Summary
Actin filament nucleation and turnover are interdependent processes in migrating cells, but the mechanisms coordinating these events are unknown. Coronin 1B influences motility, lamellipodial dynamics and actin filament architecture at the leading edge of Rat2 cells in a manner consistent with a role in coordinating filament formation and turnover. Coronin 1B interacts simultaneously with both Arp2/3 complex and Slingshot (SSH1L) phosphatase, two regulators of actin filament formation and turnover, respectively. Coronin 1B inhibits filament nucleation by Arp2/3 complex and this inhibition is attenuated by phosphorylation of Coronin 1B on Serine 2, a site targeted by SSH1L. Coronin 1B directs SSH1L to lamellipodia where it likely regulates Cofilin. Accordingly, depleting Coronin 1B increases phospho-Cofilin levels and expressing activated Cofilin partially suppresses the effects on lamellipodia dynamics of Coronin 1B depletion. Thus, Coronin 1B coordinates filament nucleation via Arp2/3 complex and turnover by Cofilin at the leading edge of migrating cells.
Introduction
Coronins are highly-conserved F-actin-binding proteins (Uetrecht and Bear, 2006). Functional studies in Dicytostelium amoeba, fibroblasts and thymocytes indicate that Coronins play an important role in lamellipodial protrusion, whole cell motility and chemotaxis (Cai et al., 2005; de Hostos et al., 1993; Foger et al., 2006; Mishima and Nishida, 1999), but the mechanism(s) by which Coronins influence motility are unknown. In yeast, one mechanism may be through inhibition of Arp2/3 complex (Humphries et al., 2002; Rodal et al., 2005), but the effects of mammalian Coronins on actin nucleation activity by Arp 2/3 complex are unknown. Mammalian Coronin 1B is ubiquitously expressed and localizes to the leading edge of migrating fibroblasts (Cai et al., 2005). The interactions of Coronin 1B or Coronin 1A with Arp2/3 complex are regulated by phosphorylation of Serine 2 via PKC, where phosphorylation of Ser2 reduced the interaction with Arp2/3 and diminished cell motility (Cai et al., 2005; Foger et al., 2006).
ADF/Cofilin proteins (hereafter referred to as Cofilin) control actin filament turnover at the leading edge and at other cellular locations (Bamburg, 1999). Mechanistically, Cofilin severs and potentially enhances depolymerization of filaments by cooperatively binding along the sides of actin filaments and inducing conformational changes in filament structure (Bamburg et al., 1999). In vivo, Cofilin regulates the dynamics of actin-based structures such as stress fibers, dendritic spines and lamellipodia (Dawe et al., 2003; Hotulainen et al., 2005; Zhou et al., 2004). The activity of Cofilin is regulated in a variety of ways including phosphorylation, PIP2 binding, intracellular pH changes and interactions with binding partners such as AIP1 (Bamburg, 1999).
Phosphorylation of Cofilin-serine 3 by LIM Kinase or TESK leads to decreased F-actin binding and inactivation of Cofilin (Stanyon and Bernard, 1999). Dephosphorylation of Serine 3 on Cofilin enhances F-binding and activates its severing/depolymerization activity (Agnew et al., 1995). Two classes of phosphatases act on Cofilin - the Slingshots and Chronophin (Huang et al., 2006). Slingshot is a family of atypical Serine/Threonine protein phosphatases that in mammals includes Slingshot-1, -2 and –3; Slingshot-1 and –2 exist as long and short isoforms (Niwa et al., 2002; Ohta et al., 2003). The long isoform of Slingshot-1 (SSH1L) functions during chemotaxis of hematopoeitic cells (Nishita et al., 2005). Regulation of SSH1L activity may occur via phosphorylation of serine residues in its C-terminus and binding of inhibitory 14-3-3 proteins (Nagata-Ohashi et al., 2004). In addition, SSH1L activity is greatly enhanced by its interaction with F-actin (Nagata-Ohashi et al., 2004; Soosairajah et al., 2005). The other known Cofilin phosphatase, Chronophin, is a HAD-type serine phosphatase that plays an important role in cytokinesis (Gohla et al., 2005).
Considerable evidence suggests that the activities of Arp2/3 complex and Cofilin are coordinately regulated at the leading edge of motile cells. Studies using correlative microscopy (Svitkina and Borisy, 1999) suggest an array treadmilling model for actin assembly and turnover within lamellipodia in which Arp2/3-dependent filament nucleation at the front of the lamellipodia was balanced by Cofilin-dependent depolymerization of filaments at the rear. Arp2/3 complex and Cofilin also work together to promote lamellipodial protrusion in quiescent adenocarcinoma cells stimulated with growth factors, albeit via a different mechanism (DesMarais et al., 2005). MTLn3 cells stimulated with EGF initiate protrusion when Cofilin severs filaments creating new barbed ends that are preferred sites for Arp2/3-dependent filament nucleation (Ichetovkin et al., 2002). Together, filament severing and filament nucleation synergize to produce a burst of new barbed ends at the leading edge that support lamellipodial protrusion (DesMarais et al., 2004). Finally, studies using quantitative fluorescent speckle microscopy in epithelial cells suggest that Arp2/3 complex and Cofilin activities within lamellipodia may be coupled (Ponti et al., 2005), but very little is known about how this occurs. In this work, we describe a molecular connection between the Arp2/3 complex and Cofilin activities at the leading edge of motile cells that involves Coronin 1B.
Results
Depletion of Coronin 1B reduces whole cell motility and modulates lamellipodial dynamics
To test the role of Coronin 1B in cellular motility, we depleted Coronin 1B in Rat2 cells and monitored the effects on whole cell migration and lamellipodial dynamics. An shRNA that selectively targets mouse and rat, but not human, Coronin 1B (Fig. 1A, B) decreased the level of Coronin 1B in mouse or rat cells (Fig. 1C, 4J) (Rubinson et al., 2003). Depletion of Coronin 1B leads to ~33% decrease in cell speed relative to uninfected cells, cells infected with a control shRNA (NS) or cells expressing the Coronin 1B shRNA and human Coronin 1B-GFP that is refractory to the shRNA (Fig. 1D). Since the decreased rate of cell motility was rescued by expressing human Coronin 1B-GFP, this effect is specifically due to loss of Coronin 1B and not due to off-target silencing. Thus, Coronin 1B is required for normal whole cell motility.
Figure 1. Depletion of Coronin 1B slows whole cell migration and alters lamellipodial dynamics.
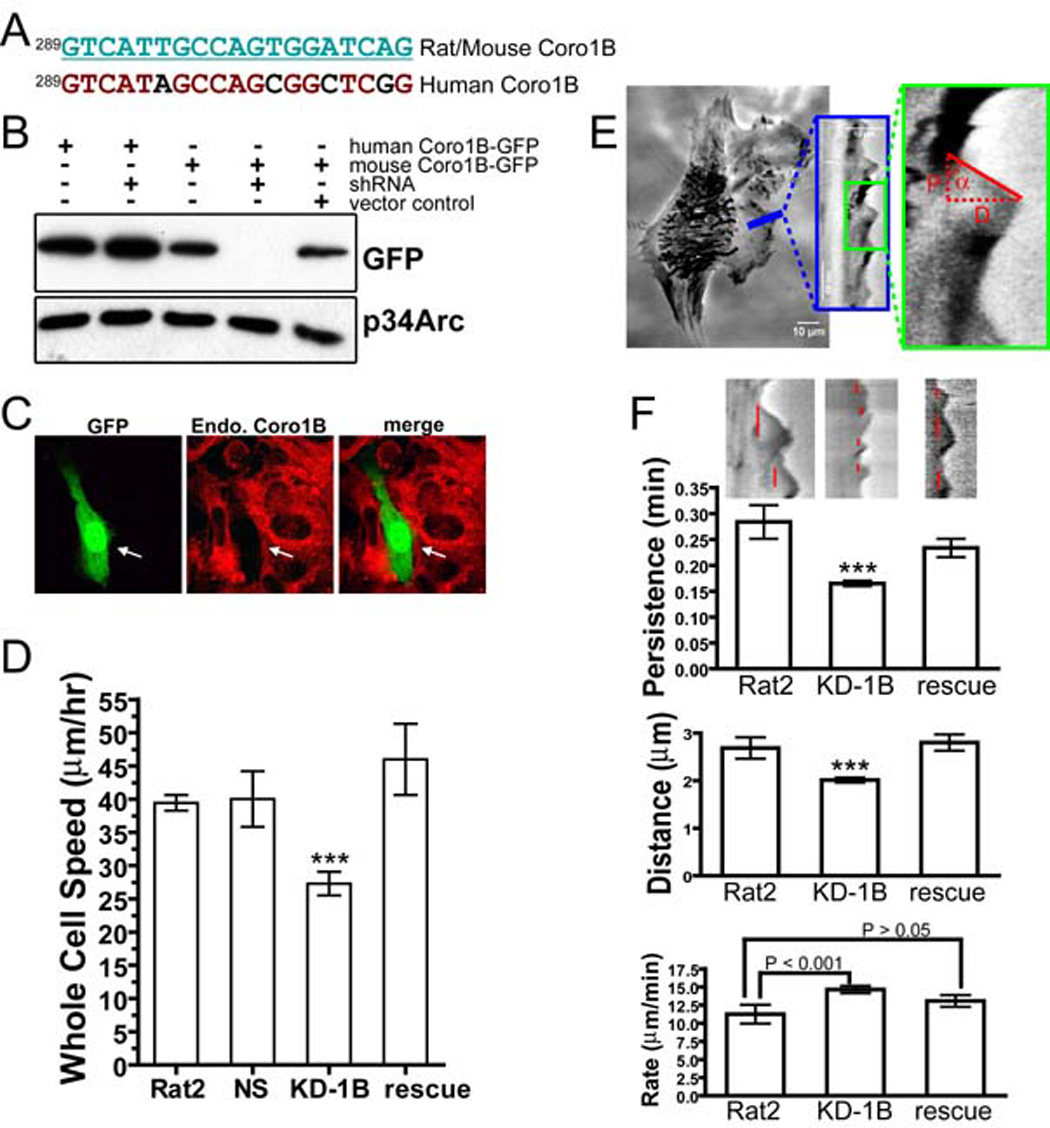
A- Target sequence of an shRNA designed to knock down expression of mouse or rat, but not human, Coronin 1B.
