Summary
The perception of pathogen or microbe-associated molecular pattern molecules by plants triggers a basal defense response analogous to animal innate immunity, and is defined in part by the deposition of the glucan polymer callose at the cell wall at the site of pathogen contact. Transcriptional and metabolic profiling in Arabidopsis mutants, coupled with the monitoring of pathogen triggered callose deposition, have identified major roles in pathogen response for the plant hormone ethylene and the secondary metabolite 4-methoxy-indol-3-ylmethylglucosinolate. Two genes, PEN2 and PEN3, are also necessary for resistance to pathogens and are required for both callose deposition and glucosinolate activation, suggesting that the pathogen triggered callose response is required for resistance to microbial pathogens. Our study shows that well-studied plant metabolites, previously identified as important in avoiding damage by herbivores, are also required as a component of the plant defense response against microbial pathogens.
Although plants are in continual contact with potential pathogens, a successful infection is rare. The ability of a particular plant species to prevent the successful colonization by a given pathogen species is referred to as non-host resistance (1–3). The molecular basis of non-host resistance is poorly understood, but presumably relies on both constitutive barriers and inducible responses. These layers are comprised of a constitutive defense layer and multiple pattern recognition receptors that respond to highly conserved microbe-associated molecular pattern (MAMP) molecules such as bacterial flagellin or peptidoglycan (4, 5). MAMP recognition triggers the activation of serine/threonine-specific protein kinases (MAPKs) and various hormone signaling pathways (6). This signaling starts a cascade that activates a variety of defense responses including callose deposition, programmed cell death, production and accumulation of antimicrobial reactive oxygen species, and induction of phytoalexins and other secondary metabolites such as the indolic antimicrobial compound camalexin (3-thiazol-2′yl-indole) and glucosinolates (1-thio-beta-D-glucosides)(3). Interestingly, plants also constitutively synthesize and store glucosinolates, which are converted by endogenous S-glycosyl hydrolases (myrosinases) into compounds that function as insect feeding and/or oviposition stimulants or deterrents (7). However, glucosinolates and their breakdown products have also been identified as potential antimicrobials (8, 9)
Despite detailed characterization of MAMP recognition and various hormone-mediated signaling pathways in plant defense response, relatively little is known about the host mechanisms that connect the perception of a particular pathogen to the downstream signaling pathways that lead to activation of specific immune responses.
Flg22-induced callose response requires FLS2, PMR4, Et signaling, and MYB51
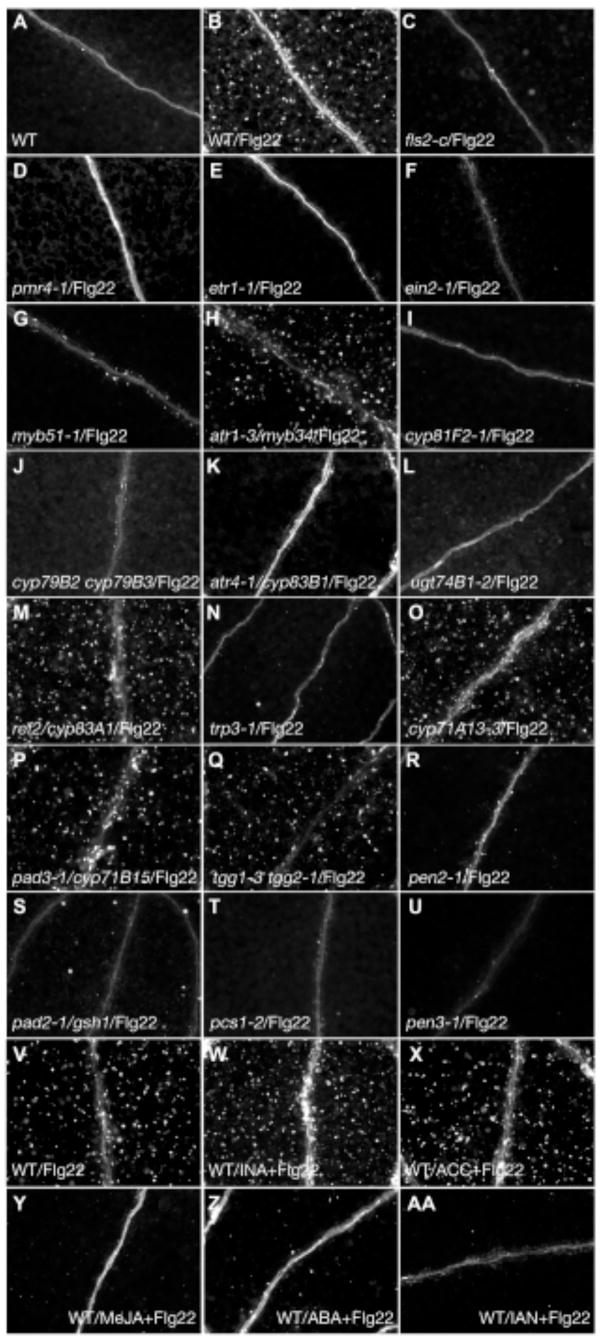
Arabidopsis genes induced in response to treatment with Flg22, a synthetic 22-amino acid polypeptide that corresponds to a highly conserved region of eubacterial flagellin, include those involved in ethylene (Et)-mediated defense hormone signaling and in indole glucosinolate biosynthesis (Table S1A). We performed a phenotypic assay for Flg22-induced callose responses in Arabidopsis seedlings under the same conditions as previous transcriptional profiling studies (10). Callose is a β(1,3) glucan polymer that strengthens and dams weak or compromised sections of plant cell walls at the site of pathogen attack. A callose-response assay was used to identify particular Flg22-induced genes involved in callose deposition. Aniline blue staining was used to detect callose, and we observed deposits on the cotyledons of Arabidopsis seedlings treated with ≥1 μM Flg22 (Fig. 1B) that were absent in water-treated plants (Fig. 1A). Mutants lacking the functional Flg22 receptor encoded by the FLS2 gene (11) or the functional callose synthase encoded by the PMR4 gene (12) did not respond to Flg22 treatment (Fig. 1C–D), demonstrating that the appearance of these fluorescent deposits was a consequence of MAMP perception and subsequent callose synthesis. A more sensitive assay of callose staining in the pmr4-1 mutant revealed faint fluorescent flecks (fig. S2A), suggesting that another callose synthase plays a minor role in response to Flg22. The callose response was also elicited by other MAMPs and was not specific to Flg22 (see SOM text; fig. S2B). The perception of Flg22 in roots requires both the Flg22 receptor FLS2 and its receptor complex partner, BAK1 (13, 14). In contrast, the Flg22-induced callose response in cotyledons does not require BAK1 (Table S1B), suggesting that the immune requirement for BAK1 in the FLS2 receptor complex is tissue-specific.
