Abstract
Dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene 1 (DAX-1) is an orphan nuclear receptor that has been demonstrated to be instrumental to the expression of the steroidogenic acute regulatory (StAR) protein that regulates steroid biosynthesis in steroidogenic cells. However, its mechanism of action remains obscure. The present investigation was aimed at exploring the molecular involvement of DAX-1 in protein kinase A (PKA)- and protein kinase C (PKC)-mediated regulation of StAR expression and its concomitant impact on steroid synthesis using MA-10 mouse Leydig tumor cells. We demonstrate that activation of the PKA and PKC pathways, by a cAMP analog dibutyryl (Bu)2cAMP [(Bu)2cAMP] and phorbol 12-myristate 13-acetate (PMA), respectively, markedly decreased DAX-1 expression, an event that was inversely correlated with StAR protein, StAR mRNA, and progesterone levels. Notably, the suppression of DAX-1 requires de novo transcription and translation, suggesting that the effect of DAX-1 in regulating StAR expression is dynamic. Chromatin immunoprecipitation studies revealed the association of DAX-1 with the proximal but not the distal region of the StAR promoter, and both (Bu)2cAMP and PMA decreased in vivo DAX-1-DNA interactions. EMSA and reporter gene analyses demonstrated the functional integrity of this interaction by showing that DAX-1 binds to a DNA hairpin at position −44/−20 bp of the mouse StAR promoter and that the binding of DAX-1 to this region decreases progesterone synthesis by impairing transcription of the StAR gene. In support of this, targeted silencing of endogenous DAX-1 elevated basal, (Bu)2cAMP-, and PMA-stimulated StAR expression and progesterone synthesis. Transrepression of the StAR gene by DAX-1 was tightly associated with expression of the nuclear receptors Nur77 and steroidogenic factor-1, demonstrating these factors negatively modulate the steroidogenic response. These findings provide insight into the molecular events by which DAX-1 influences the PKA and PKC signaling pathways involved in the regulation of the StAR protein and steroidogenesis in mouse Leydig tumor cells.
The characterization of protein kinase A- and protein kinase C-mediated steroidogenic acute regulatory (StAR) expression and steroidogenesis suggests that the orphan nuclear receptor DAX-1 is an important regulator of the steroidogenic response in Leydig cells.
Dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene 1 (DAX-1; also called Nrob1), an unusual member of the nuclear hormone receptor superfamily of transcription factors, is expressed in the hypothalamic-pituitary-adrenal/gonadal axis and is involved in adrenal and gonadal development, differentiation, and function (1,2,3,4,5). The DAX-1 gene encodes a 470-amino acid protein, whose carboxy-terminal region is homologous to the ligand-binding domain of nuclear receptors, whereas its amino terminus contains a unique 3.5 repeats of a 65- to 67-amino acid sequence that functions as a DNA-binding domain (6,7,8). Mutations in the DAX-1 gene cause adrenal hypoplasia congenita associated with hypogonadotropic hypogonadism, an X-linked inherited disorder characterized by early onset of adrenal insufficiency syndrome (1,2,9). Disruption of DAX-1 in mice results in testicular dysgenesis, which is manifested by decreased testicular size, impaired Sertoli and Leydig cell function, and infertility (10,11). We and others have demonstrated that DAX-1 expression is affected by a plethora of extracellular signals that elevate the expression of the steroidogenic acute regulatory (StAR) protein (12,13,14,15,16,17). However, the precise role of DAX-1 in regulating StAR expression and, thus steroid biosynthesis, is poorly understood.
The 30-kDa mitochondrial StAR protein controls the rate-limiting and regulated step in steroidogenesis, i.e. the mobilization of cholesterol from the outer to the mitochondrial inner membrane in which it is metabolized to pregnenolone by the cytochrome P450 cholesterol side-chain cleavage enzyme (P450scc) (18,19,20,21,22). Regulation of the StAR protein in the adrenal glands and gonads is mediated by multiple signaling pathways, including protein kinase A (PKA) and protein kinase C (PKC), and involves transcriptional and posttranslational activation (21,23,24,25). Transcriptional and/or translational inhibition of StAR expression results in a dramatic decrease in steroid biosynthesis, whereas approximately 10–15% of steroid synthesis appears to be mediated through StAR-independent mechanisms (17,19,23,26). It should be noted, however, that the PKC-dependent induction of steroid synthesis is remarkably low when compared with PKA signaling, although it potentiates gonadotropin and/or cAMP-stimulated steroidogenic responsiveness. We recently demonstrated that the inability of PKC to phosphorylate StAR accounts for the differing steroidogenic responses between PKA and PKC (25,27). Indeed, at least two phosphorylation sites, Ser56/57 and Ser194/195 (in mouse and human, respectively), have been identified in the StAR protein, and mutation of these sites (Ser→Ala) demonstrated that the phosphorylation of Ser194/195 by PKA is necessary to render StAR fully active (28,29,30).
