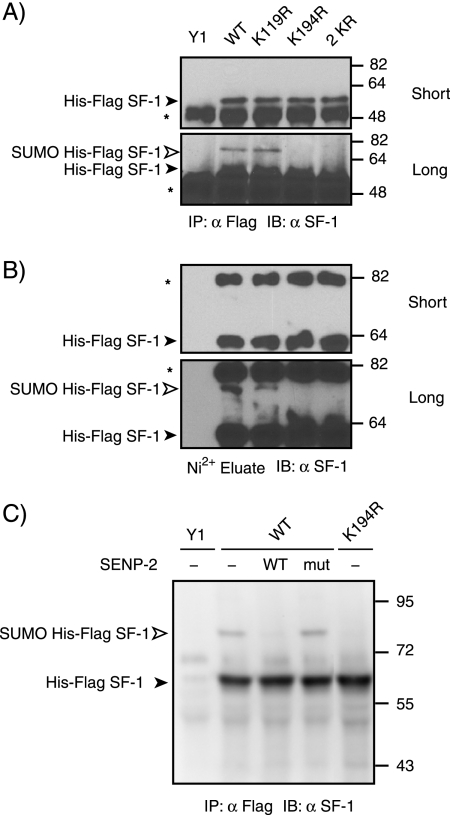
To determine which lysine residues in SF-1 are modified by endogenous SUMO in Y1 cells, we probed SF-1 preparations isolated by immunoprecipitation with anti-FLAG antibodies with a specific anti-SF-1 antibody. As can be seen in Fig. 2A, a slowly migrating species was detected in cells expressing WT SF-1. We interpret this form as being SF-1 modified by endogenous SUMO, since it is similar to the bona fide SUMO-modified form shown in Fig. 1B (the small mobility difference is attributable to the epitope tag, which is absent in the endogenous material). Moreover, this species is not observed in cells expressing an SF-1 form with mutations in both SUMOylation motifs (2KR). Interestingly, disruption of the first motif (K119R) produced no significant reduction in SF-1 SUMOylation. In contrast, mutation of K194 led to a complete loss of detectable SUMOylation. Similar results were obtained using SF-1 preparations isolated by Ni2+ chelate chromatography under denaturing conditions (Fig. 2B). To confirm that the slowly migrating species corresponded to SF-1 modified by endogenous SUMO, we treated immunopurified SF-1 with a recombinant catalytic domain of the SUMO protease SENP2. As can be seen in Fig. 2C, the slowly migrating species is sensitive to WT SENP2 but not to a catalytically inactive mutant. This form is not observed in cells expressing the K194R mutant. Taken together, these results indicate that, unlike heterologous systems, in which both motifs can serve as sites for SUMO conjugation (11, 42), only modification of K194 can be detected in the orthologous Y1 adrenocarcinoma cells.
FIG. 2.
Lysine residue 194 is the major SUMO site in SF-1. (A and B) Lysates of Y1 cells (1 × 106) stably expressing HIS-FLAG-tagged WT SF-1 (WT) or SF-1 in which lysine 119 (K119R), lysine 194 (K194R), or both lysines (2 KR) were mutated to arginine were subjected to either anti-FLAG immunoprecipitation (IP) followed by anti-SF-1 immunoblotting (IB) (A) or Ni2+ bead pulldown followed by anti-SF-1 immunoblotting (B). The asterisks indicate nonspecific bands. (C) FLAG immunoprecipitates from stably transfected HIS-FLAG-tagged WT and K194R Y1 cells were treated with 30 nM WT SENP2 or the C548S mutant form of SENP2 at 23°C for 30 min. Samples were resolved by SDS-PAGE and processed for immunoblotting using anti-SF-1 antibody. The open arrowhead indicates the position of slowly migrating SUMO-modified forms of HIS-FLAG-tagged SF-1, and the closed arrowhead indicates the position of free HIS-FLAG SF-1.
SUMOylation limits the transcriptional activity of SF-1 in Y1 cells without alterations in subcellular localization.
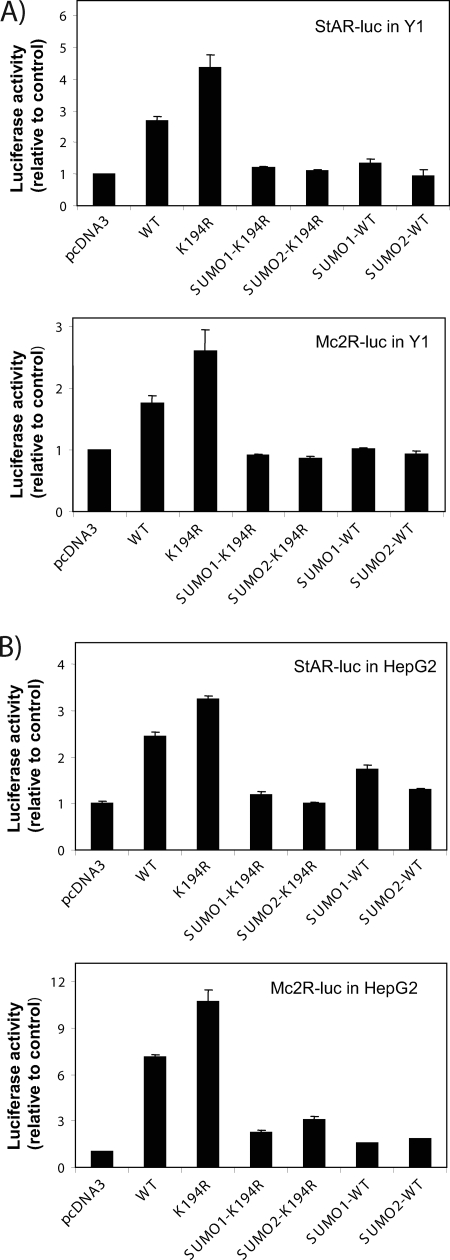
To gain insight into the role of SUMO modification of SF-1, we assessed the effect of this modification on SF-1-dependent transcription using two well-characterized SF-1-responsive regions derived from the StAR and Mc2R genes. These regions harbor multiple SF-1 binding sites and are therefore likely to be subject to SUMOylation-dependent synergy control. As can be seen in Fig. 3A, expression of WT SF-1 leads to an enhancement in the activity of a StAR promoter-driven luciferase reporter. Notably, expression of the SUMOylation-deficient K194R mutant yielded nearly twofold-higher activity. The enhanced activity of this mutant, however, is readily reversed by colinear fusion of either SUMO1 or SUMO2 at the N terminus of SF-1. This manipulation mimics a persistently SUMOylated state. Analysis of the Mc2R promoter yielded similar results. Furthermore, the functional effects of SF-1 SUMOylation are also evident in HepG2 cells (Fig. 3B). These findings indicate on one hand that the enhanced activity of K194R is likely due to loss of SUMOylation and on the other that recruitment of SUMO to SF-1 has significant inhibitory effects on its activity.
FIG. 3.