Fig. 1.
IGS biosynthesis and hydrolysis are required for Flg22-induced callose formation. (A–U) Cotyledons of seedlings treated with water (A) or Flg22 (B–U) and stained with aniline blue. (A–B) Wild type. (C) fls2-c. (D) pmr4-1; traditional staining method. (E) etr1-1. (F) ein2-1. (G) myb51-1. (H) atr1-3/myb34. (I) cyp81F2-1. (J) cyp79b2 cyp79b3. (K) atr4-1/cyp83b1. (L) ugt4b1-2. (M) ref2/cyp83a1. (N) trp3-1. (O) cyp71a13-3. (P) pad3-1/cyp71b15. (Q) tgg1-3 tgg2-1. (R) pen2-1. (S) pad2-1/gsh1. (T) pcs1-2. (U) pen3-1. (V-AA) Cotyledons pretreated with water (V), INA (W), ACC (X), MeJA (Y), or ABA (Z) for 24 hours and then treated with Flg22 (V–Z), or co-treated with IAN and Flg22 (AA). Shown are representative examples of 40 to 60 cotyledons from two independent experiments per genotype.
We screened a collection of Arabidopsis defense hormone-related mutants as well as mutants defective in various Flg22-inducible transcription factors (see Table S1B for all mutants screened), and found that etr1-1, etr1-3, ein2-1 and ein2-5 mutants in the Et signaling pathway, as well as two transcription factor mutants myb51-1 and myb51-2, were impaired in the Flg22-induced callose response (Figs. 1E–G; S2C–E). ETR1, an Et receptor, and EIN2, a membrane protein, are required for Et perception and signaling, respectively (15, 16). MYB51 is involved in the transcriptional activation of indole glucosinolate (IGS) biosynthetic genes (17).
MYB51 is downstream of Et signaling and upstream of IGS biosynthesis
Fig. 1A–G shows that the Et signaling pathway and the MYB51 transcription factor may function in the callose formation pathway. We examined the expression of candidate genes in Flg22-treated wild type and mutant seedlings and observed that the expression of the Et-responsive transcription factor ERF1 (which is up-regulated by Flg22; Table S1A) is significantly reduced in etr1-1 and ein2-1 mutants but not in myb51-1 and myb51-2 mutants (Table 1A). This indicates that Et signaling is intact in myb51 mutants. In contrast, Flg22-induced expression of MYB51 is reduced in etr1-1 and ein2-1 mutants (Table 1A), indicating that Et signaling is required for the full induction of MYB51 and ERF1 in response to Flg22 and that MYB51 is downstream of Et signaling for the callose response. Furthermore, Flg22-induced expression of all known IGS biosynthetic genes (CYP79B2, CYP79B3, CYP83B1/SUR2, SUR1, UGT74B1, and AtST5a; fig. S1) was significantly reduced in myb51 mutants (Table 1A), indicating that MYB51 up-regulates IGS biosynthesis in response to Flg22. A homolog of MYB51, ATR1/MYB34, also regulates IGS levels (18), but Flg22-elicited callose deposition was not affected in atr1-3/myb34 mutant seedlings (Fig. 1H), and when treated with Flg22, ATR1/MYB34 expression was down-regulated (Table S1A)(6).
Table 1.
Expression analysis of IGS biosynthetic and hydrolytic genes. (A) MYB51 is downstream of Et signaling and upstream of IGS biosynthesis. Ratio of mRNA levels in mutant relative to wild type plants after Flg22 treatment as determined by qRT-PCR. Data from three replicate samples; pair-wise comparisons to wild type plants; HAT, hours after treatment with Flg22; nd, not determined. (B) pen2 and pcs1 mutants exhibit feedback inhibition at the level of MYB51 expression. Ratio of mRNA levels in mutant and/or INA-pretreated relative to water-pretreated wild type plants after Flg22 treatment. Experimental design is as in (A); apair-wise comparisons to water-pretreated wild type plants; bpair-wise comparisons to INA-pretreated wild type plants. (C) Expression levels upon treatment with suppressors of Flg22-induced callose response relative to water-treated plants. Experimental design is as in (A); pair-wise comparisons to water pretreatment; *p-value<0.01 and false discovery rate<0.05, 2-tailed t-test; HAP, hours after pretreatment with ABA, IAN, or MeJA.
