Abstract
Argonaute (Ago) 2 is the catalytic engine of mammalian RNA interference, but little is known concerning the regulation of Ago2 by cell-signaling pathways. In this study we show that expression of Ago2, but not Ago1, Ago3, or Ago4, is elevated in estrogen receptor (ER) α-negative (ERα−) vs. ERα-positive (ERα+) breast cancer cell lines, and in ERα− breast tumors. In MCF-7 cells the low level of Ago2 was found to be dependent upon active ERα/estrogen signaling. Interestingly, the high expression of Ago2 in ERα− cells was severely blunted by inhibition of the epidermal growth factor (EGF) receptor/MAPK signaling pathway, using either a pharmacological MAPK kinase inhibitor, U0126, or a small interfering RNA directed against EGF receptor. Half-life studies using cycloheximide indicated that EGF enhanced, whereas U0126 decreased, Ago2 protein stability. Furthermore, a proteosome inhibitor, MG132, blocked Ago2 protein turnover. The functional consequences of elevated Ago2 levels were examined by stable transfection of ERα+ MCF-7 cells with full-length and truncated forms of Ago2. The full-length Ago2 transfectants displayed enhanced proliferation, reduced cell-cell adhesion, and increased migratory ability, as shown by proliferation, homotypic aggregation, and wound healing assays, respectively. Overexpression of full-length Ago2, but not truncated forms of Ago2 or an empty vector control, reduced the levels of E-cadherin, β-catenin, and β-actin, as well as enhanced endogenous miR-206 activity. These data indicate that Ago2 is regulated at both the transcriptional and posttranslational level, and also implicate Ago2 and enhanced micro-RNA activity in the tumorigenic progression of breast cancer cell lines.
Argonaute-2 is elevated in ERα- breast cancer cells due to epidermal growth factor receptor/MAPK signaling, and overexpression of this gene induces a more transformed phenotype in ERα+ MCF-7 cells.
Micro-RNAs (miRNAs) are small (19–24 nucleotide) noncoding RNAs that mediate posttranscriptional gene silencing through specific base pairing with target mRNAs. Once precursor miRNAs are processed and exported to the cytosol, one strand of the resulting miRNA duplex (guide strand) is incorporated into Argonaute (Ago)-containing ribonucleoprotein complexes (1,2,3). These complexes are then directed to the 3′-untranslated region (UTR) of target mRNAs where imperfect base pairing to the respective miRNA induces gene silencing via translational repression or mRNA cleavage/destabilization (4,5,6). The process of mRNA cleavage is mediated by the endonuclease activity of Ago2 containing ribonucleoprotein complexes (7,8,9).
Ago2 is a member of a family of eight proteins in mammals, four of which are germ line specific (10). Ago proteins contain a P-element induced wimpy testis (PIWI) domain that can adopt a ribonuclease H fold with potentially innate endonuclease activity (11,12,13,14). However, Ago2 is the only Ago protein shown to mediate miRNA-dependent cleavage/degradation of target mRNAs in mammals. Recently, studies have also implicated functional roles for Ago2 independent of its endonuclease activity (15,16,17,18,19). Ago2 has been observed as diffuse within the cytoplasm, and localized to both processing bodies (P bodies) and the nucleus (20).
Recent studies indicate that Ago2 is required for a variety of developmental processes that occur in a tissue-specific manner. For instance, Ago2-null mice are embryonic lethal, potentially due to neural tube defects (12,21). Disruption of Ago2 leads to gastrulation arrest and uncontrolled mesoderm expansion in mice through overactive Fgf8 signaling (22). Ago2 also plays a role in hematopoiesis because bone marrow conditional Ago2−/− mice have defective B-cell differentiation (23). Given these findings, the fact that no functional redundancy exists between Ago2 and other Ago paralogs highlights the importance of Ago2-miRNA complexes within various developmental processes (24).
Recently, microarray expression studies have indicated that genes involved in miRNA biogenesis are dysregulated in various breast tumor types (25,26). In particular, expression of Ago2 is elevated in the estrogen receptor (ER) α-negative (ERα−) ERBB2-negative [human epidermal growth factor receptor 2-negative (HER2−)] epidermal growth factor (EGF) receptor-positive (EGFR+) basal-like breast tumor subtype. Furthermore, Ago2 expression also negatively correlates with ERα status in gene profiling studies performed on various breast tumor samples (27). Although these studies implicate Ago2 in more aggressive breast tumor types, to date no studies have determined whether Ago2 is involved specifically in the tumorigenic process.
Breast cancer is the second-leading cause of cancer-related deaths among women in the United States, and approximately one third of invasive breast carcinomas will be diagnosed as ERα− (28,29). In patients with ERα− breast tumors, treatment options are limited because established antihormone adjuvant therapies (i.e. tamoxifen and CYP19-aromatase inhibitors) are ineffective at ablating tumor growth (30). In addition, this tumor type is more transformed and proliferates independent of estrogenic-stimulated signaling, leading to poor clinical outcomes (29,30). Transcriptional profiling has allowed for the categorization of breast cancer into several subtypes. The two main ERα− subtypes are designated as either “basal-like” or “HER2,” which frequently overexpress the EGFR or the related ERRB2/HER2 protein, respectively. In this study we demonstrate that Ago2 is up-regulated by an EGFR/MAPK signaling pathway in ERα− breast cancer cell lines, whereas overexpression of Ago2 is sufficient to drive breast tumor progression in the ERα+ MCF-7 human breast adenocarcinoma cell line.
Materials and Methods
Cell lines and tissue samples
Tissue culture reagents were purchased from Invitrogen Corp. (Carlsbad, CA) and Sigma-Aldrich Corp. (St. Louis, MO). All breast cancer cell lines were obtained through American Type Culture Collection (Manassas, VA). Cell lines were maintained in DMEM/F12 supplemented with 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin for standard culture (serum). Cells cultured in hormone-depleted media (HDM) were grown with media supplemented with 10% charcoal-treated dextran-stripped FBS (HyClone, Logan, UT). Stably transfected cells were maintained in DMEM/F12, 10% FBS, and 250 μg/ml G418.
Human primary breast carcinomas (≥1.5 cm) were obtained from consenting patients under an institutional review board approved protocol within 5 min of surgical resection, and frozen in optimal cutting temperature media at −80 C. Cryosections were evaluated for areas of high tumor cell density via hematoxylin and eosin stain. Tissue cores were obtained with a 2-mm dermal punch, harvested for RNA via the RNAeasy Kit from QIAGEN, Inc. (Valencia, CA), and processed for reverse transcription and hybridization to Illumina’s Human-8 cDNA array according to the manufacturer’s protocol (Illumina Inc., San Diego, CA). Gene expression analysis was performed with Bead Studio software from Illumina using Rank invariant normalization. Individual gene values were used only if multiple probe signals had P values less than 0.05.
