Abstract
Cancer results from somatic alterations in key genes, including point mutations, copy number alterations and structural rearrangements. A powerful way to discover cancer-causing genes is to identify genomic regions that show recurrent copy-number alterations (gains and losses) in tumor genomes. Recent advances in sequencing technologies suggest that massively parallel sequencing may provide a feasible alternative to DNA microarrays for detecting copy-number alterations. Here, we present: (i) a statistical analysis of the power to detect copy-number alterations of a given size; (ii) SegSeq, an algorithm to identify chromosomal breakpoints using massively parallel sequence data; and (iii) analysis of experimental data from three matched pairs of tumor and normal cell lines. We show that a collection of ∼14 million aligned sequence reads from human cell lines has comparable power to detect events as the current generation of DNA microarrays and has over two-fold better precision for localizing breakpoints (typically, to within ∼1 kb).
INTRODUCTION
Copy-number alterations represent a substantial category of genetic variation. Germline copy-number variants can be used for phenotypic mapping in genome-wide association studies, and have been linked to various diseases1-3. During carcinogenesis, tumor genomes often acquire somatic chromosomal alterations that can alter the dosage or structure of oncogenes and tumor suppressor genes. A powerful way to find cancer genes is to identify genomic regions with recurrent copy-number alterations (gains and losses) in tumor genomes4. Ideally, such characterization should include both the precise identification of the chromosomal breakpoints of each alteration and the accurate estimation of copy numbers in each chromosomal segment. Indeed, hybridization of genomic DNA to oligonucleotide microarrays can reveal genome-wide copy number changes5,6.
In principle, a simple and powerful approach to assessing copy-number alterations is to perform ‘digital karyotyping’. For instance, analyses of whole-genome shotgun sequencing data can delineate germline copy-number variations among individuals7-9. One can use a similar approach to detect copy-number alterations that arise somatically in tumor genomes. In essence, one performs shotgun sequencing of short sequence tags from tumor and normal DNA. The number of sequences aligning to each genomic region should be proportional to its copy number10-13. In practice, however, the high cost of DNA sequencing has greatly limited the practical application of this approach. Recently, a new generation of DNA sequencers has enabled massively parallel sequencing of millions of short sequence reads at dramatically lower costs8,14.
In this paper, we present a detailed analysis of the issues involved in identifying cancer copy-number alterations using massively parallel sequencing. First, we analyzed the statistical power to detect copy-number alterations and to map their boundaries accurately. Second, we developed SegSeq, a computational algorithm to detect these alterations and map their boundaries, taking advantage of the high density of sequence reads. Third, we applied these results to actual sequencing data from Illumina 1G Genome Analyzer, with reads length of 32 or 36 bp. With over 10 million aligned sequence reads per sample, we found that copy-number estimates from massively parallel sequencing achieved greater sensitivity, higher dynamic range and greater precision for mapping breakpoints than similar estimates based on microarray hybridization.
RESULTS
Statistical power: Copy-number alterations in fixed windows
We first studied the power to detect a copy-number alteration of a given size. Assuming that sequence reads are randomly chosen from the genome, the number of reads aligning to a region will follow a Poisson distribution with mean directly proportional to the size of the region and to the copy number. With 10 million aligned reads, for example, a region of 50 kb in the alignable portion of the human genome (A = 2.2 × 109) would be expected to have 50,000 × 107/A = ∼230 reads for 2 copies, ∼115 reads for one copy or ∼345 reads for three copies (Supplementary Methods online). In practice, one cannot hit repetitive sequences with uniquely aligning reads. Throughout, here we refer to the ‘uniquely aligning’ portions of a region.