SUMOylation of SF-1 represses SF-1's transcriptional activity. Fifty thousand Y1 cells (A) or HepG2 cells (B) were plated on 24-well plates. After 24 h, the cells were transfected with 200 ng of HIS-FLAG-tagged SF-1 expression plasmids as indicated, as well as either 500 ng StAR-LUC or 500 ng Mc2R-LUC plasmids. Approxi-mately 40 to 48 h after transfection, the cells were harvested. Luciferase activity was measured with a Dual Luciferase Assay System (Promega) and normalized to Renilla activity. All experiments were performed three times in triplicate. SUMO1 (SUMO2)-WT (K194R) are SUMO fusion SF-1 proteins. The error bars indicate standard errors.
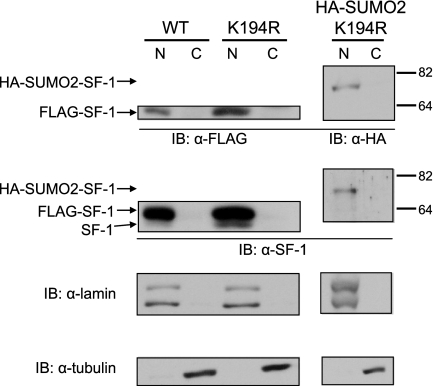
Given that the regulatory effects of SUMOylation can be accompanied by alterations in the subcellular localization of the modified protein (24), we examined whether SUMO conjugation to SF-1 is associated with modulation of its subcellular localization. As can be seen in Fig. 4, subcellular fractionation revealed that WT SF-1 is observable only in the nuclear fractions of Y1 cells. Moreover, this distribution was not visibly altered in cells expressing the K194R SUMOylation-deficient SF-1 or in the case of the N-terminal SUMO2 fusion to K194R SF-1. These results indicate that the transcriptional effects of SF-1 SUMOylation are unlikely to be due to alterations in subcellular localization and argue in favor of an intranuclear action for this modification.
FIG. 4.
SUMOylated SF-1 exhibits WT nuclear localization. Nuclear (N) and cytoplasmic (C) fractions of Y1 cells (1 × 106) stably expressing WT HIS-FLAG SF-1, K194R SF-1, or HA-SUMO2-K194R SF-1 were subjected to anti-FLAG, anti-HA-11, anti-SF-1, anti-lamin, and anti-tubulin immunoblotting (IB).
Nonreciprocal interaction between SF-1 SUMOylation and phosphorylation.
In contrast to the inhibitory effects of SUMOylation, phosphorylation at S203 is a strong stimulatory signal for the transcriptional activity of SF-1 (26). The opposing effects of these two modifications suggest that they may be coordinated. To investigate the interplay between phosphorylation and SUMOylation of SF-1, we first determined if the phosphorylation status of SF-1 influences its endogenous SUMOylation. To this end, we isolated SF-1 preparations by Ni2+ chelate chromatography under denaturing conditions from stable Y1 cells expressing the WT, as well as single and compound phosphorylation and SUMOylation mutants. As can be seen in Fig. 5A, probing with an anti-SF-1 antibody revealed SUMO-modified forms of both the phosphorylation-deficient (S203A) and phosphomimic (S203D) mutant forms of SF-1. SUMOylation of these mutants appears to occur at the same sites as for WT SF-1, since no SUMO modification was observed in the cases of mutants bearing the K194R mutation irrespective of the identity of the residue at position 203. These data indicate that SF-1 can be SUMOylated independently of the phosphorylation status of S203.
FIG. 5.
Nonreciprocal interaction between SF-1 SUMOylation and phosphorylation. (A) Lysates of Y1 cells (1 × 106) stably expressing the indicated forms of SF-1 were subjected to Ni2+ bead pulldown, followed by anti-SF-1 immunoblotting. The asterisks indicate nonspecific bands. (B) Whole-cell lysates of stable Y1 cells were subjected to anti-phospho-SF-1 (top) and anti-SF-1 (bottom) immunoblotting (IB). (C) Whole-cell lysates of Y1 cells stably expressing SF-1 K194R and HA-SUMO2-K194R were subjected to immunoblotting with the indicated antibodies. Lysates from cells transiently expressing SF-1 S203A were included as a nonphosphorylated control.
If the two modifications are independent, we anticipated a reciprocal relationship and that phosphorylation at S203 would be independent of SUMOylation. To our surprise, we observed a dramatic increase in S203 phosphorylation of the SUMOylation-deficient forms of SF-1 (K194R and 2KR), as revealed by a phospho-specific antibody (Fig. 5B). In contrast, the K119R mutation, which does not significantly alter SF-1 SUMOylation (Fig. 2), did not result in enhanced phosphorylation. As anticipated, the antibody did not detect the S203A and S203D mutants of SF-1. Because colinear fusion of SUMO to SF-1, which mimics a persistently SUMOylated state, counteracts the transcriptional effects of disrupting SF-1 SUMOylation (Fig. 3), we examined whether this extended to the phosphorylation status of SF-1. Indeed, as can be seen in Fig. 5C, the phosphorylation of the K194R mutant is diminished upon the colinear addition of SUMO2 (SUMO2 K194R). Quantitative analysis from multiple experiments indicated that, after normalization to the level of total SF-1, the extent of phosphorylation of the SUMO fusion is less than 25% of that observed for the K194R mutant alone. This indicates that the enhanced phosphorylation of K194R is due to loss of SUMOylation and that recruitment of SUMO to SF-1 limits the extent of its phosphorylation at S203. Taken together, these data demonstrate a nonreciprocal interaction between K194 SUMOylation and S203 phosphorylation. Whereas SUMOylation clearly limits phosphorylation, the converse does not appear to be true, since the phosphorylation status of SF-1 has no detectable influence on its SUMOylation.
The ACTH signaling cascade does not alter the SUMOylation of SF-1.