Luciferase reporter assays
Firefly luciferase reporter assays for miR-206 activity were performed on breast cancer cell lines essentially as described (31). Luciferase activity generated from the pIS-ERα-1 constructs were measured using the dual-luciferase reporter assay system in combination with a Renilla luciferase construct (pRL-TK), to control for transfection efficiency, from Promega Corp. (Madison, WI). Results were reported as the average relative luciferase activity from three independent experiments normalized to Renilla activity ± sem.
Chemical inhibitor and small interfering RNA (siRNA) treatments
Breast cancer cell lines were treated with various hormones and selective inhibitors, purchased from Tocris Bioscience (Ellisville, MO). Cells at 80% confluence were washed and switched to drug-containing media, which included 17β-estradiol (E2), propyl pyrazole triol (PPT), diarylpropionitrile (DPN), U0124, U0126, and/or EGF (Sigma-Aldrich). After 24 h cells were washed and retreated with the appropriate drug so cumulative effects could be determined at 48 h. Treatment duration was 12 h during studies using the MG132 compound. For siRNA experiments, MDA-MB-231 cells were transfected with EGFR siRNA (Dharmacon, Inc., Lafayette, CO), and protein lysates were isolated after 24 h.
Cycloheximide (CHX) stability studies
MDA-MB-231 cells at 80% confluency were treated with 200 μm CHX (Sigma-Aldrich) for 30 min (zero time point). Cells were then treated with 100 ng/ml EGF, 10 μm U0126, 10 μm PD153035, or any combination of the aforementioned, plus 200 μm CHX. At each time point (0, 0.5, 3, and 6 h) after drug treatments, cells were harvested for protein in 250 μl radioimmunoprecipitation assay (RIPA) buffer/well. Western blot analysis was performed on 15 μl sample, and densitometry readings of the Ago2-specific band were reported. These values were normalized to amount loaded on the gel, which is equivalent to total cell number, and reported as the mean of three independent experiments ± sem.
Quantitative real-time PCR detection assay
For mRNA and miRNA detection, real-time PCR assays were performed on 100 ng/μl total RNA isolated from various cell lines and treatment conditions, using TRIzol reagent from Invitrogen, as previously described (31). Primers used in this study are described in supplemental Table S1, which is published as supplemental data on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org. Data were analyzed using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA) and Microsoft Excel (Microsoft Corp., Redmond, WA). All experiments were performed in quadruplicate and normalized to either RPL-19 or 5s RNA. In some instances end-point RT-PCR assays were performed. For these assays the PCR products were resolved on a 2% agarose gel and detected by ethidium bromide.
Western blot analysis
Western blot analyses were performed essentially as described (31). Antibodies to Ago2 (1:2000) and ERα (1:000) from Millipore Corp. (Billerica, MA), ERβ (1:000) and EGFR (1:1500) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), E-cadherin (1:1000) and β-catenin (1:1000) from Transduction Laboratories (Lexington, KY), Ago1 (1:1000), β-actin (1:1000), and c-myc (1:500) from Abcam, Inc. (Cambridge, MA), α-tubulin (1:1000) from AbD Serotec (Raleigh, NC), and Ki67 (1:1000) from Dako Corp. (Carpinteria, CA) were used in conjunction with either antimouse, antirat, or antirabbit IgG horseradish peroxidase-conjugated secondary antibody, from Santa Cruz Biotechnology. Protein expression was detected via ECL Plus from GE Healthcare Bio-Sciences Corp. (Piscataway, NJ) and chemiluminescent film from Eastman Kodak Co. (Rochester, NY). Resultant band intensities were quantified using ImageJ software from National Institutes of Health (Bethesda, MD).
Generation of the MCF-7-Ago2 transfectants
All basic molecular biology and transfection reagents used to make the MCF-7-Ago2 transfectants were obtained from Invitrogen. MCF-7 cells were cultured to 90% confluency and transfected with pcDNA 3.1 expression constructs harboring either full-length Ago2 [MCF7-Ago2-wild type (WT)] or N-terminal-truncated mutant Ago2 (MCF-7-Ago2_167C and MCF-7-Ago2_408C) myc-tagged cDNAs (32), or pcDNA 3.1 alone (MCF-7-empty) as a control. After 24 h, cells were washed and cultured in medium containing 850 μg/ml G418 every 48 h for 3 wk. After selection, each pool of cells, five from each cell type/transfectant, were trypsinized and cultured in a six-well plate containing medium plus 250 μg/ml G418. The pooled transfectants were screened for Ago2 and myc-tagged expression by real-time PCR and Western blot analyses. Transfectants with 2-fold or more enrichment in Ago2 over parental MCF-7 cells (MCF-7-Par) were classified as Ago2high and characterized.
Trypan blue proliferation assays
The Trypan blue viability assays were performed on the five MCF-7 cell transfectants over the course of 96 h. After seeding in 12-well culture plates at 3 × 104 cells per well, the number of viable cells were determined every 24 h by trypsinization of cells, resuspension in growth medium, 1:4 dilution with 0.4% Trypan blue stain, and hemocytometer counts. Values generated are representative of three independent experiments and are reported as the average viable cell number ± sem.
Homotypic aggregation assay
Breast cancer cells and transfectants (1 × 105) were transferred to 1.5-ml microcentrifuge tubes containing 1 ml culture medium. Each tube was incubated at 37 C for 0.5 h with 0.5 μl 2 mg/ml Calcein-AM (Invitrogen). Samples were centrifuged at 460 × g for 10 min, washed with PBS, resuspended in 1 ml culture medium, and placed on a rocker for 1.5 h at 37 C. Five hundred microliters of 3.75% formaldehyde were added to each tube, and the cell mixture was transferred to three wells within a 12-well culture dish. Fluorescent and phase-contrast images of cell aggregates were obtained using a Zeiss Stemi SV11 stereomicroscope with an AttoArc HBO 100W power source (Carl Zeiss MicroImaging, Inc., Thornwood, NY), whereas quantification of aggregates was measured using a hemocytometer, where all nine grids were counted.
Wound healing/migration assay
Various breast cancer cells and transfectants were seeded into 24-well plates. Once cells reached confluence, a wound was made in the center of each well using a 200-μl pipette tip. At each time point after wound formation, cells were washed with PBS, fixed in 100% methanol, rinsed with PBS, and stained with 1% crystal violet. Images of each wound were captured on a Zeiss SV11 stereomicroscope from Carl Zeiss MicroImaging and imported into Adobe Photoshop 8.0 (Adobe Systems, Inc., San Jose, CA). Using the measure tool within the software, five measurements were taken of the distance between the two migrating fronts. Data were recorded as width of wound (mm) ± sem.
