Abstract
The basis for the extensive variability seen in the reconstitution of CD4+ T cell counts in HIV-infected individuals receiving highly active antiretroviral therapy (HAART) is not fully known. Here, we show that variations in CCL3L1 gene dose and CCR5 genotype, but not major histocompatibility complex HLA alleles, influence immune reconstitution, especially when HAART is initiated at <350 CD4+ T cells/mm3. The CCL3L1-CCR5 genotypes favoring CD4+ T cell recovery are similar to those that blunted CD4+ T cell depletion during the time before HAART became available (pre-HAART era), suggesting that a common CCL3L1-CCR5 genetic pathway regulates the balance between pathogenic and reparative processes from early in the disease course. Hence, CCL3L1-CCR5 variations influence HIV pathogenesis even in the presence of HAART and, therefore, may prospectively identify subjects in whom earlier initiation of therapy is more likely to mitigate immunologic failure despite viral suppression by HAART. Furthermore, as reconstitution of CD4+ cells during HAART is more sensitive to CCL3L1 dose than to CCR5 genotypes, CCL3L1 analogs might be efficacious in supporting immunological reconstitution.
HIV-1 infection is characterized by extensive variability in the degree of depletion or recovery in CD4+ T cell (abbreviated as CD4+) counts before or after receipt of HAART, respectively. Variability in CD4+ depletion cannot be attributed solely to the extent of viral replication (as reflected by the viral load)1,2, as host factors can influence HIV disease course independently of viral load2,3. Similarly, variability in the depth of HAART-induced suppression of viral replication cannot be the sole factor underlying intersubject differences in recovery of CD4+ counts. For example, the majority of HIV-positive subjects who initiate therapy with a low CD4+ nadir during untreated disease (<200 cells/mm3) do not fully recover peripheral or mucosal CD4+ counts, despite HIV-suppressive HAART4-12. Conversely, some HIV-positive individuals show sustained recovery of CD4+ counts in the face of incomplete viral suppression5,10,13-20. However, the host factors that mediate this variable CD4+ recovery are largely unknown.
Here we tested the hypothesis that CD4+ depletion and recovery reflect a dynamic balance between pathogenic and reparative processes integrally linked by specific common genetic pathways programmed during early stages of HIV disease. The importance of events occurring within the initial phases of HIV disease is highlighted by the observation that the magnitude of early immune damage determines the development and durability of virus-specific immunity, which dictates, in part, steady-state viral load, initial CD4+ loss and, subsequently, HIV disease course1,3,21-23. However, once host genetic factors skew the balance toward pathogenic processes (CD4+ depletion) during early untreated infection, it is unclear whether this imbalance can be fully reversed, even in the presence of HAART-induced viral suppression.
To test our hypothesis, we determined whether specific genetic factors that skew the balance toward pathogenic or reparative processes during early HIV disease also exert a negative or positive influence, respectively, on recovery of CD4+ counts during HAART. We focused on polymorphisms in the genes encoding CCR5, the major HIV co-receptor, and CCL3L1, its most potent HIV-suppressive ligand24, for three reasons. First, CCR5 genotypes and CCL3L1 gene copy number (that is, gene dose) influence HIV-AIDS susceptibility3,25-29. Second, CCL3L1-CCR5 genetic risk groups (GRGs), defined according to different inherited combinations of CCL3L1 dose and CCR5 polymorphisms, influence the extent of early immune damage3. Third, CCL3L1-CCR5 GRGs affect HIV disease progression by affecting viral load and parameters independent of the viral load (for example, cell-mediated immunity)3.
To provide a context for our hypothesis, we considered two additional issues. First, we assessed whether the impact of CCL3L1-CCR5 GRGs on CD4+ recovery is mitigated by HAART-induced suppression of viral replication during acute or early infection. Second, HLA alleles are crucial in determining the viral load set-point, degree of CD4+ depletion and HIV disease course29,30, but whether those HLA alleles that influence early immune damage also affect CD4+ recovery is unknown; thus we studied the effect of HLA alleles on recovery of CD4+ counts during HAART.
RESULTS
CCL3L1, CCR5, GRG and CD4+ dynamics during HIV infection
During the pre-HAART era, the overall CD4+ counts within the HIV+ Wilford Hall Medical Center (WHMC) cohort declined progressively, reaching ∼300 cells/mm3 (Fig. 1a). During the HAART era, CD4+ counts increased, peaked in 2001, and remained at ∼500 cells/mm3 thereafter (Fig. 1a).
Figure 1.
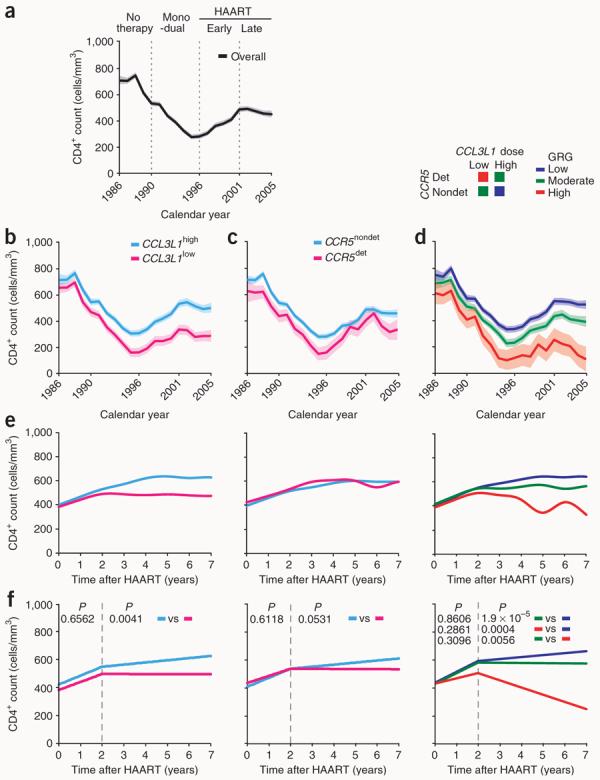
Influence of CCL3L1-CCR5 genotypes on CD4+ T cell dynamics in the WHMC HIV+ cohort. (a–d) Trajectories of CD4+ counts in the WHMC cohort across calendar years of cohort membership computed before (a) and after accounting for CCL3L1 dose (b), CCR5 genotype (c) and CCL3L1-CCR5 GRGs (d). Dotted lines in a demarcate the indicated therapy eras, and mono and dual reflect therapy with one or two antiviral agents that were prevalent during the pre-HAART era. The time trends shown were estimated by nonlinear generalized estimating equations (GEE) modeling of CD4+ counts and depicted as mean (thick lines) and 95% confidence bands (shaded regions). The genetic components that comprise CCL31-CCR5 GRGs are shown above d. (e) Trajectories of CD4+ counts (by nonlinear GEE) from time of initiation of HAART according to (from left to right) CCL3L1 dose, CCR5 genotype and CCL3L1-CCR5 GRG status. (f) Trajectories of CD4+ counts during HAART according to CCL3L1 dose, CCR5 genotype and GRG status during the first 2 years after initiation of HAART and in the years thereafter (modeled by linear GEE in two segments demarcated by the dashed lines). P values indicate differences in rate of change in CD4+ counts for the indicated groups (derived by the Student's t-test) during the first 2 years after initiation of HAART and in the years thereafter. Rate of change in CD4+ cell counts per month (with s.e.m. in parentheses) during the first 2 years of HAART in those with a high or low CCL3L1 dose, nondetrimental or detrimental CCR5 genotype, and low, moderate or high GRG were 5.23 (0.72), 4.73 (1.12), 5.29 (0.57), 4.18 (2.11), 6.36 (0.57), 6.20 (0.98), and 3.12 (2.86), respectively. Rate of change in CD4+ cell counts/month (s.e.m.) after 2 years of HAART in those with a high or low CCL3L1 dose, nondetrimental or detrimental CCR5 genotype, and low, moderate or high GRG were 1.27 (0.23), −0.04 (0.39), 1.20 (0.21), −0.05 (0.61), 0.96 (0.17), −0.09 (0.17), and −3.54 (1.22), respectively. The color code for plots shown in e and f are shown above b–d.