| A | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN2 | PCS1 | PEN3 | CYP81F2 | ERF1 | MYB51 | ASA1 | CYP79B2 | CYP79B3 | CYP83B1 | SUR1 | UGT74B1 | AtST5a | ||
| etr1-1 | 3 | 0.73 | 1.01 | 0.67 | 0.56 | 0.07* | 0.25* | 0.08* | 0.06* | 0.77 | 0.14* | nd | nd | nd |
| 6 | 0.97 | 1.10 | 0.96 | 0.31* | 0.10* | 0.17* | 0.10* | 0.04* | 0.88 | 0.19* | nd | nd | nd | |
| ein2-1 | 3 | 1.04 | 1.07 | 0.91 | 0.80 | 0.11* | 0.31* | 0.20* | 0.13* | 1.12 | 0.54 | nd | nd | nd |
| 6 | 0.85 | 0.80 | 1.29 | 0.36* | 0.05* | 0.13* | 0.13* | 0.09* | 0.75 | 0.40* | nd | nd | nd | |
| myb51-1 | 3 | 1.24 | 1.16 | 1.39 | 1.41 | 0.99 | 0.00* | 0.25* | 0.26* | 0.07* | 0.09* | 0.37* | 0.18* | 0.12* |
| 6 | 0.86 | 1.31 | 0.81 | 0.62 | 0.83 | 0.06* | 0.24* | 0.18* | 0.06* | 0.18* | 0.36* | 0.27* | 0.19* | |
| myb51-2 | 3 | 1.24 | 1.18 | 1.61 | 1.62 | 0.86 | 0.13* | 0.32 | 0.29* | 0.12* | 0.13* | 0.43* | 0.22* | 0.19* |
| 6 | 1.09 | 0.83 | 0.91 | 0.74 | 0.86 | 0.00* | 0.17* | 0.07* | 0.05* | 0.08* | 0.28* | 0.14* | 0.12* | |
| B
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype/ |
PAMP-inducible Gene |
|||||||||||||
| Chemical | H | PEN2 | PCS1 | PEN3 | CYP81F2 | ERF1 | MYB51 | ASA1 | CYP79B2 | CYP79B3 | CYP83B1 | SUR1 | UGT74B1 | AtST5a |
| A | ||||||||||||||
| T | ||||||||||||||
| pen2-1 | 3 | 0.17*a | 0.77 | 0.89 | 1.16 | 1.06 | 0.48*a | 0.25*a | 0.30 | 0.10*a | 0.20*a | 0.39*a | 0.21*a | 0.17*a |
| 6 | 0.13*a | 1.10 | 0.60 | 0.52*a | 0.42 | 0.40*a | 0.19*a | 0.15*a | 0.34*a | 0.28*a | 0.37*a | 0.19*a | 0.24*a | |
| pen2-1/INA | 3 | 0.19*b | 0.55 | 0.98 | 0.96 | 1.05 | 0.52*b | 0.51*b | 0.46*b | 0.09*b | 0.31*b | 0.60*b | 0.29*b | 0.28*b |
| 6 | 0.22*b | 1.45 | 0.54* | 0.74*b | 0.51*b | 0.59*b | 0.40*b | 0.43*b | 0.38*b | 0.35*b | 0.66 | 0.33*b | 0.41*b | |
| pcs1-2 | 3 | 1.12 | 0.08*a | 0.89 | 1.49*a | 0.81 | 0.67*a | 0.32*a | 0.19*a | 0.29*a | 0.32*a | 0.48*a | 0.30*a | 0.24*a |
| 6 | 1.00 | 0.22*a | 0.94 | 1.21 | 0.63 | 0.80 | 0.49*a | 0.41*a | 0.17*a | 0.50 | 0.64*a | 0.54*a | 0.51*a | |
| pcs1-2/INA | 3 | 0.89 | 0.12*b | 0.98 | 1.97 | 1.26 | 0.77*b | 0.76*b | 1.04*b | 0.27*b | 0.46*b | 0.79*b | 0.43*b | 0.46*b |
| 6 | 0.86 | 0.25*b | 0.91 | 1.08 | 1.10 | 0.88 | 0.79 | 0.49*b | 0.23*b | 0.67 | 0.89 | 0.62 | 0.56*b | |
| WT/INA | 3 | 0.78 | 0.86 | 0.98 | 1.57*a | 0.90 | 1.85*a | 2.69*a | 6.12*a | 1.29 | 1.80*a | 2.04*a | 2.05*a | 1.81*a |
| 6 | 0.72 | 1.31 | 0.83 | 0.99 | 0.88 | 1.17 | 1.33 | 1.64 | 1.69 | 1.14 | 1.14 | 1.00 | 0.97 | |
|
| ||||||||||||||
| pen3-1 | 3 | 1.19 | 1.04 | 1.09 | 0.82 | 1.40 | 0.40*a | 0.32*a | 0.14*a | 0.19*a | 0.30*a | 0.42*a | 0.26*a | 0.24*a |
| 6 | 1.20 | 1.46 | 1.15 | 1.10 | 1.21 | 0.85 | 0.46 | 0.53 | 0.39 | 1.30 | 0.86 | 0.97 | 0.96 | |
| C
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
PAMP-inducible Gene |
||||||||||||||
| Pretreatment | HAP | HAT | PEN2 | PCS1 | PEN3 | CYP81F2 | ERF1 | MYB51 | ASA1 | CYP79B2 | CYP83B1 | |||
| ABA | 24 | 3 | 1.33 | 0.97 | 1.42 | 1.06 | 1.81 | 0.61 | 0.65 | 0.61 | 0.83 | |||
| 24 | 6 | 1.08 | 0.81 | 0.60 | 0.35* | 0.32* | 0.33* | 0.15* | 0.22* | 0.29* | ||||
| IAN | 0 | 3 | 0.81 | 1.17 | 0.93 | 0.76 | 0.86 | 0.57* | 0.56* | 0.14* | 0.56* | |||
| 0 | 6 | 0.99 | 1.13 | 1.28 | 0.25* | 1.03 | 0.44* | 0.38* | 0.02* | 0.14* | ||||
| MeJA | 24 | 3 | 0.66 | 0.65 | 0.77 | 0.65* | 1.64 | 0.47* | 0.71 | 0.24* | 0.56 | |||
| 24 | 6 | 0.87 | 0.90 | 0.95 | 0.96 | 1.55* | 0.82 | 1.86* | 1.01 | 0.60 | ||||
Indole glucosinolates are derived from tryptophan, the biosynthesis of which is defense-regulated by Et-induced expression of ASA1. ASA1 catalyzes the first and rate-limiting step in the tryptophan biosynthetic pathway (19, 20). ASA1 gene expression was also induced by Flg22 treatment (Table S1A), and this up-regulation was dependent on MYB51 (Table 1A). Since ASA1 expression is Et-inducible, these data suggest that MYB51 functions downstream of Et signaling and that MYB51 most likely mediates at least some transcriptional responses to Et signaling.
IGS biosynthesis is required for Flg22-induced callose response
The drastically reduced expression of IGS biosynthetic genes in myb51 mutants suggested that IGS biosynthesis may be required for Flg22-induced callose deposition. Therefore, mutants defective in IGS biosynthesis or accumulation were tested for the callose response to Flg22. The IGS biosynthetic double mutant cyp79B2 cyp79B3, which completely lacks IGS (21), as well as mutants atr4-1/cyp83B1 and ugt74B1-2, were impaired in the induction of the callose response (Fig. 1J–L). Furthermore, tfl2 mutants, which have reduced IGS levels in their leaves (22, 23), were also impaired in Flg22-induced callose response (Table SB1 and fig. S2F).
In contrast to mutations that affect the biosynthesis of indole glucosinolates, the aliphatic glucosinolate biosynthetic mutant ref2/cyp83A1 (24) exhibited a wild type response to Flg22 (Fig. 1M), indicating that aliphatic glucosinolates do not play a major role in callose accumulation. The hypothesis that biosynthesis and accumulation of indole glucosinolates are specifically required for the Flg22-induced callose response is consistent with the facts that CYP83A1 expression is down-regulated in response to Flg22 (Table S1A)(6), as is the expression of MYB28 and MYB29 (Table S1A)(6), which regulate the aliphatic glucosinolate biosynthetic pathway (25–27). Finally, the tryptophan biosynthetic mutant trp3-1 (28) was also found to have a greatly reduced callose response to Flg22 (Fig. 1N), consistent with the fact that IGS biosynthesis requires tryptophan.