Whereas DAX-1 plays key roles in adrenal and gonadal development and function, it appears to do so by inhibiting the expression of several enzymes and genes that are functionally related to steroidogenesis, including StAR, steroidogenic factor 1 (SF-1), cytochrome P450scc, 3β-hydroxysteroid dehydrogenase, 17α-hydroxylase, Müllerian-inhibiting substance, and high-density lipoprotein (HDL) receptor (6,10,24,31,32,33,34,35). In most of these cases, the inhibitory action of DAX-1 is mediated indirectly rather than by directly binding to these genes. A substantial body of evidence indicates that SF-1 orchestrates the expression of several steroidogenic genes. SF-1 has been demonstrated to colocalize with DAX-1 in the hypothalamus, pituitary gonadotroph cells, adrenal glands, and gonads, and notably SF-1 and DAX-1 are both known to play significant roles in adrenal and gonadal development and function (36,37,38,39,40). Likewise, the nuclear receptor Nur77 (also called nerve growth factor-induced clone B) binds to the nerve growth factor-induced clone B response element (NBRE; 5′-AAAGGTCA-3′), a variant of the nuclear receptor half-site, an element that is similar to, but distinct from, the SF-1 binding motif (5′-CAAGGTCA-3′) and is involved in a large array of biological processes including StAR expression and steroidogenesis (41,42,43,44,45,46). Herein we report that DAX-1 binds, both in vivo and in vitro, to a putative DNA hairpin structure in the mouse StAR promoter. Additionally, we delineate novel mechanisms by which DAX-1 represses both PKA- and PKC-mediated expression of the StAR protein and steroidogenesis as well as serving to antagonize the effects of Nur77 and SF-1 in mouse Leydig tumor cells.
Materials and Methods
Plasmids, transfections, and luciferase assays
The 5′-flanking −151/−1- and −68/−1-bp regions of the mouse StAR promoter were synthesized using a PCR-based cloning strategy and inserted into the Xho1 and HindIII cloning sites of the pGL3 basic vector (Promega, Madison, WI) that contains firefly luciferase as a reporter gene (29,47). Using the −151/−1 StAR-pGL3 as template, mutation in the DAX-1 binding site was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The sense strand of the oligonucleotide sequence (mutated bases underlined lowercase letters) used was the following: 5′-GCACAGCCTTgCggccGcAGCATTTAAGGC-3′. Mutations generated in three SF-1 binding sites within the −151/−1-bp region of the mouse StAR promoter, SF-1/1 (−138/−133 bp), SF-1/2 (−46/−40 bp) and SF-1/3 (−102/−96 bp), have been described previously (48). All clones were verified by restriction mapping and confirmed by automated sequencing on a PE Applied Biosystems 310 genetic analyzer (ABI PRISM 310; PerkinElmer, Foster City, CA) at the Texas Tech University Biotechnology Core Facility. The pRL-SV40 plasmid was obtained from Promega. DAX-1, Nur77, and SF-1 expression plasmids have been previously described (24,32,42).
MA-10 mouse Leydig tumor cells (49), a generous gift from Dr. M. Ascoli (Department of Pharmacology, University of Iowa College of Medicine, Iowa City, IA), were maintained in Waymouth MB 752/1 medium supplemented with 15% horse serum and antibiotics (29,47). These cells were transfected using FuGENE HD-transfection reagent (Roche Diagnostics Corp., Indianapolis, IN) under optimized conditions (29,47,48). In brief, the −151/−1 StAR reporters (wild type and mutant) were transfected with different expression plasmids (1:1). The amount of DNA used in transfections was equalized with empty vector. Transfection efficiency was normalized by cotransfecting 10–20 ng pRL-SV40 vector (a plasmid that constitutively expresses renilla luciferase).
Transfection with small interfering RNAs (siRNAs) was performed using X-tremeGENE siRNA transfection reagent (Roche) under optimized conditions (50). Silencer negative control, and the DAX-1/NROB1 siRNAs (no. 1, 5′-CCAACACGACGCAGGAAAU-3′, and no. 2, 5′-GGGACCGUGCUCUUUAACC-3′) were obtained as annealed oligos (Ambion Inc., Austin, TX). The siRNAs were transfected using final concentrations ranging from 50 to 100 nm.
Luciferase activity in the cell lysates was determined by the dual-luciferase reporter assay system (Promega), as described previously (29,47,48). After treatments, cells were washed with 0.01 m PBS, and 300 μl of the reporter lysis buffer were added to the plates. Cellular debris was pelleted by centrifugation at 12,000 × g at 4 C, and the supernatant was measured for relative light units using a TD 20/20 luminometer (Turner Designs, Sunnyvale, CA).
