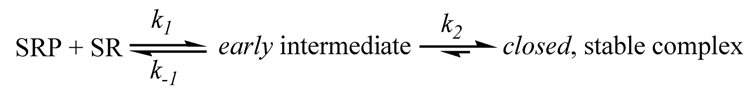Abstract
Two GTPases in the signal recognition particle (SRP) and its receptor (SR) control the delivery of newly synthesized proteins to the ER or plasma membrane. During the protein targeting reaction, the 4.5S SRP RNA accelerates the association between the two GTPases by 400-fold. Using fluorescence resonance energy transfer (FRET), we demonstrate here that formation of a stable SRP•SR complex involves two distinct steps: a fast initial association between SRP and SR to form an early, GTP-independent complex, followed by a GTP-dependent conformational rearrangement to form the stable, final complex. We also found that the 4.5S SRP RNA significantly stabilizes the early, GTP-independent intermediate. Further, mutational analyses show that there is a strong correlation between the ability of the mutant SRP RNAs to stabilize the early intermediate and their ability to accelerate SRP•SR complex formation. We propose that the SRP RNA, by stabilizing the transient early intermediate, can give this intermediate a longer dwell time and therefore a higher probability to rearrange to the final, stable complex. This provides a coherent model that explains how the 4.5S RNA exerts its catalytic role in SRP•SR complex assembly.
INTRODUCTION
To maintain proper cellular function, a cell needs to efficiently and accurately deliver all its proteins to the different subcellular organelles. The signal recognition particle (SRP) and its receptor (SR) constitute a universally conserved machinery to deliver newly synthesized proteins from the cytoplasm to the eukaryotic endoplasmic reticulum (ER) membrane, or the bacterial plasma membrane 1–3. The protein targeting reaction consists of several ordered steps that ensure the efficiency and fidelity of this process 4,5. At the beginning of the targeting cycle, the SRP recognizes translating ribosome that carries a signal sequence on the nascent chain. The SRP then forms a complex with SR localized on the target membrane; this process brings the ribosome•nascent chain complex (RNC) to the membrane surface. Upon arrival at the membrane, conformational changes in the SRP•SR complex drive the release of the RNC from the SRP to a protein conducting channel composed of the sec61p (or secYEG in bacteria) complex 6. Once the RNC is released, the SRP and SR dissociate into free components, allowing a new round of the protein targeting reaction. Thus, the ordered assembly and disassembly of the SRP•SR complex control the delivery of proteins to their proper cellular destinations.
In eukaryotes, SRP is a universally conserved ribonucleoprotein complex consisting of six proteins and an SRP RNA 7–9. The protein targeting function of the SRP requires only two components: the conserved SRP54 protein in complex with the SRP RNA. SRP54 (called Ffh in E. coli) is composed of two structurally and functionally distinct domains: a methionine-rich M domain and an NG domain. The M domain recognizes the signal sequences and binds the SRP RNA 10–14. A GTPase, G-domain and an N-terminal four helix bundle (the N-domain) together form a structural and functional unit called the NG domain, which binds and hydrolyzes GTP and forms a complex with SR (called FtsY in bacteria) 15–18. The NG domain was also suggested to play a role in signal peptide recognition 19. The SRP and SR GTPases use a regulatory mechanism distinct from that of classical signaling GTPases such as Ras, Rho and Ran 20. The structure of both GTPases are similar regardless of whether GTP or GDP is bound 21–24. Thus, the SRP and SR do not switch between active and inactive states depending on whether GTP or GDP is bound. Moreover, these GTPases bind nucleotides weakly and exchange nucleotides quickly, so that no external nucleotide exchange factors are required to switch these GTPases from the GDP- to the GTP-bound state 25. In addition, the SRP and SR reciprocally stimulate each other’s GTPase activity upon formation of the SRP•SR complex 17. Therefore, no external GTPase activating proteins are required to regulate the switch of these GTPases from the GTP- to the GDP-bound state. Instead, recent biochemical and biophysical analyses suggest that several discrete conformational changes occur during the binding and reciprocal activation between the two proteins, and each of these conformational change may provide a potential point for regulation during the protein targeting reaction 5,6.
The SRP RNA has been shown to play an indispensable role in protein targeting both in vitro and in vivo 26–30. The size of the SRP RNA varies widely from bacteria to yeast and mammalian cells; nevertheless, the most phylogenetically conserved region of the SRP RNA, domain IV, has been maintained in all three kingdoms of life 31,32. The role of SRP RNA may involve recognition and binding of the ribosome and signal sequences 11,26,33, and stabilization of the folding of the M-domain34. In addition, it was also proposed to bind and stabilize the NG domain of Ffh 34,35. Intriguingly, kinetic analyses of the role of the 4.5S SRP RNA on the GTPase cycles of Ffh and FtsY showed that the RNA also plays a critical role in the interaction between the two GTPases 17,25. In the absence of the SRP RNA, Ffh-FtsY association is extremely slow, with a rate constant of 5 × 103 M−1s−1, and the SRP RNA accelerates their association kinetics by 400-fold 17,25,36. An additional step, GTP hydrolysis after the complex is formed, is also enhanced 8-fold by the 4.5S RNA 25. Thus, the presence of the SRP RNA brings the interaction kinetics between the SRP and SR to an appropriate range for their biological functions. The SRP RNA contains a highly conserved GNRA tetraloop that was shown to be essential for the interaction between the SRP and SR. Tetraloop mutants were reported to impair the binding between SRP and SR, cause a reduction in the GTPase activity of the SRP•SR complex, as well as fail to support normal cell growth in vivo 27,30. A site directed hydroxyl radical probing study further suggest that the tetraloop is located close to the heterodimer interface of the SRP•SR GTPase complex 37.
To probe the conformational dynamics during the SRP-SR interaction and to elucidate how the SRP RNA exerts its catalytic role on SRP•SR complex assembly, we developed a highly sensitive FRET assay to monitor the interaction between the SRP and SR in real time. This new assay led to the discovery of a new SRP•SR complex that forms independently of GTP. This GTP-independent complex has been observed only once in a surface-resonance experiment using mammalian SRP and SR 38. Further characterization identifies this GTP-independent complex as an early intermediate during the initial stage of the SRP-SR interaction. Formation of the early intermediate is substantially stabilized by the 4.5S RNA, and 4.5S RNA tetraloop mutants that fail to stabilize this intermediate also fail to accelerate SRP•SR complex assembly. We propose that the catalytic role of 4.5S RNA on complex assembly can be explained by its stabilizing effect on the early intermediate, which increases its probability to rearrange to the final, GTP-stabilized complex.
RESULTS
To characterize the conformational dynamics during the SRP-SR interaction, we developed a real time assay based on fluorescence resonance energy transfer (FRET). The basic strategy is to engineer a single cysteine residue on cysteine-less Ffh and FtsY proteins, and label the single cysteines with thio-reactive fluorescent probes (Fig. 1a). In Ffh, the intrinsic cysteine at position 406 can be replaced with serine without disrupting its function (37,39 and data not shown). A cysteine was introduced at position 153 of cysteine-less Ffh and labeled with maleimide-coumarin (DACM) as the FRET donor. FtsY does not contain any cysteine residue; thereby a cysteine was introduced at position 345 and labeled with maleimide-BODIPY-fluorescein (BODIPY-FL) as the FRET acceptor. These probes are close to the nucleotide binding pocket in the G domains of both proteins, and are 31 Å apart as estimated from the crystal structure of the Thermus aquaticus Ffh•FtsY complex (Figure 1a; 16). The cysteine mutation and fluorescence labeling do not alter the ability of Ffh and FtsY to bind and activate each other’s GTPase activity (Supplementary Fig. 1), nor do they affect their ability to translocate model SRP substrates into ER microsomal membranes (data not shown).