Statistical analysis
Values reported in all analyses were expressed as the mean ± sem, and densitometry readings from Western blots were reported as averages. Differences between treatments and/or groups were analyzed by ANOVA with either a Dunnett or Bonferroni multiple comparison after test using GraphPad Instat Software 3.0. Statistical significance was accepted at P < 0.05.
Results
Ago2 mRNA levels and protein expression are elevated in ERα− breast cancer cell lines
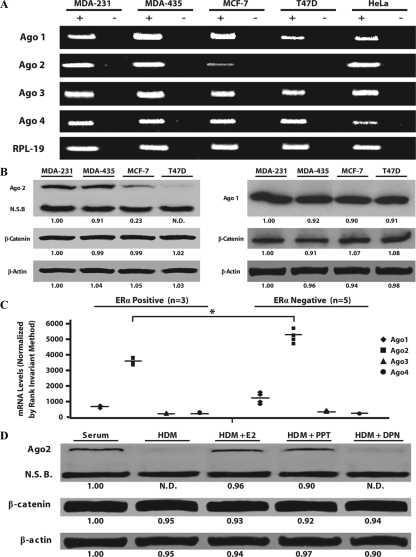
Ago2 expression was initially compared between ERα+ and ERα− breast cancer cell lines. Ago2 mRNA and protein levels were significantly higher in ERα− cell lines (MDA-MB-231 and MDA-MB-435) than ERα+ lines (T47D and MCF-7), whereas Ago1 protein levels remained unchanged in all cell lines tested (Fig. 1, A and B). In particular, MCF-7 cells had 4.4 ± 0.21-fold lower Ago2 protein levels, when compared with either of the ERα− cell lines. The T47D cells had undetectable levels of Ago2 expression, as assayed by Western blot and end-point PCR analyses.
Figure 1.
Ago2 expression is highly abundant in ERα− breast cancer cell lines. A, End-point RT-PCR of endogenous Ago1-4 and RPL-19 mRNA levels in the four breast cancer cell types, as well as HeLa cell lines. After 25 cycles (Ago1, Ago2, RPL-19) or 30 cycles (Ago3, Ago4) of amplification, the resultant band intensities for each cell type are depicted. Plus (+)/minus (−) symbols indicate plus or minus reverse transcriptase reactions. B, Western blot analysis of Ago2 protein expression in ERα+ (MCF-7 and T47D) and ERα− (MDA-MB-231 and MDA-MB-435) breast tumor cell lines. Values below each blot depict mean Ago2, β-catenin, and β-actin expression from four independent experiments, relative to MDA-MB-231 samples. C, Distribution plot of Ago1-4 mRNA levels in ERα+ and ERα− human breast tumors. Each point represents levels of a particular Ago within an individual sample after Rank invariant normalization. Horizontal bars denote mean Ago1-4 transcript levels within a data set (*, P < 0.005). D, Western blot analysis of Ago2, β-catenin, and β-actin expression in MCF-7 cells cultured in serum, HDM, or HDM with either 1.5 nm E2, 10 nm PPT, or 10 nm DPN for 48 h. Values below each blot represent mean band intensity from four independent experiments, and are reported as relative to the serum control. N.D., Nondetectable densitometric reading; N.S.B., 85-kDa nonspecific band detected by the Ago2 antibody.
To determine whether the inverse correlation between Ago2 levels and ERα positivity occurs in human breast tumors, we obtained data from a human-8 cDNA array performed on total RNA isolated from three ERα+ and five ERα− human breast tumors (Fig. 1C). Although the sample size was small, ERα− tumor samples had significantly higher Ago2 levels when compared with ERα+ tumor tissue (1.5 ± 0.05-fold; P < 0.005). Furthermore, expression of Ago1, Ago3, and Ago4 did not change within these tumors. Collectively, these findings demonstrate that Ago2 is selectively dysregulated between two breast tumor subtypes.
One hypothesis formed from these findings was that ERα/estrogen signaling suppressed Ago2 expression. However, E2 and the ERα-selective agonist, PPT, enhanced Ago2 levels in hormone-depleted MCF-7 cells (Fig. 1D).
Inhibition of EGFR/MAPK signaling reduces Ago2 expression in ERα− breast cancer cells
Two mechanisms by which Ago2 could be up-regulated in the ERα− breast cancer cells are through gene amplification and/or constitutive activation of a cell-signaling cascade. Semiquantitative end-point PCR performed on genomic DNA isolated from the four breast cancer cell lines revealed no evidence indicative of Ago2 gene amplification (data not shown).
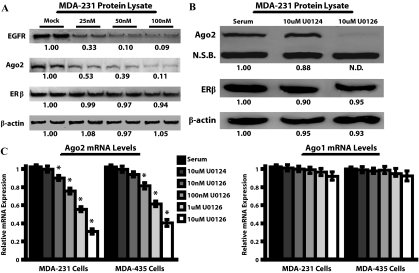
EGFR and the MAPK signaling cascade represent a major class of signal transduction pathways in ERα− breast cancers (33,34,35,36). To determine whether EGFR is required for Ago2 protein expression, Western blot experiments were performed on MDA-MB-231 cells treated with EGFR siRNA in the presence of serum-containing growth medium. After 24 h 100 nm small interfering EGFR (siEGFR) treatment, Ago2 expression was reduced 9.1 ± 0.25-fold, whereas β-actin and ERβ expression remained constant (Fig. 2A). Similarly, 100 ng/ml EGF for 48 h elevated Ago2 mRNA and protein levels approximately 1.5-fold (Fig. 3A).
Figure 2.
Abrogating MAPK signaling reduces Ago2 expression in MDA-MB-231 cells. A, MDA-MB-231 cells were transfected with 0–100 nm siEGFR for 24 h, and analyzed for Ago2, EGFR, ERβ, and β-actin protein expression via Western blot. The numbers below each band represent the mean densitometric readings from four independent experiments, relative to the serum control, which was set at 1.00. B, Western blot analysis of Ago2, ERβ, and β-actin protein expression in MDA-MB-231 cells treated with U0124, U0126, or serum alone for 48 h. N.D., Nondetectable densitometric reading; N.S.B., 85-kDa nonspecific band detected by the Ago2 antibody. C, MDA-MB-231 and MDA-MB-435 cell lines cultured in normal serum conditions were treated with 0–10 μm U0126, or 10 μm U0124 for 48 h, and Ago2 and Ago1 mRNA levels were assayed by real-time PCR. Values were normalized to RPL-19 and are reported as the mean + sem of three independent experimenters with five replicates per experiment, relative to serum-treated samples (*, P < 0.01, compared with serum control).