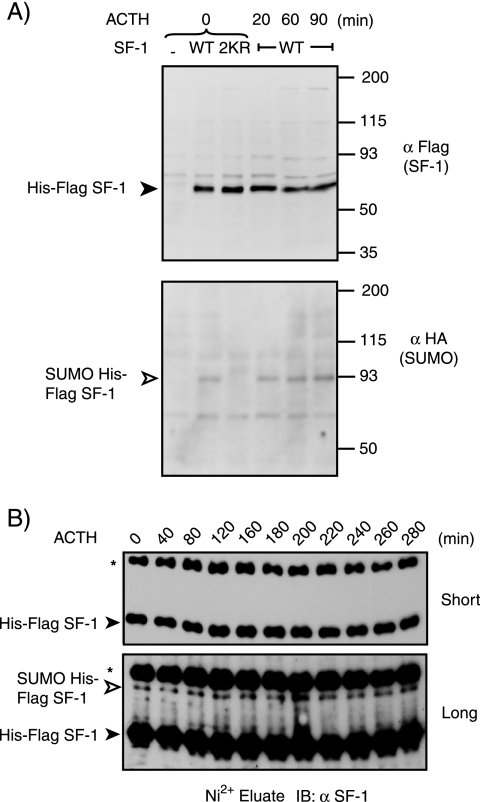
SF-1 is a major mediator of the transcriptional effects of ACTH in adrenocortical cells. ACTH treatment induces S203 phosphorylation and sets in motion a cyclical pattern of cofactor recruitment to SF-1 target genes (70). Given that SUMOylation exerts an important inhibitory effect on SF-1 function and limits S203 phosphorylation, we examined whether ACTH influences the SUMO modification of SF-1. We initially examined the SUMOylation of transiently expressed SF-1 in Y1 cells after release from α-amanitin to synchronize the transcriptional response of the cells. As can be seen in Fig. 6A, ACTH treatment did not significantly alter the levels of SF-1 SUMOylation. More detailed time course analysis in Y1 cells stably expressing WT SF-1 did not reveal significant temporal regulation of SF-1 SUMOylation in response to ACTH (Fig. 6B). These results indicate that, in contrast to S203 phosphorylation, K194 SUMOylation is not under direct regulation by ACTH, and consequently, the increase in S203 phosphorylation in response to ACTH is unlikely to be due to loss of K194 SUMOylation.
FIG. 6.
The level of SUMOylated SF-1 is not altered by ACTH treatment. (A) Y1 cells (1 × 106) were seeded in 10-cm plates and 24 h later were serum deprived in DMEM supplemented with 0.05% bovine serum albumin, followed by transfection with 3 μg HA-SUMO3 expression vector alone (−) or together with the indicated SF-1 or mutant SF-1 expression vector. Twenty-four hours after transfection, the cells were treated with 2.5 μM α-amanitin for 2 h. The cells were washed twice with PBS, and fresh serum-free medium was added 30 min prior to ACTH (10 nM) stimulation for the indicated times. The cells were harvested, and the lysates were subjected to Ni2+ bead pulldown, followed by anti-FLAG (top) or anti-HA (bottom) immunoblotting. The empty arrowhead indicates SUMOylated SF-1. The solid arrowhead indicates non-SUMOylated SF-1. (B) Lysates of Y1 cells (1 × 106) stably expressing HIS-FLAG WT-SF-1 and incubated for the indicated times with ACTH were subjected to Ni2+ bead pulldown, followed by anti-SF-1 immunoblotting (IB).
Loss of SF-1 SUMOylation increases the mRNA and protein levels of multiple SF-1 target genes responsible for steroidogenesis.
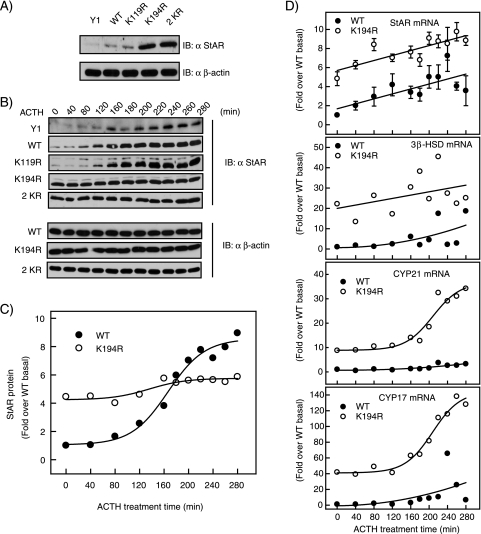
The robust steroid hormone synthesis capacity of adrenocortical cells depends on the expression of a battery of genes encoding multiple enzymes involved in steroidogenesis. As a major regulator of this process, SF-1 directly influences the expression of many such genes. We therefore examined the functional consequences of abrogating SF-1 SUMOylation on the regulation of endogenous SF-1 target genes in the steroidogenic pathway. Expression of the StAR protein, which is the rate-limiting factor in the steroidogenesis pathway, is directly regulated by SF-1 in response to ACTH. Thus, we first examined whether loss of SF-1 SUMOylation and its accompanying increase in phosphorylation resulted in greater expression of StAR protein. As depicted in Fig. 7A, basal StAR protein levels in the absence of ACTH were indeed increased in K194R and 2KR SF-1 stable Y1 cells. Notably, our ability to detect such an increase also argues that the endogenous levels of SF-1 do not mask the effects of the introduced variants. As expected, and consistent with previous reports, ACTH treatment led to a time-dependent increase in StAR protein in Y1 cells expressing WT SF-1 (Fig. 7B and C). Surprisingly, we found that ACTH did not further upregulate StAR protein levels in Y1 cells stably expressing the SUMOylation-deficient K194R and 2KR forms of SF-1 (Fig. 7B and C). In contrast, the K119R substitution, which affects neither SF-1 SUMOylation nor S203 phosphorylation, displayed a pattern indistinguishable from that of WT SF-1. Consistent with a transcriptional mechanism and with the results obtained using reporter constructs, the basal mRNA levels of StAR were markedly higher in cells expressing the K194R SF-1 mutant. In contrast, for cells expressing WT SF-1, StAR mRNA levels reached comparable levels only after prolonged stimulation with ACTH (Fig. 7D).
FIG. 7.
Loss of SF-1 SUMOylation increases the mRNA and protein levels of multiple steroidogenic-enzyme genes. (A) Lysates of Y1 cells (1 × 106) stably expressing the indicated forms of SF-1 (without ACTH treatment) were subjected to anti-StAR (top) and anti-β-actin (bottom) immunoblotting (IB). (B) Stable Y1 cell lines (1 × 106) were serum deprived for 48 h and treated with ACTH (10 nM) for the indicated times. Whole-cell lysates were prepared and subjected to anti-StAR immunoblotting or anti-β-actin immunoblotting. (C) The WT and K194R immunoblots for anti-StAR shown in panel B were quantified using a Bio-Rad phosphorimager. (D) Y1 cells (1 × 106) stably expressing WT HIS-FLAG SF-1 or K194R were treated with ACTH for the indicated times. Total RNAs were extracted from the cells, reverse transcribed, and amplified by quantitative PCR with GAPDH as an internal control. The data are presented as amounts over the RNA levels in WT HIS-FLAG SF-1 cells at time zero. Each point represents the average of three experiments, each with triplicate samples. After a 280-min incubation with ACTH, WT-HIS-FLAG SF-1-expressing cells displayed 3.5-, 18.8-, 3.2-, and 6.5-fold increases in StAR, 3β-HSD, CYP21, and CYP17 mRNAs, respectively. The error bars indicate standard errors.
Analysis of mRNA levels for other SF-1-regulated steroidogenesis genes, including 3β-HSD, CYP17, and CYP21, showed that in all cases basal levels were elevated in the cells expressing K194R mutant SF-1 cells (Fig. 7D). Interestingly, in cells expressing K194R SF-1, ACTH stimulation led to only modest further stimulation in the cases of StAR and 3β-HSD, whereas it led to significant time-dependent stimulation of CYP17 and CYP21 mRNA levels. Taken together, these results demonstrate that loss of SF-1 SUMOylation leads to enhanced expression of endogenous SF-1 target genes.