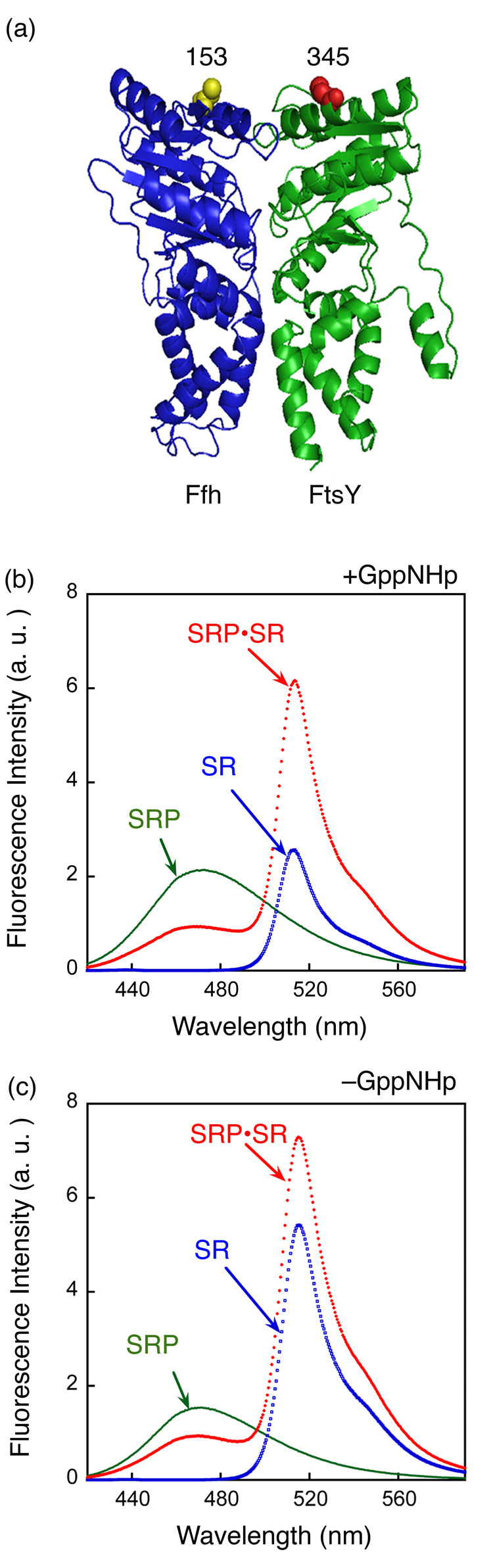
Figure 1. SRP and SR can form a complex independently of GTP.
(a) Position of FRET donor (●) and acceptor (●) probes on the SRP (Ffh) and SR (FtsY) on a co-crystal structure of the Ffh•FtsY complex 16. (b) Fluorescence emission spectrum of SRP•SR complex in the presence of 100 µM GppNHp. 0.5 µM SRP and 2 µM SR were incubated for 10 minutes at 25 °C to form the SRP•SR complex (red). SRP- and SR-only spectra (green and blue, respectively) were obtained by incubating fluorescently labeled SRP (or SR) with unlabeled SR (or SRP). (c) Fluorescence emission spectrum of SRP•SR complex in the absence of GppNHp. 5 µM SRP and 15 µM SR were incubated at 25 °C for 10 minutes. SRP or SR-only spectra were obtained as in part (b).
A GTP-independent complex is detected by the FRET assay
Previous studies have shown that SRP and SR form a stable complex in the presence of GTP or non-hydrolyzable GTP analogues such as 5’-guanylylimido-diphosphate (GppNHp), with dissociation constants of 16 – 30 nM 17. As expected, a significant amount of FRET was observed upon assembly of the SRP•SR complex in the presence of GppNHp (Figure 1b). At saturating protein concentrations, the FRET efficiency was 0.80 (Fig. 2a), in good agreement with the distance between the two residues in the crystal structure and the Förster radius of this donor-acceptor pair. To our surprise, when GppNHp was either removed from the reaction mix or replaced by GDP, significant FRET was also observed (Fig. 1c), suggesting that an SRP•SR complex can be formed independent of GTP.
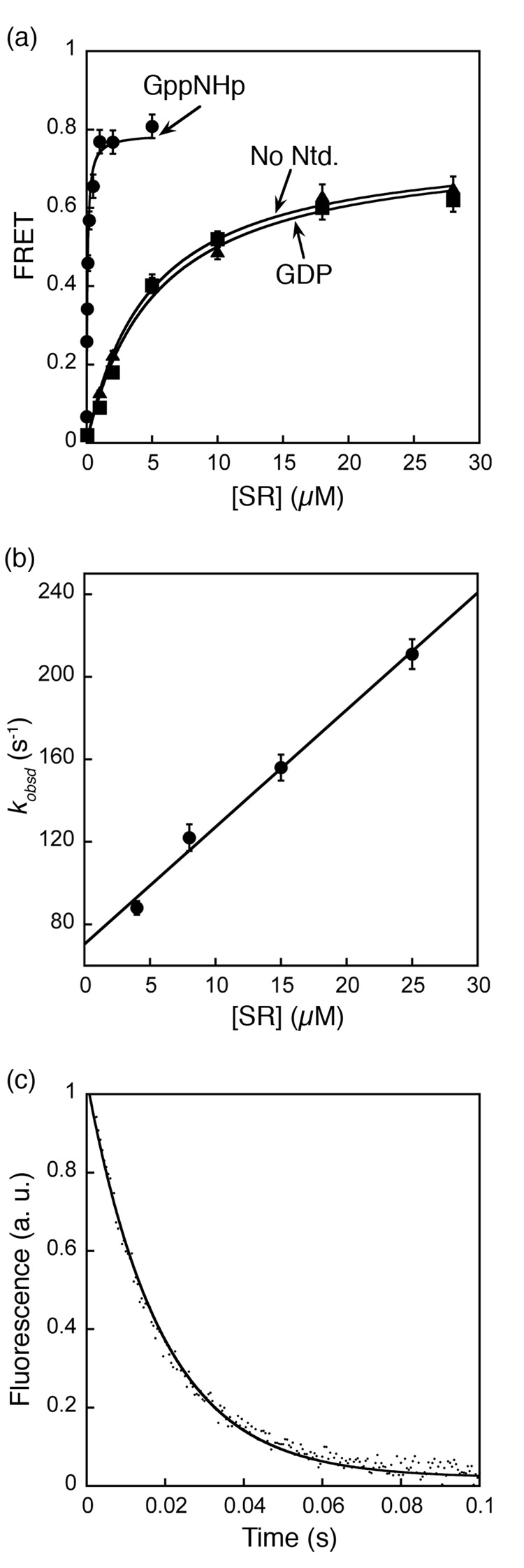
Figure 2. Stability and kinetics for formation of the GTP-independent complex.
(a) Equilibrium titration of SRP•SR complex with GppNHp (●), GDP (■), and without nucleotide (▲). The data were fit to a single binding equation and gave dissociation constants of 16 nM (GppNHp), 4 µM (GDP) and 4.2 µM (no nucleotide). (b) Association kinetics of GTP-independent complex was measured as described in Methods. Values of observed rate constants were plotted against SR concentration and a linear fit of the data gave an association rate constant of 5.6 × 106 M−1 s−1. (c) Dissociation kinetics was determined in a pulse-chase experiment described in Methods. The data were fit to a single exponential equation and gave a dissociation rate constant of 60 s−1.
The affinities of the GTP-dependent and GTP-independent complexes were measured by equilibrium titration. The dissociation constant of the complex formed in the presence of GppNHp was determined to be 16 nM using this FRET assay (Fig. 2a, circles), consistent with previous studies 17. In contrast, a dissociation constant of 4 – 10 µM was observed for the complex assembled in the presence of GDP or no nucleotide (Fig. 2a, squares and triangles, respectively). Thus, the γ-phosphate of GTP contributes over 250-fold to the stability of the SRP•SR complex. In these titration experiments, the FRET value at saturating protein concentrations represent the FRET efficiency of the two probes in their respective complexes: the GTP-independent complex has a FRET efficiency of 0.62, which is ~ 25% lower than that of the GTP-dependent complex (0.80). The different FRET values suggest that these two complexes have different conformations in which the donor and acceptor fluorophores are positioned or oriented differently. Similar results were observed when another FRET pair was engineered near the N-domain of each protein (Supplementary Fig. 2).
In addition to equilibrium measurements, we also determined the kinetics for assembly and disassembly of the GTP-independent complex by following fluorescence emission from the FRET donor over time. The time course for assembly of the GTP-independent complex fits well to single exponential kinetics (Fig. 3, blue and data now shown); plots of the observed rate constant against the concentration of SR gave an association rate constant kon of 5.7 ± 0.5 × 106 M−1s−1 (Fig. 2b). This is over 50 times faster than the association kinetics for formation of the GTP-dependent complex previously determined 17. The dissociation rate constant of the GTP-independent complex is 62 ± 6 s−1 (Fig. 2c), which is 2 × 104-fold faster than that of the GTP-dependent complex 17. Thus in addition to the lower equilibrium stability, the GTP-independent complex is also kinetically much less stable than the GTP-dependent complex previously characterized 17. This explains why this complex was not observed previously based on gel filtration analyses 16, which can only detect kinetically stable complexes.
