Abstract
Basal levels of C-reactive protein (CRP) have been associated with disease, particularly future cardiovascular events. Twin studies estimate 50% CRP heritability, so the identification of genetic variants influencing CRP expression is important. Existing studies in populations of European ancestry have identified numerous cis-acting variants but leave significant ambiguity over the identity of the key functional polymorphisms. We addressed this issue by typing a dense map of CRP single nucleotide polymorphisms (SNPs), and quantifying serum CRP in 594 unrelated African Americans. We used Bayesian model choice analysis to select the combination of SNPs best explaining basal CRP and found strong support for triallelic rs3091244 alone, with the T allele acting in an additive manner (Bayes factor >100 vs. null model), with additional support for a model incorporating both rs3091244 and rs12728740. Admixture analysis suggested SNP rs12728740 segregated with haplotypes predicted to be of recent European origin. Using a cladistic approach we confirmed the importance of rs3091244(T) by demonstrating a significant partition of haplotype effect based on the rs3091244(C/T) mutation (F=8.91, P=0.006). We argue that weaker linkage disequilibrium across the African American CRP locus compared with Europeans has allowed us to establish an unambiguous functional role for rs3091244(T), while also recognising the potential for additional functional mutations present in the European genome.
Keywords: C-reactive protein, African Americans, quantitative trait analysis, Bayesian statistics, racial admixture, genetics
Introduction
C-reactive protein (CRP) is a key component of the human acute-phase response. It is also expressed at low levels in apparently healthy individuals, although the factors regulating basal expression are unknown. CRP binds a range of exogenous and endogenous ligands and interacts with other components of the immune system, particularly C1q, resulting in activation of the classical complement cascade (Pepys et al. 2003). CRP probably evolved as a defence against bacterial infection, although its ability to bind apoptotic cells and nuclear components suggests an additional role in protecting against anti-nuclear autoimmunity (Chang et al. 2002; Mold et al. 1999; Volanakis 1982). Efforts to understand CRP regulation have been stimulated by the observation that basal CRP levels are associated with disease. Most widely studied is the link between CRP and atherosclerosis. Basal CRP is a modest independent predictor of future cardiovascular morbidity and mortality and is also elevated in the presence of a number of cardiovascular risk factors including obesity, diabetes mellitus and cigarette smoking (Danesh et al. 2004; Pepys et al. 2003). The current literature on the functional role of CRP in atherosclerosis is complex; authors variously propose that CRP is directly atherogenic, that it is simply a marker of other proatherogenic processes, or even that it has an atheroprotective role (Singh et al. 2008). CRP is also a candidate gene for systemic lupus erythematosus (SLE), both through its functional properties and through its location in an SLE susceptibility locus defined by linkage analysis (chromosome 1q22-24) (Moser et al. 1998). Three studies have shown an association between CRP polymorphism and SLE itself (Edberg et al. 2008; Jonsen et al. 2007; Russell et al. 2004).
Twin studies suggest basal serum CRP is approximately 50% heritable (MacGregor et al. 2004; Wessel et al. 2007). Levels are also longitudinally stable in any individual, at least over a period of several years, making it particularly amenable to genetic analysis (Kivimaki et al. 2007; Pepys et al. 2003). Existing studies in cohorts of European ancestry have identified cis-acting single nucleotide polymorphisms (SNPs) associated with basal CRP, but the identification of true functional candidates remains problematic due to strong linkage disequilibrium (LD) across CRP and the probability that at least three SNPs are functional (Carlson et al. 2005; Crawford et al. 2006; Hage et al. 2007; Kathiresan et al. 2006; Lange et al. 2006; Miller et al. 2005; Rhodes et al. 2008a; Verzilli et al. 2008). In this study we aimed to identify key functional SNPs within the CRP locus in African Americans: our rationale for doing this is outlined below.
The study of non-European populations may provide a solution to the problem of strong LD across CRP. Since the divergence of ethnic groups from a common ancestral population, the independent evolution of genetic loci may result in different SNPs, altered haplotype structure or novel genetic recombination events, each of which may be informative in terms of identifying functional SNPs. Theoretically the more ancient African genome (and the related African-American genome) should offer the most genetic diversity, and this is illustrated by the LD architecture of Yoruba subjects from Nigeria, which is more diverse and shows more evidence of recombination than reference populations of European and East Asian ancestry (Gabriel et al. 2002). Existing studies looking at the CRP locus in African American cohorts have not yet provided a comprehensive analysis (Carlson et al. 2005; Crawford et al. 2006; Lange et al. 2006; Szalai et al. 2005).
Variation within trans-acting loci, particularly genes associated with acute-phase signalling (interleukin-1B (IL1B), interleukin-6 (IL6) and interleukin-1 receptor antagonist (IL1RN)) have also been associated with basal CRP levels.(Berger et al. 2002; Eklund et al. 2005; Latkovskis et al. 2004; Paik et al. 2007; Reiner et al. 2008; Shin et al. 2007; Vickers et al. 2002). Existing CRP analyses have adjusted for clinical and environmental phenotype data but have generally not considered effects from these trans-acting loci: we attempt to address this.
Finally, we adopted a Bayesian strategy for our analysis. Classically, model choice in an analysis is achieved through a stepwise regression procedure, but the final model is often highly dependant on the starting model and the score parameters used to include or reject variables. In a Bayesian approach each model is evaluated by its posterior probability. We are able to incorporate prior information on the basis of existing results, and our final choice of model does not depend on arbitrary scoring parameters.