Western blotting
Western blot analyses were carried out using total cellular protein (15,17,30). Briefly, cells were homogenized in lysis buffer, and the supernatant was assayed for total protein. Equal amounts of protein (20–30 μg) were solubilized in sample buffer and loaded onto either 10 or 12% SDS-PAGE (Mini Protean II system, Bio-Rad Laboratories, Inc., Hercules, CA) and electrophoresed as previously described (15,17,30). The proteins were electrophoretically transferred onto Immuno-Blot polyvinyl difluoride membranes (Bio-Rad). Membranes were probed with specific antibodies (Abs) that recognize DAX-1 (3), total StAR (51), phosphorylated StAR (P-StAR) (30), SF-1 (52), Nur77 (Cell Signaling Technology, Inc., Beverly, MA), P450scc (Chemicon International Inc., Temecula, CA), and actin (Santa Cruz Biotechnology, Santa Cruz, CA). After overnight incubation with primary Abs, the membranes were washed and incubated with appropriate secondary Abs for 1 h, washed again, and detection of the different proteins was determined with the chemiluminescence imaging Western Lightning kit (PerkinElmer). Membranes were exposed to x-ray films (Marsh Bio Products Inc., Rochester, NY), and the intensity of immunospecific bands was quantified using a computer-assisted image analyzer (Visage 2000; BioImage, Ann Arbor, MI). Detection of different proteins was assessed using identically processed membranes (in some cases StAR and DAX-1 were detected together); however, where appropriate, the same membranes were also analyzed by stripping and reprobing with additional antibodies.
Quantitative RT-PCR
Total RNA was extracted from different treatment groups using Trizol reagent (GIBCO-BRL, Grand Island, NY). Mouse StAR (18) and DAX-1 (GenBank accession no. NM_007430) were amplified using the following primer pairs: StAR sense, 5′-GACCTTGAAAGGCTCAGGAAGAAC-3′ and StAR antisense, 5′-TAGCTGAAGATGGACAGACTTGC-3′; DAX-1 sense, 5′-GCCGAGGGCCCCCTGGTGGGAC-3′ and DAX-1 antisense, 5′-TCCAGCATCATATCATCCATGCTGAC-3′, as described previously (17,24,32). The variation in RT-PCR efficiency was normalized using an L19 ribosomal protein gene, which was amplified using the following primer pairs: the sense, 5′-GAAATCGCCAATGCCAACTC-3′ and the antisense, 5′-TCTTAGACCTGCGAGCCTCA-3′. Using 2 μg of total RNA, reverse transcription and PCR were run sequentially in the same assay tube containing [α32P]dCTP in the deoxynucleotide triphosphate mixture (24,32). The cDNAs generated were further amplified by PCR using the primer pairs listed above. The molecular sizes of StAR, DAX-1, and L19 were determined on 1.2% agarose gels, which were then vacuum dried and exposed to x-ray film (Marsh Bio Products) for 1–5 h. The levels of StAR, DAX-1, and L19 signals were quantified using a computer-assisted image analyzer (Visage 2000).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using a kit (Upstate/Chemicon, Temecula, CA) as per the manufacturer’s instructions, and carried out under optimized conditions (53). In brief, cells were incubated with formaldehyde (1%) for 10 min at 37 C to cross-link DNA and its associated proteins. Cells were collected, resuspended in lysis buffer, and then sonicated with seven to nine cycles of 10-sec pulses using a Tekmar sonic disruptor (Fisher Scientific, Pittsburgh, PA). The supernatant containing chromatin was cleared with protein A agarose/salmon sperm DNA 50% slurry for 30 min at 4 C with agitation. After centrifugation, the supernatant was immunoprecipitated with IgG, 4–5 μg of Abs to DAX-1 (as above), or phosphorylated cAMP response element-binding protein (P-CREB; Cell Signaling Technology) for 14 h at 4 C, and followed by incubation with protein A agarose/salmon sperm for an additional 1 h. The chromatin-antibody-protein A agarose complexes were subsequently washed with low salt, high salt, LiCl, and Tris/EDTA buffers. Protein-DNA complexes were eluted with freshly made elution buffer (1% sodium dodecyl sulfate, 0.1 m NaHCO3). NaCl was added (to a final concentration of 20 mm) to the eluates that were then incubated at 65 C for 4 h to reverse the formaldehyde cross-linking. The resulting samples were treated with proteinase K for 1 h at 45 C, and the purified DNA samples were analyzed by a radioactive PCR approach using [α32P]dCTP in the deoxynucleotide triphosphate mixture (53). PCR was performed with 75–100 ng of DNA and the proximal mouse StAR promoter primers (forward, 5′-CTGGTCCTCCCTTTACACAGTC-3′ and reverse, 5′-GGCGCAGATCCAGTGCGCTGC-3′), spanning bases −170/−149 and −21/−1, respectively (54). A second primer combination [forward, 5′-GTGAGGACAGCTCATACGTGCAC-3′ (bases −3596/−3574) and reverse, 5′-GAACAGGCTTAAGTTAAGACTCC-3′ (bases −3450/−3428)] recognizing the distal region of the mouse StAR promoter was used as a negative control. PCR products were separated on 2% agarose gels. Gels were vacuum dried, exposed to x-ray film (Marsh Bio Products) for 1–5 h, and the resulting signals quantified (Visage 2000).