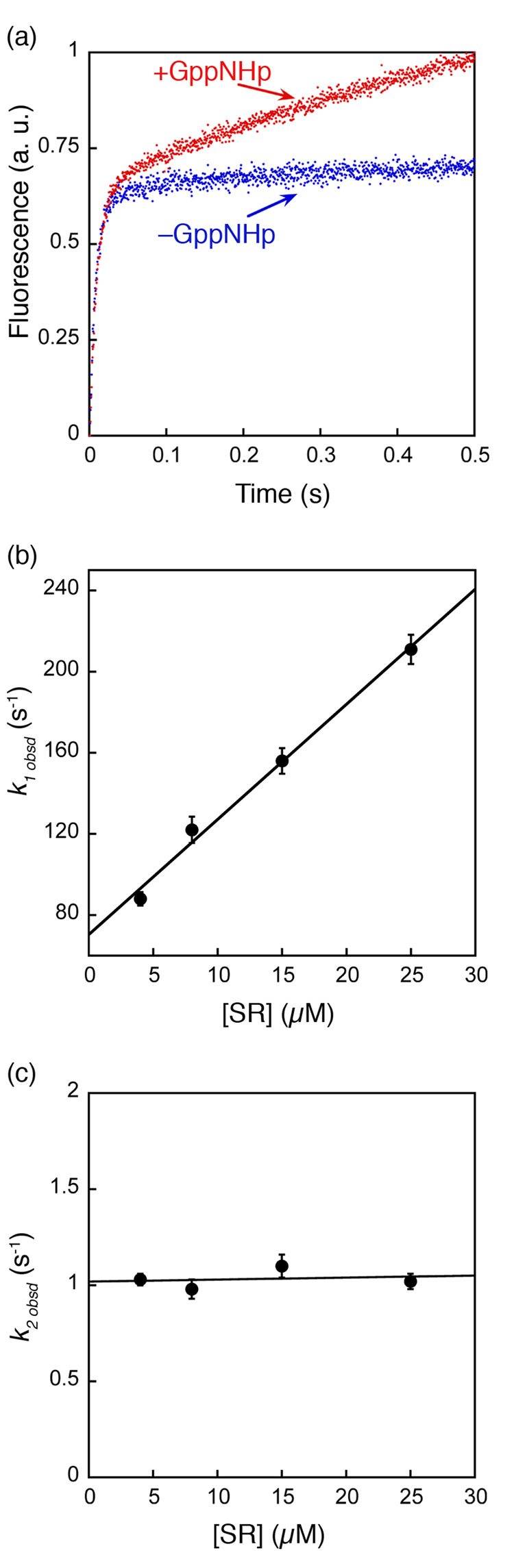
Figure 3. Formation of an SRP•SR complex in the presence of GppNHp involves two discrete steps.
(a) Comparison of the time courses for complex formation in the absence (blue) and presence of 100 µM GppNHp (red). Data were obtained with 4 µM SRP and 8 µM SR. (b) The observed rate constants of the first kinetic phase during SRP-SR association in the presence of GppNHp were plotted against SR concentration. A linear fit of the data gave an association rate constant of 5.8 × 106 M−1 s−1 (k1 in Scheme I). (c) The observed rate constants of the second kinetic phase during SRP•SR association in the presence of GppNHp are independent of SR concentration. The average of these rate constants is 1.03 s−1 (k2 in Scheme I).
The following observations strongly suggest that the GTP-independent complex is not an artifact introduced by dye labeling: (1) the FRET value is dependent on protein concentration and is saturable, suggesting that the FRET signal arises from complex formation, rather than nonspecific interactions between the dyes; (2) FRET from the GTP-independent complex can be competed away by unlabeled protein (Fig. 2c and data not shown); and (3) SR labeled with an environmentally sensitive probe (acrylodan) on position 242 also showed a fluorescence change when the complex was formed in GDP (unpublished results). Thus, FRET provides a robust and highly sensitive assay that allows us to detect, for the first time, a transient GTP-independent SRP•SR complex that has a different conformation than that observed previously for the GTP-dependent complex.
The GTP-independent complex represents a transient intermediate on the pathway for formation of the GTP-stabilized complex
In this section we provide two lines of evidence that strongly suggest that the GTP-independent complex is an on-pathway intermediate preceding the formation of the GTP-dependent complex: (1) an intermediate can be directly detected in the time course for formation of the GTP-dependent complex, and the kinetics for formation of this intermediate agrees with the kinetics for assembly of the GTP-independent complex, and (2) stabilization of the GTP-independent intermediate by the SRP RNA also accelerates the rate for formation of the final, GTP-dependent complex, consistent with the notion that the GTP-independent complex in an on-pathway intermediate.
The first piece of evidence was obtained from comparison of the kinetics of complex formation in the presence or absence of GppNHp. To ensure that low affinity intermediates can accumulate and be detected, we used a high concentration of SR during complex assembly, and fluorescence emission from the FRET donor was followed over time. The time course for complex formation in the presence of GppNHp exhibits biphasic kinetics (Fig. 3a, red), indicating that there are at least two steps involved in the assembly of the GTP-dependent complex. The first kinetic phase is dependent on SR concentration (Fig. 3b), and therefore represents fast, bi-molecular association between SRP and SR to form an intermediate that has a lower FRET value. The second kinetic phase is concentration independent (Fig. 3c) and thus represents the unimolecular rearrangement of this intermediate to a complex that has a higher FRET value. Remarkably, the rate constant of the first kinetic phase coincides very well with that for formation of the GTP-independent complex (Fig. 3a, blue), with observed rate constants of 118 and 122 s−1 at 8 µM SR (Fig. 3a). This strongly suggests that the GTP-independent complex is the intermediate observed in the first kinetic phase during complex assembly in the presence of GppNHp. In contrast to the bi-phasic kinetic behavior during assembly of the GTP-dependent complex, formation of the GTP-independent complex does not have a second kinetic phase (Fig. 3a, blue), suggesting that the rearrangement represented in the second kinetic phase is strictly GTP-dependent.
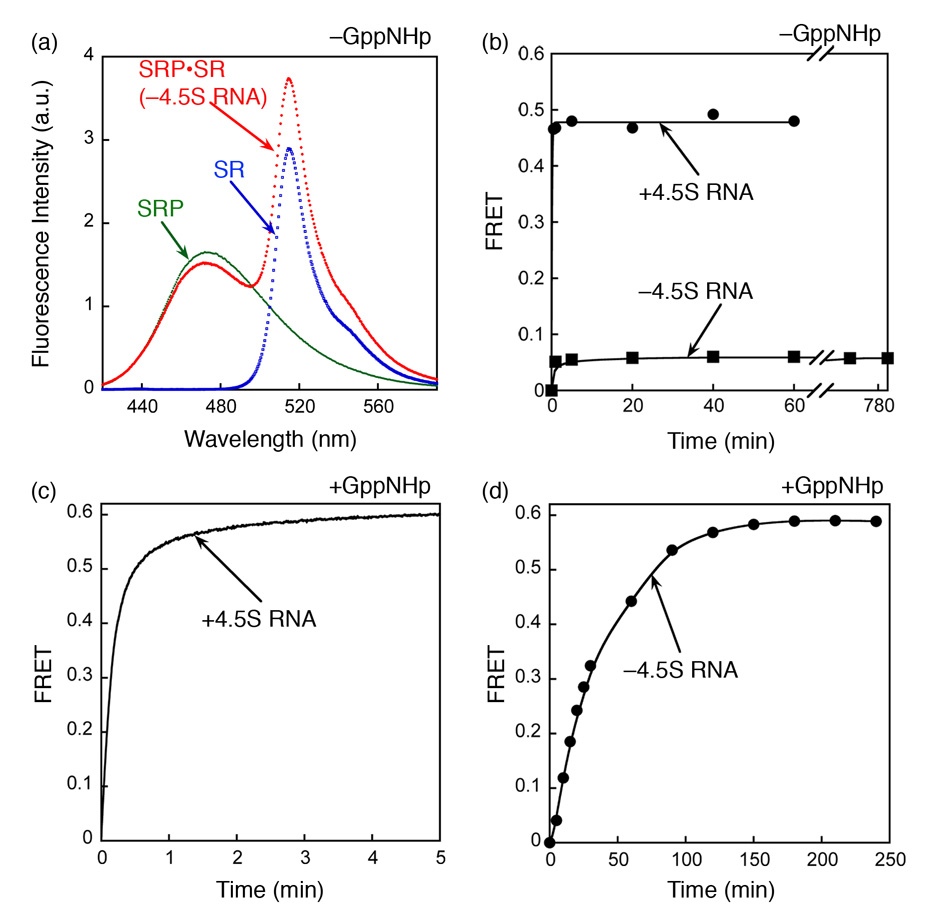
A classical criterion for an on-pathway intermediate is that stabilization of the intermediate accelerates the reaction to form the final product. This criterion was satisfied by the effects of the 4.5S SRP RNA on the GTP-independent and GTP-dependent complexes. The GTP-independent complex could not be formed in the absence of the 4.5S RNA (Fig. 4a), even after long periods of incubation when equilibrium had been reached (Fig. 4b). Thus, the 4.5S RNA increases the equilibrium stability of the GTP-independent complex. In contrast, it was shown that a stable GTP-dependent Ffh•SR complex can be formed with or without the 4.5S RNA, but the RNA accelerates the association rate of this complex by 200-fold (17 and cf. Figures 4c and 4d). The results presented here and in the next section show that there is a strong correlation between the ability of the 4.5S RNA to stabilize the GTP-independent complex and its ability to accelerate formation of the GTP-dependent complex. This provides independent evidence that the GTP-independent complex is an on-pathway intermediate. If the GTP-independent complex were off the pathway, then its stabilization by the 4.5S RNA would compromise formation of the native complex in the presence of GppNHp.