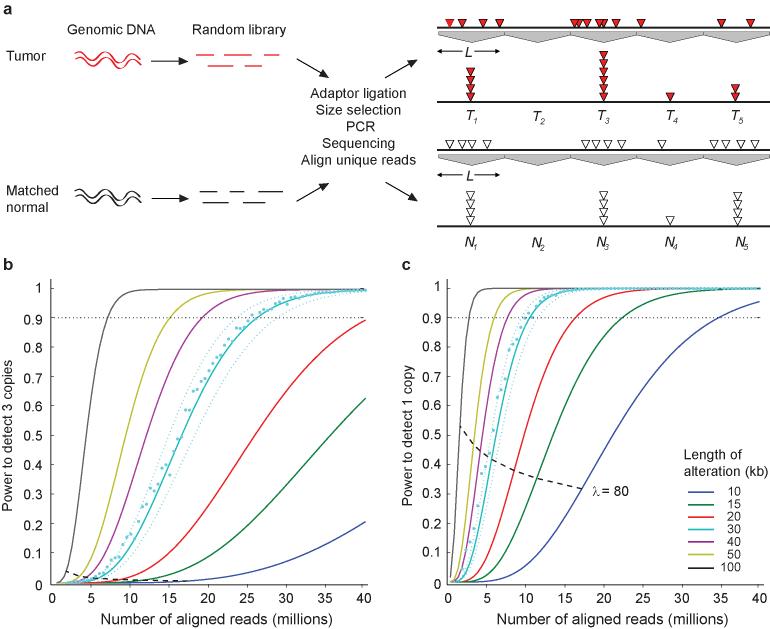
For any genomic region, its copy-number ratio equals the number of aligning reads from a tumor sample, divided by the number from the corresponding matched normal sample. One detects a copy-number alteration in regions where the copy-number ratio deviates from 1. In order to calculate the power to detect a significant alteration at a fixed genome-wide false-positive rate, we artificially partitioned the genome into non-overlapping windows of equal size (Fig. 1a). Then, we used a log-normal approximation for the logarithm of differences in copy-number ratios to calculate the total number of aligning reads required to have 90% power to discriminate between copy number 1, 2, or 3 for regions of various sizes at a stringency of a single false positive in the entire genome. To detect a 50 kb region of a single-copy gain, at this stringency, one requires ∼15 million aligned reads (Fig. 1b); for a single-copy loss one needs ∼6 million aligned reads (Fig. 1c).
Fig. 1. Theoretical coverage required to detect single copy gains and losses.
(a) Schematic overview for detecting copy-number alterations by sequencing. (b,c) Power calculations to detect copy-number alterations for a single copy gain and loss. We considered fixed windows L ranging from L = 10 kb to L = 100 kb. Lines indicate approximated power based on the distribution of ratios of normally-distributed random variables. For L = 30 kb we plotted simulation results for ratios of Poisson-distributed random variables (cyan dots). The approximation is accurate to within 10% (cyan dotted lines) for windows with average number of reads λ greater than 80 (dashed black line).
Algorithm: Detecting and localizing copy-number alterations
We developed a computational algorithm, called SegSeq, to detect and localize copy-number alterations from massively parallel sequence data. A simple approach would be to partition the genome into windows of fixed size, estimate the tumor-normal ratios for each window, and use segmentation algorithms to decompose the genome into regions of equivalent copy number15. The disadvantage of this approach, however, is that the breakpoints could not be localized more finely than the boundaries of the windows. Instead, we developed an approach with the ability to identify breakpoints at any read position. Our approach is thus not constrained to a window of a pre-specified size nor to fixed marker locations (as in microarray hybridization).