Loss of SUMOylation, but not phosphorylation, enhances SF-1 occupancy at target promoters.
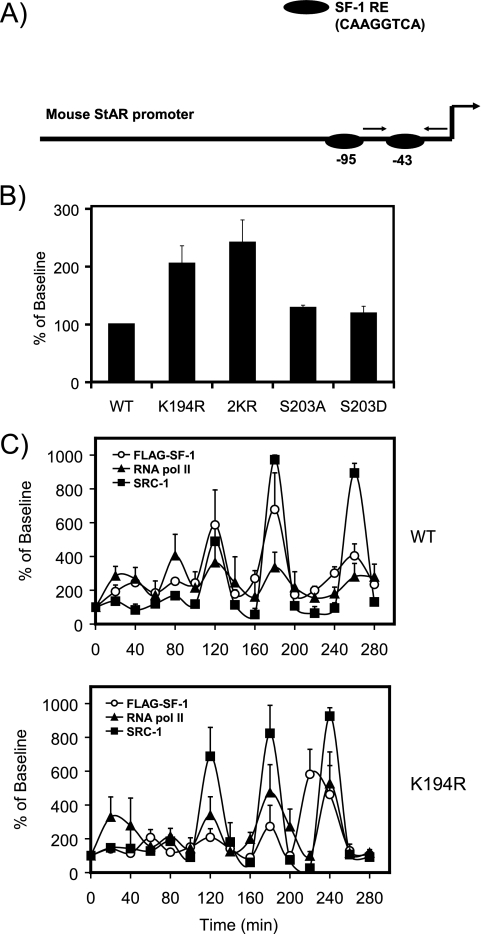
We previously determined that ACTH-induced phosphorylation of SF-1 at S203 regulates SF-1-dependent cyclic recruitment of coregulator proteins that culminates in the engagement of the RNA polymerase II transcriptional machinery. Given the significant functional effects of SF-1 SUMOylation, we examined the role of this modification in the regulated recruitment and clearance of SF-1 from endogenous target genes. Based on our observations that basal StAR expression is highly sensitive to SF-1 SUMOylation, we examined the recruitment of HIS-FLAG-SF-1 to the proximal promoter of the StAR gene by ChIP. This region of the promoter harbors two SF-1 response elements (Fig. 8A). Consistent with the mRNA and protein data, we observed enhanced occupancy for the SUMOylation-deficient (K194R and 2KR) forms of SF-1 under basal conditions (Fig. 8B). In contrast, and as we have previously demonstrated (26), neither the phosphorylation-deficient (S203A) nor the phosphomimicking (S203D) form of SF-1 displayed altered promoter occupancy (Fig. 8B).
FIG. 8.
Loss of SUMOylation, but not phosphorylation, enhances SF-1 promoter occupancy. (A) Diagram of the mouse StAR promoter showing the two SF-1 response elements. The primer pairs indicated by the arrows were used for ChIP analysis and amplified the proximal element. (B) ChIP assays were performed on HIS-FLAG-tagged SF-1 stable Y1 cell lines (1 × 106; serum deprived for 48 h and synchronized with α-amanitin) using anti-FLAG antibodies. The immunoprecipitates were analyzed by quantitative PCR using primers designed against the proximal mouse StAR promoter. The data were normalized to values obtained for 1% input controls, and the results are presented as percentage of baseline values. The error bars indicate standard errors. (C) ChIP assays were performed on HIS-FLAG-tagged WT and K194R SF-1 stable Y1 cell lines (1 × 106; serum deprived for 48 h and synchronized with α-amanitin) after ACTH (10 nM) treatment at specific time points using anti-FLAG antibodies. Anti-FLAG-immunoprecipitated lysates were re-ChIPed with anti-polymerase II or anti-SRC-1 antibodies as a function of time. The immunoprecipitates were analyzed by quantitative PCR using primers designed against the proximal mouse StAR promoter. The data were normalized to values obtained for 1% input controls, and the results are presented as percentages of baseline values.
To determine whether loss of SUMOylation alters the cyclical pattern of SF-1 and cofactor recruitment and release upon ACTH stimulation, we carried out ChIP assays in Y1 cells stably expressing WT SF-1 or the SUMOylation-deficient K194R mutant. We examined the time course of recruitment of SF-1 by performing ChIP assays using anti-FLAG antibodies. We also determined the extent of corecruitment of SRC1 and RNA-Pol II by re-ChIP with the corresponding antibodies. As shown in Fig. 8C, ACTH stimulation led to the cyclic recruitment of SF-1. Notably, although the basal level of SF-1 occupancy was higher, we did not observe significant changes in the dynamic pattern of SF-1 recruitment in cells expressing the SUMOylation-deficient form of SF-1. These results indicate that SUMOylation plays an important role in regulating the magnitude, but not the cyclical nature, of SF-1 recruitment to the StAR promoter and provide a plausible mechanism for the associated increase in the transcription of SF-1 target genes.
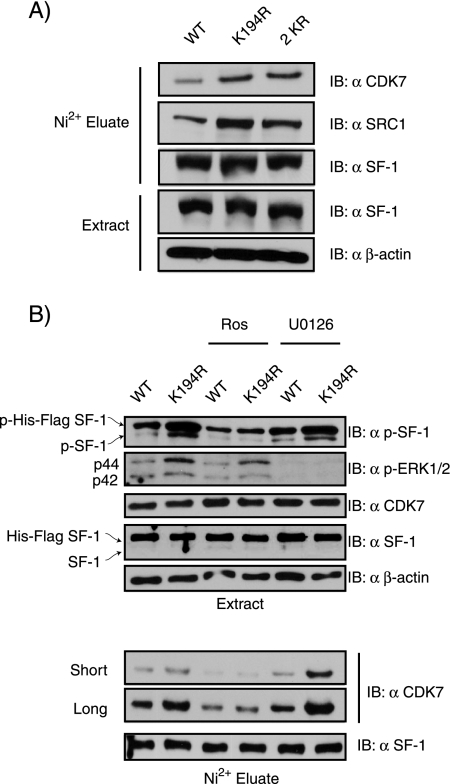
Loss of SF-1 SUMOylation enhances interaction with CDK7, a kinase required for S203 phosphorylation.