Flg22 also induces the expression of CYP71A13 and PAD3 (Table S1A)(29, 30), which are genes involved in the biosynthesis of the indole phytoalexin, camalexin. Like IGS, camalexin requires CYP79B2 and CYP79B3 for biosynthesis; however, the camalexin biosynthetic mutants cyp71A13-1, cyp71A13-3 and pad3-1 exhibited a wild type callose response to Flg22 (Fig. 1O–P and fig. S2G), showing that camalexin is not required for the callose response.
4-methoxy-I3G is required for the Flg22-induced callose response
Our data indicate that the synthesis of IGS is required for callose deposition in response to Flg22, and suggest that IGS functions as a signal or co-activator downstream of MAMP responses. Because of the large number of cytochrome P450s involved in IGS biosynthesis, we searched for other Flg22-inducible cytochrome P450 genes and found one with unassigned function, CYP81F2 (Table S1A). The mutants cyp81F2-1 and cyp81F2-2 showed a complete loss of callose deposition in response to Flg22 (Fig. 1I and fig. S2H), suggesting that CYP81F2 might also be involved in IGS biosynthesis. Unlike the characterized IGS biosynthetic genes CYP79B2, CYP79B3, CYP83B1/ATR4, SUR1, UGT74B1, and AtST5a (fig. S1), the induced expression of CYP81F2 after 3 hours of Flg22 treatment was independent of Et signaling and MYB51 (Table 1A). At 6 hours of Flg22 treatment, however, continued CYP81F2 expression became dependent on Et signaling but remained independent of MYB51 (Table 1A).
To further ascertain the biochemical function of CYP81F2, IGS metabolic profiling experiments were carried out on a variety of mutants (see below) to look for correlations between callose-deficient phenotypes and levels of the three known IGS species (indol-3-ylmethylglucosinolate (I3G), 4-methoxy-I3G, and 1-methoxy-I3G). For all tested genotypes, Flg22 treatment caused a significant reduction in I3G (Fig. 2A), a counterintuitive result given that Flg22 treatment activates the expression of IGS biosynthetic genes. This result suggested that Flg22 also activates the expression of myrosinase enzyme(s) (S-glycosyl hydrolases) that catalyze the hydrolysis of IGS (see below).
Fig. 2.
Blocking IGS biosynthesis or hydrolysis depletes or elevates 4-methoxy-I3G levels, respectively. Upon Flg22 elicitation, I3G levels (A) are reduced in wild type and mutant seedlings, and 4-methoxy-I3G levels (B) are reduced in ein2-1, myb51-1, and cyp81F2-1 mutants, and elevated in pen2-1, cad1-3 (a null pcs1 allele), and pen3-1 mutants. Mean ±S.D., n=4 replicate samples. Different letters above the bars denote statistically significant differences, P<0.01, 2-tailed t-test.
In wild type seedlings, Flg22 treatment produced no effect on 4-methoxy-I3G levels; however, in Flg22-treated myb51-1 and ein2-1 mutants, 4-methoxy-I3G levels were significantly reduced compared to those in wild type (Fig. 2B). These results suggest that the perception of Flg22 triggers both the biosynthesis and subsequent hydrolysis of 4-methoxy-I3G. Furthermore, 4-methoxy-I3G levels were extremely low in the cyp81F2-1 mutant in control (water) and Flg22 treatments (Fig. 2B), suggesting that CYP81F2 may produce 4-methoxy-I3G via the 4-methoxylation of I3G. No consistent effect on 1-methoxy-I3G levels was observed after Flg22 treatments (fig. S3), suggesting that it is not required for the Flg22-induced callose response. Therefore, we hypothesize that Flg22 perception activates the biosynthesis of MYB51 and CYP81F2-dependent 4-methoxy-I3G, accompanied by a Flg22-triggered hydrolysis of 4-methoxy-I3G into callose-eliciting compound(s).
PEN2 is the putative myrosinase involved in Flg22-triggered IGS breakdown
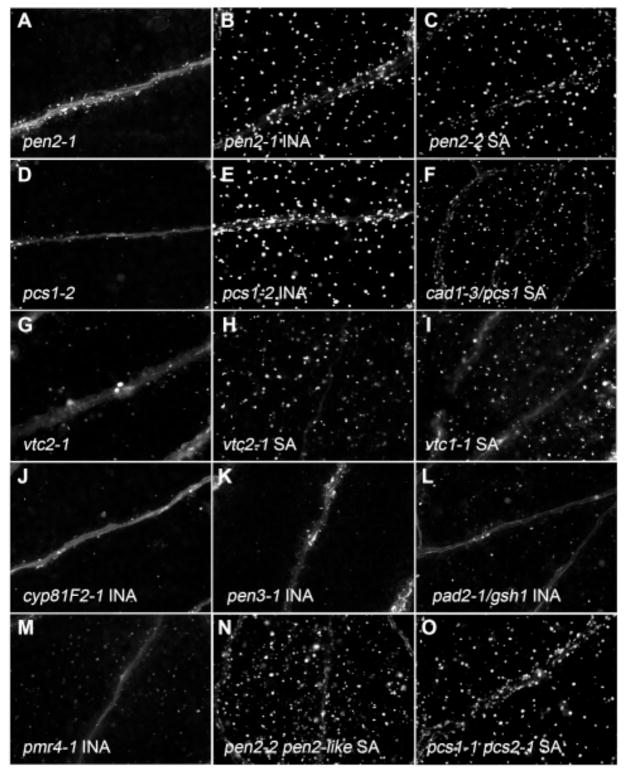
The Flg22-elicited reduction in I3G levels suggests that the Flg22-triggered IGS biosynthesis may also be accompanied by IGS hydrolysis, and that IGS hydrolytic products may be required for callose synthesis. IGS hydrolysis is brought about by myrosinase enzymes and their associated modifiers, and ascorbate is an essential cofactor for myrosinases (31). Consistent with the hypothesis that IGS hydrolysis is required for Flg22-induced callose deposition, the ascorbate-deficient mutants vtc1-1 and vtc2-1, which contain 10–30% wild type levels of ascorbic acid (32), exhibited a greatly reduced callose response to Flg22 (see Figs. 4G, S2I). However, the double mutant tgg1-3 tgg2-1, carrying lesions in the two characterized foliar myrosinases in Arabidopsis (33), exhibited a wild type callose response to Flg22 (Fig. 1Q). A search for predicted glycosyl hydrolases with a Flg22-inducible expression profile identified PEN2 (Table S1A). Corresponding mutants exhibit enhanced penetration by the non-adaptive fungal pathogen Blumeria graminis f. sp. hordei (34, 35). Both pen2-1 and pen2-2 mutants exhibited a loss of the callose response to Flg22 (Figs. 1R, S2J), indicating that PEN2 is the putative myrosinase enzyme involved in the Flg22-induced hydrolysis of IGS (see also the accompanying article by Bednarek et al. (36) for direct biochemical evidence supporting this hypothesis).