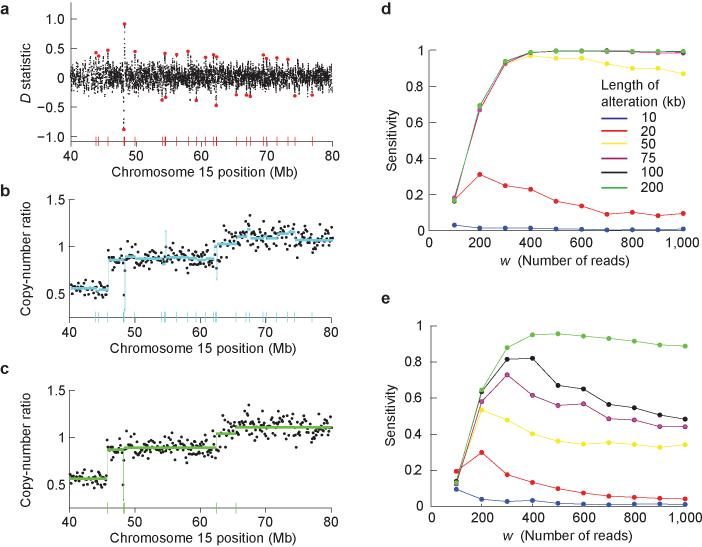
Our algorithm is a hybrid of local change-point analysis with a subsequent merging procedure that joins adjacent chromosomal segments (Fig. 2a–c). There are three user-defined parameters: w, the number of consecutive reads from the normal sample that defined the local windows for breakpoint initialization; pinit, the p-value cutoff for the initial list of candidate breakpoints; and pmerge, the p-value cutoff for merging adjacent segments.
Fig. 2. Segmentation algorithm for aligned sequenced reads.
(a-c) Schematic overview of segmentation algorithm. (a) Candidate breakpoints (red dots) correspond to tumor read positions (black dots) whose local log-ratio statistic, D, passes a lenient significance threshold. (b) These candidate breakpoints define the boundaries of the initial copy-number segments (blue lines). Each point represents the estimated copy-number ratio for a 100 kb window. (c) A merging procedure yields the final list of copy-number segments (green lines) obtained for 10 genome-wide false positives. (d-e) Sensitivity to detect copy-number alterations as a function of the local window size parameter, w. A copy-number alteration of a particular size is introduced into a diploid genome sampled by 12 million aligned reads. Each line represents the fraction of 1000 spike-in simulations for which (d) a copy-number gain or (e) a copy-number loss was correctly identified by the segmentation algorithm.
In the first step, we hyper-segmented the genome by generating a list of candidate breakpoints based on read counts in local windows. At each tumor read position, we extended a window to the left and to the right to include a fixed number of reads, w, in the normal sample. Then, we calculated the significance (p-value) of a copy-number change based on the log-ratio between the number of tumor reads contained in both windows (Supplementary Fig. 1 online). Positions which passed a lenient genome-wide significance threshold (p-value < pinit) were declared as candidate breakpoints; these positions demarcated the initial list of segments. In the next step, we iteratively joined segments by eliminating the breakpoint between them, starting from the least significant and continuing as long as its p-value was above merge. In this step, p- values are calculated based on the number of reads in the tumor and normal in the entire segments. Since these segments were typically larger than the local windows, the increased number of aligned reads enables more accurate estimation of statistical significance.
We optimized the user-defined parameters based on replicate sequencing lanes of a normal sample. The preferred values for these parameters were set as follows: (i) The p-value cutoffs, pinit and pmerge, controlled the genome-wide false positive rates and were set such that we generated ∼1,000 false positive initial breakpoints and ∼10 false positive final segments (Supplementary Methods). (ii) The local window size, w, was set to maximize the sensitivity to detect alterations, as assessed via spike-in simulations using actual sequence reads obtained from a tumor cell line and its matched normal (Fig. 2d,e). We tested single-copy alterations varying from 10 kb to 500 kb, assuming ∼12 million aligned reads in both the tumor and normal samples. At this sequencing depth, we found that w = 400 provided the best sensitivity for single-copy gains at least 50 kb in size (Fig. 2d) and w = 300 provided the best sensitivity for single-copy losses at least 75 kb in size (Fig. 2e).