Figure 3.
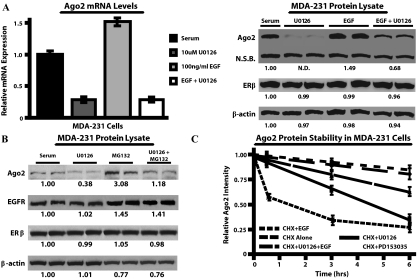
EGF enhances Ago2 protein stability through active EGFR/MAPK signaling. A, MDA-MB-231 cells were treated with 10 μm U0126 and/or 100 ng/ml EGF for 48 h, and Ago2 mRNA levels were assayed by real-time PCR (left panel). Values (mean + sem) were obtained from three experiments with five replicates per experiment, normalized to RPL-19, and depicted as relative to serum-treated samples. Western blot analysis of Ago2, ERβ, and β-actin protein expression in MDA-MB-231 cells under the same conditions as noted previously (right panel). N.D., Nondetectable densitometric reading; N.S.B., 85-kDa nonspecific band detected by the Ago2 antibody. B, MDA-MB-231 cells were treated with serum alone plus 10 μm MG132, or 10 μm U0126 plus 10 μm MG132, and analyzed for the expression of the proteins noted previously. For all Western blots, the numbers below the band represents the mean densitometric readings from four experiments, relative to the serum control. C, CHX stability studies were performed in MDA-MB-231 cells. Each point within a line graph represents the mean + sem for Ago2 expression from four independent experiments, normalized to cell number, and reported as relative to CHX alone at 0 h. At 3 h, PD153035 or U0126 significantly (P < 0.05) reduced Ago2 levels compared with U0126 plus EGF or CHX alone.
To ascertain whether EGF stimulated Ago2 expression through the MAPK pathway, ERα− breast cancer cells were treated with U0126, a MAPK kinase (MEK) inhibitor, or U0124, an inactive U0126 analog. Western blot analyses indicated that the U0124 compound had no effect on Ago2 protein levels, whereas, 48 h 10 μm U0126 treatment decreased Ago2 expression to nondetectable levels (Fig. 2B). Real-time PCR analysis of dose-response experiments indicated that 10 μm U0126 significantly reduced Ago2 transcript levels after 48 h by 3.3 ± 0.15-fold and 2.5 ± 0.21-fold in MDA-MB-231 and MDA-MB-435 cells, respectively (Fig. 2C). The effect of U0126 was specific, in that RPL-19 and Ago1 mRNA levels remained unchanged. In MDA-MB-231 cells, Ago2 mRNA levels were the same after treatment with either 100 ng/ml EGF plus 10 μm U0126, or with U0126 alone (Fig. 3A, left panel). These data indicate that the MAPK pathway serves as the primary mediator of EGF-stimulated induction of Ago2 gene expression in ERα− cells.
EGF enhances Ago2 protein stability through MAPK signaling in MDA-MB-231 cell lines
Results from Western blot analyses of the experiments described previously revealed that Ago2 protein expression did not correlate with transcript levels under certain conditions. Specifically, 100 ng/ml EGF plus 10 μm U0126 enhanced Ago2 expression at the protein level, but not at the mRNA level, compared with U0126 alone (Fig. 3A). Previous studies have shown that EGF, via EGFR, can promote protein stability through phosphorylation and/or the prevention of ubiquitin-mediated proteasomal degradation (37,38,39,40,41). To test the notion that Ago2 protein levels are regulated by the proteasome, we treated MDA-MB-231 cells with 10 μm MG132, a proteasomal inhibitor (Fig. 3B). Twelve hours after MG132 treatment, Ago2 protein levels were enhanced approximately 3.1-fold in cells pretreated with or without 10 μm U0126. These data indicate that in ERα− cells, EGF enhances Ago2 stability posttranslationally.
To test further that EGF-EGFR signaling increases Ago2 protein stability, CHX half-life studies were performed on EGF and/or U0126 treated MDA-MB-231 cells. After 6 h, Ago2 was reduced 2.5 ± 0.14-fold in CHX plus U0126 treated samples when compared with the CHX alone control (Fig. 3C). This reduction was ablated by 1.7 ± 0.12-fold in CHX, U0126, and EGF treated cells. The use of 10 μm PD153035, an EGFR inhibitor, confirmed that the loss of EGFR signaling leads to an immediate reduction, 1.7 ± 0.11-fold by 0.5 h, in Ago2 stability. These findings indicate that EGFR signaling, through an as of yet unidentified pathway, is essential for maintaining Ago2 protein expression and stability in MDA-MB-231 cells.
Generation and characterization of the Ago2 overexpressing (MCF-7-Ago2-WT) cell types
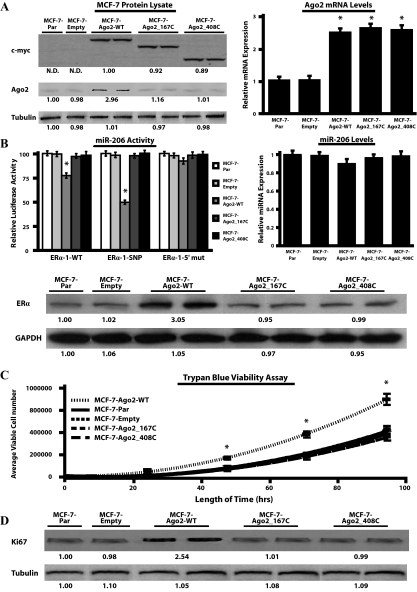
Because Ago2 is regulated by an oncogenic signaling cascade in ERα− cells, we examined whether increased Ago2 expression induced a transformed phenotype. Ago2 was stably overexpressed in the less transformed ERα+ MCF-7 cell line as described in Materials and Methods. For each of the Ago2 cell types generated, three of the five pooled MCF-7 cell transfectants were classified as having Ago2high expression (Fig. 4A). Specifically, MCF-7-Ago2-WT transfectants had 2.5 ± 0.11-fold and 2.9 ± 0.18-fold greater Ago2 mRNA and protein levels, respectively, compared with MCF-7-Par cells. Because the Ago2 antibody used in these screening experiments recognized the N-terminal region of Ago2, expression of the Ago2 N-terminal deletion mutants in the MCF-7 cells were determined by c-myc epitope-tag expression and Ago2 mRNA levels. Our analysis indicated that MCF-7-Ago2 mutants expressed Ago2 at levels comparable to the MCF-7-Ago2-WT cell types.
Figure 4.