However, the time trends for the CD4+ counts differed according to a person's CCL3L1 and CCR5 genetic makeup, that is, low or high CCL3L1 dose (Fig. 1b) and detrimental or non-detrimental CCR5 genotype27 (Fig. 1c). Both CCL3L1low and CCR5det genotype conferred rapid CD4+ loss and impaired CD4+ recovery during the pre- and post-HAART eras, respectively (Fig. 1b,c). However, the differences in CD4+ recovery associated with CCL3L1low and CCL3L1high (Fig. 1b) were more prominent than those associated with CCR5det and CCR5nondet (Fig. 1c). Concordantly, among subjects of European and African descent, a stepwise increase in CCL3L1 copy number was associated with a stepwise increase in CD4+ recovery rates during HAART (Supplementary Fig. 1a online). Analyses of the joint effects of CCL3L1 and CCR5 revealed that going from a low to moderate to high CCL3L1-CCR5 GRG was associated with accelerating depletion during the pre-HAART era or increasingly impaired recovery during the HAART era (Fig. 1d).
CCL3L1, CCR5, GRG and CD4+ recovery during chronic infection
Time trends of CD4+ counts indexed from the time of initiation of HAART contrasting CCL3L1 dose, CCR5 genotype and GRG (Fig. 1e> and Table 1) were similar to those detected by calendar-era analysis in the WHMC HIV+ cohort during the HAART era (Fig. 1b–d). We next determined whether suppression of viral replication by HAART overshadowed the effects of variations in CCL3L1 and CCR5 on CD4+ recovery. Those HIV-positive individuals who had HAARt-induced viral load suppression had robust increases in CD4+ counts and slower AIDS progression rates compared with those who were non–viral load suppressors (Supplementary Fig. 1b,c). Additionally, although HIV-infected individuals with transient or sustained viral load suppression did not differ in their rates of disease progression, gains in CD4+ counts were significantly greater in those with sustained viral load suppression (Supplementary Fig. 1b,c). However, even among subjects with sustained viral load suppression, CCL3L1-CCR5 variations affected CD4+ recovery (Table 1 and Supplementary Fig. 1d). Additionally, these effects were also evident when complementary statistical methods (mixed models) were used (Supplementary Fig. 2 online).
Table 1.
Influence of variations in CCL3L1 and CCR5 on CD4+ recovery rates
| Viral load suppressors |
Viral load suppression |
||||
|---|---|---|---|---|---|
| Genotype | Overall | Sustained | Transient | ||
| CCL3L1 | |||||
| CCL3L1high | 2.80 (0.14) | 3.43 (0.17) | 5.41 (0.32) | 2.61 (0.19) | |
| CCL3L1low | 0.71 (0.24) | 0.73 (0.24) | 2.61 (0.42) | −0.24 (0.29) | |
| Plow versus high | 6.9 × 10−13 | 2.0 × 10−17 | 8.1 × 10−7 | 2.4 × 10−13 | |
| CCR5 | |||||
| CCR5nondet | 2.38 (0.37) | 2.76 (0.14) | 5.18 (0.98) | 1.97 (0.16) | |
| CCR5det | 1.77 (0.13) | 2.29 (0.48) | 4.40 (0.26) | 1.35 (0.43) | |
| Pdet versus nondet | 0.1229 | 0.3495 | 0.4474 | 0.1786 | |
| GRG status | |||||
| Low | 2.89 (0.13) | 3.45 (0.15) | 5.08 (0.28) | 2.68 (0.16) | |
| Moderate | 1.42 (0.20) | 1.67 (0.22) | 3.68 (0.35) | 0.62 (0.27) | |
| High | −0.85 (0.76) | −0.77 (0.70) | 2.29 (0.85) | −1.74 (0.69) | |
| Plow versus moderate | 1.7 × 10−9 | 2.8 × 10−10 | 0.0023 | 4.4 × 10−9 | |
| Pmoderate versus high | 4.3 × 10−3 | 1.2 × 10−3 | 0.1357 | 2.3 × 10−3 | |
| Plow versus high | 3.1 × 10−6 | 6.5 × 10−8 | 0.0086 | 7.0 × 10−8 | |
Data are from subjects from the WHMC HIV+ cohort who received HAART during chronic infection. Values indicate the mean (s.e.m.) rate of change of CD4+ cells per month after initiation of HAART, as estimated by regression coefficients computed by linear GEE. A negative number indicates a decrease in the rate of CD4+ cell counts. Data for analyses conducted by using square-root–transformed CD4+ T cell counts, as well as segmented regressions to determine effects of genotypes on CD4+ recovery during and after the first 2 years of HAART, are shown in Supplementary Table 1 online.
CCL3L1, CCR5, GRG and durability of CD4+ recovery
Inspection of CD4+ trajectories indicated that during the first 2 years of HAART, CD4+ counts increased regardless of genotype, but differed thereafter according to variations in CCL3L1 and CCR5 (Fig. 1e). Analyses of CD4+ recovery rates before and after 2 years of HAART substantiated these observations (Fig. 1f).