Methods
Study Cohort
594 individuals of African American origin were sourced from two cohorts. Cohort 1 consisted of 294 subjects from the Chicago Health and Aging Project (CHAP), which is a population based study of community-dwelling adults aged 65 years old and over. Details are given elsewhere (Bienias et al. 2003). The mean age was 78.3 years (standard deviation (s.d.) 7.3), 37.4% were male. Phenotypic data were available for Body Mass Index (BMI) (mean 28.24kg/m2, SD 6.3), smoking status (10.5%) and history of stroke (11.6%), cancer (23.8%), hypertension (81.0%) ischaemic heart disease (12.6%) and diabetes mellitus (30.3%). Cohort 2 consisted of 150 multiple sclerosis (MS) cases and 150 controls from the UCSF Multiple Sclerosis Genetics Group. Ascertainment protocols, clinical and demographic characteristics have been summarised elsewhere (Cree et al. 2004; Oksenberg et al. 2004). The mean age was 43.5 years (s.d. 11.3), 39.5% were male; no other phenotypic data were available. Recruitment to both cohorts received institutional review board approval and written informed consent was obtained.
SNP selection and genotyping
Three approaches were used to select SNPs. Firstly the HapMap Yoruba trio families were used to identify SNPs predicted to tag African CRP haplotypes (using Haploview tagger with an r2=0.8 threshold) (Barrett et al. 2005). Secondly we aimed to type all CRP SNPs on which there is published data, whether or not this provided redundant genetic information. Finally, to address the question of CRP admixture, rs1255606, rs11585798 and rs17459580 were selected from the HapMap datasets as being monomorphic in African populations but polymorphic in Caucasians. For both IL1B and IL6 the HapMap data suggest LD between SNPs is relatively weak. Variation at these genes was not the main focus of this paper so, rather than aiming to capture all possible variants, we genotyped a selection of previously reported SNPs. Genotyping was by Matrix-Assisted Laser Desorption and Ionization-Time Of Flight (MALDI-TOF) mass spectrometry (Sequenom, Hamburg, Germany) using the iPLEX Gold assay (Ross et al. 1998). Genotyping on cohort 1 participants was performed at the Broad Institute Center for Genotyping and Analysis (Cambridge, MA, USA). Genotyping on cohort 2 was performed at the Genome Centre, Imperial College London (London, UK). Using predefined quality control exclusions we excluded any individual genotyped at <90% of markers, any marker genotyping in <90% individuals, any marker with a minor allele frequency <5% and any marker out of Hardy-Weinberg equilibrium (P<0.05 calculated by χ2).
Serum CRP quantification
CRP quantification on cohort 1 was performed at the University of Vermont Laboratory for Clinical Biochemistry Research using a particle enhanced immunonephelometric assay (BNII nephelometer, Dade Behring, Deerfield, IL, USA). CRP quantification on cohort 2 was performed at the Department of Clinical Chemistry, Hammersmith Hospital using a latex-enhanced immunoturbidimetric assay (Olympus AU2700 analyser, Olympus Diagnostics, Southall, UK).
Bayesian Model Selection
CRP levels were transformed to the natural log scale and phenotype/genotype effects were fitted using a Bayesian linear model. Bayesian model selection was performed using the association studies toolkit for WinBUGS, employing a reverse jump algorithm on the model space, in the Markov Chain Monte Carlo (MCMC) framework (Lunn et al. 2000; Lunn et al. 2006). We made simple prior assumptions based on existing literature; firstly that the magnitude of genetic effect would be relatively small, and secondly that the genetic model would be most likely to have one or two genetic effects but much less likely to have more than three effects (regression parameters β and γ, normal distribution, mean=0, variance=0.25; number of genetic effects specified by a Poisson (2) distribution). A duplicate analysis using uniform prior assumptions (no prior belief in the number of genetic effects or their magnitude) is presented in the supplementary material. The presence of highly correlated variables can cause problems fitting and interpreting linear models, and may even give the impression no variable is associated with the outcome, even if a strong association is seen for each variable individually. At the outset it was therefore decided not to simultaneously enter SNPs into our genetic models that were correlated with r2>0.8. For pairs, or clusters of SNPs with a higher degree of correlation a single tagSNP was therefore selected for analysis.
In the primary analysis SNPs were evaluated assuming an additive allele effect. Trialleleic rs3091244 was encoded twice, assuming increasing copies of either T or A relative to the commonest C allele. Simultaneously entering both allowed the identification of the allele best explaining the data, with the C allele as baseline. In secondary analyses, to assess whether additive, dominant or recessive effects best explained the CRP associations, SNPs coded accordingly were entered simultaneously into the analysis with an informative prior belief for the regression parameter but with equal prior probabilities given to all potential models.
Haplotype analysis
Each individual was assigned a phased haplotype using PHASE v2.1 (Stephens et al. 2001; Stephens et al. 2003). Population haplotype structure was inferred using Haploview and the Gabriel algorithm (default settings), having excluded individuals predicted to carry haplotypes of European origin (see below) (Barrett et al. 2005; Gabriel et al. 2002). Clade analysis was by TCS: Phylogenetic network association using statistical parsimony v1.21 (Clement et al. 2000). Haplotype association was modelled by ANOVA using Treescan v1.0: a bioinformatic application to search for genotype/phenotype associations using haplotype trees (available at http://Darwin.uvigo.es) (Templeton et al. 2005). Classical P-values were corrected for multiple hypothesis testing by step-down permutation testing, using 10,000 permutations of the dataset (Westfall et al. 1993). Permuted P-values are presented.
Missing Data
Where phenotypic data were available in both cohorts only a small amount of data were missing: 0.18% for age and none for sex or multiple sclerosis affection. Data on BMI and other disease states were not collected for cohort 2 and hence were missing for 49.2% of the combined data set. In order to maximise use of this available information without a dramatic reduction in the total size of the dataset we used a Bayesian approach to inferring missing data by inputting a prior distribution for missing variables, with information from existing data updating these priors. For example, existing lnBMI data has mean 3.32 kg/m2, s.d. 0.22, so a normal prior distribution with these parameters was used for missing values. A Bernoulli prior distribution was used for binary phenotype data. To ensure the use of these data did not dramatically alter our findings all significant models were also re-evaluated without the use of inferred values. Missing SNP data was estimated using FastPhase (Scheet et al. 2006).