Application: Copy-number alterations in tumor cell lines
To test the methodology, we generated and analyzed massively parallel sequence data on the Illumina 1G Genome Analyzer from three tumor cell lines (HCC1954, HCC1143 and NCIH2347) and their matched normal cell lines (Supplementary Methods). For each of the six cell lines, we obtained 10-19 million uniquely aligned reads (Supplementary Table 1 online). We noted that the number of observed counts in both normal and tumor cell lines depended on the local G+C content (Supplementary Fig. 2,3, Supplementary Table 2 online), which may reflect inherent biases in the sample preparation or sequencing procedures. These biases were mitigated by our approach to analyze the ratio of the number of reads seen in tumor DNA and its paired normal DNA, processed at the same time.
We used our segmentation algorithm with these optimized parameters to parse the genome into intervals of constant copy number. After filtering for segments with copy-number ratios greater than 1.5 or less than 0.5, we found 194 copy-number alterations in the HCC1954 cell line, 126 alterations in the HCC1143 cell line, and 15 alterations in the NCI-H2347 cell line (Table 1, Supplementary Fig. 4-6, Supplementary Data online). There were six high-level amplifications (copy-number ratios greater than 8), all of which matched previously reported loci16,17. We also found seven regions of homozygous deletion ranging in size from ∼29 kb to ∼582 kb (Supplementary Table 3, Supplementary Fig. 7 online).
Table 1.
Summary of copy-number alterations in tumor cell lines
| Massively parallel sequencing | Affymetrix SNP 6.0 array | |||||
|---|---|---|---|---|---|---|
| HCC1954 | HCC1143 | NCI- H2347 |
HCC1954 | HCC1143 | NCI- H2347 |
|
| Number of segments with predicted gains | ||||||
| Copy-number ratio 1.5 - 2.0 | ||||||
| 63 | 61 | 5 | 57 | 43 | 5 | |
| Copy-number ratio 2.0 - 4.0 | 78 | 38 | 0 | 62 | 29 | 1 |
| Copy-number ratio 4.0 - 8.0 | 27 | 2 | 0 | 20 | 3 | 0 |
| Copy-number ratio > 8.0 | 5 | 1 | 0 | 1 | 0 | 0 |
| Number of segments with predicted losses | ||||||
| Copy-number ratio < 0.25 | 0 | 3 | 3 | 0 | 2 | 4 |
| Copy-number ratio 0.25 - 0.50 |
21 | 21 | 7 | 13 | 16 | 8 |
| Total number of predicted alterations |
194 | 126 | 15 | 153 | 93 | 18 |
We then compared the results obtained by massively parallel sequencing to the results obtained from hybridization of the same samples to oligonucleotide arrays (Affymetrix SNP Array 6.0). After merging segments that spanned fewer than 8 consecutive probe sets, we found 153 copy-number alterations in the HCC1954 cell line, 93 alterations in the HCC1143 cell line, and 18 alterations in the NCI-H2347 cell line.
In general, the copy-number segments detected by both approaches were highly concordant with respect to identifying the existence of a copy-number alteration, while massively parallel sequencing had somewhat better resolution for localizing the breakpoints (Supplementary Fig. 8 online). Notably, sequencing achieved a higher dynamic range for estimating copy-number alterations. For instance, we considered the high-level amplification of the ERBB2 locus in the HCC1954 cell line. We estimated a 16-fold increase in copy-number ratio by microarrays, comparied to a 55.6-fold increase estimated by sequencing (Supplementary Figs. 8 and 9 online). Quantitative PCR measurement confirmed the higher extent of amplification16(at ∼70-fold). This saturation effect of microarray hybridization at high copy numbers could be explained by a Langmuir adsorption model18(Supplementary Fig. 8, Supplementary Methods online).