Recent findings support the view that SF-1 S203 phosphorylation is mediated by CDK7, since the kinase interacts with SF-1 and CDK7 inhibitors block both phosphorylation of SF-1 and its transactivation capacity (44). The enhanced S203 phosphorylation of SUMOylation-deficient SF-1 may thus involve CDK7. We therefore used Y1 cells stably expressing WT or SUMOylation-deficient K194R SF-1 to examine SF-1-associated proteins. As can be seen in Fig. 9A, SF-1 preparations isolated via Ni2+ chelate chromatography under nondenaturing conditions contained larger amounts of associated CDK7 in the case of the mutant SF-1 than in that of the WT (1.78- and 2.04-fold for the K194R and 2KR forms of SF-1, respectively). Consistent with the enhanced S203 phosphorylation, we also detected increased recovery of the coactivator SRC1 relative to WT SF-1 (1.87- and 1.86-fold for the K194R and 2KR forms of SF-1, respectively). This indicates that loss of SF-1 SUMOylation facilitates its interaction with CDK7. By using pathway-specific inhibitors, we found that the enhanced phosphorylation of the SUMOylation-deficient SF-1 appeared to be mediated by CDK7, since it was markedly reduced by treatment with the CDK7 inhibitor roscovitine. In contrast, the MAPK pathway inhibitor U0126 does not alter S203 phosphorylation, even though it effectively reduces phospho-ERK levels (Fig. 9B). Interestingly, roscovitine, but not U0126, also reduced the association of CDK7 with SF-1, suggesting that the catalytic activity of CDK7 is required for its association with SF-1 (Fig. 9B). Taken together, these novel findings suggest that the enhanced phosphorylation of the SUMOylation-deficient SF-1 in adrenocortical carcinoma cells is mediated, at least in part, by CDK7 and likely involves alterations in the association of the proteins.
FIG. 9.
Loss of SF-1 SUMOylation enhances interaction with CDK7, a kinase required for S203 phosphorylation. (A) Lysates of Y1 cells (1 × 106) stably expressing WT-SF-1, K194R, or 2KR were subjected to Ni2+ bead pulldown, followed by anti-CDK7, anti-SRC-1, or anti-SF-1 immunoblotting (IB). Whole-cell lysates from these cell lines were subjected to anti-SF-1 and anti-β-actin immunoblotting as controls. (B) Y1 cells (1 × 106) stably expressing WT-SF-1 or K194R were treated with either roscovitine (Ros) or U0126 for 1 h. The cell lysates were harvested and directly subjected to immunoblotting or Ni2+ bead pulldown, followed by immunoblotting with the indicated antibodies.
DISCUSSION
Here, we have shown for the first time that phosphorylation of SF-1 is altered by its SUMOylation status. Removal of SUMO conjugation from SF-1 upregulates the level of S203 phosphorylation, and this effect depends on CDK7 and is likely due to alterations in CDK7 recruitment. Our findings highlight how posttranslational modifications within the hinge region of SF-1 play significant roles in regulating SF-1's transcriptional activity.
Previous reports have demonstrated that SF-1 is a target of SUMO1 modification in heterologous systems (11, 40, 42). Whether other SUMO isoforms can modify SF-1 and whether this modification occurs in a more orthologous cellular system had not been examined. Our data indicate that SF-1 can be modified by SUMO3 and, importantly, that the endogenous machinery in adrenocortical cells can conjugate SUMO to SF-1. Whereas in heterologous systems species attributable to modification at both K119 and K194 can be detected (11, 40, 42), SF-1 SUMOylation in Y1 cells is sensitive only to disruption of K194. This is consistent with the reported preferential modification of K194 (11, 40, 42). Previous studies, as well as the present one, have indicated that replacement of K194 by an arginine residue prevents SUMOylation and leads to enhanced transcriptional activity. This is consistent with the reported inhibitory function of the underlying SC motif (40). Lysine residues, however, can be the targets of multiple modifications. Our finding that fusion of SUMO to the N terminus of SF-1 mimics the effects of the SC motif argues that the normal inhibitory function of the motif is indeed mediated by SUMO. By examining the regulation of endogenous genes, our findings indicate that SUMOylation is an important regulatory mechanism to control the overall activity of SF-1.
Studies of multiple sequence-specific transcription factors indicate that phosphorylation of residues in the vicinity of SUMOylation sites can alter SUMOylation (17, 29, 37, 59, 63, 66, 67). Thus, a subset of SUMOylation motifs in factors such as HSFs (29) and ERR α and γ (63, 67) conform to the recently described phosphorylation-dependent SUMOylation motif, in which an adjacent proline-directed phosphorylation site lies downstream of the core SUMOylation motif (PsiKXEXXSP). In this regard, phosphorylation may provide an additional negative charge, which appears to favor SUMOylation in certain contexts (72). Notably, whereas phosphorylation at S727 in STAT1 enhances SUMOylation at K703 (58), tyrosine phosphorylation at Y701 is mutually exclusive with K703 SUMOylation. In this regard, our findings indicate that SF-1 K194 SUMOylation is not affected by S203 phosphorylation, and thus, that this motif is not a phosphorylation-dependent SUMOylation motif. The substantially larger separation between the sites may be the basis for this observation.
On the other hand, the relationship between phosphorylation and SUMOylation is, surprisingly, not reciprocal, since we found that preventing SUMOylation enhances the phosphorylation level at S203. This is accompanied by an increased ability of SF-1 to upregulate endogenous target genes. The enhanced phosphorylation at S203 depends on CDK7, and its interaction with SF-1 is enhanced when SUMOylation is prevented. This indicates that CDK7 plays a key role as a mediator of S203 phosphorylation in response to loss of SUMOylation. Given that SUMOylation of SF-1 is a negative influence on its phosphorylation, exploring the exact mechanism of this antagonism will be very informative. One possibility is that SUMOylation sterically hinders access to kinases such as CDK7. The low stoichiometry of steady-state SUMOylation, however, argues that such a mechanism would have to be highly compartmentalized or dynamic. Determining whether SUMOylation and phosphorylation are mutually exclusive, however, will require examining the phosphorylation state of the small SUMO-modified pool of SF-1. Since S203 phosphorylation is a strong positive signal for SF-1, our findings also provide insight into the mechanism of SUMO-dependent transcriptional inhibition. Thus, in addition to the direct inhibitory role of SUMO in transcription (14, 30), our data indicate that SUMOylation can play an inhibitory role by preventing positively acting posttranslational modifications. In addition, the enhanced promoter occupancy of the SUMOylation-deficient SF-1 indicates that SUMOylation may act by limiting promoter occupancy. The time scale of such an effect, however, is likely different from that of the cyclical pattern initiated by ACTH treatment, since SUMOylation does not appear to alter such dynamics (70). To our knowledge, this is the first instance in which the SUMOylation status of a transcription factor governs its own phosphorylation. We anticipate, however, that this is likely to be a prevalent mechanism for other transcription factors or cell cycle regulators. Further studies in this area seem warranted.