Additive effects of CCL3L1 and CCR5
Although a low CCL3L1 dose is a feature common to both high and moderate GRGs (Fig. 1d), subjects in a high GRG had significantly lower rates of CD4+ gains as compared with those in a moderate GRG (Fig. 1f and Table 1). Furthermore, among all subjects who had half of the average CCL3L1 dose of their ethnic or racial group, those who also had a detrimental CCR5 genotype lost CD4+ cells at a faster rate than those who had a nondetrimental CCR5 genotype (Supplementary Fig. 1e). These findings indicate that the effects of a detrimental CCR5 genotype and a low CCL3L1 dose on CD4+ recovery are additive, and that a nondetrimental CCR5 genotype attenuates the negative impact of a low CCL3L1 dose on CD4+ gains.
CCL3L1, CCR5, GRG and CD4+ recovery during early infection
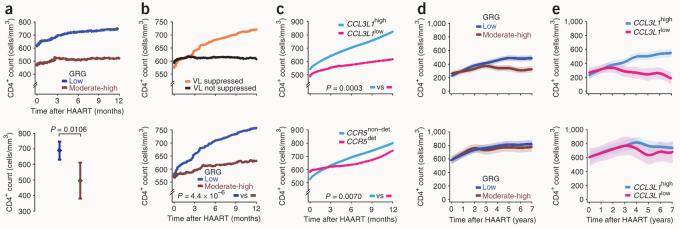
HIV-positive subjects in the Acute Infection and Early Disease Research Program (AIEDRP) were stratified according to whether they received HAART during acute or early infection, and we analyzed the effects of CCL3L1 dose and CCR5 genotype on CD4+ recovery in these two groups of subjects separately. Among subjects who initiated HAART during acute infection and who attained viral load suppression, a low GRG was associated with both a higher baseline CD4+ count at time of enrollment into the cohort (619.10 cells/mm3 versus 440.44 cells/mm3 in those with low versus moderate-high GRG, respectively; P = 0.0253), and an increase of ∼150 cells/mm3 in mean CD4+ counts through 12 months of follow-up (Fig. 2a). In contrast, in subjects with a moderate or high GRG, post-HAART CD4+ counts remained similar to those detected at baseline (∼500 CD4+ cells/mm3; Fig. 2a). The higher baseline CD4+ counts observed in those with a low GRG than in those with a moderate or high GRG is consistent with our previous report showing that possession of a low GRG blunts the extent of the initial loss in CD4+ cells3.
Figure 2.
Influence of CCL3L1-CCR5 on CD4+ recovery in subjects initiating HAART during acute or early infection (AIEDRP cohorts, a–c) and impact of the timing of HAART on effects of CCL3L1-CCR5 on CD4+ recovery in the WHMC cohort (d,e). (a) Loess curves depict trajectories of CD4+ counts in those who initiated HAART during acute infection and who attained viral load suppression according to GRG status (top). Difference in average CD4+ counts by GRG in the first year of HAART is also shown (bottom). (b) Loess curves showing CD4+ count changes in subjects according to those who did or did not achieve viral load suppression during early infection (top) and according to GRG status in those who achieved HAART-induced viral load suppression during early infection (bottom). For those initiating HAART during early infection, rate of change in CD4+ cell counts/month (s.e.m.) in those with low or moderate-high GRG were 21.62 (1.09) and 12.52 (1.59), respectively. (c) Loess curves for CD4+ count changes according to CCL3L1 dose (top) or CCR5 genotype (bottom) in subjects with HAART-induced viral load suppression during early infection. Rate of change in CD4+ counts/month (s.e.m.) in those with a high or low CCL3L1 dose and nondetrimental or detrimental CCR5 genotypes were 20.30 (1.02), 12.71 (1.79), 20.18 (0.97), and 12.56 (2.63), respectively. (d) Trajectories of CD4+ counts (mean (bold lines) and 95% confidence bands by nonlinear GEE) according to GRG status for subjects in WHMC who initiated HAART at <350 (top) or ≥350 (bottom) CD4+ cells/mm3. The data are for all subjects who received HAART. (e) Trajectories of CD4+ counts according to CCL3L1 dose in subjects who were therapy-naive and initiated HAART at <350 (top) or ≥350 CD4+ cells/mm3 (bottom). The HAART-initiating CD4+ T cell count is the average CD4+ T cell count during the year preceding initiation of therapy. Where indicated, the moderate and high GRGs were combined into a single category (brown).
Similar findings were detected in those who received HAART during early infection. Viral load suppression during early HIV infection was associated with robust gains (19.8 CD4+ cells/month) in CD4+ counts (Fig. 2b). However, even among those who initiated HAART during early infection and who attained viral load suppression, the CD4+ recovery rate was greatest in those with a low GRG (Fig. 2b). Of note, the individual components of the GRGs—CCL3L1 and CCR5—influenced CD4+ recovery (Fig. 2c).
GRG effects on CD4+ recovery according to timing of HAART
We next determined whether CD4+ recovery was sensitive to both the CCL3L1-CCR5 GRG (or CCL3L1 dose alone) and the immunological status of a subject (as reflected by CD4+ count at the time of initiation of HAART). Among individuals from the WHMC cohort who initiated HAART at <350 CD4+ cells/mm3, a low GRG was associated with CD4+ gains, whereas recovery was highly muted in those with a moderate or high GRG (Fig. 2d and Table 2). Notably, each 50-cell decrease in the CD4+ count at the time at which HAART was initiated was associated with a step-wise decline in recovery rates in those possessing a moderate or high GRG and, consequently, a progressively widening difference in CD4+ recovery rates for those with a low versus moderate or high GRG (Table 2). Notably, the latter associations persisted even when the high GRG (prevalence of 8%) was removed from analyses (Table 2), indicating that the associations observed are not solely due to a high GRG and that a moderate GRG (prevalence of 42%) also has a significant negative impact on CD4+ recovery rates. In subjects who initiated HAART at ≥350 CD4+ cells/mm3, the time trends in CD4+ counts did not reveal significant differences in CD4+ recovery (Fig. 2d). Similar patterns of CD4+ recovery were observed in therapy-naive and therapy-experienced subjects (Supplementary Fig. 3a online). Notably, the differential impact of genotype in subjects who differed according to the CD4+ count at HAART initiation was even more prominent when subjects were categorized according to CCL3L1 dose (Fig. 2e).
Table 2.