Figure 4. The GTP-independent complex is stabilized by the 4.5S RNA.
(a) Spectrum of the GTP-independent complex in the absence of 4.5S RNA. The experiment setup is the same as in Fig. 1c except that the 4.5S RNA was not included. (b) Formation of the GTP-independent complex was monitored in the presence (●) and absence (■) of the 4.5S RNA. (c and d) The time course for formation of the GTP-dependent complex was monitored in the presence (c) and absence (d) of 4.5S RNA. In (c), 0.5 µM SRP and 2 µM SR were used. In (d), 2 µM Ffh and 10 µM SR were used to obtain a faster reaction rate. Note the difference in time scales in (c) and (d).
Taken together, these results demonstrate that formation of the GTP-stabilized SRP•SR complex involves at least two steps (Scheme I): (1) GTP-independent bimolecular association between the SRP and SR to form a transient intermediate (referred to as the early intermediate); and (2) GTP-dependent rearrangement of the early intermediate to form the stable complex previously observed. As demonstrated previously, additional conformational stages are present even after the stable complex is formed (5 and see Fig. 8 in Discussion). Thus, the interaction between the SRP and SRP receptor is a dynamic process involving multiple conformational changes during complex assembly and activation.
Scheme 1.
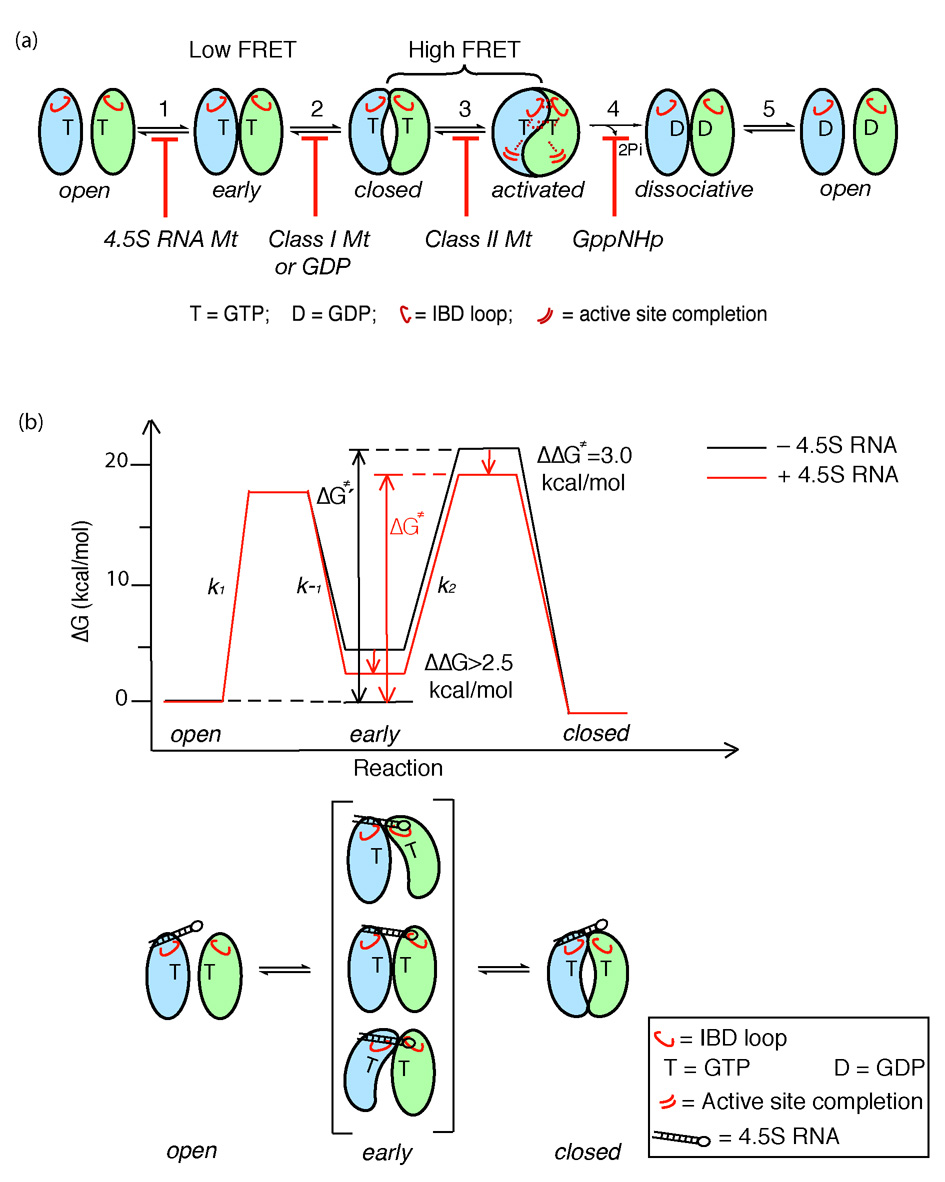
Figure 8. Multiple conformational changes during SRP-SR complex formation and activation.
(a) SRP and SR GTPases form an early GTP-independent intermediate that exhibits a low FRET (step 1). In the presence of GTP, early rearranges to a more stable, closed complex that exhibits a high FRET (step 2). Additional rearrangements in the catalytic loops activate GTP hydrolysis (step 3). GTP hydrolysis drives the dissociation of the SRP•SR complex (steps 4 and 5). Each step can be blocked using specific mutants or nucleotides. 4.5S RNA tetraloop mutants block formation of the early intermediate. Class I mutants of SR 5 or GDP blocks formation of a closed complex. Class II mutants on SRP or SR 5, 6 block the rearrangement that activates GTP hydrolysis. GppNHp blocks the chemical step. (b) top panel: free energy profile for the SRP-SR interaction in the absence (black) and presence (red) of the 4.5S RNA for a standard state of 200 nM that mimics the in vivo protein concentrations in bacteria. Activation energies were calculated from the observed association and dissociation rate constants using ΔG = −RT ln(kh/kBT), where R = 1.987 cal K−1 mol−1, h = 1.58 × 10−37 kcal s−1, kB = 3.3 × 10−27 kcal K−1, and T = 298K. The relative energies of the different complexes were calculated from the observed equilibrium stabilities using ΔG = − RT lnK. The 4.5S RNA stabilizes the early intermediate (in bracket) by > 2.5 kcal mol−1, and the overall activation energy is subsequently lowered by ~3 kcal mol−1. ΔG≠ and ΔG≠′ defines the overall activation energy for forming the GTP-stabilized complex with and without RNA, respectively. The bottom panel depicts a physical picture of how the 4.5S RNA exerts its effect on the SRP-SR interaction as described in the text.