Overexpression of Ago2 induces alterations in the phenotype of MCF-7 cells. A, MCF-7-Ago2 transfectants (MCF-7-Ago2-WT, MCF-7-Ago2_167C, and MCF-7-Ago2_408C) were screened for myc-epitope and Ago2 expression by Western blot analysis (left panel). The densitometry readings depicted below each Western blot represent the mean myc-epitope, Ago2, and tubulin expression. Real-time PCR was performed to monitor Ago2 transcript levels in the MCF-7 transfectants (right panel). Values were normalized to RPL-19 and reported as relative to MCF-7 parental (MCF-7-Par) cells. B, Luciferase assays using the pIS-ERα-1 constructs were performed to monitor miR-206 activity in the MCF-7-Ago2 transfectants (left panel), whereas real-time PCR was performed to determine mature miR-206 levels (right panel). Values from the luciferase and real-time PCR assays were normalized to Renilla luciferase or 5s RNA, respectively, and reported as relative to the levels in MCF-7-Par cells. Western blots were also performed to measure endogenous ERα expression in the various MCF-7 transfectants (bottom panel). C, The number of viable MCF-7 transfectants was determined by Trypan blue growth curves as described in Materials and Methods. D, Western blot analysis of Ki67 expression levels in the MCF-7 transfectants. Numbers below bands represent the average densitometric readings for Ki67 and tubulin expression relative to MCF-7-Par cells. For all assays three independent experiments were performed in quadruplicate, and values were reported as the mean + sem. Data generated from the Ago2 transfectants are representative of three stably selected pools of cells with equivalent WT or mutant Ago2 expression (*, P < 0.05, compared with MCF-7-Par control). GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; N.D., nondetectable densitometric reading; SNP, single nucleotide polymorphism.
We previously showed that miR-206 targets two sites (ERα-1 and ERα-2) within the 3′-UTR of the ERα mRNA (31). This was done by insertion of each predicted site into the 3′-UTR of the pIS-0 luciferase reporter construct. We also observed that miR-206 induced degradation of pIS-ERα-1 luciferase mRNA only in cells that contained endogenous Ago2 (supplemental Fig. S1). Based on these findings, cells that overexpress Ago2 protein would presumably have enhanced miR-206 activity. To test this, pIS-ERα-1 constructs were transiently transfected into the five MCF-7 cell types for 24 h to determine their miR-206 activity (Fig. 4B, left panel). In the MCF-7-Ago2-WT cells, miR-206 activity on pIS-ERα-1-WT and pIS-ERα-1-single nucleotide polymorphism constructs were enhanced by 1.3 ± 0.12-fold and 2.1 ± 0.13-fold, respectively, when compared with MCF-7-Par cells. No change in miR-206 activity was observed in cells transfected with the pIS-ERα-1-5′ mutant construct, or with any construct used in the MCF-7-empty and MCF-7-Ago2 mutant transfectants. Real-time PCR experiments on the five MCF-7 cell types confirmed that these results were due to changes in Ago2 expression and not miR-206 levels (Fig. 4B, right panel). Thus, MCF-7-Ago2 cells express elevated levels of functional Ago2, which correlates with enhanced miR-206 activity.
Elevated Ago2 levels did not result in reduced ERα expression (Fig. 4B, bottom panel). In fact, MCF-7-Ago2-WT transfectants had elevated ERα protein levels. This is discordant with the finding that Ago2 overexpression resulted in elevated activity of a miRNA, miR-206, that targets ERα, and indicates that there is a greater concentration of miRNAs that target proteins involved in ERα suppression than those that target ERα itself.
Trypan blue proliferation assays performed on the various MCF-7 transfectants indicated that after 96 h, the number of viable MCF-7-Ago2-WT cells was 1.9 ± 0.14-fold greater than the other four MCF-7 cell types (Fig. 4C). Further evidence of enhanced proliferation was obtained by Western blot analysis for the expression of Ki67 antigen, a cell-cycle related nuclear protein (Fig. 4D). The levels of Ki67 were enhanced 2.5 ± 0.12-fold in the MCF-7-Ago2-WT transfectants when compared with the other MCF-7 cell types. These findings suggest that the expression of functional Ago2 protein (i.e. miRNA-responsive) stimulates cell cycle progression in MCF-7 cells.
Ago2 expression correlates with a scattered phenotype and promotes loss of cell adhesion
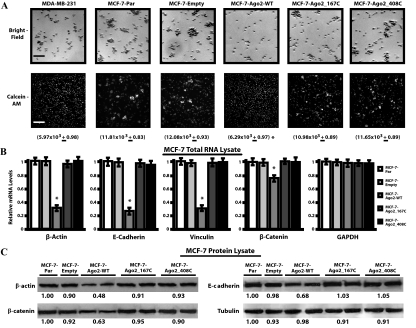
In addition to enhanced proliferation, the MCF-7-Ago2-WT transfectants displayed a distinct “scattered” phenotype not shared by any other MCF-7 cell type. Because MCF-7 cells normally grow in tight colonies, this “scattered” phenotype is indicative of reduced cell-cell adhesion. Homotypic aggregation assays performed to measure this reduction of functional cell-cell adhesion revealed that parental MCF-7, MCF-7-Ago2 mutant, and MCF-7-empty cell types readily formed cell aggregates, whereas the MCF-7-Ago2-WT cells mimicked the phenotype of the nonadherent MDA-MB-231 cells (Fig. 5A, top panel). This finding was verified using Calcein-AM, a fluorescent dye retained in metabolically active cells (Fig. 5A, bottom panel). To quantify the extent of cell aggregation, the number of aggregates was measured via Trypan blue exclusion and counted on a hemocytometer. On average, MCF-7-Ago2-WT cells generated half the number of aggregates compared with parental, empty vector control, or Ago mutant cells (6.3 × 103 ± 0.98 × 103 vs. ∼11.6 × 103 ± 0.88 × 103; P < 0.05), respectively. These results indicate the loss of cell-cell adhesion molecules in the MCF-7-Ago2-WT transfectants, which is an indicator of tumorigenic transformation.
Figure 5.
Ago2 reduces cell adhesion molecule expression and homotypic aggregation. A, Bright-field (top panel) and fluorescent Calcein-AM (bottom panel) images of fixed cell aggregates from a representative experiment are shown. Quantification of the homotypic aggregation assay was obtained from three independent experiments performed in duplicate, and values under each picture represent the average number of aggregates per ml of sample + sem. Scale bars represent a length of approximately 350 μm. B, Total RNA from the cell types mentioned previously was analyzed via real-time PCR for β-actin, E-cadherin, vinculin, β-catenin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. Values were normalized to RPL-19 and reported as the mean + sem, relative to the MCF-7-Par samples. C, Western blot analyses on the cell types noted previously. Each blot indicates the representative expression of β-actin, E-cadherin, β-catenin, and tubulin in each of the respective cell types. Values under each band represent the average densitometric intensity of protein expression relative to the MCF-7-Par cell line. Data collected from the Ago2 transfectants are representative of three stably selected pools of cells with equal WT or mutant Ago2 expression (*, P < 0.05, compared with MCF-7-Par control).