Admixture analysis
Admixture analysis suggests that a significant proportion of the African American genome (perhaps 10-30%) may arise from recent admixture with European populations (Wassel Fyr et al. 2007). At the level of a single locus this may complicate analyses by distorting the inferred haplotype structure if the pattern of LD differs between the background and the admixed genetic elements. It may also affect our ability to detect an indirect genetic association. We therefore identified three SNPs predicted to be polymorphic in European populations but monomorphic in populations of an African origin. We used PHASE v.2.1 to explicitly infer a haplotype pair for each individual and we hypothesised that haplotypes containing these SNPs would have a European origin.
Results
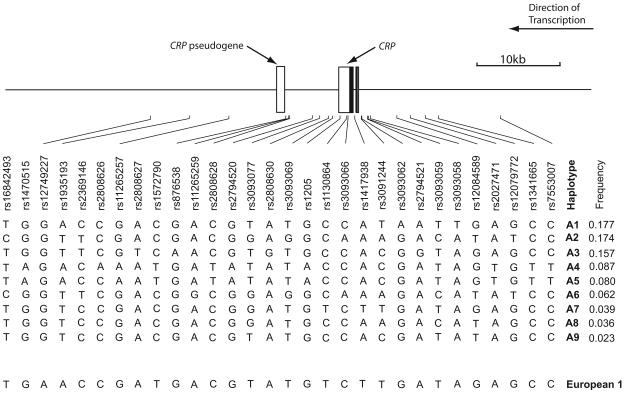
The 44 SNPs genotyped in both cohorts are listed in table 1. Eight of these SNPs failed genotyping according to predetermined criteria. We also had available typing on an additional five SNPs from one cohort only (supplementary table 1), which were used in the inference of population haplotypes, but were not included for any association analyses. Figure 1 demonstrates the position of SNPs alongside the inferred haplotype structure.
Table 1.
SNPs typed in all subjects
| rs number | Location | alleles | minor allele frequency (%) |
HWPa | missing data (%) |
|---|---|---|---|---|---|
| CRP SNPs | |||||
| rs11265263* | 5′ flank | T>G | 3.26 | 0.428 | 1.08 |
| rs12728740 | 5′ flank | G>T | 9.58 | 0.964 | 0.90 |
| rs7553007 | 5′ flank | C>T | 20.77 | 0.197 | 2.51 |
| rs1341665 | 5′ flank | C>T | 18.83 | 0.237 | 4.84 |
| rs12079772 | 5′ flank | G>T | 20.15 | 0.092 | 1.25 |
| rs2027471 | 5′ flank | A>T | 20.49 | 0.559 | 0.72 |
| rs12084589 | 5′ flank | G>T | 28.23 | 0.800 | 2.87 |
| rs3093058 | 5′ flank | A>T | 15.65 | 0.943 | 5.56 |
| rs3093062 | upstream promoter | G>A | 16.18 | 0.641 | 0.90 |
| rs3091244 | upstream promoter | C>T>A | 30.55(T)/25.41(A) | 0.480 | 1.07 |
| rs3093063* | upstream promoter | C>T | 2.53 | 0.266 | 0.90 |
| rs1417938 | intron 1 | A>T | 14.16 | 0.776 | 0.00 |
| rs1800947* | exon 2 (synonymous) | G>C | 1.80 | 0.665 | 0.54 |
| rs3093066 | 3′-UTR | C>A | 22.03 | 0.259 | 1.97 |
| rs1205 | 3′-UTR | G>A | 18.39 | 0.924 | 0.90 |
| rs3093069 | 3′ flank | T>G | 20.40 | 0.124 | 1.61 |
| rs2808630 | 3′ flank | A>G | 17.95 | 0.574 | 0.18 |
| rs3093077 | 3′ flank | T>G | 28.79 | 0.545 | 0.72 |
| rs2794520 | 3′ flank | G>A | 20.68 | 0.754 | 0.36 |
| rs2808628 | 3′ flank | C>T | 20.55 | 0.838 | 1.43 |
| rs11265259 | 3′ flank | A>G | 5.70 | 0.266 | 8.78 |
| rs1572970 | 3′ flank | C>T | 35.00 | 0.760 | 1.43 |
| rs12755606 | 3′ flank | G>C | 9.95 | 0.823 | 0.90 |
| rs11265257 | 3′ flank | G>A | 20.27 | 0.956 | 0.54 |
| rs2808626 | 3′ flank | C>A | 7.99 | 0.797 | 0.18 |
| rs2369146 | 3′ flank | C>T | 37.12 | 0.843 | 5.91 |
| rs17459580* | 3′ flank | T>G | 3.25 | 0.429 | 0.72 |
| rs1935193 | 3′ flank | A>T | 46.30 | 0.764 | 0.72 |
| rs10465953* | 3′ flank | C>T | 1.80 | 0.667 | 0.18 |
| rs12749227 | 3′ flank | A>G | 11.55 | 0.562 | 0.72 |
| rs16842502* | 3′ flank | G>T | 4.03 | 0.781 | 15.59 |
| rs1470515 | 3′ flank | G>A | 24.50 | 0.805 | 1.25 |
| rs16842493 | 3′ flank | T>C | 20.34 | 0.200 | 0.90 |
| rs12093699 | 3′ flank | C>T | 33.36 | 0.429 | 1.97 |
| rs12744244 | 3′ flank | G>T | 5.18 | 0.679 | 1.43 |
| rs11585798* | 3′ flank | T>A | 4.05 | <0.001 | 2.69 |
| IL6 SNPs | |||||
| rs1554606 | 5′ flank | G>T | 35.93 | 0.707 | 2.62 |
| rs1800796 | 5′ flank | G>C | 7.52 | 0.888 | 0.17 |
| rs1800795 | 5′ flank | G>C | 8.42 | 0.637 | 0.87 |
| rs2069837 | intron 2 | A>G | 13.99 | 0.162 | 2.09 |
| rs2069840 | intron 3 | C>G | 15.15 | 0.771 | 0.35 |
| IL1B SNPs | |||||
| rs3917368 | 3′ flank | G>A | 19.73 | 0.558 | 2.27 |
| rs4849124* | 3′ flank | A>G | 2.45 | <0.001 | 10.99 |
| rs1143634 | exon 5 | C>T | 14.05 | 0.092 | 1.22 |
excluded from analysis by pre-determined quality control criteria.