Defects of mutant 4.5S RNAs in formation of the early intermediate correlates with defects in accelerating SRP•SR complex formation
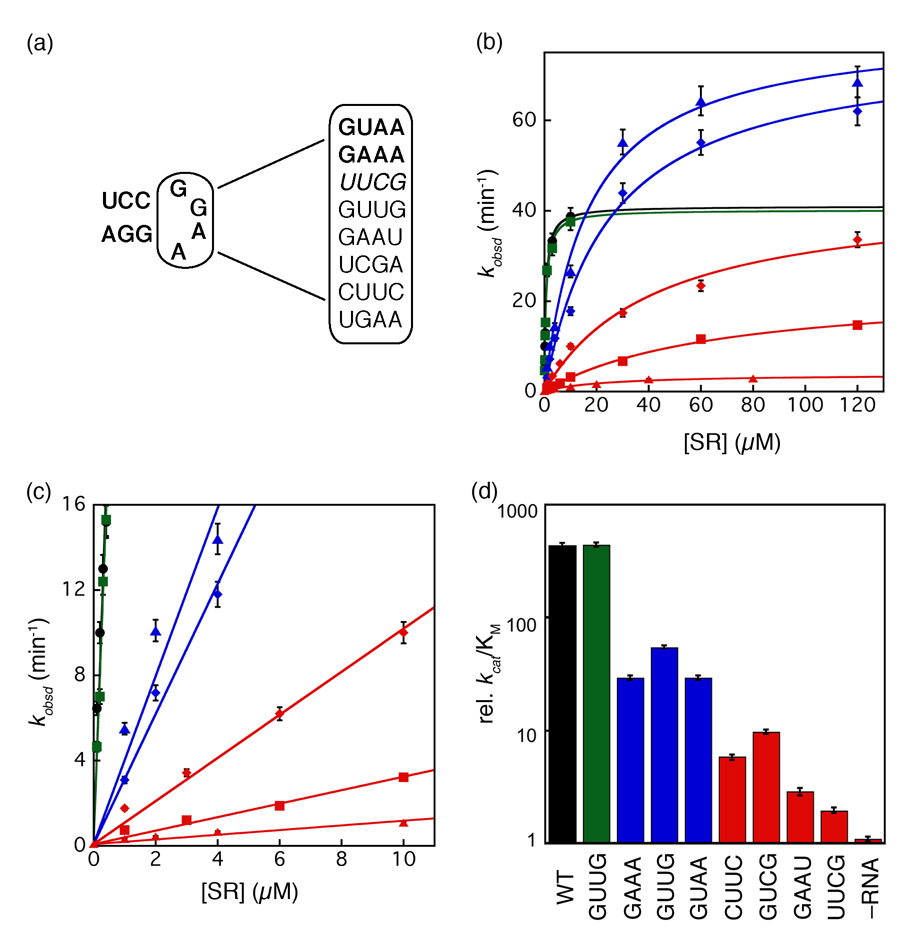
The observation that the 4.5S RNA can stabilize the early intermediate suggests that the RNA may exert its catalytic effect on SRP•SR complex formation by prolonging the lifetime of the intermediate, thereby increasing its probability to rearrange to the final stable complex. If this were true, then mutant RNAs that are defective in accelerating SRP•SR complex formation would also be predicted to be defective in stabilizing the early intermediate. To test this model, we re-examined mutations in the universally conserved GGAA tetraloop of SRP RNA (Fig. 5a) that have previously been shown to impair formation of the SRP•SR complex 27,30.
Figure 5. Tetraloop mutants in 4.5S RNA slows down the assembly rate of an active SRP•SR complex.
(a) List of tetraloop mutants studied in this work. GAAA and GUAA form GNRA type tetraloops (shown as bold); UUCG forms a UNRG type tetraloop (shown as italics); GUUG, GAAU, UCGA, CUUC and UGAA do not form a tetraloop (shown as normal). (b and c) Tetraloop mutants in the 4.5S RNA were classified into three classes based on the severity of defects in SRP-SR association (refer to the classification and color-coding in table 1). The GTPase reaction rate constants were measured and analyzed as described in Methods using 100 nM SRP and 100 µM GTP (wild-type (●), GUUG (■), UGAA (▲), GAAA (◆), GUCG (◆), UUCG (■), and no RNA (▲)). The initial linear portion of (b) are expanded in (c) to show the difference in kcat/KM of the various RNA mutants. The values of kcat/KM and kcat for each RNA are listed in Table 1. (d) Comparison of kcat/KM values for the various RNA mutants. Data were from Fig. 5c.
To this end, eight tetraloop mutants were constructed with various base substitutions: GNRA-type, UNCG-type and mutations that do not form a tetraloop (Fig. 5a). Mutant RNAs were assembled into SRPs with Ffh under the same conditions as wild-type 4.5S RNA, as previous results have shown that mutations in the RNA tetraloop does not affect its ability to bind Ffh 27,30. Although the effects of these mutations on SRP•SR complex have been characterized before, the earlier study described these effects as a deficiency in forming a stable SRP•SR complex 30. However, kinetic analyses subsequently showed that a stable Ffh•SR complex can be formed without the SRP RNA; the role of RNA is to accelerate the kinetics of complex formation 17. Therefore, we re-characterized these RNA tetraloop mutants to test whether the defects arise from altered kinetics or stability of complex formation.
We first analyzed SRP•SR complex formation using the well-characterized GTPase assay; stimulation of the GTPase activity in the SRP•SR complex provides a convenient assay for protein-protein interactions. In this assay, the rate constants of two molecular events can be measured. First, at low concentrations of SR, the reaction is rate-limited by SRP-SR association to form an activated SRP•SR complex. Therefore the slope of the initial linear portion of the concentration dependence, which represents the reaction: SRP + SR → products (kcat/KM), is equal to the rate constant for formation of a stable, active complex. Secondly, at saturating protein concentrations, the reaction is rate-limited by a different step, the activated GTP hydrolysis after a stable SRP•SR is formed. Therefore, the rate constant at saturating FtsY concentrations, kcat, represents the rate constant of GTP hydrolysis from the activated SRP•SR complex. Most of the tetraloop mutants show defects in the rate of complex formation (kcat/KM, Fig. 5b–d and Table 1). Moderate mutants GAAA, UGAA, and GUAA exhibit 8 – 15-fold defects (blue) and severe mutants CUUC, GUCG, GAAU, and UUCG exhibit 45 – 224-fold defects (red). GUUG is the only neutral mutant that exhibits no functional defect in this assay (green). In contrast, most of the mutant RNAs do not significantly impair the activated GTPase reaction in the SRP•SR complex (kcat, Fig. 5b and Table 1), with some mutants exhibiting even higher GTPase activity than wild-type SRP. Only the most severe mutants GAAU and UUCG showed a modest reduction (1.8- and 1.2-fold, respectively) in the stimulated GTPase activity. These data showed that the primary defect of the RNA tetraloop mutants is the slower kinetics to form the SRP•FtsY complex.
Table 1.
Summary of mutational effects of tetraloop mutants in the 4.5S RNA. Three classes of mutants are classified based on the severity of the defect as defined in the text.
| Tetraloop Mutant | kcat/KM, rela | Translocation Efficiency | kcat (min−1) | FRETb |
|---|---|---|---|---|
| Wild Type | 439 | 55% | 40.9 | 0.40 |
| GUUG | 439 | 46% | 38.7 | 0.34 |
| GAAA | 29.2 | 37% | 76.4 | 0.12 |
| UGAA | 54.9 | 38% | 81.9 | 0.15 |
| GUAA | 29.2 | 38% | 80.6 | 0.11 |
| CUUC | 5.8 | 12% | 35.8 | 0.06 |
| GUCG | 9.8 | 14% | 44.3 | 0.07 |
| GAAU | 2.9 | 11% | 23.0 | 0.08 |
| UUCG | 1.9 | 8% | 33.8 | 0.05 |
| No RNA | 1 | 8% | 3.8 | 0.05 |
Relative value of kcat/KM compared to that of the no RNA reaction.
FRET values were determined in the presence of 10 µM SR.
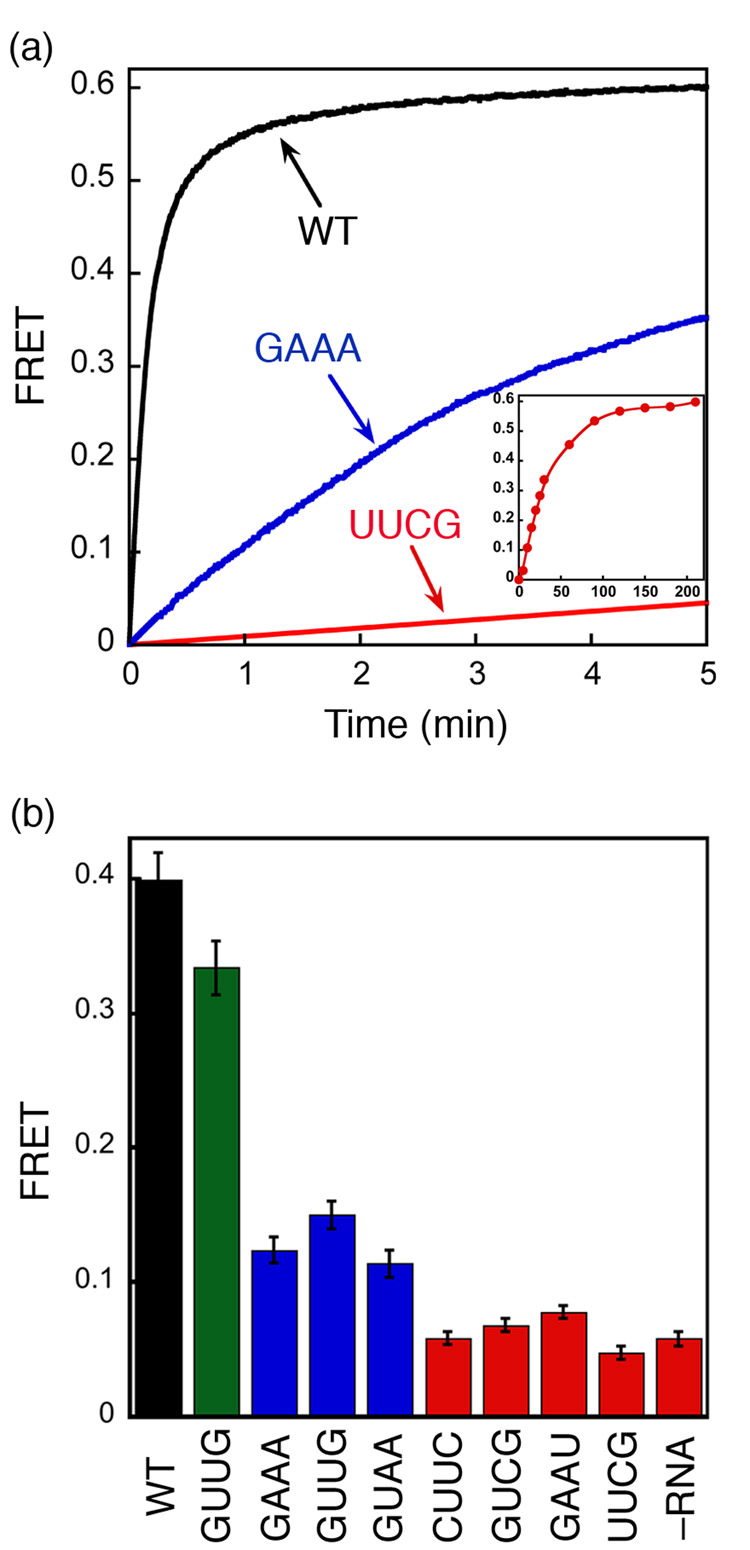
We also used the FRET assay to independently determine the effect of mutant RNAs on formation of the GTP-dependent SRP•SR complex. Consistent with the results from the GTPase assay, mutant SRPs form GTP-dependent complexes with SR much more slowly than wild-type SRP (Fig. 6a). In addition, the FRET assay directly demonstrates that SRP•SR complexes can be formed with the mutant RNAs, given that sufficient time is provided to allow complex formation.