B- HEK293 cells were co-transfected with the Coronin 1B shRNA and GFP-tagged human or mouse Coronin 1B. Cell lysates were immunoblotted with anti-GFP to verify the efficiency and species specificity of knockdown. p34Arc protein was detected as a loading control.
C- NIH3T3 fibroblasts were transfected with Coronin 1B shRNA (indicated by GFP fluorescence) for 72 hours, and immunostained for Coronin 1B (red) to verify knockdown of the endogenous gene.
D- Mean cell speeds of Rat2 fibroblasts infected with lentivirus to express Coronin 1B shRNA (KD-1B, without or with (rescue) expression of GFP-tagged human Coronin 1B) or control shRNA (NS). Error bars represent the 95% confidence intervals. Newman-Keuls multiple comparison test was used after one-way ANOVA to generate the P values (P<0.001, ***).
E- Method for kymography analysis. Minimal intensity projection of a 300-frame 1-second-interval movie was presented on the right. Pixel intensities along a 1-pixel wide line (blue) were used to generate the kymograph presented in the blue box; a magnified region (outlined in green) is displayed on the right. Red dashed lines indicate the parameters for one protrusion. D is protrusion distance, P is protrusion persistence, and tanα is protrusion rate.
F- Protrusion parameters of Rat2 cells infected with lentivirus expressing the Coronin 1B shRNA without or with the rescue construct for expression of GFP–tagged human Coronin 1B (rescue). Sample kymographs corresponding to each treatment are shown above each bar; red lines indicate persistence time for each protrusion. The mean value for each parameters is presented; error bars indicate the 95% confidence intervals. Newman-Keuls multiple comparison test was used after one-way ANOVA to generate the P values (***P<0.001 when compared to Rat2 cells).
Figure 4. Interactions between Coronin 1B and Slingshot 1L.
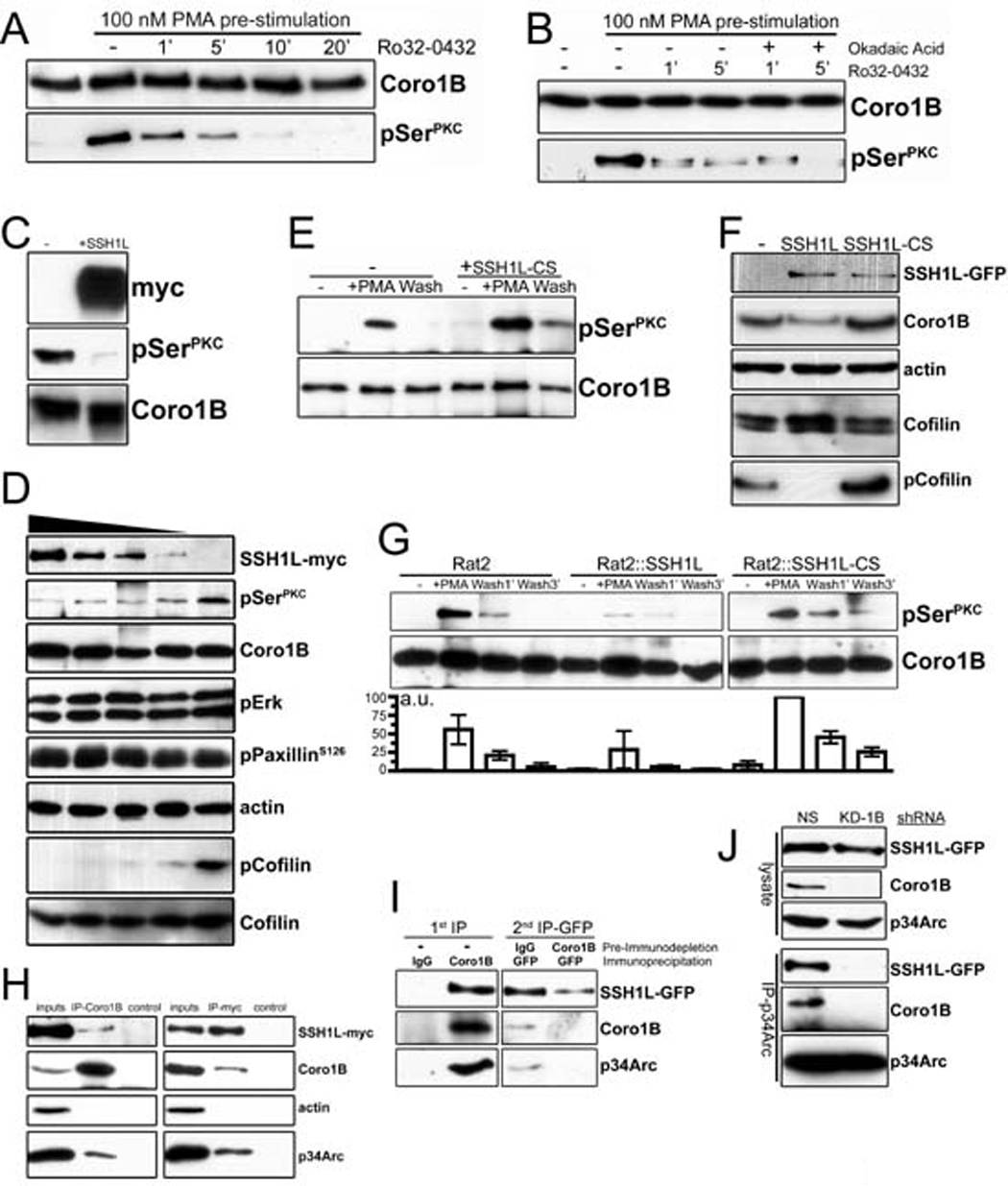
A- Rat2 fibroblasts stably expressing Coronin 1B-GFP were treated with 100 nM PMA for 30 min to stimulate Coronin 1B phosphorylation. Cells were washed with fresh media and incubated with the pan-PKC inhibitor Ro32-0432 (1 µM) to inhibit further phosphorylation. Coronin 1B was immunoprecipitated from lysates at the indicated times and blotted with anti-pSerPKC to monitor dephosphorylation kinetics.
B- Dephosphorylation assay carried out as in panel A with or without okadiac acid (100 nM).
C- Recombinant Coronin 1B was phosphorylated in vitro by PKC and subjected to in vitro dephosphorylation with SSH1L-myc immunoprecipitated from transfected HEK293 cells. Blots were reacted with anti-pSerPKC to detect phospho-Coronin 1B.
D- Serum starved Rat2 fibroblasts were treated with 100 nM PMA for 30 min and lysates were subjected to dephosphorylation in vitro by increasing concentrations of SSH1L-myc. Phospho-Coronin 1B was detected following immunoprecipitation as described above. Phosphorylation of PaxillinS126, Erk1/2 and Cofilin were monitored by immunoblotting the unbound fraction after Coronin 1B immunoprecipitation using phospho-specific antibodies. Blots shown are representative of at least three independent experiments.
E- Phospho-Coronin 1B was detected in control or HEK293 cells expressing myc-tagged dominant negative mutant SSH1L C393S (CS) as described in panel B.
F- Lysates of control Rat2 cells or cells expressing either WT SSH1L-GFP or SSH1L-CS-GFP were blotted with the indicated antibodies.
G- Rat2 cells stably expressing either WT SSH1L-GFP or SSH1L-CS-GFP were subjected to the in vivo dephosphorylation assay. Phospho-Coronin 1B was detected by anti-pSerPKC and quantified by densitometry relative to total Coronin 1B. Results from three independent experiments are presented as means with error bars indicating standard error of the means.
H- Lysates from HEK293 cells transiently expressing SSH1L-myc were immunoprecipitated with antibodies to endogenous Coronin 1B or SSH1L-myc and blotted with the indicated antibodies. Immunoprecipitations with rabbit or mouse IgG were performed as negative controls.
I- Endogenous Coronin 1B was immunodepleted from lysates of Rat2 cells stably expressing SSH1L-GFP. Control rabbit IgG was used for mock depletion. Residual SSH1L-GFP was immunoprecipitated from the immunodepleted lysates using anti-GFP antibody and blotted with the indicated antibodies.
J- Rat2 cells stably expressing SSH1L-GFP were infected with lentivirus expressing Coronin 1B shRNA (KD-1B) or a control shRNA (NS). Arp2/3 complex was immunoprecipitated using anti-p34Arc and blotted for SSH1L using anti-GFP. Similar results were obtained using anti-GFP to immunoprecitate SSH1L-GFP (not shown).
Since Coronin 1B is concentrated at the leading edge (Fig. S1), we reasoned that the effects of Coronin 1B depletion on whole cell motility might arise from defects in lamellipodial dynamics. We used kymography to quantify the effects of Coronin 1B depletion on lamellipodial dynamics (Fig. 1E). Three parameters of lamellipodial behavior were analyzed: protrusion rate, protrusion persistence and protrusion distance (Hinz et al., 1999). Depletion of Coronin 1B increased the protrusion rate and decreased the protrusion persistence and the distance protruded (Fig. 1F). Thus, Coronin 1B modulates lamellipodial dynamics, but is not absolutely required for protrusions to form.
Depletion of Coronin 1B slows retrograde actin flow, influences barbed end distribution and density, and actin architecture at the leading edge
The assembly and disassembly of actin filament networks underlie the dynamic behavior of lamellipodia. Since Coronin 1B depletion affects lamellipodia and Coronins bind F-actin, we tested if Coronin 1B depletion affects actin dynamics at the leading edge. The network of actin filaments assembling at the cell margin moves rearwards towards the cell body via retrograde flow. Using kymography of cells expressing GFP-actin to visualize actin, the rate of retrograde actin flow in Coronin 1B-depleted cells was reduced to ~50% the rate in control cells (Fig. 2A, B), supporting the idea that Coronin 1B modulates the dynamics of the actin network at the leading edge.
Figure 2. Depletion of Coronin 1B slows retrograde actin flow, influences barbed end distribution and filament architecture at the leading edge.