Hardy-Weinburg probability P-value calculated by χ2
Figure 1.
African American CRP haplotypes. The position of typed SNPs relative to CRP is demonstrated in the top part of the diagram. Individuals predicted to carry haplotypes of a recent European origin have been excluded from this analysis (see methods), although the commonest of these haplotypes is shown at the bottom of the diagram for reference. SNP rs12728740 is located upstream of this main block while rs12744344 and rs12093699 are located downstream.
Median (95th percentile) CRP was 3.38 mg/L (0.18, 27.50) in cohort 1, 1.79 mg/L (0.13, 16.81) in cohort 2, and 2.52 mg/L (0.24, 22.75) in the combined dataset. Bayesian model choice analysis was used to evaluate the combination of phenotypic covariates that explained the CRP data. The highest posterior probability (P(M/D)) was observed for a model incorporating age and BMI only (P(M/D)=0.98, representing overwhelming support). There was little support for models including sex or a diagnosis of multiple sclerosis (MS) and the marginal probabilities (Pmarg) for these covariates were small (Pmarg=0.002 and Pmarg=0.0003 respectively). There was even less support for other diseases including ischaemic heart disease and diabetes mellitus, although given the small numbers of affected individuals and the absence of data from cohort 2, this was not surprising. We looked for additional cohort specific effects and found strong evidence for no effect (age, BMI, sex and MS adjusted: P(M/D)=0.008 for model with cohort effect, P(M/D)=0.992 for model with no cohort effect, Bayes factor >100)
Genetic models
Of the 29 CRP SNPs passing quality control, some formed highly correlated clusters. As outlined in the methods section rs2808628 was selected to tag rs2794520, rs2027471, rs1341665, rs7553007 and rs1205 (mean pairwise r2=0.89); rs3093066 to tag rs3093069, rs12079772 and rs16842493 (mean pairwise r2=0.90); and rs3093062 to tag rs3093058 (r2=1.00). A full comparison of markers (with chromosomes of predicted European origin excluded) is presented in supplementary table 2. Age, BMI, sex and MS affection were specified as covariates (sex and MS affection were thought important to include despite the lack of evidence for an effect). Models were also run with the addition of “cohort” as a fixed phenotype effect, but this made negligible difference to the overall results (data not shown).
Table 2 shows the results of Bayesian model choice analysis incorporating all genetic markers. Highest posterior probability was attached to a model incorporating rs3091244(T) alone, representing overwhelming evidence for a genetic effect (Bayes factor >100 vs. null model (no SNPs)). A secondary model incorporating both rs3091244(T) + rs12728740 had a smaller posterior probability. Evidence was weak for models using the alternative codings for rs3091244 [rs3091244(A) P(M/D)<1×10−4; rs3091244(A) + rs3091244(T) P(M/D)=0.003].
Table 2.
Bayesian choice models with more than 5% posterior probability.
| Model | P(M/D) | Pmarg | Effect estimate (95% CI) |
|---|---|---|---|
| rs3091244(T) | 0.14 | 0.65 | 0.37 (0.21, 0.53) |
| rs3091244 (T)+ | 0.07 | 0.65 | 0.45 (0.29,0.63) |
| rs12728740 | 0.20 | −0.32(−0.59,−0.05) | |
| null | <0.01 |
P(M/D) - Posterior probability of the model given the data, Pmarg – Marginal probability associated with each SNP in the model, Effect estimate (95% CI) – Effect estimate with 95% credible intervals.
Secondary models
Additional analyses were performed to substantiate our findings. Firstly, having identified rs3091244 as the key SNP we investigated whether additive, dominant or recessive effects best explained this CRP association. We observed substantial evidence in favour of an additive model (Bayes factor 5.7 compared with dominant model). A boxplot demonstrating the relationship between CRP (adjusted for age, sex, BMI and MS affection) and rs3091244 genotype is shown as supplementary figure 1, and confirms graphically that an additive genotype affect is most likely. Secondly, we performed an analysis using uniform prior assumptions. This did not differ in its conclusion that rs3091244(T) was the key genetic effect (supplementary table 3). Thirdly, to re-examine the possibility of cohort effects we re-ran our analysis with the inclusion of cohort*genotype interactive terms. All the top models selected across these terms included genotype at rs3091244(T) +/− rs12728740, but there was evidence to support a cohort*genotype interaction with genotype exerting a smaller effect in cohort 1 (supplementary table 4). Consistent with this was an analysis of each cohort separately, with rs3091244 being the most significant SNP marginally in both cohorts, but with lower significance in cohort 1 (Pmarg = 0.58 in cohort 2 and Pmarg = 0.10 in cohort 1). Finally, to ensure that our use of inferred phenotype data was not distorting the results we re-ran our analysis excluding BMI as a covariate (BMI being the only important phenotypic effect with substantial missing data). In these models the significance attached to both the key SNPs actually increased [for rs3091244(T) Pmarg = 0.90 and for rs12728740 Pmarg = 0.26]. The posterior probability of the two top models also increased [rs3091244(T) alone P(M/D)=0.19 and for rs3091244(T) + rs12728740 P(M/D)=0.11].