Application: Mapping breakpoints in tumor cell lines
We next studied our ability to map breakpoints accurately. For this purpose, we considered interstitial homozygous deletions, whose boundaries can be mapped to single-nucleotide resolution by sequencing across the deletion. We detected 3 homozygous deletions in the NCI-H2347 cell line: a novel 44-kb deletion at the UTRN locus, as well as previously reported deletions at the PTPRD and HS3ST3A1 loci19,20 (Supplementary Table 3, Supplementary Fig. 10-12 online). After confirming that these deletions were absent in the paired normal cell line, we mapped their breakpoints by the conventional sequencing of PCR products spanning each deletion.
Our segmentation algorithm (using ∼14 million tumor reads) predicted breakpoints that were extremely close to the actual breakpoints (the differences for the six breakpoints being 2, 52, 226, 527, 829 and 1,007 bp, with a mean of 440 bp) (Fig. 3a—c, Supplementary Table 3 online). Since short sequence reads cannot uniquely align to repeat regions, the presence of Alu repeats flanking three of the six breakpoints limited the precision of mapping. Segmentation of data from microarrays had a mean error of 1,068 bp; it missed the actual breakpoints by +2718 bp and -1,262 bp for the UTRN locus, by -491 bp and -1,242 bp for the PTPRD locus and by +608 bp and -86 bp for the HS3ST3A1 locus (Fig. 3d–f).
Fig. 3. Mapping the chromosomal breakpoints of homozygous deletions.
(a-c) Breakpoint mapping with aligned sequence reads at: (a) the UTRN locus; (b) the PTPRD locus; or (c) the HS3T3A1 locus. Each point represents the location of a sequence read aligning to the NCI-H2347 (blue) tumor cell line or its matched normal, NCI-BL2347 (black). Vertical green lines indicate the exact chromosomal breakpoints mapped by sequencing of a PCR product spanning each homozygous deletion. For each breakpoint, we report the difference between the predicted and actual breakpoint positions. (d-f) Breakpoint mapping with an Affymetrix SNP 6.0 Array, where each point represents the log2copy-number ratio interrogated by an array probeset in: (d) the UTRN locus; (e) the PTPRD locus; or (f) the HS3T3A1 locus. The minimum value for log2copy-number ratios was set to -7. Horizontal blue lines represent copy-number segments inferred by the circular binary segmentation algorithm28.
DISCUSSION
With the advent of powerful new technologies, massively parallel sequencing will provide increasingly high-resolution analyses of copy-number alterations in cancer genomes. We found that a collection of ∼14 million sequence reads had over two times higher resolution than the current generation of DNA microarrays (median spacing ∼700 bp) to localize breakpoints. Our analysis of sequence data from three tumor-normal cell line pairs provided experimental confirmation of our statistical analyses. Although the sequencing of 14 million reads is currently more expensive than microarray hybridization, relative costs may change with higher sequencing throughput.
Cancer genome analysis will benefit considerably from these improvements in measurement accuracy. A common approach to localizing key cancer-related genes relies on pinpointing a ‘common region of overlap’ among overlapping gains or losses across hundreds of samples4,21,22. The increased precision of mapping chromosomal breakpoints in individual samples will identify more precise coordinates for the aggregate overlapping region. Even more importantly, improvements in sequencing will enable the detection of extremely small intragenic events, especially homozygous deletions. For example, we identified four intragenic homozygous deletions ranging in size from 44 kb to 582 kb that affected between one and 15 coding exons. The higher precision of breakpoint mapping may thus help to identify recurrent alterations in tumor suppressor genes that have been previously missed by other genome characterization technologies.
Massively parallel sequencing technologies offer three other key advantages relative to microarray-based hybridization approaches. First, smaller copy-number alterations could be detected by simply increasing the depth of sequencing. Second, one could compensate for stromal admixture in tumor samples by performing deeper sequencing. Third, paired-end sequence reads provide information about structural rearrangements that cannot easily bedetected by array-based methods14. Future improvements to our method would evaluate the statistical significance for detecting structural rearrangements from paired-end reads.