Figure 6. FRET measurement shows the deficiency in SRP-SR complex formation caused by 4.5S RNA tetraloop mutants.
(a) Time course for formation of the GTP-dependent complex in the presence of different RNA mutants. The inset shows the data over a longer time course with the UUCG mutant (time scale in minutes). 0.5 µM SRP (2 µM SRP for UUCG mutant) and 2 µM SR (10 µM SR for UUCG) were used in the experiment in the presence of 100 µM GppNHp. (b) FRET measurement of the extent of formation of the GTP-independent complex with various 4.5S RNA mutants. 4 µM SRP and 16 µM SR were incubated without GppNHp.
We then tested whether the mutant RNAs can allow formation of the GTP-independent early intermediate using the FRET assay (Fig. 6b). The severe mutants GAAU, CUUC, GUCG and UUCG, which cause the most deleterious defect on the assembly rate of the GTP-dependent SRP•SR complex, also severely block the formation of the GTP-independent early intermediate, with the observed FRET efficiency similar to that in the absence of 4.5S RNA (Fig. 6b, red). Slightly higher FRET efficiencies are observed with moderate mutants UGAA, GUAA, and GAAA (blue), indicating partial formation of the GTP-independent early intermediate at the concentration used in this experiment. In contrast, the neutral mutant GUUG (green) formed the GTP-independent complex as efficiently as the wild type SRP. Due to the very weak affinity of the GTP-independent complex formed by the mutant RNAs (>50 µM), saturation could not be reached in equilibrium titration experiments to measure the stabilities of these complexes (not shown). Nevertheless, the results in Figure 6b show that the GTP-independent complex is substantially destabilized by mutations in the tetraloop of the 4.5S RNA. Further, there is a strong correlation between the defects of RNA mutants in stabilizing the GTP-independent early intermediate and their defects in accelerating the assembly rate of the GTP-stabilized, final SRP•SR complex (cf Fig. 6b vs Fig. 5d).
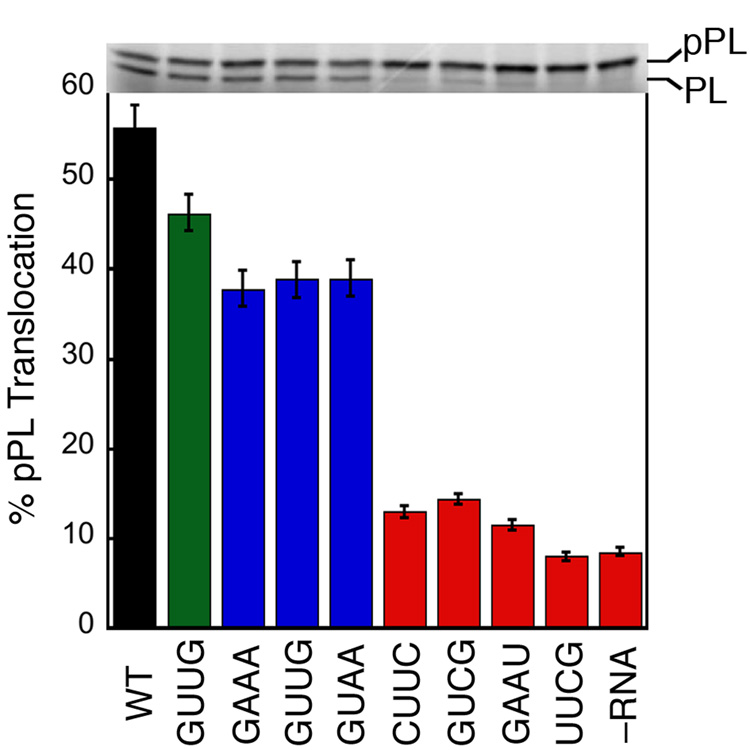
If stabilization of the early intermediate and efficient SRP•SR complex formation are essential for protein targeting, then the mutant RNAs would be predicted to also impair the protein targeting reaction. To test this notion, we measured the efficiency of protein targeting mediated by the mutant RNAs using a heterologous, co-translational protein targeting assay based on the model SRP substrate preprolactin (pPL) 6,40. As shown in Figure 7, most of the mutant RNAs also exhibit translocation defects. The severe mutants (red), which impair complex formation by over 50-fold, completely block pPL translocation. The moderate mutants (blue), which reduce the SRP-SR interaction kinetics by about 15-fold, caused a more modest (~20%) reduction in translocation efficiency. The small translocation defect caused by the moderate mutants is presumably due to the limited sensitivity of this targeting assay, as it can detect translocation defect only when the SRP-SR interaction is reduced by more than 20-fold 6. In contrast, the neutral mutant GUUG does not significantly affect protein translocation. Thus there is also a good correlation between the translocation defect and the degree to which complex formation is blocked by each mutant RNA (cf. Fig 5d, Fig. 6b, and Fig. 7; see also Table 1).
Figure 7. Tetraloop mutants impair the co-translational translocation of pre-prolactin.
The translocation efficiencies were determined and analyzed as described in Methods. Top panel shows the SDS-PAGE analysis of the translocation of 35S-labeled prolactin. pPL and PL indicate the precursor and mature form of prolactin.
DISCUSSION
In this study, we developed a highly sensitive, real time FRET assay that allows us to detect a transient, GTP-independent early intermediate during assembly of a stable SRP•SR complex. This demonstrates that SRP•SR complex assembly is a complex multi-step process. Intriguingly, this early intermediate is substantially stabilized by the 4.5S SRP RNA, and there is a strong correlation between the abilities of mutant RNAs to stabilize this early intermediate and their abilities to accelerate the assembly of the stable SRP•SR complex. This led us to propose a new model in which the SRP RNA exerts its catalytic effect on SRP•SR complex assembly through stabilizing a transient intermediate, thereby allowing it more dwell time to rearrange into the GTP-stabilized final complex. The presence of this additional conformational step provides another potential point for regulation in the protein targeting reaction.
Previous studies have established that GTP or non-hydrolyzable GTP analogues are required for formation of a stable SRP•SR complex, but no complexes have been observed in the absence of GTP 17,25,41–43. In this study, FRET provides a highly sensitive assay that allows us to observe an unstable SRP•SR complex in solution that can be formed independently of GTP. Only Mandon et al. have reported a mammalian SRP•SR complex formed in GDP in surface-resonance measurements 38. This complex was not observed in solution previously, presumably because previous studies have relied on gel filtration analysis 16 or the use of tryptophan fluorescence 17,25,44. Gel filtration chromatography can only observe kinetically stable complexes but will not be able to detect a more transient complex. Tryptophan fluorescence relies on a late conformational change in FtsY that accompanies complex formation 17, but could miss earlier steps. In contrast, the FRET assay is able to detect transient complexes, because FRET signal relies only on the distance approximation and relative orientation of the donor and acceptor fluorophores on the two proteins. We also showed that the FRET value is different for the GTP-independent complex from the stable, GTP-dependent complex; thus these two complexes have different conformations. Finally, this assay allows us, for the first time, to quantitatively evaluate the contribution of the γ-phosphate group to complex stability. The presence of the γ-phosphate of GTP stabilizes the SRP•SR complex over 250-fold; the actual interaction energy of the proteins with the γ-phosphate group is presumably larger, as a significant amount of the interaction energy has to be used to induce conformational changes in the complex5,16.