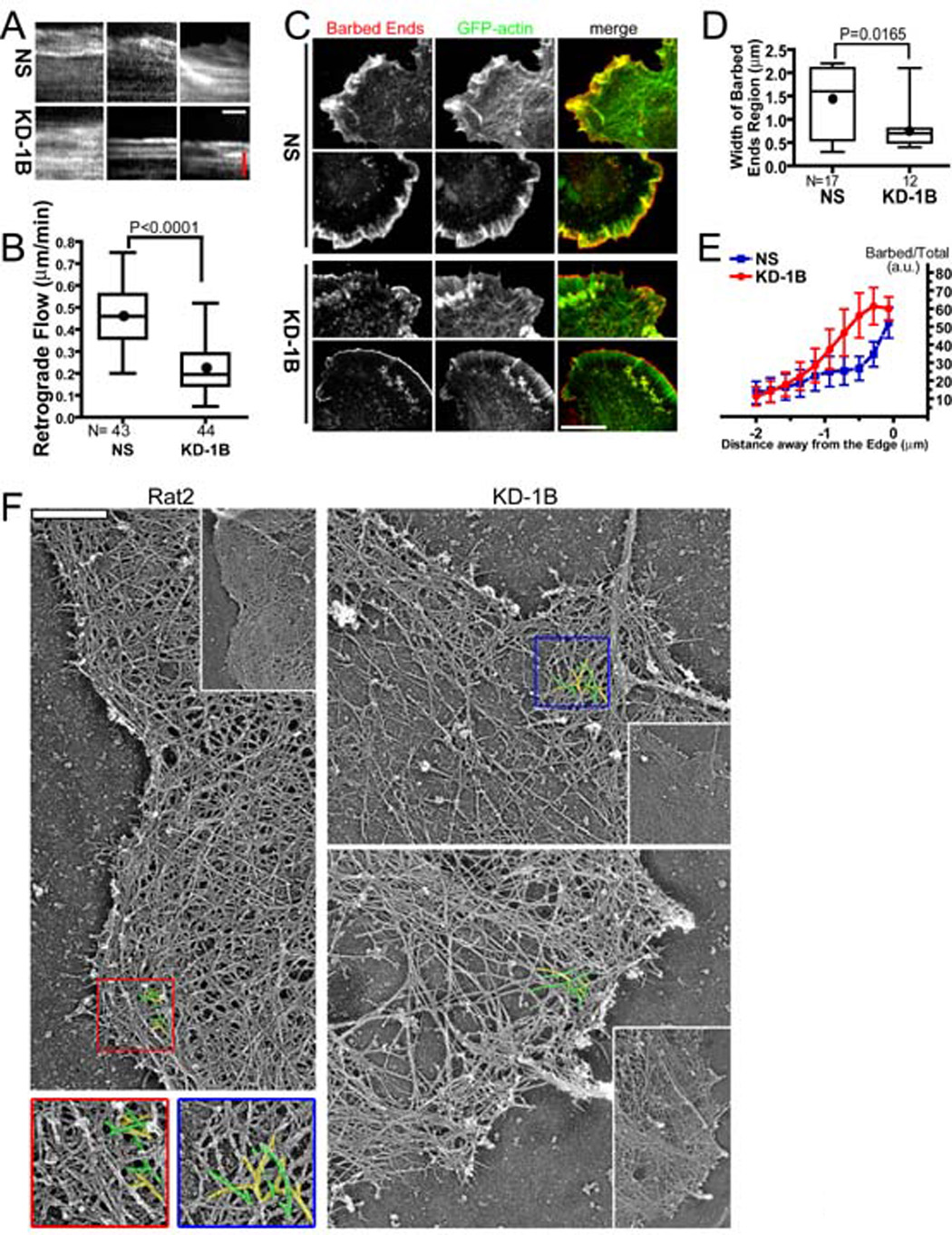
GFP-actin in Rat2 cells infected with Lentivirus expressing Coronin 1B (KD-1B) or control (NS) shRNA and GFP-actin was imaged for at 1 sec intervals for 2 min.
A- Three representative kymographs showing retrograde actin flow are presented for each condition. Red bar = 1.14 µm; white bar = 30 sec.
B- Average actin retrograde flow rate in KD-1B and NS expressing cells (3 measurements/cell, n = 40 cells) for each condition are presented as box and whisker plots (Dot = mean, middle line = median, top & bottom of box = 75% and 25%, whiskers = full data range). Unpaired student t-test indicates a signification difference between samples (P<0.0001).
C- KD-1B- and NS-expressing Rat2 cells were subjected to the barbed end assay using Alexa Fluor 568-labeled G-actin. Two cells for each condition are shown. Scale bar = 10 µm.
D- Quantification of free barbed ends in KD-1B and NS cells. Pixel intensities of Alexa-568-actin around the leading edge were plotted as described in Fig. S1. The region encompassing the top 50% of the barbed end signal is defined as the width of the barbed end zone (see Fig. S2). Width of this zone in KD-1B and NS cells is presented as a box and whisker plot. Unpaired student t test indicates a significant difference between the samples (P=0.0165).
E- The normalized ratio of barbed ends (Alexa-568 actin signal) to total actin (AlexaFluor647-phalloidin signal) was determined as a function of distance from the cell edge (see Fig. S2). Data were from cells shown in D and presented as mean with error bars indicating standard errors of the mean. Paired student t-test indicates a significant difference in barbed end density relative to F-actin between Coronin 1B-depleted and control cells (P=0.013 for the region 0.3µm - 1.4µm from the edge).
F- Platinum replica electron micrographs of lamellipodia in Coronin 1B-depleted and control Rat2 fibroblasts. Expanded views of each cell is presented as an inset. Branched actin filaments are pseudo colored with yellow and green. Scale bar = 500 nm.
A zone of rapidly growing filament barbed ends is a hallmark of the actin network at the leading edge (Condeelis et al., 1988). To measure the distribution and density of barbed ends in Coronin 1B depleted cells, we used an established assay to monitor barbed ends in situ (Symons and Mitchison, 1991). Depletion of Coronin 1B leads to a striking narrowing of the zone of barbed ends near the cell edge compared to control cells (Fig. 2C, D). In addition to altering the spatial distribution of barbed ends, Coronin 1B depletion increased the density of barbed ends relative to total F-actin (Fig. 2E). Thus, Coronin 1B inhibits the generation of barbed ends at the leading edge and alters their spatial distribution.
To examine the underlying actin filament architecture at the leading edge of Coronin 1B depleted cells, we used platinum replica electron microscopy. Rat2 cells have a robust and uniform dendritic network of actin filaments at the leading edge that is approximately 2µm wide (Fig. 2F). Cells depleted of Coronin 1B have an abnormal actin network characterized by densely branched filaments at the cell margin and a relative paucity of actin filaments at the rear of the lamellipodium (Fig. 2F). These changes in the organization of actin filaments are not observed in cells expressing a control shRNA or in Coronin 1B-depleted cells rescued with human Coronin 1B-GFP (Fig. S3). Thus, Coronin 1B appears to plays a role in coordinating assembly of actin filaments at the cell edge and disassembly of actin filaments at the rear of the lamellipodium.
Coronin 1B inhibits Arp2/3 complex activity in a phosphorylation-dependent manner
Yeast Coronin inhibits actin filament nucleation by Arp2/3 complex in vitro (Humphries et al., 2002). To determine if human Coronin 1B inhibits Arp2/3 complex nucleation activity, we added recombinant Coronin 1B to pyrenyl actin polymerization reactions. Coronin 1B had no effect on the rates of spontaneous actin assembly or of assembly nucleated from Spectrin-F-actin seeds (Fig. S6). However, in reactions containing Coronin 1B and GST-VCA-activated Arp2/3 complex, the rate of actin polymerization was reduced (Fig. 3A, B). To determine if phosphorylation at Ser2 regulates Coronin 1B’s inhibition of Arp2/3 complex, we compared wild-type Coronin 1B (WT), phosphorylated Coronin 1B (p-WT) and a phosphomimetic S2D mutant of Coronin 1B. In contrast to WT Coronin 1B, phosphorylated Coronin 1B (p-WT) and the S2D mutant Coronin 1B weakly inhibit Arp2/3 complex nucleation activity at all doses tested (Fig. 3B, Fig. S7). Furthermore, purified Arp2/3 complex bound directly to wild-type Coronin 1B, but did not bind to the phosphomimetic S2D Coronin 1B mutant (Fig. 3C), which corroborates previous immunoprecipitation experiments (Cai et al., 2005). Thus, Coronin 1B inhibits Arp2/3 complex nucleation in vitro and phosphorylation of Coronin 1B on Ser2 regulates this activity.
Figure 3. Coronin 1B inhibits actin nucleation by Arp2/3 complex in vitro.