GRG effects on CD4+ recovery in those initiating HAARTat <350 CD4+ cells/mm3
| Pre-HAART CD4 count (cells/mm3)a |
GRG status |
Low versus moderate-high GRG CD4 difference (s.e.m.), P |
Low versus moderate GRG CD4 difference (s.e.m.), P |
||
|---|---|---|---|---|---|
| Low | Moderate-high | Moderate | |||
| <350 | 3.23 (0.21)b | 0.83 (0.22) | 0.80 (0.24) | 2.40 (0.29), 5.8 × 10−13 | 2.43 (0.31), 2.9 × 10−12 |
| <300 | 3.52 (0.21) | 0.28 (0.26) | 0.23 (0.28) | 3.24 (0.33), 5.0 × 10−16 | 3.29 (0.35), 6.2 × 10−12 |
| <250 | 3.47 (0.25) | −0.26 (0.27) | −0.33 (0.30) | 3.73 (0.37), 1.9 × 10−15 | 3.80 (0.39), 1.5 × 10−14 |
| <200 | 4.51 (0.24) | −0.23 (0.33) | −0.22 (0.33) | 4.74 (0.41), 3.6 × 10−16 | 4.73 (0.41), 8.1 × 10−16 |
| <150 | 4.60 (0.30) | −0.16 (0.37) | −0.16 (0.37) | 4.76 (0.47), 1.1 × 10−11 | 4.76 (0.47), 1.1 × 10−11 |
| <100 | 4.82 (0.34) | −0.06 (0.41) | −0.06 (0.41) | 4.88 (0.54), 1.5 × 10−9 | 4.88 (0.54), 1.5 × 10−9 |
| <50 | 4.59 (0.52) | −0.47 (0.90) | −0.47 (0.90) | 5.06 (1.04), 2.0 × 10−4 | 5.06 (1.04), 2.0 × 10−4 |
Data are for subjects from the WHMC HIV+ cohort who received HAART during chronic infection.
Pre-HAART CD4+ T cell count was the average of all CD4+ T cell counts during the 3 months before initiation of HAART.
Values are defined as in Table 1. Similar analyses were also conducted by using square-root-transformed CD4+ T cell counts (Supplementary Fig. 1f).
Specificity of GRG effects on CD4+ recovery
The influence of CCL3L1-CCR5 GRGs on CD4+ recovery rates could not be explained by significant inter-GRG differences in viral load suppression or other parameters such as the number of subjects who initiated HAART at <350 or ≥350 CD4+ cells/mm3 according to GRG status, the age at which HAART was initiated, or the pre-HAART CD4+ cell count or viral load (Supplementary Methods and Supplementary Note online). We found that even after adjustment for these covariates (potential confounders), a moderate or high GRG remained as a significant independent determinant of impaired CD4+ recovery rate after 2 years of HAART in those initiating HAART at <350 CD4+ cells/mm3 (Table 3). A trend toward a negative effect of a moderate-high GRG on CD4+ recovery rates was also observed in those who initiated HAART at ≥350 cells/mm3 (Table 3). These results, together with the findings discussed below, indicate that the effect of GRG status on CD4+ recovery is independent of other parameters that are also known to influence immune reconstitution during HAART.
Table 3.
Independent effects of CCL3L1-CCR5 GRGs on CD4+ recovery
| Pre-HAART CD4 | Low GRG | Moderate-high GRG | P | ||
|---|---|---|---|---|---|
| Model 1: unadjusted | |||||
| During first 2 years of HAART | |||||
| ≥ 350 | 7.07 (1.00)a | 6.87 (1.44) | 0.9093 | ||
| < 350 | 6.05 (0.76) | 5.13 (0.89) | 0.4332 | ||
| After 2 years of HAART | |||||
| ≥ 350 | 0.65 (0.40) | −0.65 (0.62) | 0.0808 | ||
| < 350 | 1.90 (0.35) | 0.12 (0.34) | 0.0004 | ||
| Model 2: adjusted for covariatesb | |||||
| During first 2 years of HAART | |||||
| ≥ 350 | 5.16 (1.19) | 4.58 (1.49) | 0.7611 | ||
| < 350 | 6.03 (0.69) | 4.49 (0.62) | 0.1008 | ||
| After 2 years of HAART | |||||
| ≥ 350 | 1.01 (0.50) | −0.45 (0.59) | 0.0626 | ||
| < 350 | 2.07 (0.40) | 0.11 (0.38) | 0.0007 | ||
| Model 3: adjusted for covariatesb and HLA-A*68, HLA-B*57 and HLA-C*16 alleles | |||||
| During first 2 years of HAART | |||||
| ≥ 350 | 4.77 (1.21) | 5.15 (1.71) | 0.8565 | ||
| < 350 | 6.36 (0.76) | 4.97 (0.33) | 0.0971 | ||
| After 2 years of HAART | |||||
| ≥ 350 | 1.02 (0.48) | −1.55 (0.68) | 0.0028 | ||
| < 350 | 2.44 (0.46) | 0.20 (0.36) | 0.0003 | ||
| Model 4; adjusted for covariatesb and HLA allele frequency score | |||||
| During first 2 years of HAART | |||||
| ≥ 350 | 4.72 (0.17) | 5.15 (1.57) | 0.8258 | ||
| < 350 | 6.38 (0.78) | 4.97 (0.43) | 0.1206 | ||
| After 2 years of HAART | |||||
| ≥ 350 | 1.01 (0.49) | −1.56 (0.90) | 0.0033 | ||
| < 350 | 2.45 (0.46) | 0.19 (0.36) | 0.0003 | ||
Data are for subjects from the WHMC HIV+ cohort and indicate the mean (s.e.m.) rate of change of CD4+ cell counts per month after initiation of HAART, as estimated by regression coefficients computed by linear GEE.
Values are defined as in Table 1.
Covariates were calendar year of HAART initiation, age at HAART initiation, pre-HAART viral load, achievement of viral load suppression and prior receipt of antiretroviral therapy. Pre-HAART CD4+ T cell count is as defined in Table 2. Similar results for models 1 and 2 were obtained in subjects who were naive for HAART (Supplementary Table 2 online).
Influence of disease-modifying HLA alleles on CD4+ recovery
To test further the generalizability of our hypothesis, we focused on those HLA alleles that influence early immune damage as reflected by steady-state viral load, initial CD4+ loss and delayed-type hypersensitivity (DTH) skin test responses, an in vivo marker of cell-mediated immunity. We addressed this by adopting two unbiased approaches. First, we determined which of the HLA class I alleles at the A, B, and C loci and class II alleles at the DRB1 locus independently influenced AIDS progression rates in a multivariate model. In the combined analyses of European and African Americans from the WHMC, HLA-A*68, HLA-B*57 and HLA-C*16 were the only three HLA alleles that predicted altered disease progression rates, with HLA-A*68 and HLA-C*16 associating with disease acceleration and HLA-B*57 associating with disease retardation (Fig. 3a). Of these three alleles, only the HLA-B*57 allele also influenced all three markers of early immune damage, as well as the cumulative CD4+ cell count (Fig. 3b and Supplementary Fig. 3b). Although possession of the HLA-B*57 allele was associated with a striking reduction in disease progression rates independently of other covariates such as steady-state viral load and GRGs (Fig. 3a), there were no differences in the overall CD4+ recovery according to HLA-B*57 status (Fig. 3c). Thus, an overall protective effect of the HLA-B*57 allele was not evident during HAART, and it was noteworthy that during the first 2–3 years after starting HAART possession of this allele was associated with impaired CD4+ cell recovery (Fig. 3c).