Functional cell-cell adhesion requires the coordinated regulation of a variety of adhesion molecules and associated structural, scaffolding, and signaling proteins. Therefore, Ago2 may be involved in directly silencing the E-cadherin cell-cell adhesion complex in MCF-7 cells. Real-time PCR assays indicated that β-actin, E-cadherin, vinculin, and β-catenin mRNA levels were reduced between 1.5- to 4.1-fold in MCF-7-Ago2-WT cells when compared with the other MCF-7 cell types (Fig. 5B). These values were normalized to RPL-19, which remained constant in all five cell types. Western blot analyses performed on the five MCF-7 cell types indicated that β-actin, E-cadherin, and β-catenin expression were reduced by 2.1 ± 0.12, 1.5 ± 0.14, and 1.6 ± 0.13-fold in the MCF-7-Ago2-WT cells, respectively, whereas tubulin remained unchanged (Fig. 5C). These data support the notion that functional Ago2 can dysregulate functional protein complexes by silencing the key proteins within a given pathway.
Overexpression of Ago2 in MCF-7 cell lines enhances their migratory/invasive properties
Although MCF-7 cells generally have poor invasive capabilities, the MCF-7-Ago2-WT cells had a transformed phenotype similar to the MDA-MB-231 cell line. To test whether Ago2 might enhance the migratory/invasive capabilities of the MCF-7 cell type, wound healing assays were performed over the course of 24 h (Table 1). MDA-MB-231 cells quickly occupied the wound by 72.8 ± 8.12% over the course of 24 h, whereas parental MCF-7 cells invaded the wound by only 14.1 ± 2.31%. However, the MCF-7-Ago2-WT transfectants were able to infiltrate significantly the wound area by 35.9 ± 4.56%, compared with the MCF-7-empty transfectants, which only had 17.9 ± 3.54% wound closure. This migratory phenotype was not observed in the MCF-7-Ago2_167C and MCF-7-Ago2_408C mutant transfectants. These data indicate that Ago2 enhances the migratory capabilities of MCF-7 breast cancer cells, potentially by enhancing the miRNA activity within this cell type.
Table 1.
Quantification of wound healing in stably transfected MCF-7-Ago2 cells
| Cell type | Time point | Width of the wound (mm)
|
Total wound closure (%) | |
|---|---|---|---|---|
| Average (n = 6) | ± sem | |||
| MDA-MB-231 | 0 h | 188.98 | 6.528 | |
| 8 h | 124.97 | 7.061 | ||
| 16 h | 90.42 | 7.315 | ||
| 24 h | 51.31 | 5.639 | 72.8 | |
| MCF-7-Par | 0 h | 189.48 | 10.52 | |
| 8 h | 184.41 | 11.63 | ||
| 16 h | 174.24 | 10.29 | ||
| 24 h | 162.81 | 15.01 | 14.1 | |
| MCF-7-empty | 0 h | 191.52 | 8.001 | |
| 8 h | 184.66 | 4.191 | ||
| 16 h | 172.47 | 2.845 | ||
| 24 h | 157.23 | 2.642 | 17.9 | |
| MCF-7-Ago2-WT | 0 h | 189.48 | 12.42 | |
| 8 h | 160.27 | 5.537 | ||
| 16 h | 137.92 | 4.267 | ||
| 24 h | 121.49 | 4.343 | 35.9 | |
| MCF-7-Ago2_167C | 0 h | 185.32 | 11.13 | |
| 8 h | 168.52 | 4.325 | ||
| 16 h | 161.73 | 3.451 | ||
| 24 h | 155.12 | 3.327 | 16.3 | |
| MCF-7-Ago2_408C | 0 h | 190.37 | 10.17 | |
| 8 h | 175.19 | 8.924 | ||
| 16 h | 165.23 | 6.174 | ||
| 24 h | 156.48 | 5.052 | 17.8 | |
The table depicts ″total wound closure″ for each cell type, as determined by relative distance encroached upon by invasive cells after 24 h. Each assay was performed three times with duplicates within each experiment and reported as the mean ± sem. Values from various Ago2 transfectants are representative of three pools of stable transfectants with equivalent WT or mutant Ago2 expression. At 16 h, wound closure was significantly (P < 0.05) greater in MCF-7-Ago2-WT as compared with MCF-7-Par cells.
Discussion
miRNAs have emerged as critical factors in the regulation of tumorigenesis. Some miRNAs (e.g. miR-21) are oncogenic, whereas others (e.g. let-7) function as tumor suppressors (42). Numerous miRNAs are differentially expressed in breast tumors. Some of these have been linked to general aspects of breast cancer biology (e.g. cell cycle traverse, metastasis, and apoptosis), and a smaller percentage has been identified as regulators of specific mRNAs in breast cancer cells (e.g. ERα, programmed cell death 4 protein, and HER2) (31,43,44,45). Despite the broad range of miRNA-induced phenotypes, all miRNAs studied to date assemble into Ago-miRNA-containing ribonucleoprotein complexes (7). It has also been shown that a subset of miRNA/mRNA hybrids containing imperfect base pairing can lead to the degradation of the targeted mRNA (46,47). Due to its innate catalytic slicer activity, the cleavage/degradation of target mRNAs by a miRNA requires Ago2 (7,12). As such, this unique action confers a centrally important role upon Ago2 in the biology of normal and transformed cells. Nevertheless, the study of whether the expression of Ago2 itself is regulated, and how changes in Ago2 expression impact normal physiology, development, and tumorigenesis, has largely remained unstudied.
The first major finding of this study was that mRNA levels of Ago2, but not of Agos 1, 3, or 4, were markedly elevated in ERα− breast cancer cells when compared with ERα+ cells, and Ago2 mRNA levels paralleled this change in protein expression. In addition, microarray analysis on a small panel of human breast tumors revealed a selective increase in Ago2 expression in ERα− tumors vs. ERα+ tumors. Our findings are consistent with a recent report on expression of miRNA biogenesis machinery in various breast cancer samples (26). In this microarray study, Ago2 expression was differentially up-regulated in human ERα− vs. ERα+ breast tumors, whereas other components such as Drosha and other Agos remained unchanged. Thus, elevated Ago2 expression correlates with a transformed phenotype in breast cancer cells, and raises the possibility that elevated Ago2 might augment tumorigenic potential by enhancing miRNA activity.