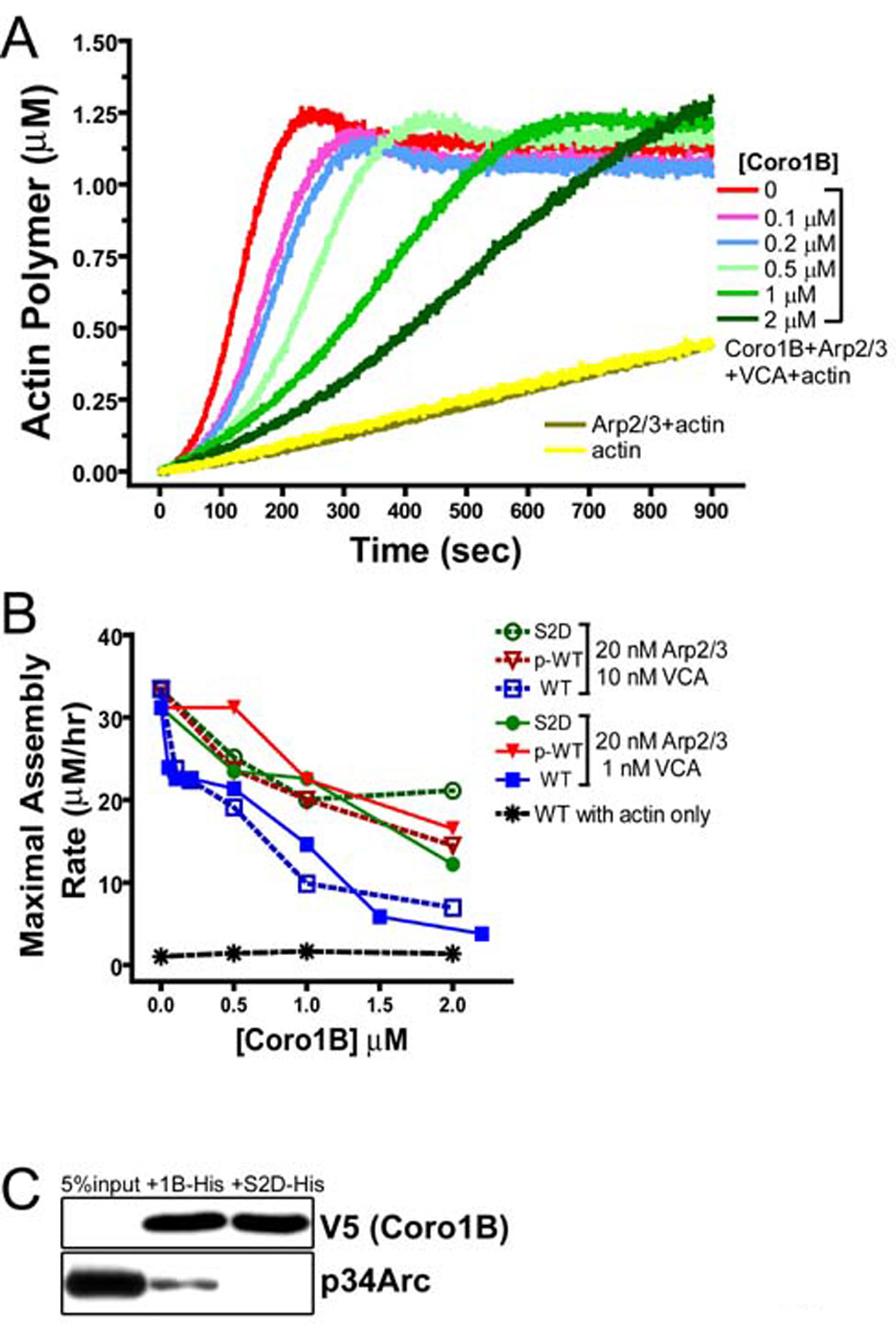
A- Plotted are actin polymer concentrations versus time in reactions containing 1.5 µM actin (5% pyrene labeled), 20 nM Arp2/3 complex, 10 nM GST-VCA, and Coronin 1B as indicated in figure. The curves labeled actin (yellow) and Arp2/3+actin (tan) contained neither Coronin 1B nor GST-VCA.
B- Phospho-Coronin 1B is less potent at inhibiting Arp2/3 complex. Plotted are the maximal rates of actin polymerization at varying Coronin 1B concentrations. WT, wild-type Coronin 1B (no detectable phosphorylation); p-WT, wild type Coronin 1B purified from PMA-stimulated cells (~75% phosphorylated); S2D, a mutant Coronin 1B with a phospho-mimetic aspartate substitution at Ser2. Maximal assembly rates were calculated from a linear fit to the rate of actin polymerization where 0.5–1.0 µM F-actin formed during each reaction. Data are derived from experiments presented in Fig. S7.
C- Coronin 1B binds Arp2/3 complex directly. His-tagged Coronin 1B or Coronin 1B (S2D) was bound to Ni-NTA beads and mixed with 20 nM bovine Arp2/3 complex for 1 hour. Immunoblotting detected Arp2/3 complex bound to the beads.
Coronin 1B is rapidly dephosphorylated by an okadiac acid-insensitive phosphatase
The phosphatase that dephosphorylates and activates Coronin 1B is unknown. To identify this 1B phosphatase, we developed an assay in which cells were first treated with PMA to stimulate maximal phosphorylation, followed by PMA washout in the presence of a pan-PKC inhibitor (Ro32-0432) to block further phosphorylation. Using this regime, dephosphorylation of Coronin 1B was detected within one minute and phospho-Coronin 1B returned to basal levels by 10 minutes (Fig. 4A). We examined the sensitivity of this phosphatase to the Ser/Thr phosphatase inhibitor okadaic acid, which potently inhibits PP1 and PP2A (Cohen et al., 1990). Okadaic acid at concentrations from 100nM (Fig. 4B) to 1µM (data not shown) had no effect on the rate of Coronin 1B dephosphorylation, making it unlikely that phospho-Coronin 1B is a substrate of either PP1 or PP2A.
Coronin 1B is a substrate of the Slingshot-1L phosphatase
We considered whether the Coronin 1B phosphatase might be Slingshot, which acts on Cofilin. Slingshots are among a small number of Ser/Thr phosphatases resistant to okadaic acid (Niwa et al., 2002). To test this hypothesis, we performed in vitro and in vivo dephosphorylation assays using Slingshot-1L (SSH1L). Recombinant Coronin 1B phosphorylated in vitro with purified PKCα was efficiently dephosphorylated by immunoprecipitated SSH1L-myc (Fig 4C). Since phosphatases often exhibit promiscuous activity on purified proteins, we tested whether SSH1L dephosphorylates Coronin 1B in cell lysates. Coronin 1B was phosphorylated by stimulating cells with PMA prior to lysis and increasing amounts of SSH1L-myc were then added; the phosphorylation status of Coronin 1B and other substrates was monitored by immunoblotting. Coronin 1B was dephosphorylated by SSH1L in a dose-dependent manner (Fig. 4D). Cofilin was also efficiently dephosphorylated and may be a preferred substrate. In contrast, no detectable dephosphorylation of phospho-Erk1/2 or phospho-Paxillin was detected, suggesting that the SSH1L phosphatase activity is highly specific under these conditions.
To determine if Coronin 1B is a substrate of SSH1L in vivo, we performed dephosphorylation assays in two different cell types. First, HEK293 cells were transiently transfected with dominant negative mutant form of SSH1L (SSH1L-CS), which harbors a mutation (C393S) in the phosphatase domain that renders it catalytically inactive (Ohta et al., 2003). In the presence of SSH1L-CS, phosphor-Coronin 1B was elevated after PMA stimulation and the return to basal levels after washing out PMA was delayed (Fig. 4E). Since ectopic SSH1L expression is relatively high in HEK293 cells, we confirmed these observations using Rat2 cell stably expressing lower levels of either wild-type (WT) SSH1L-GFP or dominant negative SSH1L-CS-GFP (Fig. 4F). As expected, GFP-tagged SSH1L localized to stress fibers, focal adhesions and the leading edge (Ohta et al., 2003) (Fig. S1A) in Rat2 cells and the level of phosopho-Cofilin decreased in lysates from Rat2 cells expressing WT SSH1L-GFP and increased in lysates from Rat2 cells expressing CS SSH1L-GFP. We conclude that, like Cofilin, Coronin 1B is a substrate of the SSH1L phosphatase in vitro and in vivo.
Coronin 1B, Slingshot 1L and Arp2/3 form a complex in vivo that is bridged by Coronin 1B
Coronin 1B and Arp2/3 complex interact in vivo (Cai et al., 2005). To determine if Slingshot 1L is part of this complex, we immunoprecipitated SSH1L and probed for Coronin 1B and Arp2/3 complex. SSH1L-myc interacted with endogenous Coronin 1B using reciprocal co-immunoprecipitations (Fig. 4H). Arp2/3 complex (as reported by the p34 subunit) was detected in both the Coronin 1B and SSH1L immunoprecipitates. Notably, actin was not detected, despite the fact that Coronin 1B, SSH1L and Arp2/3 complex all bind F-actin, indicating that the interaction among these components is not bridged by residual F-actin.
While these results are consistent with the existence of a ternary complex containing Coronin 1B, SSH1L and Arp2/3 complex, they do not exclude the possibility of two bivalent complexes (a Coronin 1B-Arp2/3 complex and a SSH1L-Arp2/3 complex). To identify a ternary complex, we used a two-step immunoprecipitation protocol in which Coronin 1B was first immuno-depleted from the lysates derived from Rat2:SSH1L-GFP cells followed by a second immunoprecipitation of SSH1L-GFP from the depleted lysate (Fig 4I). In the first step, anti-Coronin 1B (but not control IgG) immunoprecipitated both SSH1L-GFP and Arp2/3 complex. When SSH1L-GFP was immunoprecipitated from the Coronin 1B-depleted lysate, no additional Arp2/3 complex was co-immunoprecipitated. In contrast, SSH1L-GFP co-immunoprecipitated both Coronin 1B and Arp2/3 complex from a mock-depleted lysate.
To confirm this result, we tested for an interaction between SSH1L and Arp2/3 complex in Coronin 1B-depleted cells. No SSH1L was detected when Arp2/3 complex was immunoprecipitated from lysates derived from Coronin 1B-depleted, SSH1L-GFP expressing Rat2 (Fig. 4J). We conclude that SSH1L, Coronin 1B and Arp2/3 exist in a ternary complex that is bridged by Coronin 1B.
Depletion of Coronin 1B inhibits SSH1L-induced membrane ruffling
Expression of SSH1L-GFP in Rat2 cells induces constitutive hyper-ruffling at the cell periphery (Fig. 5A). Lamellipodia in SSH1L-expressing cells protrude at approximately twice the rate and protrusions are short-lived compared to control cells, with no difference in protrusion distance. Remarkably, depletion of Coronin 1B completely suppressed SSH1L-induced hyper-ruffling. One possible explanation is that Coronin 1B generally influences lamellipodial dynamics by a mechanism unrelated to SSH1L activity. To address this possibility, we investigated lamellipodia in Coronin 1B-depleted cells expressing FP4-CAAX, which induces hyper-ruffling by targeting Ena/VASP proteins to the plasma membrane (Bear et al., 2000; Bear et al., 2002). As expected, expression of FP4-CAAX increased protrusion rate and shortened protrusion persistence, similar to the effects of SSH1L expression, but depletion of Coronin 1B had no effect on the ruffling induced by FP4-CAAX. Thus, the suppression of the SSH1L-induced ruffling in Coronin 1B–depleted cells is a specific, rather than a general, effect on lamellipodial dynamics.