Figure 3.
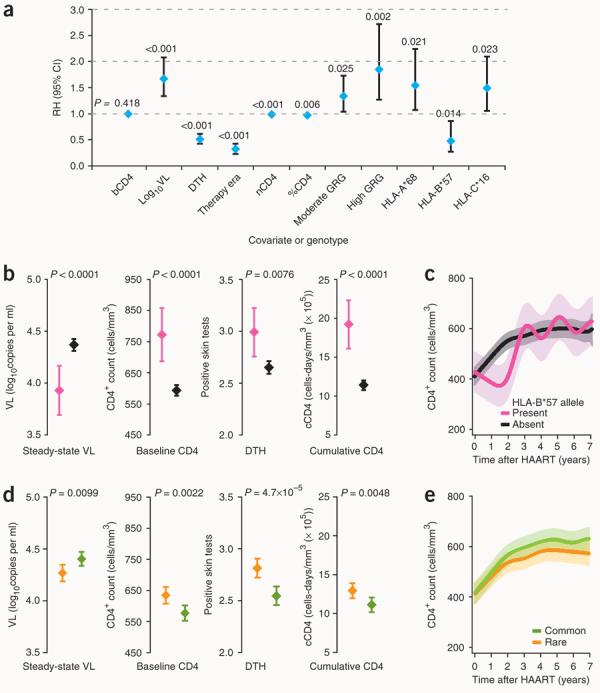
Influence of HLA alleles on early events and CD4+ T cell dynamics in subjects from the WHMC HIV+ cohort. (a) Independent disease-modifying effects of HLA alleles. The plot shows the relative hazards (RH, diamonds) and 95% confidence interval (CI, error bars) for rates of progression to AIDS for the variables listed along the x-axis. Results are derived from the final model using stepwise multivariate Cox proportional hazards regression analyses for time to AIDS (1987 Centers for Disease Control criteria) and are based on data from 719 European- and African-American subjects from the WHMC HIV+ cohort. bCD4, baseline CD4+ T cell count; VL, steady-state viral load; nCD4, nadir CD4+ T cell count during HIV disease course; %CD4, percentage of baseline CD4+ T cell counts; these parameters as well as the therapy era and DTH skin test reactions are as described previously3. The RH (95% CI) for bCD4, nCD4 and %CD4 are 1.00 (0.99 – 1.00), 0.99 (0.99 – 0.99) and 0.97 (0.96 – 0.99), respectively. (b) Association of the HLA-B*57 allele (pink, present; black, absent) with parameters that reflect early immune damage (steady-state VL, baseline CD4, DTH responses) and cumulative CD4+ T cell counts. Diamonds and error bars represent the mean and 95% CI, and the P values were obtained using the Student's t-test. (c) Recovery of CD4+ T cell counts after initiation of HAART on the basis of possession of a HLA-B*57 allele (by nonlinear GEE). (d) Association of HLA class I allele frequency score groups (low score representing rare (orange) and high score representing common (green) class I HLA alleles) with the indicated parameters. (e) Recovery of CD4+ T cell counts after initiation of HAART on the basis of the HLA class I allele frequency score groups representative of rare and common HLA class I alleles (by nonlinear GEE).
In an alternative approach, we sought to capture the contribution of all HLA alleles, rather than focusing on specific alleles. This approach takes into account the prevailing viewpoint that a common class I allele confers a detrimental on HIV disease course, whereas a rare class I allele confers a beneficial effect31. We assigned each HIV+ subject an HLA allele frequency score that provided a measure of whether they possessed a common (high score) or rare (low score) HLA allele. In agreement with the prevailing viewpoint, a high score was associated with greater early immune damage and lower cumulative CD4+ counts than a low score (Fig. 3d). Despite this, after initiation of HAART, the CD4+ time trends according to HLA allele frequency score were similar (Fig. 3e), indicating that HLA alleles that mitigate early immune damage (for example, lower initial loss of CD4+ cells) do not necessarily confer salutary effects on CD4+ recovery during HAART.
Effects of GRGs on CD4+ recovery are independent of HLA
Although a direct effect of HLA alleles on CD4+ recovery was not detected, we could not exclude the possibility of gene-gene interactions between two biological systems (that is, CCL3L1-CCR5 and HLA) that influence early immune damage and, consequently, these interactions affecting CD4+ recovery. We therefore statistically adjusted the effects of GRGs on CD4+ recovery for the three HLA alleles that affected HIV disease progression and for HLA allele frequency score. These analyses showed that after 2 years of HAART, the influence of GRG on CD4+ recovery rates was independent of HLA alleles (Table 3).
DISCUSSION
By studying subjects who received HAART during chronic infection, we demonstrate that although both CCL3L1-CCR5 genotype and HLA alleles affect the extent of early immune damage and HIV disease progression independently of viral load (on the basis of results shown here and previously3), only variations in the CCL3L1-CCR5 axis influence recovery of CD4+ counts. This supports our hypothesis that particular genetic pathways (for example, the CCL3L1-CCR5 axis) have dominant effects on both pathogenic and reparative processes during HIV disease.
We also investigated subjects who received HAART during acute or early infection with the following supposition: if there is no relationship between CCL3L1-CCR5 genotype–dependent differences in early immune damage and the subsequent recovery of CD4+ counts during HAART, then initiation of therapy during this early phase would overshadow the impact of CCL3L1-CCR5 genotype on CD4+ gains. However, this was not the case. Hence, collectively, the results of this study emphasize that certain CCL3L1-CCR5 genotypes promote HIV pathogenesis despite viral load suppression by HAART during acute, early or chronic HIV disease; that is, they have an effect on pathogenesis that is independent of the viral load. This inference of a viral load–independent effect on HIV pathogenesis by CCL3L1-CCR5 genotypes amplifies and extends the conclusions of our previous study3. However, given the known effects of CCL3L1 dose and CCR5 genotypes on viral entry, we cannot definitively exclude the possibility that they also influence CD4+ reconstitution by affecting the viral infection–target cell component of the host-pathogen relationship, that is, by influencing the extent and rate of suppression of viral replication during HAART.