Fig. 4.
SA pretreatment rescues Flg22-induced callose formation in pen2 and pcs1 mutants. (A–O) Cotyledons of seedlings pretreated with water (A, D), INA (B, E, J–M), or SA (C, F, H–I, and N–O) for 24 hours, treated with Flg22, and stained with aniline blue. (A–B) pen2-1. (C) pen2-2. (D–E) pcs1-2. (F) cad1-3/pcs1. (G–H) vtc2-1. (I) vtc1-1. (J) cyp812-1. (K) pen3-1. (L) pad2-1/gsh1. (M) pmr4-1. (N) pen2-2 pen2-like. (O) pcs1-1 pcs2-1. Shown are representative of 40 to 60 cotyledons from two independent experiments.
In contrast to the myb51-1 and cyp81F2-1 mutants, the IGS profile of the pen2-1 mutant shows increased accumulation of 4-methoxy-I3G upon Flg22 treatment (Fig. 2B), supporting the hypothesis that PEN2 acts as a myrosinase catalyzing the hydrolysis of 4-methoxy-I3G. Furthermore, in the absence of PEN2, Flg22-treated plants demonstrate up-regulation of IGS biosynthesis and 4-methoxylation, and the substrate 4-methoxy-I3G accumulates. The correlation between reduced and/or depleted levels of 4-methoxy-I3G in the cyp81F2-1 mutant and accumulation of 4-methoxy-I3G in the pen2-1 mutant and the callose-deficient phenotypes of both the cyp81F2-1 and pen2-1 mutants suggest that a hydrolytic product of 4-methoxy-I3G functions as a signaling molecule or co-activator for callose deposition.
Interestingly, our results not only suggest that the IGS biosynthetic pathway is required for callose deposition but that the IGS biosynthetic pathway may be under feedback inhibition by elevated levels of 4-methoxy-I3G and that this feedback regulation occurs at the level of MYB51 expression, consistent with previous reports that glucosinolates regulate their biosynthesis by feedback inhibition of relevant transcription factors (37): in the pen2-1 mutant, Flg22-induced expression of MYB51 and IGS biosynthetic genes was greatly reduced after Flg22 elicitation (Table 1B). PEN2 expression is independent of Et signaling and MYB51 (Table 1B), indicating that Flg22-elicited activation of IGS breakdown is independent of IGS biosynthesis. Mutants of the modifiers of myrosinase-catalyzed reactions, esp and esm1 (38, 39), as well as the Col-0 ESP-overexpressing transgenic line (Col-0 is a natural esp mutant), exhibited a wild type callose response to Flg22 (fig. S2K–L; Table S1B), suggesting that other as yet unidentified associated modifier proteins may function to catalyze MAMP-triggered glucosinolate activation in conjunction with PEN2. Consistent with these results, recent work has shown the presence of an ESP-independent nitrile-forming IGS activation in Arabidopsis (40).
PCS1 is required for Flg22-triggered IGS activation
Glutathione may function in IGS biosynthesis (41), and consistent with this, glutathione biosynthetic mutants (pad2-1 and cad2-1)(42, 43) exhibited reduced callose response to Flg22 (figs. 1S, S2M). The PAD2 (or GSH1) gene encodes a γ-glutamylcysteine synthetase (42), which catalyzes the first committed step in the synthesis of the tripeptide glutathione (GSH). Glutathione is a precursor for a class of heavy-metal-chelating glutathione polymers known as phytochelatins (44). Microarray data of Flg22-inducible expression profiles identified a phytochelatin synthase gene, PCS1 (Table S1A), whose corresponding mutants (pcs1-1, pcs1-2 and cad1-3) were all impaired in the callose response to Flg22 (Figs. 1T, S2N; Table S1B). More importantly, the transcriptional and IGS profiles of the pcs1 mutant resemble those of the pen2 mutant (Fig. 2B), suggesting that it too is involved in the breakdown of 4-methoxy-I3G. Like pen2, the pcs1 mutant also exhibited reduced Flg22-induced expression of MYB51 and IGS biosynthetic genes (Table 1B), supporting the conclusion that accumulation of IGS is involved in feedback inhibition of IGS biosynthesis at the level of MYB51 expression. Also, like PEN2, the Flg22-induced expression of PCS1 is independent of Et signaling and MYB51 (Table 1A).
PEN3 is also required for Flg22-induced IGS activation
Phytochelatin synthases require the cofactor cadmium for enzymatic activity (44) and may be involved in the transport and sequestration of cadmium into the vacuole (43, 45). ABC-type transporters in plants and yeast also transport and sequester cadmium into the vacuole, and may work in concert with phytochelatins (46–48). We searched the Flg22-elicited transcriptional profiling data and identified one ABC transporter, PEN3/AtPDR8 (Table S1A)(49, 50), whose corresponding mutants (pen3-1 and atpdr8-2/pen3) were also impaired in the callose response to Flg22 (Figs. 1U, S2O). Furthermore, like the pen2 and pcs1 mutants, the pen3 mutant accumulated 4-methoxy-I3G upon Flg22 treatment (Fig. 2B), suggesting that it too is involved in IGS breakdown. However, unlike the pen2 and pcs1 mutants, the pen3 mutant does not exhibit diminished MYB51 expression after 6 hours of Flg22 treatment (Table 1B). PEN3 localizes to the plasma membrane, not the vacuole (49, 50), and is involved in the extrusion of cadmium out of the cell (51), which may provide a targeting mechanism of IGS hydrolytic products towards the plasma membrane/cell wall where the callose synthase PMR4 resides. These observations may explain the lack of presumptive feedback inhibition of MYB51 expression in the pen3-1 mutant and why increased levels of 4-methoxy-I3G in the pen3-1 mutant are not as high as those in the pen2-1 and pcs1-2 mutants (Fig. 2B).