Figure 5. Depletion of Coronin 1B alters lamellipodial dynamics and inhibits SSH1L-induced membrane ruffling.
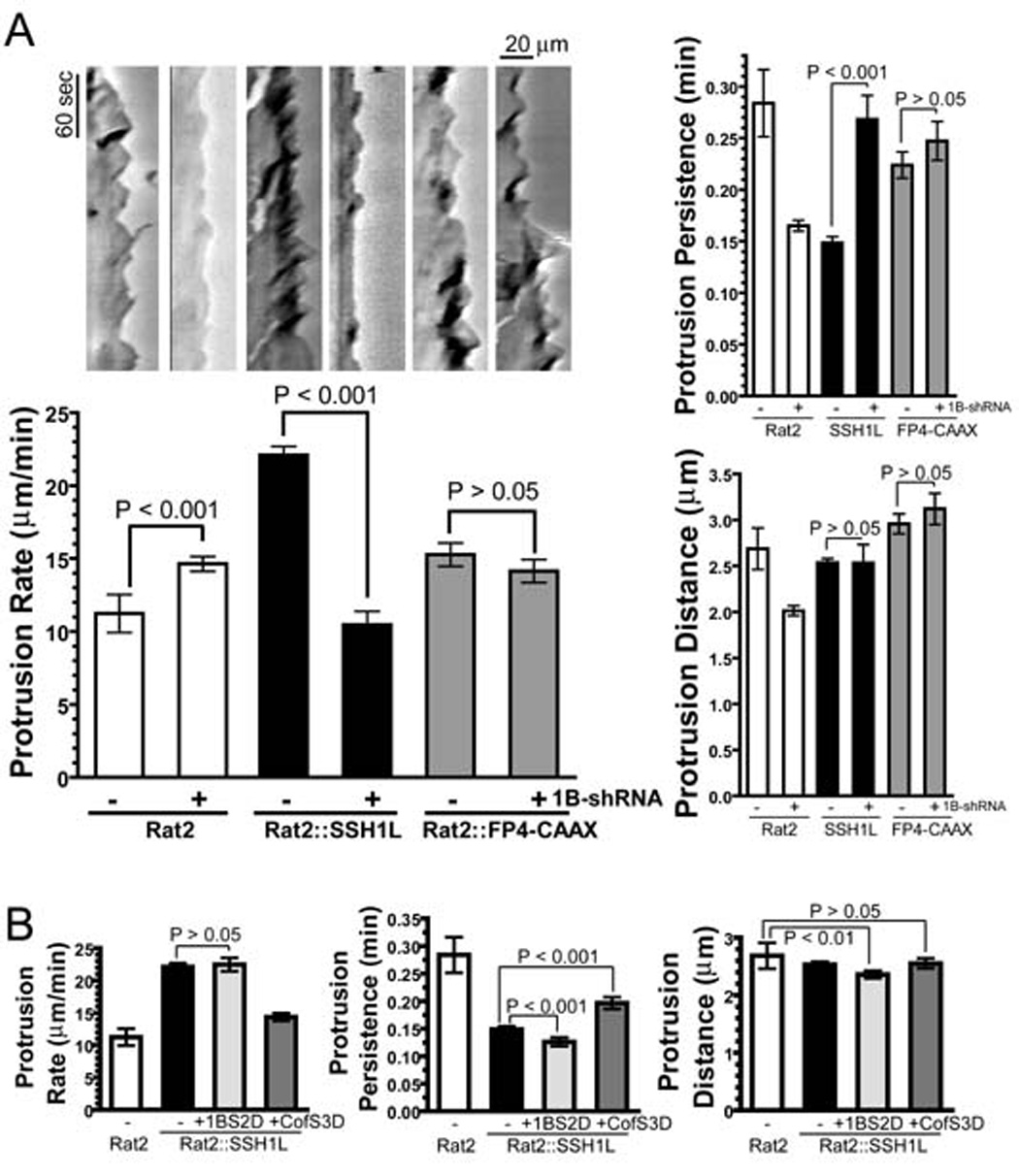
A- Depletion of Coronin 1B inhibits SSH1L-induced hyper-ruffling. Rat2 fibroblasts stably expressing SSH1L or FP4-CAAX were transfected with the Coronin 1B shRNA, and the cells were subjected to kymography analysis as described in Fig. 1E. Newman-Keuls multiple comparison test was used after one-way ANOVA to generate the P values.
B- Cofilin S3D suppresses SSH1L-induced hyper-ruffling. Phospho-mimetic mutants of either Coronin 1B (S2D) or Cofilin (S3D) were transiently expressed in Rat2 fibroblasts stably expressing SSH1L-GFP; kymography analysis was carried out as in Fig. 1E.
Suppression of SSH1L-induced ruffling upon Coronin 1B depletion may occur via dephosphorylation of Coronin 1B by SSH1L. Thus, a phosphomimetic mutant form of Coronin 1B would be predicted to suppress SSH1L-induced hyper-ruffling by competing with endogenous substrates. To test this hypothesis, lamellipodial dynamics were examined in cells expressing Coronin 1B S2D and SSH1L. To our surprise, the Coronin 1B S2D mutant is ineffective in suppressing SSH1L-induced hyper-ruffling (Fig. 5B). In contrast, expression of the phosphomimetic Cofilin S3D mutant suppresses SSH1L-induced hyper-ruffling (Fig. 5B), confirming that SSH1L-induced ruffling is due to the phosphatase activity of SSH1L. While these results do not preclude a contribution of SSH1L dephosphorylation of Coronin 1B in regulating lamellipodial behavior, they do suggest that regulation of Cofilin by SSH1L influences lamellipodial behavior. Alternately, Cofilin S3D may be a more effective competitor of SSH1L phosphatase than Coronin 1B S2D. Nonetheless, we sought another explanation for the potent suppression of the ectopic SSH1L-induced ruffling upon Coronin 1B depletion.
Depletion of Coronin 1B disrupts normal Slingshot 1L targeting to the leading edge
Coronin 1B may act upstream of SSH1L and thereby influence SSH1L-induced hyper-ruffling. For example, Coronin 1B may allow SSH1L to reach its substrate (Cofilin) and thereby influence lamellipodial dynamics. To test this idea, we compared the localization of SSH1L-GFP in control and in Coronin 1B-depleted cells. shRNAs were co-expressed with SSH1L-GFP using a single lentirviral vector (Fig. 6A), so that every cell expressing SSH1L-GFP also expressed either Coronin 1B-specific or control shRNA. SSH1L-GFP was not as enriched at the leading edge of Coronin 1B-depleted cells as in the control cells (Fig. 6B) when compared to the distribution of Cortactin (see Fig. S1). In control shRNA expressing cells, the maximal peaks of Cortactin and SSH1L-GFP were separated by an average of 0.35 µm. In Coronin 1B-depleted cells, Cortactin was distributed identically to that in control cells, but the peak of SSH1L-GFP signal was approximately twice as far rearward from the peak of Cortactin than in control cells (Fig. 6C). These data suggest that Coronin 1B is required to target SSH1L to a distinct region within lamellipodia and that proper targeting of SSH1L is required to exert an effect on lamellipodial dynamics.
Figure 6. Depletion of Coronin 1B disrupts targeting of Slingshot 1L to the leading edge.
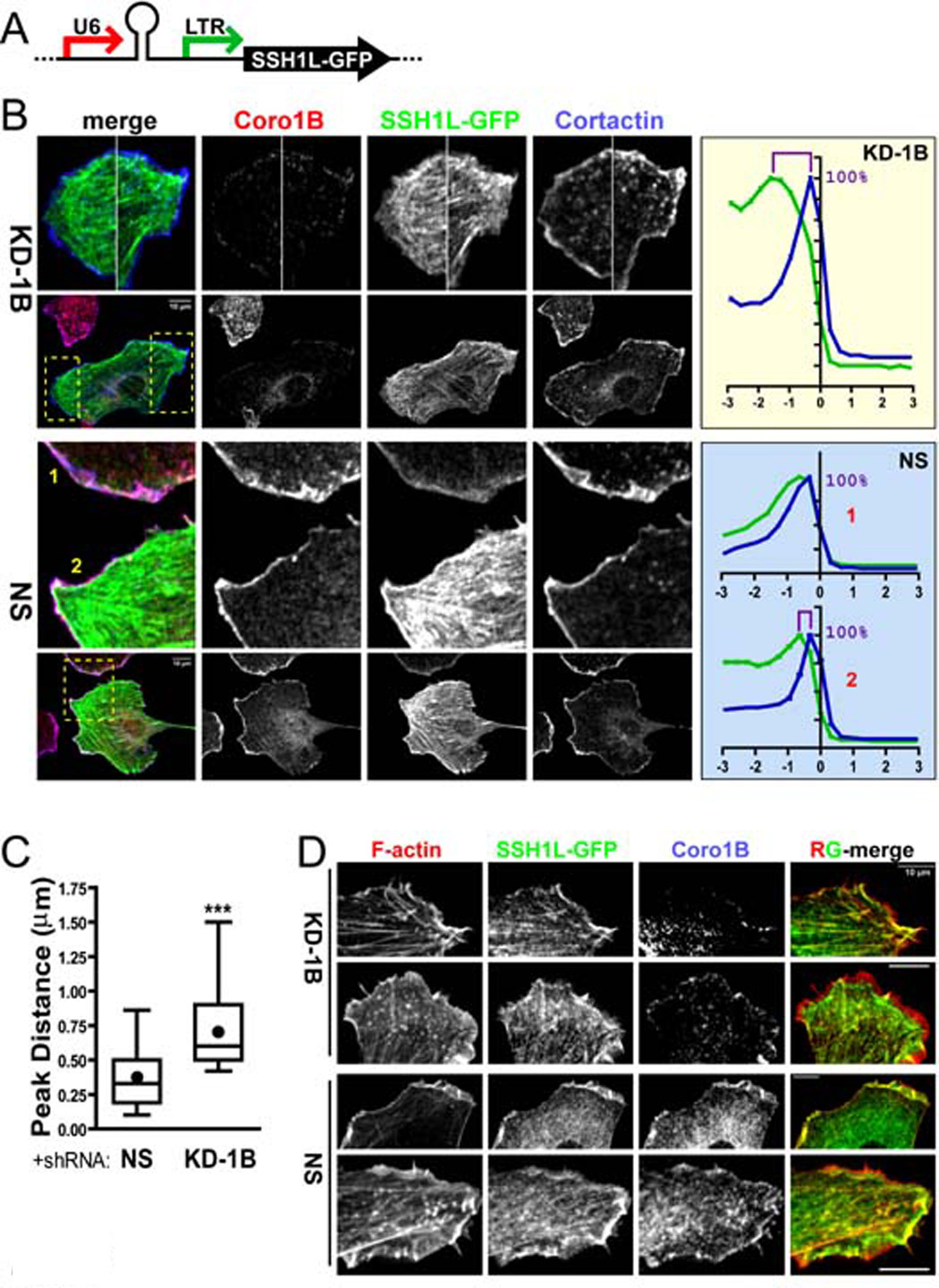
A- Diagram of the lentiviral vector combining Coronin 1B (KD-1B) or NS control shRNA expression from the Pol III U6 promoter and SSH1L-GFP expression from the MSCV 5’ LTR promoter.