Haplotype association
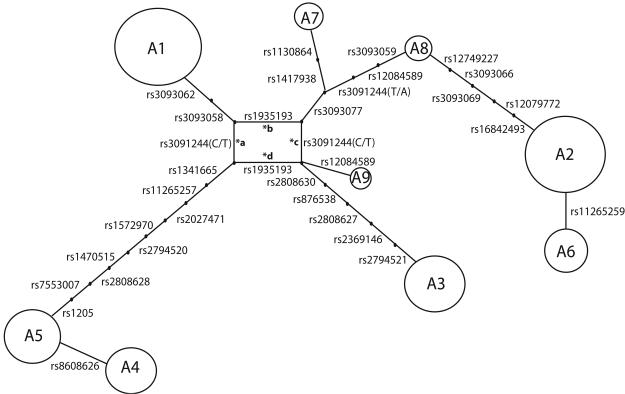
Admixture analysis predicted 10.8% of the observed haplotypes were of recent European origin. 20.2% of the cohort carried one European haplotype and 0.73% carried two. Figure 2 presents a phylogenetic relationship between haplotypes, having excluded these individuals. As explained in the figure caption there are four possible resolutions of the tree, labelled *a, *b, *c or *d. Significant partitions were only observed for trees *a and *c (those where the rs3091244 C/T mutation occurs only once). The most significant partition was defined by the rs3091244 C/T mutation, segregating haplotypes A1, A2, A6, A7 and A8 as clade one and haplotypes A3, A4, A5 and A9 as clade two (For tree *a, F=8.91, P=0.006). Clade one was associated with higher CRP (β=0.312 95%CI 0.146, 0.491), although the overall proportion of CRP variance explained by this partition was quite small (5.20%). A weakly significant effect was also observed for a partition defined by rs3093062/rs3093058, separating haplotype A1 from all the others (for tree *a F=6.385, P=0.038), although this weakly significant effect may be largely due to correlation with the rs3091244 C/T partition. The A1 haplotype was associated with higher CRP level than all the rest (β=0.448 95%CI 0.241, 0.655). No significant secondary partition was detected, although it should be noted the power to detect such an effect is dramatically reduced because of the smaller sample size consequent on the primary partition.
Figure 2.
Cladogram demonstrating a phylogenetic relationship between African American haplotypes. Each haplotype is represented by a circle whose size is roughly proportional to the population frequency. Connecting lines are annotated by the SNPs which distinguish one haplotype from another. Small dots along these lines represent haplotypes which are no longer observed but can be inferred to have existed in the population history. The loop at the centre of the tree represents uncertainty over the clade structure. Either rs3091244 (C/T) or rs1935193 may be the first mutational event, and consequently either rs3091244 (C/T) or rs1935193 must have mutated twice, from the wild type to the new allele, and then back to the wild type. These four explanations may be envisaged by removing either of the four branches labelled *a, *b, *c or *d.
Discussion
Our data suggest a single key variant, rs3091244 (or a variant in strong LD with rs3091244), regulates CRP expression in African Americans. In particular the T allele acting in an additive manner explained most of the genetic component of variation in serum CRP levels. Consistent with previous reports this effect was modest, explaining only 5.20% of CRP variance. The C allele of rs3091244 is commonest in all studied human populations, is found on the Chimpanzee and Macaque genome reference sequences, and has not been identified as polymorphic in these species (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007; The Chimpanzee Sequencing and Analysis Consortium 2005). It is therefore likely the C allele is ancestral, and a key functional moment in the evolution of the human CRP locus was the mutation of rs3091244 C to T, thus increasing basal serum CRP expression. We can only hypothesise how this enhanced expression may have conferred an advantage to early humans. Perhaps basal CRP has a surveillance role in protecting against the early stages of bacterial infection; alternatively higher basal CRP may enhance clearance of apoptotic debris and thus protect against developing autoimmunity. Another explanation, although untested, is that rs3091244 also alters the magnitude or time-course of the acute-phase CRP response, and this may have additional functional consequences.
A model incorporating both rs3091244 and rs12728740 should also be kept in consideration. The posterior probability of this two-SNP model was smaller than the model with rs3091244 alone but, because there are more possible two-SNP combinations, the Bayes factor gave it slightly more weight (Bayes factor = 6 in favour of two-SNP model). The incorporation of rs12728740 into a model is interesting. Although not originally selected to tag the admixed European genome it became apparent that 96% of the minor alleles of rs12728740 were segregating with haplotypes of inferred European origin. In addition it was predicted to lie outside the main CRP haplotype block in our population. By reference to the HapMap CEPH panel it is clear that in Europeans rs1272870 is in much stronger LD with the main CRP SNPs, in particular with rs1417938 (r2=0.857), which tags the commonest European haplotype, and has been associated with higher CRP production in existing European studies (Kathiresan et al. 2006; Lange et al. 2006; Miller et al. 2005). This does not downplay the importance of rs3091244 as the key regulator of CRP, but it does provide some evidence for a separate functional effect of recent European origin. On the other hand, we failed to demonstrate any role for SNPs at the IL6 and IL1B loci. Some of these have been associated with serum CRP levels in other populations, but these associations are modest (Berger et al. 2002; Eklund et al. 2005; Latkovskis et al. 2004; Paik et al. 2007; Rhodes et al. 2008b; Shin et al. 2007; Vickers et al. 2002). Given our sample size, we are not able to definitively rule out a role for these other loci in CRP expression.
Of the existing studies of the CRP locus in African Americans, the first concentrated on CRP levels associated with rs3091244 and rs3093062 only (Szalai et al. 2005). The authors' key argument hinged on an interaction between rs3091244(T) and rs3093062 such that haplotype rs3091244(T)/rs3093062(G) was associated with high CRP, while haplotype rs3091244(T)/rs3093062(A), the equivalent to our haplotype A1, was associated with lowest CRP. There are therefore differences between this paper and our data; we found little evidence for a role for both rs3091244 and rs3093062. Furthermore we found the rs3093062(A) haplotype to be associated with high rather than low CRP.