Our observations also underscore that HLA alleles that retard HIV disease course might have contrasting effects during HAART, conferring no effects on CD4+ cell recovery or possibly, as observed for the HLA-B*57 allele, a detrimental effect during the initial years of HAART. Recent findings by others concur with these inferences32,33. The reasons for why the beneficial effects of HLA alleles observed during untreated HIV disease are lost during HAART are unclear, but might relate in part to the following conceptual model. During untreated HIV disease, the magnitude of viral replication contributes to CD4+ cell depletion, and, in this scenario, HLA-dependent restriction of viral replication helps mitigate CD4+ cell loss. However, when viral replication is suppressed by HAART, the ability of a person's HLA alleles to recognize HIV peptides and mount an effective cytotoxic Tlymphocyte response may not be so consequential. Thus, in the latter scenario, HLA alleles that have protective effects on CD4+ cells in the context of viremia during untreated HIV disease might have minimal beneficial effects on CD4+ recovery during HAART. In contrast, by influencing viral replication, cell-mediated immunity, and processes that are independent of the viral load and cell-mediated immunity3, CCL3L1-CCR5 genotypes may affect the CD4+ cell loss observed before initiation of HAART and the CD4+ cell gain during HAART.
The differing effects of low and moderate or high CCL3L1-CCR5 GRGs on reconstitution of CD4+ counts were evident in subjects in whom HAART-induced suppression of viral replication was sufficient to sustain robust gains in CD4+ counts and was also present after accounting for potential gene-gene interactions with HLA alleles and other parameters that influence HIV disease course (for example, age, pre-HAART CD4+ cell count or viral load). These divergent effects suggest that a moderate or high CCL3L1-CCR5 GRG might serve as a genetic basis for the phenotype of incomplete immunologic reconstitution despite sustained viral suppression, whereas a low GRG might underlie the converse phenotype of partial immunologic-clinical stability or recovery in the face of incomplete or transient viral suppression. The clinical importance of identifying genetic factors that contribute to the former phenotype is underscored by the finding that impaired immune reconstitution despite HAART-induced viral load suppression is associated with an increased risk of both AIDS-related and non–AIDS-related morbidity and mortality10,11,34-38.
The genotype-phenotype associations reported here are robust because of several features of the study. First, it includes a large number of HIV-positive subjects representative of the general US population. Second, in our study cohorts, confounding factors such as unequal access to health care and loss of subjects to follow-up were minimized. Third, we found agreement between CCL3L1-CCR5 genotype and phenotype at multiple levels: similar genotype-dependent effects were observed whether HAART was given during acute, early or chronic infection; genotypes linked with robust recovery of CD4+ cell numbers also associate with improved CD4+ cell function during HAART3,27; and there is concordance in the genotypes that mitigate or promote CD4+ depletion during the pre-HAART era and those genotypes that associate with robust or impaired CD4+ recovery during the HAART era. Substantiating the last point, previous studies have found that an HIV disease–retarding genotype (heterozygosity for the CCR5-Δ32–containing HHG*2 haplotype25) has beneficial effects on CD4+ recovery, whereas an HIV disease–accelerating CCR5 genotype (homozygosity for CCR5 HHE/HHE25) has detrimental effects on CD4+ recovery during HAART39-42. However there is an important caveat: as the findings in these reports and our study are derived from cohorts from the US, the genotype-phenotype relationships observed herein might pertain specifically to clade B HIV-1–host interactions, and thus it it is possible that similar relationships might not be detected within the context of other host–non-B clade HIV settings.
Although CCL3L1 and CCR5 genotype affect CD4+ recovery across a spectrum of both high and low CD4+ counts at which HAART was initiated, it is remarkable that the greatest impact of these genotypes appears to be on sustaining the initial CD4+ gains after 2 years of HAART, especially in those individuals initiating HAART at <350 CD4+ cells/mm3. This is noteworthy because the GRG-dependent inflections in CD4+ gains between 2 and 3 years could be missed in clinical trials, which are typically of a shorter duration. The prominent beneficial effects associated with a low CCL3L1-CCR5 GRG on CD4+ recovery in HIV-positive individuals initiating HAART at <350 CD4+ cells/mm3 suggests that the magnitude of immune reconstitution may be more sensitive to the genetically determined (that is, CCL3L1-CCR5 genetic status) immunologic, rather than virologic, status of the infected host.
On the basis of these findings and those from our previous studies3,25,26, we propose that very soon after exposure to virus, a person's CCL3L1-CCR5 genotype serves as a rheostat, setting the balance between pathogenic and reparative processes that contribute, in part, to the observed wide heterogeneity in the dynamics of CD4+ cell counts and function not just during untreated HIV infection, but also during therapy. As this phenotypic pathway is genetically programmed during early stages of the disease3,25,26 and can persist during HAART, CCL3L1-CCR5 genotype–dependent HIV pathogenesis may continue in spite of therapy, resulting in impaired immune reconstitution. Thus, the current debate about when to initiate antiretroviral therapy43 might need to be redirected toward first assessing for whom therapy should be considered, on the basis of the host genotype. For HIV-positive individuals with ≥350 CD4+ cells/mm3, the current treatment guidelines44 recommend ‘consideration’ of therapy, and these guidelines provide some general criteria regarding which people might benefit the most from earlier initiation of HAART. We suggest that, given the poor immunologic response in individuals with a moderate or high CCL3L1-CCR5 GRG, this genetic parameter may also be a useful guide in deciding whether to initiate therapy in those with ≥350 CD4+ cells/mm3. Furthermore, our findings suggest that, for subjects with a moderate or high GRG in whom therapy is delayed, in addition to HAART, immunologically based adjunctive therapies (for example, recombinant interleukin-2 or recombinant interleukin-7)45 might need to be developed to diminish the risk of impaired durability of CD4+ responses. The observation that recovery of CD4+ counts during HAART is more sensitive to the copy number of CCL3L1 than to CCR5 genotype raises the possibility that CCL3L1 mimetics might be efficacious in promoting immune reconstitution. Finally, it may be useful to evaluate whether a subject's CCL3L1 gene dose and CCR5 genotype will affect their response to CCR5 blockers used in HIV therapy.
METHODS
Study cohorts
The characteristics of the WHMC cohort, a prospective observational cohort that is a component of the United States Military's Tri-Service AIDS Clinical Consortium Natural History Study, and the AIEDRP cohort are described previously3,27,46 and in the Supplementary Methods. Subjects within the WHMC and AIEDRP cohorts received HAART during chronic and acute-early infection, respectively. Subjects gave informed consent for these studies, and the study was approved by the Institutional Review Boards at the participating institutions (WHMC, University of California at San Diego and University of California at San Francisco).