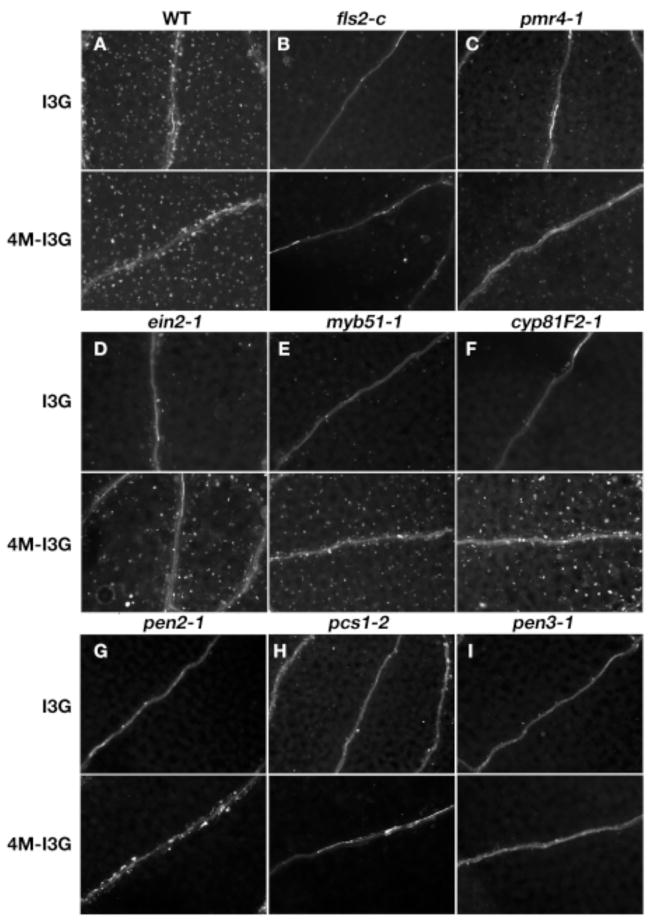
4-methoxy-I3G rescues callose formation
Callose formation was induced by 100 μM 4-methoxy-I3G in conjunction with Flg22 treatment in mutants defective in IGS biosynthesis or 4-methoxylation of I3G (Fig. 3D–F) but not in mutants defective in IGS hydrolysis (Fig. 3G–I), Flg22 perception (Fig. 3B), or callose synthesis (Fig. 3C). This supports the hypothesis that 4-methoxy-I3G is involved as a signaling molecule in or a potential co-activator of callose deposition. I3G had no effect on callose-deficient mutants (Fig. 3), indicating that it is not involved in Flg22-induced callose response. Identifying breakdown products of 4-methoxy-I3G associated with the Flg22-induced callose response will be technically challenging as these compounds may be unstable and it is not feasible to purify large quantities of them from Arabidopsis. A presumed IGS hydrolytic product, 4-methoxy-indole-3-acetonitrile (4-methoxy-IAN), has been purified from Chinese cabbage, a close relative of Arabidopsis, but neither 4-methoxy-IAN nor the endogenous IGS hydrolytic products 4-methoxy-indole-3-carboxylate and methyl-4-methoxy-indole-3-carboxylate rescued callose-deficient mutants (Table S1B), suggesting that they are most likely not directly involved in callose deposition.
Fig. 3.
4-methoxy-I3G induces callose formation in IGS biosynthetic mutants. (A–I) Cotyledons of seedlings simultaneously treated with Flg22 and I3G or 4-methoxy-I3G (4M–I3G), and then stained with aniline blue. (A) Wild type. (B) fls2-c. (C) pmr4-1. (D) ein2-1. (E) myb51-1. (F) cyp81F2-1. (G) pen2-1. (H) cad1-3/pcs1. (I) pen3-1.
The parallel SA-dependent IGS hydrolytic pathway
Microbes can trigger callose formation in Arabidopsis leaves via the plant hormone salicylic acid (SA)-dependent pathway (52), suggesting that there are multiple signaling pathways in MAMP-induced callose formation. We added hormones to rescue callose-deficient mutants and found that pretreatment with SA or the functional SA analog 2,6-dichloro-isonicotinic acid (INA)(53) rescued the callose-deficient phenotype of the pen2, pcs1, and ascorbate-deficient vtc mutants (Fig. 4B–C, E–F and H–I, respectively). This suggests that SA compensates for deficient PEN2 myrosinase activity and that PCS1 activity is necessary for PEN2 function. This SA-mediated rescue was not seen with any other described callose mutants (Fig. 4J–M), and interestingly, the transcriptional and IGS profiles of pen2 and pcs1 mutants were unchanged (Table 1B; fig. S4A). No other hormones were found to rescue callose-deficient mutants (Table S1B). Because the double mutants pen2-2 pen2-like (PEN2-like (at3g60120; Table S1A)) and pcs1-1 pcs2-1 (PCS2 (54)) exhibited a Flg22-triggered callose response in the presence of SA (Fig. 4N–O), this removes the possibility of functional redundancy. An SA-mediated pathway apparently can bypass the requirement of PEN2 and PCS1 in IGS hydrolysis through an as yet unknown mechanism.
Glucosinolate-dependent callose deposition restricts bacterial growth
A published report has shown that PMR4-dependent callose deposition contributes to MAMP-induced growth suppression of the type III secretion system-deficient bacterial pathogen Pseudomomas syringae PtoDC3000hrcC (55). In our seedling growth assay, the Flg22 receptor mutant fls2-c and the IGS biosynthetic mutant ein2-1 are very susceptible, and the IGS biosynthetic mutant cyp81F2-1 and the IGS hydrolytic mutant pen2-1 are slightly but significantly more susceptible to wild-type PtoDC3000 (fig. S5), suggesting that glucosinolate-dependent callose deposition most likely contributes to MAMP-induced growth suppression of PtoDC3000. Interestingly, this growth suppression is not dependent on PMR4 (fig. S5), probably due to the increased SA levels in the mutant (12).