For the SRP-subfamily of GTPases, the structural difference between the GppNHp-, GDP- and apo-proteins is rather minimal 21,23,45–47. It is therefore reasonable to suspect that the conformation of the GTP-independent complex can also be adopted by GTP-bound SRP and SR. Here we provide several lines of evidence that strongly suggest that the GTP-independent complex represents an intermediate on the pathway to formation of the final, stable complex by GTP-bound SRP and SR. First, the time course for complex formation in the presence of GppNHp exhibits bi-phasic kinetics indicative of a two-step process, and the first kinetic phase agrees well with the kinetics for formation of GTP-independent complex. Second, the 4.5S RNA is shown to thermodynamically stabilize the GTP-independent complex and also accelerate formation of a GTP-stabilized complex. This observation is consistent with the classical criterion for an on-pathway intermediate: stabilization of an on-pathway intermediate should accelerate the reaction to form the final product. In contrast, if the GTP-independent complex were off-pathway, then stabilizing this complex would be expected to inhibit formation of the GTP-dependent complex. Together, these observations provide strong evidence that the GTP-independent complex is an early intermediate that precedes a GTP-dependent rearrangement to form the final, GTP-dependent complex. The omission of GTP provides a convenient means to isolate this intermediate by preventing the subsequent conformational rearrangements, thereby characterizing its kinetic, thermodynamic, and structural properties and its roles in the protein targeting reaction.
Previously, mutational analysis of the SRP•SR complex have isolated multiple classes of mutant GTPases that each blocks a different stage during the SRP-SR interaction: class I mutants are defective in complex formation; class II mutants primarily block reciprocal GTPase activation; class III mutants impair both steps; and class IV mutants specifically affect activation of one GTPase in the complex 5. The results with these mutants suggest that during the SRP-SR interaction, complex formation and activation of GTP hydrolysis in the individual GTPases are discrete and separable steps. Our results here further showed that assembly of a stable complex is also a multi-step process that involves an additional GTP-independent early intermediate. Together, these results demonstrate the dynamic nature of the SRP-SR interaction. The fact that this early intermediate is much less stable than the previously characterized complexes, and that the class I mutant SR (G455W), which blocks formation of a stable complex, does not affect the formation of the early intermediate (Supplemental Fig. 3), indicates that the early intermediate precedes formation of the closed complex.
The model in Figure 8a describes the multiple steps during the SRP-SR binding and activation cycle. The free SRP and SR, predominantly in an inactive, open conformation, quickly associate with one another to form a transient, GTP-independent early intermediate (Fig. 8a, step 1). Interactions of both proteins with the GTP γ-phosphate allow this complex to rearrange into a stable closed complex (step 2). Activation of GTP hydrolysis in the complex requires an additional local rearrangement of the conserved insertion box domain (IBD) loops from both SRP and SR that precisely aligns the catalytic residues in the loop with respect to both GTP molecules (step 3). GTP hydrolysis then generates a GDP-complex (step 4), which quickly disassembles due to its low kinetic stability (step 5).
Notably, the early intermediate formed in the first step is significantly stabilized by the 4.5S SRP RNA. Unlike the purely kinetic effect of this RNA on formation of the stable SRP•SR complex (i.e., both complex formation and disassembly is accelerated by the same 200 – 400-fold without affecting the equilibrium stability of the complex)17,25, the RNA thermodynamically stabilizes the early intermediate. Further, mutations in the conserved tetraloop of the 4.5S RNA are defective in stabilizing the early intermediate, and this defect strongly correlates with the defect of these RNA mutants in accelerating formation of the final, stable SRP•SR complex. Judging from the FRET efficiency of the GTP-independent intermediate in the absence of the RNA, we estimate that the RNA exerts a >60-fold stabilizing effect on this intermediate; this effect accounts for a large part of the ~200-fold acceleration of SRP-SR complex assembly by the RNA.
These data allow us to propose a new model for how the 4.5S RNA catalyzes both the association and dissociation between SRP and SR (Fig. 8b). We propose that the early intermediate, although forms quickly, does not have sufficient contacts between the two proteins and thus disassembles just as quickly. The 4.5S SRP RNA, by stabilizing the early intermediate, could provide this intermediate a longer lifetime during which each protein searches the conformational space and attempts to rearrange to the correct conformation for interacting with each other (Fig. 8b lower panel). The subsequent rearrangement of the early intermediate to the closed complex is the rate-limiting step for formation of a stable SRP•SR complex (Fig. 8b, ΔG‡ and ΔG‡′ represents the free energy barrier for formation of the final complex with and without the RNA, respectively). Even if the RNA do not provide additional transition state stabilization for the early → closed rearrangement and the same barrier remains for this rearrangement with or without the RNA, the overall energy barrier for formation of the stable complex is reduced, thus leading to an accelerated assembly rate (Fig. 8b). This model explains how the SRP RNA accelerates assembly of the Ffh•SR complex without affecting its equilibrium stability 17. Several previous models have been proposed to account for the catalytic effect of the RNA by suggesting that the RNA pre-organizes the conformation of Ffh to allow a better interaction with SR; however, such models predict that the stability of the Ffh•SR complex would also be increased by the SRP RNA and are not consistent with experimental data.
Although we provide here an energetic model to explain the catalytic role of the 4.5S RNA, the structural origin of this effect remains to be determined. Most likely, the SRP RNA provides a transient tether that holds the two GTPases together upon their initial encounter (Fig. 8B). This tether is broken after rearrangement to the final stable SRP•SR complex since the RNA does not stabilize this stable complex 17, and as such, it has been difficult to identify these transient interactions that the RNA makes with the GTPase domains. Since the thermodynamic stability of the early intermediate directly affects the overall energy barrier of the assembly reaction, instead of characterizing the transition state, we can conveniently characterize the structural and energetic properties of the early intermediate to identify molecular interactions made by the 4.5S RNA to exert its catalytic role.
The presence of the early intermediate and an additional conformational rearrangement required to form the closed complex provides an additional potential point for regulation in the protein targeting reaction. In solution, the initial collisional encounter of the SRP and SR leads to a transient and unstable early intermediate that would not accumulate under cellular conditions. In the presence of spatial and temporal cues such as cargo binding and membrane localization, it is possible that the kinetic and thermodynamic stability of this early intermediate and its subsequent rearrangement can be modulated to coordinate the proper delivery of cargo during the protein targeting reaction (X.Z. et al., manuscript in preparation).
MATERIALS AND METHODS
Material
Eschericia coli Ffh, FtsY and 4.5S RNA were expressed and purified using established procedures 25. Mutant proteins and RNAs were constructed using QuickChange procedure (Stratagene, La Jolla, CA), and were expressed and purified by the same procedure as that for wild-type proteins and RNAs. Fluorescent dyes DACM and BODIPY-FL were purchased from Invitrogen (Carlsbad, CA).
Fluorescence labeling
Single-cysteine mutants of Ffh and FtsY were labeled with maleimide derivatives of coumarin and BODIPY-FL, respectively. Protein was dialyzed in labeling buffer [50 mM KHEPES (7.0), 300 mM NaCl, 2 mM EDTA)] and treated with 2 mM TCEP to reduce the disulfide bonds. The labeling reaction was carried out using a five-fold excess of dye over protein for over 2 hours at 4 °C, and stopped by adding 2 mM DTT. Excess dye was removed by gel filtration using Sephadex G-25 (Sigma). Absorbance of DACM (ε363 = 27,000 M−1 cm−1) and BODIPY-FL (ε504 = 79,000 M−1 cm−1) was used to determine the concentration of labeled protein. The efficiency of labeling reaction was evaluated using:
| (1) |
The efficiency of labeling reaction was typically ≥95% for both probes. The background, estimated from the labeling of cysteinless Ffh and FtsY using the same procedure, are less than 3%.