Shortly after this initial report, data from the biracial CARDIA cohort was published (Carlson et al. 2005). Tag SNPs were selected on the basis of sequencing a small number of individuals and hence failed to capture all the variation we detected. The authors also analysed both European and African ethnicities together (albeit with a “race” term in their regression models), which may be a suboptimal approach given the different SNP and haplotype frequencies between groups. Two subsequent studies in different cohorts used the same combination of SNPs, but with a separate analysis of African American participants (Crawford et al. 2006; Lange et al. 2006). All these studies generated results broadly similar to ours, with haplotypes tagged by the C and T alleles of rs3091244 being associated with low and high levels of CRP respectively. Notably haplotype rs3091244(T)/rs3093058(T) (equivalent to our haplotype A1) was associated with highest overall CRP expression, as we found.
Both the initial papers produced in vitro functional evidence to support their hypotheses, but again with differences in their key findings (Carlson et al. 2005; Szalai et al. 2005). It has been noted that there are frequently discrepancies between genetic associations and in vitro functional data so this is a field that requires further study (Ioannidis et al. 2006).
Our data suggest a simple model of genetic effect at CRP based on rs3091244 alone (or a variant in strong LD with rs3091244). This contrasts with existing data from Europeans, which identifies multiple associations, and a meta-analysis, using similar methodology to us, which finds greatest evidence for models containing at least three different SNPs (Verzilli et al. 2008). There are a number of explanations for this difference. Firstly, and this was the initial rational behind our study, there is a difference in allele frequency and patterns of LD between SNPs in the two ethnic groups. Studies in Europeans, in addition to rs3091244, commonly report associations with rs1205, rs1417938 and rs1800947. The LD between these SNPs and rs3091244(T) is considerably weaker in our African American cohort than in European groups, as shown by a comparison with another cohort of 799 unrelated individuals of European Ancestry (Rhodes et al. 2008a). It should be noted that rs1800947 was rare in African Americans, so was not included in the main analysis. The degree of LD with rs3091244(T) in African Americans as measured by r2 was 0.138 for rs1205, 0.225 for rs1417938, 0.007 for rs1800947, and the equivalent in Europeans was 0.256 for rs1205, 0.990 for rs1417938, and 0.035 for rs1800947. The association with these additional SNPs in Europeans may therefore be influenced by both their higher frequency and their stronger LD with rs3091244(T), with the true effect only becoming apparent in this African American cohort. A second possibility is that there genuinely is an additional functional site that has arisen recently in European populations. Our demonstration of a possible effect from rs12728740 provides some evidence for this argument.
We appreciate there are limitations to our study. Firstly we used two cohorts whose participants had rather different phenotypic characteristics. We explored this problem using a “cohort” term as a fixed phenotype effect, using genotype*cohort interaction terms and, in addition, we examined each cohort separately. These secondary analyses suggest that there were some quantitative differences between cohorts in terms of the magnitude of SNP effect, but there were no qualitative differences in terms of which were the key SNPs or SNP combinations associated with CRP levels. The likely explanation for the less extreme SNP effects in cohort 1 (older individuals) is that with higher disease burden (both documented and occult) a greater proportion of the observed CRP level is due to stimulated acute-phase expression rather than truly basal CRP. This increase in “background noise” may decrease our power to detect CRP associations. A second potential problem was our use of inferred phenotype data. In practice the only key variable for which levels of missing data were great enough for this to be an issue was BMI. A secondary analysis, with the complete omission of BMI, still attached greatest significance to the same key SNPs. While recognising the limitations of having incomplete BMI data, we have no evidence to suggest that if these additional data were available our genetic conclusions would change.
Our conclusion, therefore, is that the most important SNP regulating CRP expression is rs3091244, and we discuss how our study helps unravel the complex genetic association seen in European populations. Identifying the genetic determinants of CRP expression remains an important question because of the link between basal CRP and disease. The use of larger cohorts in different ethnic populations may make this possible.
Supplementary Material
Acknowledgements
This work was funded by an Arthritis Research Campaign Clinical Research Fellowship awarded to B.R. and a Wellcome Trust Senior Research Fellowship awarded to T.J.V. P.L.D. is the William C. Fowler Scholar in Multiple Sclerosis Research and is supported by a National Institute of Neurological Disorders and Stroke KO8 grant (NS46341). C.F.MdL, J.F.K. and D.A.E. are supported by grants from the National Institutes of Environmental Health Sciences (ES 10902), and the National Institute of Aging (AG 11101) of the US National Institutes of Health. We would like to thank John Meek for facilitating CRP quantification in the Department of Clinical Chemistry, Hammersmith Hospital, London.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berger P, McConnell JP, Nunn M, Kornman KS, Sorrell J, Stephenson K, Duff GW. C-reactive protein levels are influenced by common IL-1 gene variations. Cytokine. 2002;17:171–174. doi: 10.1006/cyto.2001.0974. [DOI] [PubMed] [Google Scholar]