Suppressing the Flg22-triggered callose response
Defense-related plant hormones (SA, methyljasmonate (MeJA), abscisic acid (ABA) and the Et precursor 1-aminocyclopropane-1-carboxylic acid (ACC)) and the common IGS hydrolytic product indole-3-acetonitrile (IAN) did not induce callose formation in the absence of Flg22. In the presence of Flg22, pretreatment with SA or ACC had no effect on the callose response (Fig. 1W–X). In contrast, pretreatment with ABA or MeJA or co-treatment with IAN completely suppressed the Flg22-induced callose response (Fig. 1Y-AA). ABA, MeJA and IAN may differ in their modes of callose suppression. By 6 hours after Flg22 elicitation, ABA pretreatment greatly reduced Flg22-induced expression of ERF1, MYB51 and CYP81F2, which depend on Et signaling for full activation (Table 1C), suggesting that ABA antagonizes the Et signaling triggered by Flg22. This is consistent with antagonistic interactions between ABA and Et signaling pathways (56, 57).
In contrast to ABA, MeJA pretreatment transiently reduced transcript levels of MYB51 and CYP81F2, but after 6 hours of Flg22 treatment, gene expression returned to wild type levels (Table 1C). MeJA and JA induce I3G and 1-methoxy-I3G but not 4-methoxy-I3G production in mature plants (58–60), and we also observed this in seedlings (fig. S4). It is possible that MeJA blocks callose deposition by inducing the N-methoxylation pathway, which predominates over the Flg22-induced 4-methoxylation pathway, thereby reducing the production of 4-methoxy-I3G required for callose formation. However, this does not appear to be the case as 4-methoxy-I3G levels were unchanged in both the wild type and the 4-methoxy-I3G-accumulating pen2 mutant upon MeJA and Flg22 treatment (fig. S4B), suggesting that there is no significant substrate competition between the two methoxylation pathways and that the mode of MeJA suppression occurs downstream of methoxylation, probably at the level of IGS hydrolysis.
IGS biosynthetic pathway is inhibited by IAN feeding
IAN treatment immediately suppressed Flg22-induced expression of MYB51 and MYB51-regulated IGS biosynthetic genes (Table 1C). By 6 hours after Flg22 elicitation, CYP81F2 expression also decreased, but ERF1 remained at wild type levels (Table 1C), suggesting that Et signaling is not affected in IAN-treated plants. Since IAN is a common IGS hydrolytic product, the suppressive effect of exogenous IAN treatment suggests that the IGS biosynthetic pathway is under feedback inhibition by the accumulation of IGS hydrolytic products although the identity of the actual regulatory metabolite remains unknown. Interestingly, expression in IAN-treated wild type plants resemble those of putative IGS breakdown mutants pen2 and pcs1 at the level of MYB51 expression (Table 1B), suggesting that the IGS biosynthetic pathway is under feedback inhibition by IGS accumulation and its hydrolytic products. Other IGS hydrolytic products, 4-methoxy-IAN, methyl-4-methoxy-indole-3-carboxylate, and methyl-indole-3-carboxylate, also suppressed the Flg22-induced callose response (Table S1B), further supporting the conclusion that IGS biosynthesis is under feedback inhibition by its hydrolytic products.
Conclusions
Our study of the MAMP-triggered callose defense response, a classic innate immune response to both adapted and non-adapted pathogens, shows that the essential and ubiquitous compounds glutathione and ascorbate, the transported metal ion cadmium, and the secondary metabolite 4-methoxy-indol-3-ylmethylglucosinolate are all required for callose deposition. We showed that the Flg22-triggered callose response in Arabidopsis seedlings requires the concomitant induction of three pathways: Et- and MYB51-dependent I3G biosynthesis, CYP81F2-dependent 4-methoxylation of I3G, and the PEN2, PCS1, and PEN3-mediated hydrolysis of 4-methoxy-I3G (Fig. 5). The core IGS biosynthetic pathway is under feedback inhibition at the level of MYB51 expression by the accumulation of IGS and IGS hydrolytic products. The defense hormones ABA and MeJA suppress the callose response by antagonizing Et signaling and IGS breakdown, respectively, whereas an SA-dependent pathway can bypass the hydrolytic requirement of PEN2 and PCS1. The role of glucosinolate hydrolysis in insect resistance has been studied for over 100 years. Here we have identified a new role for this metabolic pathway, linking the endogenous hydrolytic products of glucosinolates to MAMP-mediated defense responses as potential signaling molecules. Further exploration of the species and genus-specific effects of these molecules, including their potential role as compounds with direct antimicrobial activity as suggested in the accompanying paper by Bednarek et al. (36), will be needed to determine their multiple functions in plant-microbe interactions.
Fig. 5.
A model of MAMP-triggered callose deposition in Arabidopsis.
Supplementary Material
Acknowledgments
We thank S. Abel for ugt74B1-2; J.L. Celenza for atr1-3, atr4-1/cyp83B1, cyp79B2 cyp79B3 and trp3-1; C.S. Cobbett for cad1-3/pcs1 and cad2-1/gsh1; P.L. Conklin for vtc1-1 and vtc2-1; R. Blum for pcs1-1, pcs2-1, and pcs1-1 pcs2-1; S.C. Somerville for pcs1-2; and ABRC for insertion lines. PhD stipend support to N.K.C. comes from National Institutes of Health (NIH) NRSA-Kirschstein Postdoctoral Fellowship. Support to A.M.A. and G.J. comes from National Science Foundation (NSF) grants IOS-0718733 and DBI-0500550. Support to F.M.A. comes from NSF Grant 06-SC-NSF-1020 and NIH Grant R37-GM48707.
Footnotes
This manuscript has been accepted for publication in Science. This version has notundergone final editing. Please refer to the complete version of record athttp://www.sciencemag.org/. The manuscript may not be reproduced or used in anymanner that does not fall within the fair use provisions of the Copyright Act without theprior, written permission of AAAS.