It is worth indicating that the lower levels of Ago2 in ERα+ cells are not due to repression by ERα. In fact, estrogen and ERα enhanced Ago2 expression. Although further work is needed to characterize fully the mechanism by which liganded ERα stimulates Ago2 expression, it is noteworthy that the Ago2 promoter harbors a putative estrogen-response element (48).
The second major finding of this study was that the elevated Ago2 expression is a response to the EGFR/MAPK signaling pathway. Overexpression and/or activation of the EGFR is a frequent finding in ERα− breast tumors (49). We observed that EGF stimulated Ago2 gene expression in MDA-MB-231 cells. Conversely, MDA-MB-231 cells treated with siEGFR or the MEK inhibitor, U0126, significantly reduced Ago2 expression. Therefore, transcriptional regulation of the Ago2 gene appears to require intact MAPK signaling downstream of EGFR. A recent transcriptional profiling study described the presence of a “MAPK signature” in breast cancer cells (50). This consisted of a subset of approximately 400 genes, whose transcription was either up-regulated or down-regulated by stable overexpression of EGFR, or constitutively active Erbb2, Raf, or MEK, in MCF-7 cells. We propose that Ago2 is a centrally important component of the “MAPK signature” in breast cancer cells, whose up-regulation is likely to affect the expression of numerous mRNAs through miRNA/Ago2-dependent mechanisms. Future studies will be aimed at the characterization of transcription factors downstream of the MAPK pathway (e.g. Ets, Jun/Fos) and their interaction with the promoter region of the Ago2 gene.
Combined U0126 and EGF treatments along with MG132 studies indicated that stabilization of Ago2 protein levels are determined, in part, by ubiquitination and proteasomal degradation. EGFR signaling has modified the stability of specific proteins via posttranslational mechanisms (51,52). Our findings suggest that a specific E3 ubiquitin ligase, acting as a tumor suppressor, targets Ago2 in an EGF-suppressible manner. Several E3 ligases have been implicated in breast cancer, both as oncogenes and tumor suppressors (53,54). Thus, Ago2 may be targeted by a breast cancer-related tumor-suppressive E3 ligase, such as BRCA1/BARD1 or CHIP, or by one of the over 500 E3 ligases currently identified.
Alternatively, EGF may increase the biological half-life of Ago2 through an effect on its subcellular localization. A recent study by Zeng et al. (55) demonstrated that Ago2 is phosphorylated on Ser-387 by p38 MAPK, which induced Ago2 localization to P bodies. If, in fact, EGF/MAPK signaling has a similar effect on Ago2, the sequestration to P bodies may contribute to an increased stability of the enzyme.
The third major finding of this study was that moderate overexpression of Ago2 produced a more transformed, less epithelial-like phenotype in MCF-7 cells. We also expressed two Ago2 mutant constructs that generate proteins incapable of mediating miRNA-dependent functions (32). Mutant MCF-7-Ago2_167C and MCF-7-Ago2_ 408C cells, expressed both myc-tagged Ago2 proteins and total (endogenous and exogenous) Ago2 mRNA levels comparable to the MCF-7-Ago-WT cells. In a previous study, we determined the efficiency of miR-206 to target ERα using heterologous luciferase reporters harboring two miR-206 target sites within the ERα 3′-UTR (31). Similar luciferase assays in the various MCF-7-Ago2 transfectants showed that the activity of miR-206 was elevated in MCF-7-Ago2-WT transfectants but remained unchanged in the MCF-7-Ago2 mutant cells. Despite the change in miR-206 activity, mature miR-206 levels remained constant within all MCF-7 cell types. This finding provides evidence that formation of Ago2-miRNA-containing ribonucleoprotein complexes are the rate-limiting factor for miR-206 function within MCF-7 cells, and emphasizes that the relative levels of a miRNA are not the sole determinant of activity.
Two hallmarks of enhanced tumorigenic progression of epithelial cells are the loss of cell-cell adhesion and enhanced cell motility. Both processes contribute to a transformed phenotype that resembles the epithelial-to-mesenchymal transition during development (56,57,58). MCF-7 cells are poorly tumorigenic in xenograft models, maintain an epithelial-like phenotype due to the presence of cell-cell adhesion molecules, grow in tight colonies, and are weakly motile. In contrast, MCF-7-Ago2-WT cells have a “scattered” phenotype reminiscent of the epithelial-to-mesenchymal transition process noted previously. This includes reduced cell-cell adhesion, lower levels of the adhesion molecules E-cadherin and β-catenin, and a greater degree of migratory capabilities as measured by the wound healing assay. Because none of the MCF-7-Ago2 mutant cells displayed these transformed phenotypes, the aforementioned findings indicate that enhanced Ago2 expression, along with increased miRNA activity, contribute to the greater degree of transformation within this cell type.
An unexpected finding was that elevated Ago2 associated with increased endogenous ERα. This suggests that miRNAs in MCF-7 cells actively target proteins involved in the suppression of ERα expression and/or promotion of the degradation of ERα protein. This enrichment in ERα may also aid in the transformation of the Ago2 transfectants.
Given the complexity of miRNA-mRNA interactions, it is surprising to observe such a clear phenotypical shift toward increased transformation in the Ago2 transfectants. One explanation may lie within the role of miRNAs to dampen transcriptional noise and stochastic variations in mRNAs and corresponding proteins that play a role in the differentiated function of a cell (59). At low levels, Ago2 would allow for such dampening and promote stability of a differentiated phenotype. However, upon elevation of Ago2, the “homeostatic” miRNAs could suppress those transcripts involved in a differentiated phenotype to an inappropriately low level, allowing oncogenic pathways to predominate. Ago2 could also promote transformation through miRNA-independent mechanisms. Further work is needed to profile miRNAs and characterize their targets/ physiological roles to test this supposition.
Supplementary Material
Acknowledgments
We thank Dr. Gregory Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for the c-myc-Argonaute-2 expression constructs, and Dr. Henry Furneaux for providing necessary reagents, and reviewing and providing insights for this manuscript. We also thank Drs. T. Hla, J. Tirnauer, L. Shapiro, and B. Graveley, Department of Cell Biology and the Translational Genomics Core at the University of Connecticut Health Center, for providing equipment, reagents, and data analysis for these studies. A special thanks to Ms. Catherine O'Conor for providing insight and ideas for the functional studies reported here.
Footnotes
This research was funded by Grant 1-R21-DK073456 from the National Institutes of Health. The costs of publication were also defrayed in part by the Carole & Ray Neag Comprehensive Cancer Center at the University of Connecticut Health Center.
Disclosure Summary: The authors have no conflicts of interest.