- Bienias JL, Beckett LA, Bennett DA, Wilson RS, Evans DA. Design of the Chicago Health and Aging Project (CHAP) J Alzheimers Dis. 2003;5:349–355. doi: 10.3233/jad-2003-5501. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Aldred SF, Lee PK, Tracy RP, Schwartz SM, Rieder M, Liu K, Williams OD, Iribarren C, Lewis EC, Fornage M, Boerwinkle E, Gross M, Jaquish C, Nickerson DA, Myers RM, Siscovick DS, Reiner AP. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet. 2005;77:64–77. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MK, Binder CJ, Torzewski M, Witztum JL. C-reactive protein binds to both oxidized LDL and apoptotic cells through recognition of a common ligand: Phosphorylcholine of oxidized phospholipids. Proc Natl Acad Sci U S A. 2002;99:13043–13048. doi: 10.1073/pnas.192399699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, Witrak L, Rieder MJ, Nickerson DA. Genetic variation is associated with C-reactive protein levels in the Third National Health and Nutrition Examination Survey. Circulation. 2006;114:2458–2465. doi: 10.1161/CIRCULATIONAHA.106.615740. [DOI] [PubMed] [Google Scholar]
- Cree BA, Khan O, Bourdette D, Goodin DS, Cohen JA, Marrie RA, Glidden D, Weinstock-Guttman B, Reich D, Patterson N, Haines JL, Pericak-Vance M, DeLoa C, Oksenberg JR, Hauser SL. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology. 2004;63:2039–2045. doi: 10.1212/01.wnl.0000145762.60562.5d. [DOI] [PubMed] [Google Scholar]
- Danesh J, Wheeler JG, Hirschfield GM, Eda S, Eiriksdottir G, Rumley A, Lowe GD, Pepys MB, Gudnason V. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350:1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- Edberg JC, Wu J, Langefeld CD, Brown EE, Marion MC, McGwin G, Jr., Petri M, Ramsey-Goldman R, Reveille JD, Frank SG, Kaufman KM, Harley JB, Alarcon GS, Kimberly RP. Genetic variation in the CRP promoter: association with systemic lupus erythematosus. Hum Mol Genet. 2008;17:1147–1155. doi: 10.1093/hmg/ddn004. [DOI] [PubMed] [Google Scholar]
- Eklund C, Lehtimaki T, Hurme M. Epistatic effect of C-reactive protein (CRP) single nucleotide polymorphism (SNP) +1059 and interleukin-1B SNP +3954 on CRP concentration in healthy male blood donors. Int J Immunogenet. 2005;32:229–232. doi: 10.1111/j.1744-313X.2005.00515.x. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Hage FG, Szalai AJ. C-reactive protein gene polymorphisms, C-reactive protein blood levels, and cardiovascular disease risk. J Am Coll Cardiol. 2007;50:1115–1122. doi: 10.1016/j.jacc.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Kavvoura FK. Concordance of functional in vitro data and epidemiological associations in complex disease genetics. Genet Med. 2006;8:583–593. doi: 10.1097/01.gim.0000237775.93658.0c. [DOI] [PubMed] [Google Scholar]
- Jonsen A, Gunnarsson I, Gullstrand B, Svenungsson E, Bengtsson AA, Nived O, Lundberg IE, Truedsson L, Sturfelt G. Association between SLE nephritis and polymorphic variants of the CRP and FcgammaRIIIa genes. Rheumatology (Oxford) 2007;46:1417–1421. doi: 10.1093/rheumatology/kem167. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Larson MG, Vasan RS, Guo CY, Gona P, Keaney JF, Jr., Wilson PW, Newton-Cheh C, Musone SL, Camargo AL, Drake JA, Levy D, O'Donnell CJ, Hirschhorn JN, Benjamin EJ. Contribution of clinical correlates and 13 C-reactive protein gene polymorphisms to interindividual variability in serum C-reactive protein level. Circulation. 2006;113:1415–1423. doi: 10.1161/CIRCULATIONAHA.105.591271. [DOI] [PubMed] [Google Scholar]
- Kivimaki M, Lawlor DA, Smith GD, Eklund C, Hurme M, Lehtimaki T, Viikari JS, Raitakari OT. Variants in the CRP gene as a measure of lifelong differences in average C-reactive protein levels: the Cardiovascular Risk in Young Finns Study, 1980-2001. Am J Epidemiol. 2007;166:760–764. doi: 10.1093/aje/kwm151. [DOI] [PubMed] [Google Scholar]
- Lange LA, Carlson CS, Hindorff LA, Lange EM, Walston J, Durda JP, Cushman M, Bis JC, Zeng D, Lin D, Kuller LH, Nickerson DA, Psaty BM, Tracy RP, Reiner AP. Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. JAMA. 2006;296:2703–2711. doi: 10.1001/jama.296.22.2703. [DOI] [PubMed] [Google Scholar]
- Latkovskis G, Licis N, Kalnins U. C-reactive protein levels and common polymorphisms of the interleukin-1 gene cluster and interleukin-6 gene in patients with coronary heart disease. Eur J Immunogenet. 2004;31:207–213. doi: 10.1111/j.1365-2370.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS - a Bayesian modelling framework: concepts, structure and extensibility. Statistics and Computing. 2000;10:325–337. [Google Scholar]
- Lunn DJ, Whittaker JC, Best N. A Bayesian toolkit for genetic association studies. Genet Epidemiol. 2006;30:231–247. doi: 10.1002/gepi.20140. [DOI] [PubMed] [Google Scholar]
- MacGregor AJ, Gallimore JR, Spector TD, Pepys MB. Genetic effects on baseline values of C-reactive protein and serum amyloid a protein: a comparison of monozygotic and dizygotic twins. Clin Chem. 2004;50:130–134. doi: 10.1373/clinchem.2003.028258. [DOI] [PubMed] [Google Scholar]
- Miller DT, Zee RY, Suk DJ, Kozlowski P, Chasman DI, Lazarus R, Cook NR, Ridker PM, Kwiatkowski DJ. Association of common CRP gene variants with CRP levels and cardiovascular events. Ann Hum Genet. 2005;69:623–638. doi: 10.1111/j.1529-8817.2005.00210.x. [DOI] [PubMed] [Google Scholar]