B- Depletion of Coronin 1B alters the distribution of SSH1L-GFP upon EGF stimulation. Rat2 fibroblasts infected with lentivirus described in panel A were serum starved and stimulated with EGF for 15 min before fixation and staining with anti-Coronin 1B and anti-Cortactin. Regions boxed by yellow dash lines are magnified in the upper panels. Pixel intensities around the cell edge were extracted and plotted as described in Fig. S1. The relative intensities of Cortactin (blue) and SSH1L-GFP (green) are plotted for Coronin 1B depleted cells (KD-1B, upper panel) and for two control cells (NS, lower panels).
C- Relative distributions of SSH1L-GFP and Cortactin in control and Coronin 1B-depleted cells. The distance between the maximal intensity of SSH1L-GFP and that of Cortactin staining (panel B) was determined for Coronin 1B-depleted or control cells (n = 27 cells). Unpaired Student’s t-test indicates a significant difference between (KD-1B) and control (NS) cells (P<0.001).
D- Cells prepared as described in panel B were stained with anti-Coronin 1B and Alexa Fluor 568-phalloidin. RG-merge shows overlap between SSH1L-GFP and F-actin.
Previous studies suggested that binding of SSH1L to F-actin targeted it to the leading edge (Nagata-Ohashi et al., 2004). To examine the role of F-actin in localizing SSH1L within lamellipodia, we stained cells expressing SSH1L-GFP (+/− Coronin 1B shRNA) with phalloidin (Fig. 6D). In control cells, SSH1L-GFP and F-actin co-localized within lamellipodia, however in Coronin 1B-depleted cells, the distribution of F-actin was indistinguishable from that in control cells at the level of light microscopy, but SSH1L-GFP was excluded from the most distal, F-actin rich region near the leading edge. This result indicates that F-actin is insufficient to target SSH1L to the leading edge, but does not exclude a contribution of F-actin-SSH1L interactions for stimulating phosphatase activity (Nagata-Ohashi et al., 2004; Soosairajah et al., 2005).
Depletion of Coronin 1B inhibits endogenous Cofilin phosphatases
To test whether Coronin 1B depletion also inhibited endogenous Cofilin phosphatase activity, we compared the phospho-Cofilin levels in cells expressing Coronin 1B-shRNA and control shRNA. Cells depleted of Coronin 1B had higher levels of phospho-Cofilin relative to controls (Fig. 7A). This effect is not due activation of Cofilin kinases because Coronin 1B-depletion does not alter the level of active LIMK 1/2. To confirm the effect of Coronin 1B depletion on phospho-Cofilin levels, we determined the ratio of active-to-total Cofilin in Coronin 1B-depleted cells using ratiometric immunofluorescent imaging (Fig. 7B). In control cells, the ratio of active-to-total Cofilin was similar to that in surrounding uninfected cells; in Coronin 1B-depleted cells, the ratio of active-to-total Cofilin is lower than in uninfected cells, particularly in lamellipodial regions (Fig. 7B). The ratio of active-to-total Cofilin across the whole cell is approximately two fold lower in Coronin 1B–depleted cells (Fig. 7C). Taken together these results indicate that Coronin 1B enhances dephosphorylation and activation of Cofilin. Moreover, these findings are consistent with the biochemical and cellular effects of ectopic expression of SSH1L-GFP and suggest Coronin 1B influences Cofilin activity via a Slingshot-dependent mechanism.
Figure 7. Depletion of Coronin 1B increases phosphorylation of Cofilin and expression of activated Cofilin (S3A) partially rescues the effects of Coronin 1B depletion on lamellipodia dynamics.
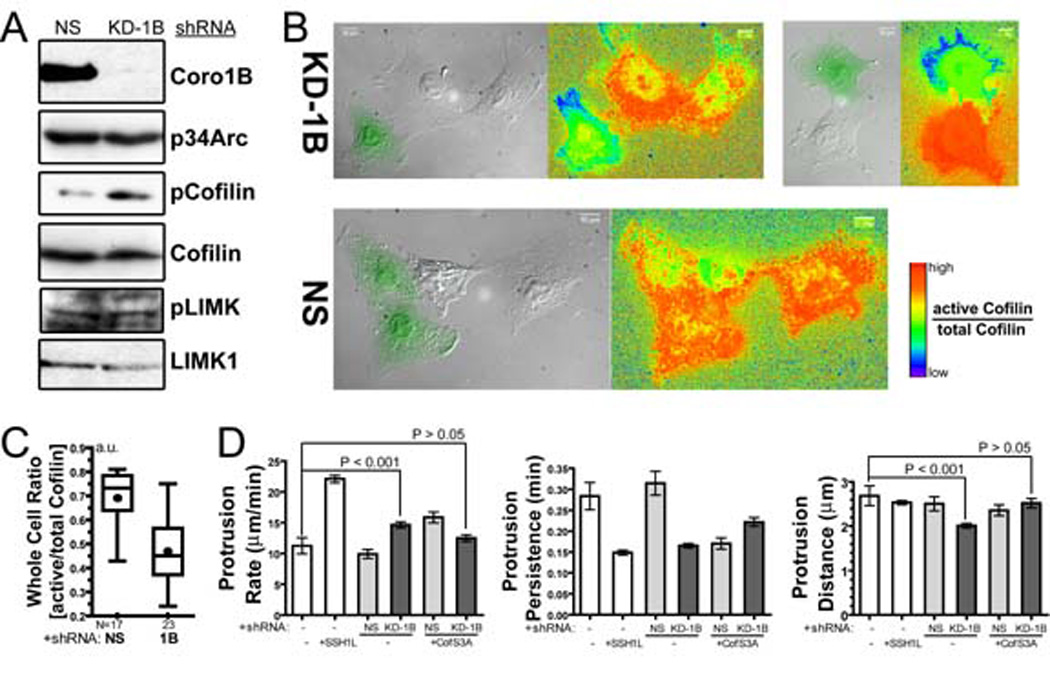
A- Depletion of Coronin 1B leads to elevated pCofilin, but not via activation of LIMK. Lysates from Rat2 cells infected with lentivirus expressing either the Coronin 1B shRNA (KD-1B) or control shRNA (NS) were blotted with the indicated antibodies.
B- Depletion of Coronin 1B decreases the level of active Cofilin in the lamellipodia. Cells generated as described in panel A were immunostained for total Cofilin (MAB22) and phospho-Cofilin (4321) and subjected to ratio imaging. GFP-positive cells in the DIC image express either Coronin 1B (KD-1B) or control (NS) shRNA. Ratios of Active/Total Cofilin were calculated from [(Total Cofilin – pCofilin) / Total Cofilin)] and presented using a rainbow look-up table.
C- Active/total Cofilin ratios in whole cells expressing Coronin 1B shRNA (KD-1B) or control shRNA (NS). Unpaired Student’s t-test indicates a significant difference between Coronin 1B-depleted and control cells (P<0.0001).
D- Expression of activated Cofilin (S3A) partially rescues the effects of Coronin 1B depletion on protrusive activity. Rat2 fibroblasts infected with Lentivirus for expressing constitutively activated Cofilin (S3A) and Coronin 1B shRNA were subjected to kymography analsysis. Protrusion parameters were determined as described in Fig. 1E. ANOVA test shows significant difference between the samples. Newman-Keuls multiple comparison tests were used to generate P values.
Activated Cofilin partially rescues the effects of Coronin 1B depletion on lamellipodial dynamics
Since Coronin 1B-depletion increases the amount of phospho-Cofilin, we posit that the effects on lamellipodial dynamics might be due, in part, to a failure to activate Cofilin at the leading edge. To test this idea, we co-expressed an activated Cofillin mutant (S3A) along with Coronin 1B or control shRNA. Lamellipodial protrusions extended faster and were shorter-lived in cells expressing Cofilin (S3A)-GFP and a control shRNA than those in either control cells expressing no Cofilin (S3A)-GFP or non-transfected cells (Fig. 7D), similar to the effects of expressing SSH1L-GFP. Co-expression of Cofilin (S3A)-GFP and Coronin 1B shRNA partially suppressed the effects on lamellipodial dynamics of Coronin 1B depletion. Lamellipodial protrusion rate decreased and protrusion persistence was modestly increased, but did not reach control levels. It is important to note that, although Cofilin S3A rescued the lamellipodial dynamics associated with depletion of Coronin 1B, it did not rescue the effects on whole cell motility (data not shown). Together, these data suggest that Coronin 1B promotes the activation of Cofilin at the leading edge.