Genotyping
Methods used to assess CCL3L1 copy number, CCR5 genotype and assign a low, moderate and high CCL3L1-CCR5 GRG are described previously3,27 and in the Supplementary Methods. Briefly, CCL3L1low indicates a CCL3L1 gene copy below the population-specific median and CCL3L1high indicates a gene copy number greater than or equal to the population-specific median27. The average gene dose in European Americans is two copies of CCL3L1, whereas it is three or four copies in HIV-positive and HIV-negative African Americans, respectively27. CCR5det indicates a genotype associated with a rapid rate of disease progression to AIDS and death, and CCR5nondet indicates all other CCR5 genotypes27. A low GRG is defined by possession of a high CCL3L1 gene dose plus nondetrimental CCR5 genotype (CCR5nondet), a moderate GRG is defined by a low CCL3L1 gene dose and nondetrimental CCR5 genotype or a high CCL3L1 gene dose and detrimental CCR5 genotype (CCR5det), and a high GRG is defined by a low CCL3L1 dose plus detrimental CCR5 genotype27 (schematic shown in Fig. 1d). The prevalence of a high and moderate GRG was 8% and 42%, respectively, in the WHMC HIV+ cohort3,27. Methods for genotyping of HLA alleles are described in the Supplementary Methods. Although several HLA allele scoring systems have been described33,47-50, we adapted one33, and the details are provided in the Supplementary Methods.
Outcomes
The primary outcome variables were changes in CD4+ T cell counts, plasma HIV-1 viral loads and time trends for these two laboratory parameters. Viral load suppression was defined as attainment of at least two viral load measurements that were below the assay-specific limits of detection on HAART. Sustained viral load suppression was defined as the attainment of at least two viral load measurements that were below the level of detection, and all subsequent viral load measurements were less than 500 copies/ml. Transient viral load suppression was defined as the attainment of at least two viral load measurements below the level of detection and at least one subsequent viral load measurement that exceeded 500 copies/ml on HAART. Other endpoints analyzed were parameters of early immune damage reflected by the baseline CD4+ T cell count, steady-state viral load and DTH skin test responses, as well as a parameter designated as the cumulative CD4+ T cell count, which reflects the extent of CD4+ T cell count changes during HIV disease; these parameters are described previously3 and in the Supplementary Methods.
Statistical analyses
Detailed statistical methods and the number of subjects and CD4+ T cell count or viral load measurements for the data shown herein are in the Supplementary Methods and Supplementary Note. The basis for conducting the linear regressions in two segments, that is, during the first 2 years after initiation of HAART and in the years thereafter, are also described in the Supplementary Methods.
Supplemental Material
ACKNOWLEDGMENTS
The people and funding agencies that made this work possible are listed in the Acknowledgements section in the Supplementary information online.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Douek DC, Picker LJ, Koup RA. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 2003;21:265–304. doi: 10.1146/annurev.immunol.21.120601.141053. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez B, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. J. Am. Med. Assoc. 2006;296:1498–1506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]
- 3.Dolan MJ, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry–independent mechanisms. Nat. Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 4.Moore RD, Keruly JC. CD4+ cell count 6 years after commencement of highly active antiretroviral therapy in persons with sustained virologic suppression. Clin. Infect. Dis. 2007;44:441–446. doi: 10.1086/510746. [DOI] [PubMed] [Google Scholar]
- 5.Grabar S, et al. Clinical outcome of patients with HIV-1 infection according to immunologic and virologic response after 6 months of highly active antiretroviral therapy. Ann. Intern. Med. 2000;133:401–410. doi: 10.7326/0003-4819-133-6-200009190-00007. [DOI] [PubMed] [Google Scholar]
- 6.Tarwater PM, et al. Increase and plateau of CD4 T-cell counts in the 3(1/2) years after initiation of potent antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 2001;27:168–175. doi: 10.1097/00126334-200106010-00012. [DOI] [PubMed] [Google Scholar]
- 7.Hunt PW, et al. Continued CD4 cell count increases in HIV-infected adults experiencing 4 years of viral suppression on antiretroviral therapy. AIDS. 2003;17:1907–1915. doi: 10.1097/00002030-200309050-00009. [DOI] [PubMed] [Google Scholar]
- 8.Moore DM, et al. Discordant immunologic and virologic responses to highly active antiretroviral therapy are associated with increased mortality and poor adherence to therapy. J. Acquir. Immune Defic. Syndr. 2005;40:288–293. doi: 10.1097/01.qai.0000182847.38098.d1. [DOI] [PubMed] [Google Scholar]
- 9.Valdez H, et al. Limited immune restoration after 3 years' suppression of HIV-1 replication in patients with moderately advanced disease. AIDS. 2002;16:1859–1866. doi: 10.1097/00002030-200209270-00002. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann GR, et al. Characteristics, determinants, and clinical relevance of CD4 T cell recovery to <500 cells/μl in HIV type 1–infected individuals receiving potent antiretroviral therapy. Clin. Infect. Dis. 2005;41:361–372. doi: 10.1086/431484. [DOI] [PubMed] [Google Scholar]
- 11.Podlekareva D, et al. Factors associated with the development of opportunistic infections in HIV-1–infected adults with high CD4+ cell counts: a EuroSIDA study. J. Infect. Dis. 2006;194:633–641. doi: 10.1086/506366. [DOI] [PubMed] [Google Scholar]
- 12.Mehandru S, et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med. 2006;3:e484. doi: 10.1371/journal.pmed.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deeks SG, Barbour JD, Martin JN, Swanson MS, Grant RM. Sustained CD4+ T cell response after virologic failure of protease inhibitor–based regimens in patients with human immunodeficiency virus infection. J. Infect. Dis. 2000;181:946–953. doi: 10.1086/315334. [DOI] [PubMed] [Google Scholar]
- 14.Ledergerber B, et al. Predictors of trend in CD4-positive T-cell count and mortality among HIV-1–infected individuals with virological failure to all three antiretroviral drug classes. Lancet. 2004;364:51–62. doi: 10.1016/S0140-6736(04)16589-6. [DOI] [PubMed] [Google Scholar]
- 15.Deeks SG. Durable HIV treatment benefit despite low-level viremia: reassessing definitions of success or failure. J. Am. Med. Assoc. 2001;286:224–226. doi: 10.1001/jama.286.2.224. [DOI] [PubMed] [Google Scholar]
- 16.Kaufmann GR, et al. CD4 T-lymphocyte recovery in individuals with advanced HIV-1 infection receiving potent antiretroviral therapy for 4 years: the Swiss HIV Cohort Study. Arch. Intern. Med. 2003;163:2187–2195. doi: 10.1001/archinte.163.18.2187. [DOI] [PubMed] [Google Scholar]