Many studies support a model whereby ACTH activates ERK-dependent phosphorylation of SF-1 S203 and perhaps facilitates the generation of phospholipid ligands for SF-1. Interestingly, a recent study (44), using proliferating cancer cell lines, indicated that SF-1 can be phosphorylated/activated on the identical S203 residue by CDK7. This enzyme functions both as a CDK-activating kinase (CAK) by phosphorylating cell cycle CDKs and also as a component of the general transcription factor TFIIH. In this context, CDK7 phosphorylates the C-terminal tail of the largest subunit of polymerase II (7, 53). While the C-terminal tail of polymerase II can be phosphorylated and hence activated by multiple kinases (including CDK8 and CKD9) in different promoter and cellular contexts, CDK7 is unique, as it participates in both cell cycle regulation and transcription. Thus, activation of CDK7 occurs only in the context of the CDK7-cyclin H-Mat1 complex in cells engaged in the cell cycle. Moreover, CDK7 is itself activated by its own targets, CDK1 and CDK2, supporting a feed-forward amplification. Such a mechanism is thus predicted to sustain proliferation induced by mitogenic stimuli (20). In addition, CDK7 is a component of a variety of nuclear receptor complexes and has been shown to phosphorylate multiple nuclear receptors to facilitate active transcription and engagement with RNA polymerase II (10). In this view, CDK7-mediated phosphorylation of nuclear receptors provides a mechanism to activate a unique subset of nuclear receptor genes that are coupled to proliferation. Our current finding that increased phosphorylation of SF-1 upon loss of SUMOylation is likely mediated by CDK7 reveals an additional mechanism by which CDK7 regulates the transcriptional activities of nuclear receptors. Future efforts will aim to characterize the molecular mechanisms by which SUMOylation regulates both CDK7 association and the phosphorylation of SF-1.
The transcriptional activities of steroidogenic enzymes regulated by SF-1 are thought to be dependent on the cell type, promoter context, and cell-signaling pathways (1, 3, 28, 41, 71). The combined observations that loss of SUMOylation enhances the SF-1 occupancy and activity of target promoters and that ACTH does not appear to regulate SF-1 SUMOylation indicate that SUMOylation exerts a tonic inhibitory effect on SF-1 activity. The consequences of the interplay between ACTH-induced alterations in SF-1 function and SUMOylation, however, appear to be promoter dependent. Thus, for genes such as StAR and 3β-HSD, loss of SUMOylation is sufficient for nearly maximal activation, whereas for CYP21 and CYP17, ACTH can further enhance the elevated basal levels caused by loss of SUMOylation. These differences support a model in which loss of SUMOylation favors promoter occupancy by SF-1 as well as CDK7-mediated phosphorylation of SF-1. For some promoters, this is sufficient for nearly complete induction, whereas for others, ACTH engagement of additional signaling cascades further enhances their transcription. In this regard, the S203D mutation in SF-1 has been shown to mimic phosphorylation and to increase transcriptional activity in numerous studies (2, 26, 73). However, our current data show that this substitution does not alter overall SF-1 promoter occupancy in the absence of ACTH stimulation. Thus, in addition to the tonic inhibitory effects of SUMOylation, other ACTH-initiated signals impinge on SF-1 through different pathways (ERK-mediated phosphorylation and/or generation of phospholipid ligands). The nature of such pathways and their significance will have to be evaluated in nontransformed cell lines and relevant in vivo systems. It is interesting that our recent studies have identified SF-1 response elements in the promoters of a number of upregulated kinases in adrenocortical cells, including the MAPK activator MAP4K2 (G. D. Hammer, unpublished observation). Moreover, additional kinases, including CDK10 and adenylate cyclase 4, are direct transcriptional targets of SF-1 in mouse Y1 adrenocortical cells (J. O. Scheys and G. D. Hammer, unpublished observation; 54). Thus, additional signaling pathways may be engaged as a result of loss of SF-1 SUMOylation. Clearly, exploring the mutual relationships between posttranslational modifications of SF-1 and signaling cascades is an important area for future research efforts.
In summary, we have identified a novel nonreciprocal relationship between posttranslational modifications within the hinge region of SF-1 that contribute to transcriptional activity in adrenocortical cells. The findings that non-SUMOylated SF-1 leads to enhanced recruitment to chromatin, increased association with CDK7, and concomitant phosphorylation, together with amplified transcriptional activity, support a general model in which SUMOylation participates in transcriptional repression in part by preventing additional activating posttranslational modifications of nuclear receptors.
Acknowledgments
We thank H. A. Ingraham for providing the phospho-specific SF-1 antibody.
This work is supported by grants NIH NIDDK DK62027 (G.D.H.) and NIH-NIDDK DK61656 (J.A.I.-L.), as well as NIH P60 DK20572 for core support and a grant from the University of Michigan Cancer Biology Training Program (T32 CA009676 to W.-H.Y.).
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Ariazi, E. A., R. J. Kraus, M. L. Farrell, V. C. Jordan, and J. E. Mertz. 2007. Estrogen-related receptor α1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol. Cancer Res. 571-85. [DOI] [PubMed] [Google Scholar]
- 2.Babu, P. S., D. L. Bavers, S. Shah, and G. D. Hammer. 2000. Role of phosphorylation, gene dosage and Dax-1 in SF-1 mediated steroidogenesis. Endocr. Res. 26985-994. [DOI] [PubMed] [Google Scholar]
- 3.Barry, J. B., J. Laganière, and V. Giguère. 2006. A single nucleotide in an estrogen-related receptor α site can dictate mode of binding and peroxisome proliferator-activated receptor γ coactivator 1α activation of target promoters. Mol. Endocrinol. 20302-310. [DOI] [PubMed] [Google Scholar]
- 4.Berta, M. A., N. Mazure, M. Hattab, J. Pouyssegur, and M. C. Brahimi-Horn. 2007. SUMOylation of hypoxia-inducible factor-1α reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 360646-652. [DOI] [PubMed] [Google Scholar]
- 5.Bohren, K. M., V. Nadkarni, J. H. Song, K. H. Gabbay, and D. Owerbach. 2004. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 27927233-27238. [DOI] [PubMed] [Google Scholar]
- 6.Borud, B., T. Hoang, M. Bakke, A. L. Jacob, J. Lund, and G. Mellgren. 2002. The nuclear receptor coactivators p300/CBP/cointegrator-associated protein (p/CIP) and transcription intermediary factor 2 (TIF2) differentially regulate PKA-stimulated transcriptional activity of steroidogenic factor 1. Mol. Endocrinol. 16757-773. [DOI] [PubMed] [Google Scholar]