Discussion
The cellular function of Coronin 1B
Our data indicate that Coronin 1B is a key regulator of whole cell motility and lamellipodial dynamics. Although Coronin 1B is not absolutely required for cells to move or lamellipodia to protrude, it enhances these processes and is a likely target of signal transduction cascades that regulate motility. Coronin 1B influences motility through control of actin filament dynamics and architecture at the leading edge. In the absence of Coronin 1B, retrograde actin flow is reduced and the dendritic actin meshwork of the lamellipodia is abnormal. These two observations are likely to be aspects of the same underlying phenomenon, as the architecture of the actin network reflects the balance of actin assembly at the front and disassembly at the back of the lamellipodia. Future studies will dissect the relative contributions of actin assembly/disassembly and myosin-based contraction to Coronin 1B’s contribution to retrograde flow.
Most striking is the different distribution and density of actin barbed ends in the lamellipodia of cells lacking Coronin 1B. Cells depleted of Coronin 1B have higher barbed end density than controls and the barbed ends are concentrated in a narrower zone at the leading edge. Increased barbed ends could arise by three mechanisms: failure to cap or uncapping of existing filaments, severing of filaments or de novo nucleation of new filaments. Since Coronin 1B has no detectable capping activity (Fig. S6), failure to cap filaments can be eliminated from consideration. The increased barbed ends observed with Coronin 1B depletion are also unlikely to result from increased filament severing since Cofilin is less active in the lamellipodia of knockdown cells. We cannot exclude an effect on Cofilin-independent severing (eg. Gelsolin), but since Cofilin is required for protrusion, it is probably the predominant severing factor in lamellipodia. We suggest that increased nucleation of filaments most likely results in increased barbed ends in lamellipodia of Coronin 1B-depleted cells.
Coronin 1B inhibits Arp2/3-mediated nucleation in a phospho-regulated manner
Consistent with the increased density of barbed ends in lamellipodia of Coronin 1B-depleted cells, recombinant Coronin 1B inhibits the nucleation activity of Arp2/3 in vitro. This inhibitory effect on Arp2/3 activity is a conserved property of Coronins from yeast to humans (Humphries et al., 2002). Other proteins such as Tropomyosin, Caldesmon and EPLIN inhibit Arp2/3 complex nucleation activity, but these proteins are thought to bind tightly to the sides of actin filaments and compete with binding of Arp2/3 complex to the mother actin filament (Blanchoin et al., 2001; Maul et al., 2003; Yamakita et al., 2003) to indirectly inhibit filament-dependent Arp2/3 nucleation activity. In contrast, inhibition by Coronin 1B appears to arise from direct binding of Coronin to Arp2/3 complex and stabilization of its “open”, inactive form (Rodal et al., 2005). Our findings support this notion since Coronin 1B S2D binds poorly to Arp2/3 and has weaker inhibitory activity. Further studies will be required to elucidate the molecular mechanism of Coronin 1B’s inhibition of Arp2/3 complex.
Our results suggest that limiting filament nucleation activity via Coronin 1B is important for efficient lamellipodial protrusion. This conclusion is consistent with other studies in which nucleation of actin filaments was rampant and lamellipodial protrusions formed in response to EGF were blocked in cells microinjected with the Arp2/3 complex activating VCA (WA) domain of SCAR or WASP (Shao et al., 2006). Coronin 1B apparently serves to temper filament nucleation by Arp2/3 complex in lamellipodia, especially close to the membrane where several WASP/SCAR co-activating factors, such as Rac, Cdc42 and PIP2, are enriched. In the absence of Coronin 1B, nucleation by Arp2/3 complex may be overactive, leading to the increased free barbed ends and the faster rate of protrusion observed in Coronin 1B depleted cells. Without a means to limit Arp2/3 complex activity, explosive generation of new filaments could result in short-lived protrusions that stall as G-actin becomes limiting, consistent with the decreased protrusion persistence and distance observed in the Coronin 1B-depleted cells. Previous models for the control of filament nucleation in lamellipodia focused on the localized activation of WASP/SCAR/Formin proteins by inositol lipids and Rho-type GTPases. Our studies suggest that nucleation control is more complex than current models predict and involves a balance between nucleation promoting factors and inhibitors.
Coronin 1B regulates SSH1L localization and Cofilin activity
Coronin 1B is required for SSH-induced lamellipodial activity and for targeting SSH1L within lamellipodia. Through these processes, Coronin 1B could significantly influence the activity of Cofilin at the leading edge. This mechanism for regulating Cofilin activity is analogous to the mechanisms regulating many kinases and phosphatases via targeting subunits and scaffolds (Cohen, 2002; Sim and Scott, 1999). It is important to note that targeting of SSH1L does not override other mechanisms of Slingshot regulation such as phosphorylation or F-actin binding (Nagata-Ohashi et al., 2004). Rather, it is likely that appropriate targeting of SSH1L works in conjunction with these other regulatory events to control Slingshot’s activity spatially and temporally.
Our studies in fibroblasts link Coronins and Cofilin activities via SSH, however there is precedence for such a connection in other systems (Goode et al., 1999). Mutations in the single yeast Coronin gene are benign except when combined with mutations in the Cofilin gene. F-actin accumulates in abnormal aggregates in double Coronin/Cofilin mutants, but not with each single mutant, suggesting that these proteins function together to promote actin turnover in this organism. Recent studies also identified Coronin 1A as a factor leading to actin depolymerization in Listeria actin tails (Brieher et al., 2006). This study suggested that Coronin 1A-bound F-actin has increased affinity for Cofilin, perhaps due to altered helical twist. One prediction of this idea is that Cofilin may not be recruited to the leading edge in the absence of Coronin. We observed no change in Cofilin distribution at the leading edge in Coronin 1B-depleted cells (Fig. S8), but future studies are needed to explicitly test whether Coronin 1B has similar activity to Coronin 1A in this regard. It is possible that Coronins direct Cofilin recruitment and activation via SSH targeting to synergistically control Cofilin activity. Regardless of the precise mechanistic details, a conserved functional connection between Coronins and Cofilin in regulating filament dynamics exists from yeast to humans.
The coordination of Arp2/3 complex and Cofilin activity in lamellipodia
Why is a factor that coordinates the activities of Arp2/3 complex and Cofilin in lamellipodia necessary? Cell motility is a system-based process that requires precise spatial and temporal control of the individual components. Chemotaxis requires localized and coordinate cytoskeletal remodeling to achieve directional movement towards a chemotactic signal. Our work identifies Coronin 1B as a link between two major components, Cofilin and Arp2/3 complex, that control cytoskeletal remodeling in many cells. Since factors that coordinate cytoskeletal dynamics are likely to be crucial for directional sensing and motility, Coronin 1B is likely regulated by chemotactic signaling pathways. This idea is strongly supported by the recent observations that thymocytes from Coronin 1A knockout mice are severely defective in chemotaxis towards chemokines (Foger et al., 2006). Future studies will focus on the role that Coronin 1B plays in directed cell motility and on elucidating the molecular mechanism of Coronin 1B action.
Experimental Procedures
Antibodies were obtained from Cell Signaling Technologies (pSerPKC, pLIMK1/2, LIMK1), Upstate Biotechnology (p34Arc, Cortactin), Cytoskeleton (Cofilin), Chemicon (actin), Biosource (pPaxillinSer126, pErk1/2), Roche (GFP), Sigma (Myc) and Jackson ImmunoResearch (Cy2, Rhodamine Red-X, Cy5, and HRP conjugated secondary antibodies). AlexaFluor phalloidins were from Molecular Probes. Cells were obtained from ATCC. Inhibitors Ro32-0432 and okadaic acid were from Calbiochem. Antibodies to Coronin 1B (4245.Exp) were affinity purified as described (Cai et al., 2005).
Cell culture and viral transduction
Cell culture, immunoprecipitation and immunoblotting were performed as described (Cai et al., 2005). Transient transfections were performed using FuGENE 6 (Roche) for HEK293 cells, PolyFect (Qiagen) for Rat2 cells and Lipofectamine 2000 (Invitrogen) for NIH3T3 cells. Retroviral packaging, infections, and fluorescence-activated cell sorting were as described (Bear et al., 2000). Lentivirus production and infection were as described (Rubinson et al., 2003).
Molecular cloning
Descriptions of the construction of plasmids are in Supplemental Experimental Procedures.
Single cell tracking and kymography analysis
Migration of Rat2 fibroblasts was analyzed as described with slight modifications (Cai et al., 2005). Cells were infected with lentivirus for expressing Coronin1B or control shRNA (with and without rescue proteins) together with GFP. GFP-expressing cells were tracked and whole cell speed calculated using Tracking Analysis software (Andor Bioimaging). GFP negative cells were analyzed as an internal control. For kymography, 300 images were captured at 1 second intervals and processed using an ImageJ plugin (http://www.embl-heidelberg.de/eamnet/html/body_kymograph.html). Kymographs were generated from protrusive areas of at least five cells per treatment and lamellipodial parameters were calculated as described (Bear et al., 2002). Data were exported to Prism for statistical analysis.
Actin retrograde flow measurement
Rat2 cells were infected with lentivirus to express shRNA against Coronin 1B or control shRNA together with GFP-actin. Images were captured at 1 sec intervals using a Nipkow-type spinning disk confocal microscope (Yokogawa CSU-10 and IX-81, Olympus) equipped with a 100x objective and a Hamamatsu CCD camera (model C4742-80-12AG). Kymographs were generated and analyzed as described above.
Analysis of free actin filament barbed ends in vivo
Free actin filament barbed ends were detected in permeabilized fibroblasts as described in Supplemental Experimental Procedures using AlexaFluor568-actin (Bryce et al., 2005). AlexaFluor568-actin incorporation was quantified from the intensity profile around the cell periphery as described for Fig. S1. The upper 50% of intensities were used to determine the width of the zone of free barbed ends (Fig. S2). The ratio of the barbed end intensity to phalloidin intensity along the edge was normalized and plotted as a function of distance from the cell margin (Fig S2).
Platinum replica electron microscopy
Rat2 cells were infected with lentivirus expressing Coronin 1B or control shRNA for 4 days. Cells were plated onto coverslips coated with 10 µg/ml fibronectin for 4 hours and samples were prepared for platinum replica electron microscopy as described (Svitkina, 2005). Images were collected at 6300X using a Philips CM12 microscope at 60 kV using Kodak SO-163 negative film, scanned using an Imacon FlexTight 848 scanner (Hasselblad AB) at resolution 2500 dpi and adjusted for publication using Photoshop (Adobe).