As sequencing and microarray technologies continue to improve, it will be important to continually benchmark their performance. We anticipate that each massively parallel sequencing platform may be susceptible to particular biases23,24(Supplementary Figs. 2 and 3 online). We propose that the trio of cancer cell lines and the sequence data reported here may provide a useful foundation for such evaluation.
METHODS
Sample preparation, sequencing and alignment
For each cell line, we prepared 3 micrograms of genomic DNA for sequencing on the Illumina 1G Genome Analyzer25 (Supplementary Methods).
Statistical analysis of tumor-normal copy-number ratios
We describe a statistical framework for observing a certain number of reads obtained from a tumor and a matched normal sample that align to a genomic window (Supplementary Methods, Supplementary Fig. 1 and 13 online).
Segmentation algorithm for the identification of copy-number alterations
We identified copy-number alterations based on changepoint detection, followed by agglomerative merging of adjacent segments. The input to this algorithm is a list of positions for aligned sequence reads from a tumor sample and a normal sample, while the output includes a list of breakpoints and copy number estimates for each inferred chromosomal segment (Supplementary Methods).
Comparison of copy-number alterations with single nucleotide polymorphism arrays
We calculated copy numbers for the Affymetrix Genome-Wide Human SNP Array 6.0 with a GenePattern pipeline26 according to methods previously described27. We optimized parameters for the Circular Binary Segmentation algorithm28 to infer chromosomal segments of constant copy number from the median of replicate arrays (Supplementary Fig. 14 online and Supplementary Methods). We determined consensus chromosomal segments from the list of breakpoints predicted by each method and evaluated the concordance between predicted copy numbers (Supplementary Fig. 8 and Supplementary Methods).
Data and software availability
National Center for Biotechnology Information (NCBI) Short Read Archive: SRP000246 (sequence reads); NCBI Gene Expression Omnibus: GSE13372 (Affymetrix SNP 6.0 array data). MATLAB code that implements the segmentation algorithm can be obtained from: http://www.broad.mit.edu/cancer/pub/solexa_copy_numbers.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Mermel, M. Berger and E. Hom for commenting on the manuscript. This work was supported by the US National Institutes of Health (grants 5U24CA126546 to M.M. and 5U54HG003-67 to E.S.L.).
REFERENCES
- 1.Freeman JL, et al. Copy number variation: New insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 2.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 3.Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 4.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat. Genet. 2005;37:S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 6.Kallioniemi A. CGH microarrays and cancer. Curr. Opin. Biotechnol. 2008;19:36–40. doi: 10.1016/j.copbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 8.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang TL, et al. Digital karyotyping. Proc. Natl. Acad. Sci. USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih I, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc. Natl. Acad. Sci. USA. 2005;102:14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leary RJ, Cummins J, Wang TL, Velculescu VE. Digital karyotyping. Nat. Protoc. 2007;2:1973–1986. doi: 10.1038/nprot.2007.276. [DOI] [PubMed] [Google Scholar]
- 13.Morozova O, Marra MA. From cytogenetics to next-generation sequencing technologies: advances in the detection of genome rearrangements in tumors. Biochem. Cell Biol. 2008;86:81–91. doi: 10.1139/O08-003. [DOI] [PubMed] [Google Scholar]
- 14.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai WR, Johnson MD, Kucherlapati R, Park PJ. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi N, et al. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- 18.Hekstra D, Taussig AR, Magnasco M, Naef F. Absolute mRNA concentrations from sequence-specific calibration of oligonucleotide arrays. Nucleic Acids Res. 2003;31:1962–1968. doi: 10.1093/nar/gkg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 20.Nagayama K, et al. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer. 2007;46:1000–10. doi: 10.1002/gcc.20485. [DOI] [PubMed] [Google Scholar]
- 21.Guttman M, et al. Assessing the significance of conserved genomic aberrations using high resolution genomic microarrays. PLoS Genet. 2007;3:e143. doi: 10.1371/journal.pgen.0030143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedemeyer R, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.