Fluorescence measurement
FRET was determined by steady-state fluorescence measurement on a Fluorolog-3 spectrofluorometer (Jobin Yvon, Edison, NJ). All measurements were carried out at 25 °C in assay buffer [50 mM KHEPES, pH 7.5, 150 mM KOAC, 2 mM Mg(OAc)2, 2 mM DTT, 0.01% Nikkol] using an excitation wavelength of 380 nm. Fluorescence emission spectra were acquired from 420 to 600 nm. Equilibrium titration or kinetic measurements using FRET were determined by monitoring the fluorescence emission at 470 nm. FRET efficiency (E) is calculated by the relative fluorescence intensities of the donor in the presence and absence of acceptor (equation 2),
| (2) |
where FDA and FD are the fluorescence intensities of the donor measured in the presence and in the absence of acceptor, respectively. FD was measured using donor-labeled Ffh and unlabeled FtsY. The Förster distance for the donor-acceptor pair coupled to the different positions was experimentally determined to be R0 ~ 47 Å 48. Fast reactions were measured on a Kintek stop-flow apparatus at 25 °C. The association rate constant for the SRP•SR complex was measured by mixing 2 µM SRP with 4, 8, 15, 25 µM SR in the presence or absence of GppNHp. The observed rate constant (kobsd) is linearly dependent on SR concentration (eq 3) and the slope of the concentration dependence gives the association rate constant, kon 17 .
| (3) |
The dissociation rate constant for the GTP-independent complex (koff) was determined by a pulse-chase experiment 25. 2 µM SRP and 8 µM SR were incubated in the absence of GppNHP for 5 minutes to form the SRP•SR complex, then the solution was mixed with equal volume of 400 µM unlabeled SR to drive irreversible dissociation of the complex. The time course of change in donor fluorescence was fit to exponential function (eq 4), where Fobsd is the observed fluorescence, Ft→∞ is the fluorescence when reaction reaches equilibrium, and ΔF is the amount of fluorescence change during the experiment.
| (4) |
Translocation assay
Mutant 4.5S RNAs were used to reconstitute SRP with Ffh, and protein targeting efficiency of the mutant SRPs were measured using a heterologous co-translational translocation assay as described6,40.
GTPase assay
The GTPase assay to measure the stimulated GTP hydrolysis reaction between SRP and FtsY were carried out and analyzed as described 25.
Supplementary Material
Acknowledgment
We thank members of the Shan laboratory for comments on the manuscript. This work was supported by NIH grant GM078024 to S.S. S.S. was supported by the Burroughs Wellcome Fund career award, the Henry and Camille Dreyfus foundation, the Beckman Young Investigator award, and the Packard and Lucile award in science and engineering. X.Z. was supported by a fellowship from the Ulric B. and Evelyn L. Bray Endowment Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic-reticulum membrane. Annual Review of Cell Biology. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 2.Matlack KES, Mothes W, Rapoport TA. Protein translocation: Tunnel vision. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- 3.Johnson AE, van Waes MA. The translocon: A dynamic gateway at the ER membrane. Annual Review of Cell and Developmental Biology. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 4.Shan SO, Walter P. Co-translational protein targeting by the signal recognition particle. FEBS Letters. 2005;579:921–926. doi: 10.1016/j.febslet.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 5.Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. Plos Biology. 2004;2:1572–1581. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its initiation of protein translocation. Journal of Cell Biology. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic-reticulum .1. Signal recognition protein (SRP) binds to in vitro-assembled polysomes synthesizing secretory srotein. Journal of Cell Biology. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter P, Blobel G. Translocation of proteins across the endoplasmic-reticulum .2. Signal recognition protein (SRP) mediates the selective binding to microsomal-membranes of in vitro-assembled polysomes synthesizing secretory protein. Journal of Cell Biology. 1981;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter P, Blobel G. Translocation of proteins across the endoplasmic-reticulum .3. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal-membranes. Journal of Cell Biology. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 11.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 12.Krieg UC, Walter P, Johnson AE. Photo-cross-linking of the signal sequence of nascent preprolactin to the 54-Kilodalton polypeptide of the signal recognition particle. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA. The signal sequence of nascent preprolactin interacts with the 54Kd polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 14.Zopf D, Bernstein HD, Johnson AE, Walter P. The methionine-rich domain of the 54 Kd protein subunit of the signal recognition particle contains an RNA-binding site and can be cross-linked to a signal sequence. EMBO Journal. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connolly T, Rapiejko PJ, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 16.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 17.Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- 18.Powers T, Walter P. Reciprocal stimulation of GTP hydrolysis by 2 directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 19.Cleverley RM, Gierasch LM. Mapping the signal sequence-binding site on SRP reveals a significant role for the NG domain. J Biological Chemistry. 2002;277:46763–46768. doi: 10.1074/jbc.M207427200. [DOI] [PubMed] [Google Scholar]
- 20.Bourne HR, Sanders DA, Mccormick F. The GTPase superfamily - conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 21.Gawronski-Salerno J, Coon JSV, Focia PJ, Freymann DM. X-ray structure of the T-aquaticus FtsY: GDP complex suggests functional roles for the C-terminal helix of the SRP GTPases. Proteins-Structure Function and Bioinformatics. 2007;66:984–995. doi: 10.1002/prot.21200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Focia PJ, Gawronski-Salerno J, Coon JS, Freymann DM. Structure of a GDP:AlF4 complex of the SRP GTPases Ffh and FtsY, and identification of peripheral nucleotide interaction site. Journal of Molecular Biology. 2006;360:631–643. doi: 10.1016/j.jmb.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freymann DM, Keenan RJ, Stroud RM, Walter P. Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+ GDP. Nature Structural Biology. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- 24.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 25.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 26.Gu SQ, Jockel J, Beinker P, Warnecke J, Semenkov YP, Rodnina MV, Wintermeyer W. Conformation of 4.5S RNA in the signal recognition particle and on the 30S ribosomal subunit. RNA. 2005;11:1374–1384. doi: 10.1261/rna.7219805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siu FY, Spanggord RJ, Doudna JA. SRP RNA provides the physiologically essential GTPase activation function in co-translational protein targeting. RNA. 2007;13:240–250. doi: 10.1261/rna.135407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sagar MB, Lucast L, Doudna JA. Conserved but nonessential interaction of SRP RNA with translation factor EF-G. RNA. 2004;10:772–778. doi: 10.1261/rna.5266504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown S, Fournier MJ. The 4.5S RNA gene of Escherichia-coli is essential for cell-growth. Journal of Molecular Biology. 1984;178:533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- 30.Jagath JR, Matassova NB, De Leeuw E, Warnecke JM, Lentzen G, Rodnina MV, Luirink J, Wintermeyer W. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA. 2001;7:293–301. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poritz MA, Strub K, Walter P. Human SRP RNA and Escherichiacoli 4.5s RNA contain a highly homologous structural domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- 32.Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An Escherichia-coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 33.Batey RT, Sagar MB, Doudna JA. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. Journal of Molecular Biology. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- 34.Zheng N, Gierasch LM. Domain interactions in E. coli SRP: stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Molecular Cell. 1997;1:79–87. doi: 10.1016/s1097-2765(00)80009-x. [DOI] [PubMed] [Google Scholar]
- 35.Hainzl T, Huang S, Sauer-Eriksson AE. Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14911–14916. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stroud RM, Walter P. Signal sequence recognition and protein targeting. Current Opinion in Structural Biology. 1999;9:754–759. doi: 10.1016/s0959-440x(99)00040-8. [DOI] [PubMed] [Google Scholar]
- 37.Spanggord RJ, Siu F, Ke AL, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nature Structural & Molecular Biology. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 38.Mandon EC, Jiang Y, Gilmore R. Dual recognition of the ribosome and the signal recognition particle by the SRP receptor during protein targeting to the endoplasmic reticulum. Journal of Cell Biology. 2003;162:575–585. doi: 10.1083/jcb.200303143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. Journal of Molecular Biology. 2005;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO Journal. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connolly T, Gilmore R. The signal recognition particle receptor mediates the GTP-dependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell. 1989;57:599–610. doi: 10.1016/0092-8674(89)90129-3. [DOI] [PubMed] [Google Scholar]
- 42.Rapiejko PJ, Gilmore R. Protein translocation across the ER requires a functional GTP binding-site in the alpha-subunit of the signal recognition particle receptor. Journal of Cell Biology. 1992;117:493–503. doi: 10.1083/jcb.117.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zopf D, Bernstein HD, Walter P. GTPase domain of the 54-Kd subunit of the mammalian signal recognition particle is required for protein translocation but not for signal sequence binding. Journal of Cell Biology. 1993;120:1113–1121. doi: 10.1083/jcb.120.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jagath JR, Rodnina MV, Wintermeyer W. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. Journal of Molecular Biology. 2000;295:745–753. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- 45.Padmanabhan S, Freymann DM. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase. Structure. 2001;9:859–867. doi: 10.1016/s0969-2126(01)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez UD, Minasov G, Focia PJ, Stroud RM, Walter P, Kuhn P, Freymann DM. Structural basis for mobility in the 1.1 angstrom crystal structure of the NG domain of Thermus aquaticus Ffh. Journal of Molecular Biology. 2002;320:783–799. doi: 10.1016/s0022-2836(02)00476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Focia PJ, Alam H, Lu T, Ramirez UD, Freymann DM. Novel protein and Mg2+ configurations in the Mg2+ GDP complex of the SRP GTPase Ffh. Proteins-Structure Function and Genetics. 2004;54:222–230. doi: 10.1002/prot.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lakowicz JR. Principles of fluorescence spectroscopy. London: Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.