- 7.Bregman, D. B., R. G. Pestell, and V. J. Kidd. 2000. Cell cycle regulation and RNA polymerase II. Front. Biosci. 5D244-D257. [DOI] [PubMed] [Google Scholar]
- 8.Burris, T. P., W. Guo, T. Le, and E. R. McCabe. 1995. Identification of a putative steroidogenic factor-1 response element in the DAX-1 promoter. Biochem. Biophys. Res. Commun. 214576-581. [DOI] [PubMed] [Google Scholar]
- 9.Carlone, D. L., and J. R. Richards. 1997. Functional interactions, phosphorylation, and levels of 3′,5′-cyclic adenosine monophosphate-regulatory element binding protein and steroidogenic factor-1 mediate hormone-regulated and constitutive expression of aromatase in bonadal cells. Mol. Endocrinol. 11292-304. [DOI] [PubMed] [Google Scholar]
- 10.Chen, D., E. Washbrook, N. Sarwar, G. J. Bates, P. E. Pace, V. Thirunuvakkarasu, J. Taylor, R. J. Epstein, F. V. Fuller-Pace, J. M. Egly, R. C. Coombes, and S. Ali. 2002. Phosphorylation of human estrogen receptor alpha at serine 118 by two distinct signal transduction pathways revealed by phosphorylation-specific antisera. Oncogene 214921-4931. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W. Y., W. C. Lee, N. C. Hsu, F. Huang, and B. C. Chung. 2004. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 27938730-38735. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T., H. Itoh, L. Subramanian, J. A. Iniguez-Lluhi, and K. Nakao. 2003. Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ. Res. 921201-1208. [DOI] [PubMed] [Google Scholar]
- 13.Chupreta, S., H. Brevig, L. Bai, J. L. Merchant, and J. A. Iniguez-Lluhi. 2007. Sumoylation-dependent control of homotypic and heterotypic synergy by the Kruppel-type zinc finger protein ZBP-89. J. Biol. Chem. 28236155-36166. [DOI] [PubMed] [Google Scholar]
- 14.Chupreta, S., S. Holmstrom, L. Subramanian, and J. A. Iniguez-Lluhi. 2005. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol. Cell. Biol. 254272-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford, P. A., C. Dorn, Y. Sadovsky, and J. Milbrandt. 1998. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol. 182949-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford, P. A., J. A. Polish, G. Ganpule, and Y. Sadovsky. 1997. The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1. Mol. Endocrinol. 111626-1635. [DOI] [PubMed] [Google Scholar]
- 17.Daniel, A. R., E. J. Faivre, and C. A. Lange. 2007. Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol. Endocrinol. 212890-2906. [DOI] [PubMed] [Google Scholar]
- 18.De Santa Barbara, P., N. Bonneaud, B. Boizet, M. Desclozeaux, B. Moniot, P. Sudbeck, G. Scherer, F. Poulat, and P. Berta. 1998. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol. Cell. Biol. 186653-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorn, C., Q. Ou, J. Svaren, P. A. Crawford, and Y. Sadovsky. 1999. Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J. Biol. Chem. 27413870-13876. [DOI] [PubMed] [Google Scholar]
- 20.Garrett, S., W. A. Barton, R. Knights, P. Jin, D. O. Morgan, and R. P. Fisher. 2001. Reciprocal activation by cyclin-dependent kinases 2 and 7 is directed by substrate specificity determinants outside the T loop. Mol. Cell. Biol. 2188-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 182046-2059. [DOI] [PubMed] [Google Scholar]
- 22.Gizard, F., B. Lavallee, F. DeWitte, E. Teissier, B. Staels, and D. W. Hum. 2002. The transcriptional regulating protein of 132 kDa (TReP-132) enhances P450scc gene transcription through interaction with steroidogenic factor-1 in human adrenal cells. J. Biol. Chem. 27739144-39155. [DOI] [PubMed] [Google Scholar]
- 23.Gong, Z., M. Brackertz, and R. Renkawitz. 2006. SUMO modification enhances p66-mediated transcriptional repression of the Mi-2/NuRD complex. Mol. Cell. Biol. 264519-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, B., S. H. Yang, J. Witty, and A. D. Sharrocks. 2007. Signalling pathways and the regulation of SUMO modification. Biochem. Soc. Trans. 351414-1418. [DOI] [PubMed] [Google Scholar]
- 25.Halvorson, L. M., M. Ito, J. L. Jameson, and W. W. Chin. 1998. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone β-subunit gene expression. J. Biol. Chem. 27314712-14720. [DOI] [PubMed] [Google Scholar]
- 26.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3521-526. [DOI] [PubMed] [Google Scholar]
- 27.Hay, R. T. 2005. SUMO: A history of modification. Mol. Cell 181-12. [DOI] [PubMed] [Google Scholar]
- 28.Herzog, B., J. Cardenas, R. K. Hall, J. A. Villena, P. J. Budge, V. Giguère, D. K. Granner, and A. Kralli. 2006. Estrogen-related receptor α is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J. Biol. Chem. 28199-106. [DOI] [PubMed] [Google Scholar]
- 29.HietaKangas, V., J. Anckar, H. A. Blomster, M. Fujimoto, J. J. Palvimo, A. Nakai, and L. Sistonen. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 10345-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmstrom, S., M. E. Van Antwerp, and J. A. Iniguez-Lluhi. 2003. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl. Acad. Sci. USA 10015758-15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hossain, A., and G. F. Saunders. 2003. Role of Wilms tumor 1 (WT1) in the transcriptional regulation of the Mullerian-inhibiting substance promoter. Biol. Reprod. 691808-1814. [DOI] [PubMed] [Google Scholar]
- 32.Iniguez-Lluhi, J. A., and D. Pearce. 2000. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 206040-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito, M., R. N. Yu, and J. L. Jameson. 1998. Steroidogenic factor-1 contains a carboxy-terminal transcriptional activation domain that interacts with steroid receptor coactivator-1. Mol. Endocrinol. 12290-301. [DOI] [PubMed] [Google Scholar]
- 34.Ito, M., R. N. Yu, and J. L. Jameson. 1997. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 171476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73355-382. [DOI] [PubMed] [Google Scholar]
- 36.Kabe, Y., M. Goto, D. Shima, T. Imai, T. Wada, K. Morohashi, M. Shirakawa, S. Hirose, and H. Handa. 1999. The role of human MBF1 as a transcriptional coactivator. J. Biol. Chem. 27434196-34202. [DOI] [PubMed] [Google Scholar]
- 37.Kang, J., C. B. Gocke, and H. Yu. 2006. Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 75-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawabe, K., T. Shikayama, H. Tsuboi, S. Oka, K. Oba, T. Yanase, H. Nawata, and K. Morahashi. 1999. Dax-1 as one of the target genes of Ad4BP/SF-1. Mol. Endocrinol. 131267-1284. [DOI] [PubMed] [Google Scholar]
- 39.Kelly, S. N., T. J. McKenna, and L. S. Young. 2004. Modulation of steroidogenic enzymes by orphan nuclear transcriptional regulation may control diverse production of cortisol and androgens in the human adrenal. J. Endocrinol. 181355-365. [DOI] [PubMed] [Google Scholar]
- 40.Komatsu, T., H. Mizusaki, T. Mukai, H. Ogawa, D. Baba, M. Shirakawa, S. Hatakeyama, K. I. Nakayama, H. Yamamoto, A. Kikuchi, and K. Morohashi. 2004. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 182451-2462. [DOI] [PubMed] [Google Scholar]