References
- 1.Thordal-Christensen H. Curr Opin Plant Biol. 2003;6:351. doi: 10.1016/s1369-5266(03)00063-3. [DOI] [PubMed] [Google Scholar]
- 2.Mysore KS, Ryu CM. Trends Plant Sci. 2004;9:97. doi: 10.1016/j.tplants.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Nürnberger T, Lipka V. Mol Plant Pathol. 2005;6:335. doi: 10.1111/j.1364-3703.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Gomez L, Boller T. Trends Plant Sci. 2002;7:251. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- 5.Nürnberger T, Kemmerling B. Trends Plant Sci. 2006;11:519. doi: 10.1016/j.tplants.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Denoux C, et al. Mol Plant. 2008;1:423. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halkier BA, Gershenzon J. Annu Rev Plant Biol. 2006;57:303. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 8.Tierens KFMJ, et al. Plant Physiol. 2001;125:1688. doi: 10.1104/pp.125.4.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedras MSC, Zheng QA, Gadagi RS. Chem Commun. 2007:368. doi: 10.1039/b615424g. [DOI] [PubMed] [Google Scholar]
- 10.Songnuan W, et al. In: Biology of Plant-microbe interactions. Lorito M, Loo SL, Scala F, editors. Vol. 6 International Society for Molecular Plant-Microbe Interactions; St. Paul, MN: 2008. [Google Scholar]
- 11.Gomez-Gomez L, Boller T. Mol Cell. 2000;5:1003. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura MT, et al. Science. 2003;301:969. doi: 10.1126/science.1086716. [DOI] [PubMed] [Google Scholar]
- 13.Heese A, et al. Proc Natl Acad Sci USA. 2007;104:12217. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinchilla D, et al. Nature. 2007;448:497. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 15.Schaller GE, Bleecker AB. Science. 1995;270:1809. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- 16.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. Science. 1999;284:2148. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 17.Gigolashvili T, et al. Plant J. 2007;50:886. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 18.Celenza JL, et al. Plant Physiol. 2005;137:253. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niyogi KK, Fink GR. Plant Cell. 1992;4:721. doi: 10.1105/tpc.4.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. Plant Cell. 2005;17:2230. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, et al. Genes Dev. 2002;16:3100. doi: 10.1101/gad.1035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haughn GW, Davin L, Giblin M, Underhill EW. Plant Physiol. 1991;97:217. doi: 10.1104/pp.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, Durrett TP, Last RL, Jander G. Plant Mol Biol. 2004;54:671. doi: 10.1023/B:PLAN.0000040897.49151.98. [DOI] [PubMed] [Google Scholar]
- 24.Hemm MR, Ruegger MO, Chapple C. Plant Cell. 2002;15:179. doi: 10.1105/tpc.006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge UI. Plant J. 2007;51:246. doi: 10.1111/j.1365-313X.2007.03133.x. [DOI] [PubMed] [Google Scholar]
- 26.Hirai MY, et al. Proc Natl Acad Sci USA. 2007;104:6478. [Google Scholar]
- 27.Sønderby IE, et al. PlosOne. 2007;2:e1322. [Google Scholar]
- 28.Radwanski ER, Barczak AJ, Last RL. Mol Gen Genetics. 1996;253:353. doi: 10.1007/pl00008602. [DOI] [PubMed] [Google Scholar]
- 29.Nafisi M, et al. Plant Cell. 2007;19:2039. doi: 10.1105/tpc.107.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuhegger R, et al. Plant Physiol. 2007;141:1248. doi: 10.1104/pp.106.082024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burmeister WP, Cottaz S, Rollin P, Vasella A, Henrissat B. J Biol Chem. 2000;275:39385. doi: 10.1074/jbc.M006796200. [DOI] [PubMed] [Google Scholar]
- 32.Conklin PL, Saracco SA, Norris SR, Last RL. Genetics. 2000;154:847. doi: 10.1093/genetics/154.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barth C, Jander G. Plant J. 2006;46:549. doi: 10.1111/j.1365-313X.2006.02716.x. [DOI] [PubMed] [Google Scholar]
- 34.Collins NC, et al. Nature. 2003;425:973. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 35.Lipka V, et al. Science. 2005;310:1180. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 36.Bednarek P, et al. 2008 Manuscript submitted. [Google Scholar]
- 37.AM W, et al. PLOS Genetics. 2007;3:1687. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J. Plant Cell. 2006;13:2793. doi: 10.1105/tpc.010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, Ober JA, Kliebenstein DJ. Plant Cell. 2006;18:1524. doi: 10.1105/tpc.105.039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wentzell AM, Kliebenstein DJ. Plant Physiol. 2008;147:415. doi: 10.1104/pp.107.115279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlaeppi K, Bodenhausen N, Buchala A, Mauch F, Reymond P. Plant J. 2008;55:774. doi: 10.1111/j.1365-313X.2008.03545.x. [DOI] [PubMed] [Google Scholar]
- 42.Parisy V, et al. Plant J. 2007;49:159. doi: 10.1111/j.1365-313X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- 43.Howden R, Andersen CR, Goldsbrough PB, Cobbett CS. Plant Physiol. 1995;107:1067. doi: 10.1104/pp.107.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grill E, Löffler S, Winnacker EL, Zenk MH. Proc Natl Acad Sci USA. 1989;86:6838. doi: 10.1073/pnas.86.18.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howden R, Cobbett CS. Plant Physiol. 1992;100:100. doi: 10.1104/pp.100.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortiz DF, Ruscitti T, McCue KF, Ow DW. J Biol Chem. 1995;270:4721. doi: 10.1074/jbc.270.9.4721. [DOI] [PubMed] [Google Scholar]
- 47.Li ZS, et al. Proc Natl Acad Sci USA. 1997;94:42. [Google Scholar]
- 48.Rea PA, Li ZS, Lu YP, Drozdowicz YM, Martinoia E. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:727. doi: 10.1146/annurev.arplant.49.1.727. [DOI] [PubMed] [Google Scholar]
- 49.Stein M, et al. Plant Cell. 2006;18:731. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobae Y, et al. Plant Cell Physiol. 2006;47:309. doi: 10.1093/pcp/pcj001. [DOI] [PubMed] [Google Scholar]
- 51.Kim DY, Bovet L, Maeshima M, Martinola E, Lee Y. Plant J. 2007;50:207. doi: 10.1111/j.1365-313X.2007.03044.x. [DOI] [PubMed] [Google Scholar]
- 52.Debroy S, Thilmony R, Kwack YB, Nomura K, He SY. Proc Natl Acad Sci USA. 2004;101:9927. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uknes S, et al. Plant Cell. 1992;4 doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cazalé AC, Clemens S. FEBS Let. 2001;507:215. doi: 10.1016/s0014-5793(01)02976-3. [DOI] [PubMed] [Google Scholar]
- 55.Kim MG, et al. Cell. 2005;121:749. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 56.Beaudoin N, Serizet C, Gosti F, Giraudat J. Plant Cell. 2000;12:1103. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghassemian M, et al. Plant Cell. 2000;12:1117. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brader G, Tas E, Palva ET. Plant Physiol. 2001;126:849. doi: 10.1104/pp.126.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kliebenstein DJ, Figuth A, Mitchell-Olds T. Genetics. 2002;161:1685. doi: 10.1093/genetics/161.4.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mikkelsen MD, et al. Plant Physiol. 2003;131:298. doi: 10.1104/pp.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.