Recombinant Coronin 1B protein production
Recombinant Coronin 1B was expressed and purified either from Drosophila S2 cells (Invitrogen) or from HEK293 cells as described in Supplemental Experimental Procedures. Phosphorylated Coronin 1B was obtained from HEK293 cells treated for 30 min with 50nM PMA prior to harvesting and sodium orthovanadate (4 mM) was included in all buffers. The phosphorylation status of Coronin 1B was verified by pSerPKC blotting, immunoprecipitation and by mass spectrometry (Fig. S5).
Actin polymerization assay
Spectrin F-actin seeds (SAS), actin, pyrene labeled actin, GST-VCA and Arp2/3 complex were prepared as described (Bryan and Coluccio, 1985; DiNubile et al., 1995; Egile et al., 1999; Higgs et al., 1999; Spudich and Watt, 1971). Seeded actin polymerization reactions were performed as described (Barzik et al., 2005) with 1 µM actin (5% pyrene labeled) and 0.2 nM SAS. VCA-induced Arp2/3 nucleation reactions were performed as follows: Coronin 1B and Arp2/3 (20 nM) were mixed in MKEI-50 Buffer and incubated at room temperature for 5 min; reactions were initiated by the simultaneous addition of 1.5 µM actin (5% pyrene labeled, primed with 1 mM EGTA and 0.1 mM MgCl2 for 90 sec) and GST-VCA (1 nM or 10 nM). The delay between mixing reactants and recording fluorescence was 20 sec. Fluorescence was converted to the molar concentration of F-actin from the fluorescence of completely polymerized (24 hours post reaction) and unpolymerized actin, assuming a critical concentration of 0.1 µM. Maximal actin assembly rates were determined from linear fits of the rate as 0.5–1 µM actin polymer formed during the reactions.
In vivo Coronin 1B dephosphorylation assay
Rat2 cells were treated with 100 nM PMA for 30 min to stimulate Coronin 1B phosphorylation. Cells were then washed with fresh media and incubated in media containing the pan-PKC inhibitor, 1 µM Ro32-4032, for the indicated time. Okadaic acid (up to 1 µM) was included in the washout media in some experiments. Reactions were stopped by lysing the cells in RIPA Buffer and Coronin 1B was immunoprecipitated and blotted with pSerPKC antibody as described (Cai et al., 2005).
In vitro Slingshot phosphatase assay
The in vitro Slingshot phosphatase assay was as described (Nagata-Ohashi et al., 2004; Niwa et al., 2002) with slight modifications; a detailed description is in the Supplemental Experimental Procedures.
Imaging and lamellipodial co-localization analysis
Immunofluorescent staining and imaging were performed as described (Cai et al., 2005). For co-localization analyses, a custom ImageJ macro was used to extract pixel intensity as a function of distance from the leading edge. A detailed explanation of this method is in the Supplemental Experimental Procedures.
Active/total Cofilin ratio imaging
To assess active/total Cofilin ratios, cells were stained with antibodies against Cofilin (MAB22; 1:100) and phospho-Cofilin (4321; 1:170) and processed for immunofluorescence imaging as described in the Supplemental Experimental Procedures. DIC images indicated the outline of the cells. The ImageJ plugin, Ratio Plus (http://rsb.info.nih.gov/ij/plugins/ratio-plus.html) was used to calculate the ratio between phospho-Cofilin and Cofilin staining. The ratio was converted into active/total Cofilin ratio by [(Total Cofilin-phosphoCofilin) / Total Cofilin], normalized and presented using a rainbow look-up table.
Supplementary Material
Acknowledgments
We are grateful to J. Bamburg (Colorado State University) for providing antibodies to pCofilin (4321) and Cofilin (MAB22) and the protocol to quantify active Cofilin by ratio imaging prior to publication, to A. Makhov and H. Mekeel for help with electron microscopy, to K. Burridge, M. Schaller, F. Gertler, J. Bamburg and D. Roadcap for critical reading of the manuscript, to E. Izaurralde, M. Wilm, Y. Durocher for reagents and to T. Kotova and F. Tariq for technical assistance. Electron microscopy work was supported by NIH grant CA-16086 to JD Griffith. This work was supported by NIH (GM067222) to DAS and funds from the V Foundation and Melanoma Research Foundation to JEB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP Proteins and Actin Filament Capping Regulates Fibroblast Motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J, Coluccio LM. Kinetic analysis of F-actin depolymerization in the presence of platelet gelsolin and gelsolin-actin complexes. J Cell Biol. 1985;101:1236–1244. doi: 10.1083/jcb.101.4.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Cai L, Holoweckyj N, Schaller MD, Bear JE. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J Biol Chem. 2005;280:31913–31923. doi: 10.1074/jbc.M504146200. [DOI] [PubMed] [Google Scholar]
- Cohen P, Holmes CF, Tsukitani Y. Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem Sci. 1990;15:98–102. doi: 10.1016/0968-0004(90)90192-e. [DOI] [PubMed] [Google Scholar]
- Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Hall A, Bresnick A, Warren V, Hock R, Bennett H, Ogihara S. Actin polymerization and pseudopod extension during amoeboid chemotaxis. Cell Motil Cytoskeleton. 1988;10:77–90. doi: 10.1002/cm.970100113. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Minamide LS, Bamburg JR, Cramer LP. ADF/cofilin controls cell polarity during fibroblast migration. Curr Biol. 2003;13:252–257. doi: 10.1016/s0960-9822(03)00040-x. [DOI] [PubMed] [Google Scholar]
- de Hostos EL, Rehfuess C, Bradtke B, Waddell DR, Albrecht R, Murphy J, Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V, Ghosh M, Eddy R, Condeelis J. Cofilin takes the lead. J Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni S, De Biase A, Yakovlev A, Finn T, Beers J, Hoffman EP, Faden AI. In vivo and in vitro characterization of novel neuronal plasticity factors identified following spinal cord injury. J Biol Chem. 2005;280:2084–2091. doi: 10.1074/jbc.M411975200. [DOI] [PubMed] [Google Scholar]
- DiNubile MJ, Cassimeris L, Joyce M, Zigmond SH. Actin filament barbed-end capping activity in neutrophil lysates: the role of capping protein-beta 2. Mol Biol Cell. 1995;6:1659–1671. doi: 10.1091/mbc.6.12.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin- based motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foger N, Rangell L, Danilenko DM, Chan AC. Requirement for coronin 1 in T lymphocyte trafficking and cellular homeostasis. Science. 2006;313:839–842. doi: 10.1126/science.1130563. [DOI] [PubMed] [Google Scholar]
- Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- Goode BL, Wong JJ, Butty AC, Peter M, McCormack AL, Yates JR, Drubin DG, Barnes G. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J Cell Biol. 1999;144:83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Hinz B, Alt W, Johnen C, Herzog V, Kaiser HW. Quantifying lamella dynamics of cultured cells by SACED, a new computer- assisted motion analysis. Exp Cell Res. 1999;251:234–243. doi: 10.1006/excr.1999.4541. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TY, Dermardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Humphries CL, Balcer HI, D'Agostino JL, Winsor B, Drubin DG, Barnes G, Andrews BJ, Goode BL. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- Maul RS, Song Y, Amann KJ, Gerbin SC, Pollard TD, Chang DD. EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J Cell Biol. 2003;160:399–407. doi: 10.1083/jcb.200212057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M, Nishida E. Coronin localizes to leading edges and is involved in cell spreading and lamellipodium extension in vertebrate cells. J Cell Sci. 1999;112(Pt 17):2833–2842. doi: 10.1242/jcs.112.17.2833. [DOI] [PubMed] [Google Scholar]
- Nagata-Ohashi K, Ohta Y, Goto K, Chiba S, Mori R, Nishita M, Ohashi K, Kousaka K, Iwamatsu A, Niwa R, et al. A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J Cell Biol. 2004;165:465–471. doi: 10.1083/jcb.200401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J Cell Biol. 2005;171:349–359. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Kousaka K, Nagata-Ohashi K, Ohashi K, Muramoto A, Shima Y, Niwa R, Uemura T, Mizuno K. Differential activities, subcellular distribution and tissue expression patterns of three members of Slingshot family phosphatases that dephosphorylate cofilin. Genes Cells. 2003;8:811–824. doi: 10.1046/j.1365-2443.2003.00678.x. [DOI] [PubMed] [Google Scholar]
- Ponti A, Matov A, Adams M, Gupton S, Waterman-Storer CM, Danuser G. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative Fluorescent Speckle Microscopy. Biophys J. 2005;89:3456–3469. doi: 10.1529/biophysj.104.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Shao D, Forge A, Munro PM, Bailly M. Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil Cytoskeleton. 2006;63:395–414. doi: 10.1002/cm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim AT, Scott JD. Targeting of PKA, PKC and protein phosphatases to cellular microdomains. Cell Calcium. 1999;26:209–217. doi: 10.1054/ceca.1999.0072. [DOI] [PubMed] [Google Scholar]
- Soosairajah J, Maiti S, Wiggan O, Sarmiere P, Moussi N, Sarcevic B, Sampath R, Bamburg JR, Bernard O. Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. Embo J. 2005;24:473–486. doi: 10.1038/sj.emboj.7600543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Stanyon CA, Bernard O. LIM-kinase1. Int J Biochem Cell Biol. 1999;31:389–394. doi: 10.1016/s1357-2725(98)00116-2. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TMaB, G G. Correlative light and electron microscopy studies of cytoskeletal dynamics. Cell Biology: A Laboratory Handbook (Ed J Celis) 2005;v. 3:277–286. [Google Scholar]
- Symons MH, Mitchison TJ. Control of actin polymerization in live and permeabilized fibroblasts. J Cell Biol. 1991;114:503–513. doi: 10.1083/jcb.114.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht AC, Bear JE. Coronins: the return of the crown. Trends Cell Biol. 2006;16:421–426. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Yamakita Y, Oosawa F, Yamashiro S, Matsumura F. Caldesmon inhibits Arp2/3-mediated actin nucleation. J Biol Chem. 2003;278:17937–17944. doi: 10.1074/jbc.M208739200. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