- Mold C, Gewurz H, Du Clos TW. Regulation of complement activation by C-reactive protein. Immunopharmacology. 1999;42:23–30. doi: 10.1016/s0162-3109(99)00007-7. [DOI] [PubMed] [Google Scholar]
- Moser KL, Neas BR, Salmon JE, Yu H, Gray-McGuire C, Asundi N, Bruner GR, Fox J, Kelly J, Henshall S, Bacino D, Dietz M, Hogue R, Koelsch G, Nightingale L, Shaver T, Abdou NI, Albert DA, Carson C, Petri M, Treadwell EL, James JA, Harley JB. Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci U S A. 1998;95:14869–14874. doi: 10.1073/pnas.95.25.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg JR, Barcellos LF, Cree BA, Baranzini SE, Bugawan TL, Khan O, Lincoln RR, Swerdlin A, Mignot E, Lin L, Goodin D, Erlich HA, Schmidt S, Thomson G, Reich DE, Pericak-Vance MA, Haines JL, Hauser SL. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JK, Kim OY, Koh SJ, Jang Y, Chae JS, Kim JY, Kim HJ, Hyun YJ, Cho JR, Lee JH. Additive effect of interleukin-6 and C-reactive protein (CRP) single nucleotide polymorphism on serum CRP concentration and other cardiovascular risk factors. Clin Chim Acta. 2007;380:68–74. doi: 10.1016/j.cca.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Wurfel MM, Lange LA, Carlson CS, Nord AS, Carty CL, Rieder MJ, Desmarais C, Jenny NS, Iribarren C, Walston JD, Williams OD, Nickerson DA, Jarvik GP. Polymorphisms of the IL1-Receptor Antagonist Gene (IL1RN) Are Associated With Multiple Markers of Systemic Inflammation. Arterioscler Thromb Vasc Biol epub ahead of print. 2008 doi: 10.1161/ATVBAHA.108.167437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rhodes B, Meek J, Whittaker JC, Vyse TJ. Quantification of the Genetic Component of Basal C-Reactive Protein Expression in SLE Nuclear Families. Ann Hum Genet. 2008a doi: 10.1111/j.1469-1809.2008.00444.x. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Rhodes B, Wong A, Navarra SV, Villamin C, Vyse TJ. Genetic determinants of basal C-reactive protein expression in Filipino systemic lupus erythematosus families. Genes Immun. 2008b;9:153–160. doi: 10.1038/sj.gene.6364459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- Russell AI, Cunninghame Graham DS, Shepherd C, Roberton CA, Whittaker J, Meeks J, Powell RJ, Isenberg DA, Walport MJ, Vyse TJ. Polymorphism at the C-reactive protein locus influences gene expression and predisposes to systemic lupus erythematosus. Hum Mol Genet. 2004;13:137–147. doi: 10.1093/hmg/ddh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KK, Jang Y, Koh SJ, Chae JS, Kim OY, Park S, Choi D, Shin DJ, Kim HJ, Lee JH. Influence of the IL-6 -572C>G polymorphism on inflammatory markers according to cigarette smoking in Korean healthy men. Cytokine. 2007;39:116–122. doi: 10.1016/j.cyto.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Singh SK, Suresh MV, Voleti B, Agrawal A. The connection between C-reactive protein and atherosclerosis. Ann Med. 2008;40:110–120. doi: 10.1080/07853890701749225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai AJ, Wu J, Lange EM, McCrory MA, Langefeld CD, Williams A, Zakharkin SO, George V, Allison DB, Cooper GS, Xie F, Fan Z, Edberg JC, Kimberly RP. Single-nucleotide polymorphisms in the C-reactive protein (CRP) gene promoter that affect transcription factor binding, alter transcriptional activity, and associate with differences in baseline serum CRP level. J Mol Med. 2005;83:440–447. doi: 10.1007/s00109-005-0658-0. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Maxwell T, Posada D, Stengard JH, Boerwinkle E, Sing CF. Tree scanning: a method for using haplotype trees in phenotype/genotype association studies. Genetics. 2005;169:441–453. doi: 10.1534/genetics.104.030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Verzilli C, Shah T, Casas JP, Chapman J, Sandhu M, Debenham SL, Boekholdt MS, Khaw KT, Wareham NJ, Judson R, Benjamin EJ, Kathiresan S, Larson MG, Rong J, Sofat R, Humphries SE, Smeeth L, Cavalleri G, Whittaker JC, Hingorani AD. Bayesian meta-analysis of genetic association studies with different sets of markers. Am J Hum Genet. 2008;82:859–872. doi: 10.1016/j.ajhg.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers MA, Green FR, Terry C, Mayosi BM, Julier C, Lathrop M, Ratcliffe PJ, Watkins HC, Keavney B. Genotype at a promoter polymorphism of the interleukin-6 gene is associated with baseline levels of plasma C-reactive protein. Cardiovasc Res. 2002;53:1029–1034. doi: 10.1016/s0008-6363(01)00534-x. [DOI] [PubMed] [Google Scholar]
- Volanakis JE. Complement activation by C-reactive protein complexes. Ann N Y Acad Sci. 1982;389:235–250. doi: 10.1111/j.1749-6632.1982.tb22140.x. [DOI] [PubMed] [Google Scholar]
- Wassel Fyr CL, Kanaya AM, Cummings SR, Reich D, Hsueh WC, Reiner AP, Harris TB, Moffett S, Li R, Ding J, Miljkovic-Gacic I, Ziv E. Genetic admixture, adipocytokines, and adiposity in Black Americans: the Health, Aging, and Body Composition study. Hum Genet. 2007;121:615–624. doi: 10.1007/s00439-007-0353-z. [DOI] [PubMed] [Google Scholar]
- Wessel J, Moratorio G, Rao F, Mahata M, Zhang L, Greene W, Rana BK, Kennedy BP, Khandrika S, Huang P, Lillie EO, Shih PA, Smith DW, Wen G, Hamilton BA, Ziegler MG, Witztum JL, Schork NJ, Schmid-Schonbein GW, O'Connor DT. C-reactive protein, an ‘intermediate phenotype’ for inflammation: human twin studies reveal heritability, association with blood pressure and the metabolic syndrome, and the influence of common polymorphism at catecholaminergic/beta-adrenergic pathway loci. J Hypertens. 2007;25:329–343. doi: 10.1097/HJH.0b013e328011753e. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based Multiple Testing: Examples and Methods for P-value Adjustment. Wiley; New York: 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.