- 41.Kraus, R. J., E. A. Ariazi, M. L. Farrell, and J. E. Mertz. 2002. Estrogen-related receptor α1 actively antagonizes estrogen receptor regulated transcription in MCF-7 mammary cells. J. Biol. Chem. 27724826-24834. [DOI] [PubMed] [Google Scholar]
- 42.Lee, M. B., L. A. Lebeveda, M. Suzawa, S. A. Wadekar, M. Desclozeaux, and H. A. Ingraham. 2005. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 251879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leers-Sucheta, S., K. Morahashi, J. I. Mason, and M. H. Melner. 1997. Synergistic activation of the human type II 3β-hydroxysteroid dehydrogenase/δ5-δ4 isomerase promoter by the transcription factor steroidogenic factor-1/adrenal 4-binding protein and phorbol ester. J. Biol. Chem. 2727960-7967. [DOI] [PubMed] [Google Scholar]
- 44.Lewis, A. E., M. Rusten, E. A. Hoivik, E. L. Vikse, M. L. Hansson, A. E. Wallberg, and M. Bakke. 2008. Phosphorylation of steroidogenic factor 1 is mediated by cyclin-dependent kniase 7. Mol. Endocrinol. 2291-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, Z., and E. R. Simpson. 1997. Steroidogenic factor 1 (SF-1) and SP1 are required for regulation of bovine CYP11A gene expression in bovine luteal cells and adrenal Y1 cells. Mol. Endocrinol. 11127-137. [DOI] [PubMed] [Google Scholar]
- 46.Mizusaki, H., K. Kawabe, T. Mukai, E. Ariyoshi, M. Kasahara, H. Yoshioka, A. Swain, and K. Morohashi. 2003. Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by wnt4 in the female developing gonad. Mol. Endocrinol. 17507-519. [DOI] [PubMed] [Google Scholar]
- 47.Monte, D., F. DeWitte, and D. W. Hum. 1998. Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. J. Biol. Chem. 2734585-4591. [DOI] [PubMed] [Google Scholar]
- 48.Nachtigal, M. W., Y. Hirokawa, D. L. Enyeart-VanHouten, J. N. Flanagan, G. D. Hammer, and H. A. Ingraham. 1998. Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell 93445-454. [DOI] [PubMed] [Google Scholar]
- 49.Nishida, T., M. Terashima, K. Fukami, and Y. Yamada. 2007. Repression of E1AF transcriptional activity by sumoylation and PIASy. Biochem. Biophys. Res. Commun. 360226-232. [DOI] [PubMed] [Google Scholar]
- 50.Ou, Q., J. F. Mouillet, X. Yan, C. Dorn, P. A. Crawford, and Y. Sadovsky. 2001. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 1569-79. [DOI] [PubMed] [Google Scholar]
- 51.Owerbach, D., E. M. McKay, E. T. Yeh, K. H. Gabbay, and K. M. Bohren. 2005. A praline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337517-520. [DOI] [PubMed] [Google Scholar]
- 52.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The Nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108109-120. [DOI] [PubMed] [Google Scholar]
- 53.Riedl, T., and J. M. Egly. 2000. Phosphorylation in transcription: the CTD and more. Gene Expr. 93-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rui, X., J. Tsao, J. O. Scheys, G. D. Hammer, and B. P. Schimmer. 2008. Contributions of specificity protein-1 and steroidogenic factor 1 to Adcy4 expression in Y1 mouse adrenal cells. Endocrinology 1493668-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rytinki, M. M., and J. J. Palvimo. 2008. SUMOylation modulates the transcription repressor function of RIP140. J. Biol. Chem. 28311586-11595. [DOI] [PubMed] [Google Scholar]
- 56.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 153088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saotoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2756252-6258. [DOI] [PubMed] [Google Scholar]
- 58.Song, L., S. Bhattacharya, A. A. Yunus, C. D. Lima, and C. Schindler. 2006. Stat1 and SUMO modification. Blood 1083237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spengler, M. L., L. W. Guo, and M. G. Brattain. 2008. Phosphorylation mediates Sp1 coupled activities of proteolytic processing, desumoylation and degradation. Cell Cycle. 7623-630. [DOI] [PubMed] [Google Scholar]
- 60.Su, H. L., and S. S. Li. 2002. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 29665-73. [DOI] [PubMed] [Google Scholar]
- 61.Subramanian, L., M. D. Benson, and J. A. Iniguez-Lluhi. 2003. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biol. Chem. 2789134-9141. [DOI] [PubMed] [Google Scholar]
- 62.Sugawara, T., J. A. Holt, M. Kiriakidou, and J. F. Strauss III. 1996. Steroidogenic factor 1-dependent promoter activity of the human steroidogenic acute regulatory protein (StAR) gene. Biochemistry 359052-9059. [DOI] [PubMed] [Google Scholar]
- 63.Tremblay, A. M., B. J. Wilson, X. J. Yang, and V. Giguere. 2008. Phosphorylation-dependent sumoylation regulates estrogen-related receptor-α and -γ transcriptional activity through a synergy control motif. Mol. Endocrinol. 22570-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tremblay, J. J., A. Marcil, Y. Gauthier, and J. Drouin. 1999. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 183431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tremblay, J. J., and R. S. Viger. 1999. Transcription factor GATA-4 enhances Mullerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol. Endocrinol. 131388-1401. [DOI] [PubMed] [Google Scholar]
- 66.Vanhatupa, S., D. Ungureanu, M. Paakkunainen, and O. Silvennoinen. 2008. MAPK-induced Ser727 phosphorylation promotes SUMOylation of STAT1. Biochem. J. 409179-185. [DOI] [PubMed] [Google Scholar]
- 67.Vu, E. H., R. J. Kraus, and J. E. Mertz. 2007. Phosphorylation-dependent sumoylation of estrogen-related receptor α1. Biochemistry 469795-9804. [DOI] [PubMed] [Google Scholar]
- 68.Wang, M., Y. Ikeda, X. Lou, K. M. Caron, T. J. Weber, A. Swain, B. P. Schimmer, and K. L. Parker. 1997. Steroidogenic factor 1 plays multiple roles in endocrine development and function. Recent Prog. Horm. Res. 52167-182. [PubMed] [Google Scholar]
- 69.Watanabe, K., T. R. Clarke, A. H. Lane, X. Wang, and P. K. Donahoe. 2000. Endogenous expression of Müllerian inhibiting substance in early postnatal rat Sertoli cells requires multiple steroidogenic factor-1 and GATA-4-binding sites. Proc. Natl. Acad. Sci. USA 971624-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winnay, J. N., and G. D. Hammer. 2006. Adrenocorticotropic hormone-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor 1-dependent transcriptional activation. Mol. Endocrinol. 20147-166. [DOI] [PubMed] [Google Scholar]
- 71.Yang, C., D. Zhou, and S. Chen. 1998. Modulation of aromatase expression in the breast tissue by ERRα-1 orphan receptor. Cancer Res. 585695-5700. [PubMed] [Google Scholar]
- 72.Yang, S. H., A. Galanis, J. Witty, and A. D. Sharrocks. 2006. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 255083-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, P., and S. H. Mellon. 1996. The orphan nuclear receptor steroidogenic factor-1 regulates the cyclic adenosine 3′,5′-monophosphate-mediated transcriptional activation of rat cytochrome P450c17 (17 alpha-hydroxylase/c17-20 lyase). Mol. Endocrinol. 10147-158. [DOI] [PubMed] [Google Scholar]