First Published Online September 11, 2008
Abbreviations: Ago, Argonaute; CHX, cycloheximide; DPN, diarylpropionitrile; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; E2, 17β-estradiol, ER, estrogen receptor; FBS, fetal bovine serum; HDM, hormone-depleted media; HER2, human epidermal growth factor receptor 2; MEK, MAPK kinase; miRNA, micro-RNA; PPT, propyl pyrazole triol; siEGFR, small interfering epidermal growth factor receptor; siRNA, small interfering RNA; UTR, untranslated region; WT, wild type.
References
- Meister G, Tuschl T 2004 Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS 2005 Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol 15:331–341 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004 The microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG 2006 MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103:4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W 2005 Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309:1573–1576 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W 2007 Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17:118–126 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004 Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15:185–197 [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G 2007 Argonaute proteins: mediators of RNA silencing. Mol Cell 26:611–623 [DOI] [PubMed] [Google Scholar]
- Parker JS, Barford D 2006 Argonaute: a scaffold for the function of short regulatory RNAs. Trends Biochem Sci 31:622–630 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ 2002 The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16:2733–2742 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L 2004 Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305:1434–1437 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004 Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441 [DOI] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ 2005 Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19:405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T 2002 Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563–574 [DOI] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Schedl P 2005 Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev 19:1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukioka H, Takahashi M, Mon H, Okano K, Mita K, Shimada T, Lee JM, Kawaguchi Y, Koga K, Kusakabe T 2006 Role of the silkworm argonaute2 homolog gene in double-strand break repair of extrachromosomal DNA. Nucleic Acids Res 34:1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM 2005 Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol 12:133–137 [DOI] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z 2005 Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA 11:849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z 2007 An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129:1141–1151 [DOI] [PubMed] [Google Scholar]
- Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G 2008 A multifunctional human Argonaute2-specific monoclonal antibody. RNA 14:1244–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I 2007 One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89:687–696 [DOI] [PubMed] [Google Scholar]
- Alisch RS, Jin P, Epstein M, Caspary T, Warren ST 2007 Argonaute2 is essential for mammalian gastrulation and proper mesoderm formation. PLoS Genet 3:e227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A 2007 A slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21:1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Natalin P, Stark A, Brennecke J, Cohen SM, Izaurralde E 2006 Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol 26:2965–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM 2006 The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7:96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavare S, Caldas C, Miska EA 2007 MicroRNA expression profiling of human breast cancer identifies new markers of tumour subtype. Genome Biol 8:R214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH 2002 Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ 2006 Cancer statistics, 2006. CA Cancer J Clin 56:106–130 [DOI] [PubMed] [Google Scholar]
- Giacinti L, Claudio PP, Lopez M, Giordano A 2006 Epigenetic information and estrogen receptor α expression in breast cancer. Oncologist 11:1–8 [DOI] [PubMed] [Google Scholar]
- Lapidus RG, Nass SJ, Davidson NE 1998 The loss of estrogen and progesterone receptor gene expression in human breast cancer. J Mammary Gland Biol Neoplasia 3:85–94 [DOI] [PubMed] [Google Scholar]
- Adams BD, Furneaux H, White B 2007 The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 21:1132–1147 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001 Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146–1150 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick SL, LaChance MP, Schultz GS 1984 Characterization of epidermal growth factor receptor and action on human breast cancer cells in culture. Cancer Res 44:3442–3447 [PubMed] [Google Scholar]
- Salomon DS, Brandt R, Ciardiello F, Normanno N 1995 Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 19:183–232 [DOI] [PubMed] [Google Scholar]
- Woodburn JR 1999 The epidermal growth factor receptor and its inhibition in cancer therapy. Pharmacol Ther 82:241–250 [DOI] [PubMed] [Google Scholar]
- McKay MM, Morrison DK 2007 Integrating signals from RTKs to ERK/MAPK. Oncogene 26:3113–3121 [DOI] [PubMed] [Google Scholar]
- Riedemann J, Takiguchi M, Sohail M, Macaulay VM 2007 The EGF receptor interacts with the type 1 IGF receptor and regulates its stability. Biochem Biophys Res Commun 355:707–714 [DOI] [PubMed] [Google Scholar]
- Daniel AR, Qiu M, Faivre EJ, Ostrander JH, Skildum A, Lange CA 2007 Linkage of progestin and epidermal growth factor signaling: phosphorylation of progesterone receptors mediates transcriptional hypersensitivity and increased ligand-independent breast cancer cell growth. Steroids 72:188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akca H, Tani M, Hishida T, Matsumoto S, Yokota J 2006 Activation of the AKT and STAT3 pathways and prolonged survival by a mutant EGFR in human lung cancer cells. Lung Cancer 54:25–33 [DOI] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, Hung MC 2006 Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol 8:1359–1368 [DOI] [PubMed] [Google Scholar]
- Bao J, Gur G, Yarden Y 2003 Src promotes destruction of c-Cbl: implications for oncogenic synergy between Src and growth factor receptors. Proc Natl Acad Sci USA 100:2438–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ 2006 Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 6:259–269 [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH 2008 Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283:1026–1033 [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY 2008 MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18:350–359 [DOI] [PubMed] [Google Scholar]
- Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S 2007 The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res 67:11001–11011 [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR 1984 Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226:409–416 [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE 2005 Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122:553–563 [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S 2004 Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol 18:1411–1427 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX 2001 Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2:127–137 [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D 2006 Activation of mitogen-activated protein kinase in estrogen receptor α-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor α-negative human breast tumors. Cancer Res 66:3903–3911 [DOI] [PubMed] [Google Scholar]
- DaSilva J, Xu L, Kim HJ, Miller WT, Bar-Sagi D 2006 Regulation of sprouty stability by Mnk1-dependent phosphorylation. Mol Cell Biol 26:1898–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenferink AE, Busse D, Flanagan WM, Yakes FM, Arteaga CL 2001 ErbB2/neu kinase modulates cellular p27(Kip1) and cyclin D1 through multiple signaling pathways. Cancer Res 61:6583–6591 [PubMed] [Google Scholar]
- Ohta T, Fukuda M 2004 Ubiquitin and breast cancer. Oncogene 23:2079–2088 [DOI] [PubMed] [Google Scholar]
- Chen C, Seth AK, Aplin AE 2006 Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol Cancer Res 4:695–707 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR 2008 Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J 413:429–436 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000 The hallmarks of cancer. Cell 100:57–70 [DOI] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C 2004 Tumor metastasis: a new twist on epithelial-mesenchymal transitions. Curr Biol 14:R719–R721 [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA 2004 Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927–939 [DOI] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J, Stark A 2006 Denoising feedback loops by thresholding—a new role for microRNAs. Genes Dev 20:2769–2772 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.