- 17.Garcia F, et al. Long-term CD4+ T-cell response to highly active antiretroviral therapy according to baseline CD4+ T-cell count. J. Acquir. Immune Defic. Syndr. 2004;36:702–713. doi: 10.1097/00126334-200406010-00007. [DOI] [PubMed] [Google Scholar]
- 18.Tenorio AR, et al. HIV-1–infected antiretroviral-treated patients with prolonged partial viral suppression: clinical, virologic, and immunologic course. J. Acquir. Immune Defic. Syndr. 2003;34:491–496. doi: 10.1097/00126334-200312150-00007. [DOI] [PubMed] [Google Scholar]
- 19.Anastos K, et al. The prognostic importance of changes in CD4+ cell count and HIV-1 RNA level in women after initiating highly active antiretroviral therapy. Ann. Intern. Med. 2004;140:256–264. doi: 10.7326/0003-4819-140-4-200402170-00007. [DOI] [PubMed] [Google Scholar]
- 20.Grabar S, et al. Response to highly active antiretroviral therapy at 6 months and long-term disease progression in HIV-1 infection. J. Acquir. Immune Defic. Syndr. 2005;39:284–292. doi: 10.1097/01.qai.0000160925.33935.72. [DOI] [PubMed] [Google Scholar]
- 21.Zhang ZQ, et al. The impact of early immune destruction on the kinetics of postacute viral replication in rhesus monkey infected with the simian-human immunodeficiency virus 89.6P. Virology. 2004;320:75–84. doi: 10.1016/j.virol.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Mellors JW, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 23.Lifson JD, et al. Containment of simian immunodeficiency virus infection: cellular immune responses and protection from rechallenge following transient postinoculation antiretroviral treatment. J. Virol. 2000;74:2584–2593. doi: 10.1128/jvi.74.6.2584-2593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nibbs RJ, Yang J, Landau NR, Mao JH, Graham GJ. LD78β, a non-allelic variant of human MIP-1α (LD78α), has enhanced receptor interactions and potent HIV suppressive activity. J. Biol. Chem. 1999;274:17478–17483. doi: 10.1074/jbc.274.25.17478. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez E, et al. Race-specific HIV-1 disease–modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. USA. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangano A, et al. Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus. J. Infect. Dis. 2001;183:1574–1585. doi: 10.1086/320705. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez E, et al. The influence of CCL3L1 gene–containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 28.Martin MP, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 29.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J. Infect. Dis. 2005;191(Suppl 1):S68–S77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 30.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu. Rev. Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen L, et al. Frequent human leukocyte antigen class I alleles are associated with higher viral load among HIV type 1 seroconverters in Thailand. J. Acquir. Immune Defic. Syndr. 2004;37:1318–1323. doi: 10.1097/01.qai.0000127059.98621.55. [DOI] [PubMed] [Google Scholar]
- 32.Rauch A, et al. HLA-Bw4 homozygosity is associated with an impaired CD4-T-cell recovery after initiation of antiretroviral therapy. Clin. Infect. Dis. doi: 10.1086/588479. in the press. [DOI] [PubMed] [Google Scholar]
- 33.Brumme ZL, et al. Effects of human leukocyte antigen class I genetic parameters on clinical outcomes and survival after initiation of highly active antiretroviral therapy. J. Infect. Dis. 2007;195:1694–1704. doi: 10.1086/516789. [DOI] [PubMed] [Google Scholar]
- 34.Moore DM, et al. Disease progression in patients with virological suppression in response to HAART is associated with the degree of immunological response. AIDS. 2006;20:371–377. doi: 10.1097/01.aids.0000196180.11293.9a. [DOI] [PubMed] [Google Scholar]
- 35.Weber R, et al. Liver-related deaths in persons infected with the human immunodeficiency virus: the D:A:D study. Arch. Intern. Med. 2006;166:1632–1641. doi: 10.1001/archinte.166.15.1632. [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez F, et al. Clinical Outcome of HIV-infected patients with sustained virologic response to antiretroviral therapy: long-term follow-up of a multicenter cohort. PLoS ONE. 2006;1:e89. doi: 10.1371/journal.pone.0000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Sadr WM, et al. CD4+ count–guided interruption of antiretroviral treatment. N. Engl. J. Med. 2006;355:2283–2296. doi: 10.1056/NEJMoa062360. [DOI] [PubMed] [Google Scholar]
- 38.Lewden C, et al. HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J. Acquir. Immune Defic. Syndr. 2007;46:72–77. doi: 10.1097/QAI.0b013e318134257a. [DOI] [PubMed] [Google Scholar]
- 39.Valdez H, Purvis SF, Lederman MM, Fillingame M, Zimmerman PA. Association of the CCR5Δ32 mutation with improved response to antiretroviral therapy. J. Am. Med. Assoc. 1999;282:734. doi: 10.1001/jama.282.8.734. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita TE, et al. Immunologic and virologic response to highly active antiretroviral therapy in the Multicenter AIDS Cohort Study. AIDS. 2001;15:735–746. doi: 10.1097/00002030-200104130-00009. [DOI] [PubMed] [Google Scholar]
- 41.Kasten S, et al. Positive influence of the Δ32CCR5 allele on response to highly active antiretroviral therapy (HAART) in HIV-1–infected patients. Eur. J. Med. Res. 2000;5:323–328. [PubMed] [Google Scholar]
- 42.O'Brien TR, et al. Effect of chemokine receptor gene polymorphisms on the response to potent antiretroviral therapy. AIDS. 2000;14:821–826. doi: 10.1097/00002030-200005050-00008. [DOI] [PubMed] [Google Scholar]
- 43.Holmberg SD, Palella FJ, Jr, Lichtenstein KA, Havlir DV. The case for earlier treatment of HIV infection. Clin. Infect. Dis. 2004;39:1699–1704. doi: 10.1086/425743. [DOI] [PubMed] [Google Scholar]
- 44.Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; Jan 29, 2008. Panel on Antiretroviral Guidelines for Adult and Adolescents; pp. 1–128. < http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGl.pdf>, accessed 29 February 2008. [Google Scholar]
- 45.Pahwa S. Role of common γ chain–utilizing cytokines for immune reconstitution in HIV infection. Immunol. Res. 2007;38:373–386. doi: 10.1007/s12026-007-0036-9. [DOI] [PubMed] [Google Scholar]
- 46.Hecht FM, et al. A multicenter observational study of the potential benefits of initiating combination antiretroviral therapy during acute HIV infection. J. Infect. Dis. 2006;194:725–733. doi: 10.1086/506616. [DOI] [PubMed] [Google Scholar]
- 47.Saah AJ, et al. Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. Multicenter AIDS Cohort Study. AIDS. 1998;12:2107–2113. doi: 10.1097/00002030-199816000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Kaslow RA, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 49.Tang J, et al. Host genetic profiles predict virological and immunological control of HIV-1 infection in adolescents. AIDS. 2002;16:2275–2284. doi: 10.1097/00002030-200211220-00007. [DOI] [PubMed] [Google Scholar]
- 50.Carrington M, Nelson G, O'Brien SJ. Considering genetic profiles in functional studies of immune responsiveness to HIV-1. Immunol. Lett. 2001;79:131–140. doi: 10.1016/s0165-2478(01)00275-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.