EMSA
EMSA reactions were performed using nuclear extracts (NE) and in vitro-transcribed/translated DAX-1 (29,47,53). The oligonucleotide probes were engineered and synthesized by heating sense and antisense primers to 65 C for 5 min in annealing buffer [10 mm Tris-HCl, 100 mm NaCl, 1 mm EDTA (pH 7.5)], followed by cooling at room temperature. The sense strands of the oligonucleotide sequences [mutated (Mut) bases in underlined lowercase letters] used were: DAX-1 wild-type (−48/−16 bp), 5′-GGGCACAGCCTTCCACGGGAAGCATTTAAGGCAGCG-3′; DAX-1 Mut, 5′-GGGCACAGCCTTgCggccGcAGCATTTAAGGCAGCG-3′; SF-1 binding site (−48/−35 bp of the mouse StAR promoter), 5′-GGCACAGCCTTCCACG-3′ (48); and consensus NBRE, 5′-GATCTTCGTGCGAAAAGGTCAAGCGCTAG-3′ (55). The specific doubled-stranded oligonucleotides were end labeled with (α32P)-dCTP (PerkinElmer Life Sciences) using Klenow fill-in reaction and purified using spin columns (29,47). NE (12–15 μg) and in vitro-translated DAX-1 (2–5 μg) were incubated for 15 min at room temperature in a 20-μl reaction buffer [25 mm Tris-HCl, 1 mm EDTA, 4% Ficoll, 10 mm dithiothreitol, 2 μg poly dIdC, 40 ng/μl BSA, and 12 mm MgCl2 (pH 7.9)] before the addition of a 32P-labeled probe either alone or in the presence of unlabeled oligonucleotide. When Abs were used, binding reactions were carried out for an additional 45 min on ice before addition of the labeled DNA. The reaction mixture was then subjected to electrophoresis at 200 V for 2 h through a 5% polyacrylamide gel in 0.5× a buffer of 90 mmol/liter Tris-borate, 2 mmol/liter EDTA (pH 8.0). The gels were dried and DNA-protein complexes were visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Statistical analysis
Statistical analysis was performed by ANOVA using Statview (Abacus Concepts Inc., Berkeley, CA) followed by Fisher’s protected least significant differences test. Data represent the mean ± se, and P < 0.05 was considered statistically significant.
Results
Involvement of the PKA and PKC pathways in DAX-1 expression and its relevance to StAR and steroid synthesis
The role of DAX-1 in PKA- and PKC-mediated regulation of StAR expression and progesterone synthesis was investigated. MA-10 cells were treated with dibutyryl cAMP [(Bu)2cAMP; 0.1 and 0.5 mm] or phorbol 12-myristate 13-acetate (PMA; 1 and 10 nm) for 6 h to activate the PKA and PKC pathways, respectively. As illustrated in Fig 1, the expression of DAX-1 protein was attenuated to approximately 50 to 70% of controls by (Bu)2cAMP and PMA. Lower doses of (Bu)2cAMP and PMA, ineffective at inducing StAR protein levels, were also capable of decreasing DAX-1 expression (Fig. 1, A and B). The decrease in DAX-1 levels by (Bu)2cAMP and PMA was found to correlate with an increase in StAR protein expression. Induction of StAR protein levels in response to (Bu)2cAMP (0.5 mm) and PMA (10 nm) was 8.6 ± 1.5 and 8.4 ± 1.3-fold over untreated cells, respectively. However, PMA had no effect on P-StAR, an indispensable requirement for optimal steroid biosynthesis (17,25,28,30). Conversely, (Bu)2cAMP (0.5 mm) strongly increased levels of P-StAR. Accumulation of progesterone in the media was 9.8 ± 1.5 and 423 ± 29-fold with (Bu)2cAMP (0.1 and 0.5 mm, respectively), and 5.4 ± 0.7-fold with PMA (10 nm) over untreated (1.8 ± 0.2) cells (Fig. 1C and inset). PMA (1 nm) had no apparent effect on progesterone synthesis. These findings show the impact of PKA and PKC activation on directing StAR expression and also the dependence of steroidogenesis on the phosphorylation of StAR by PKA.
Figure 1.
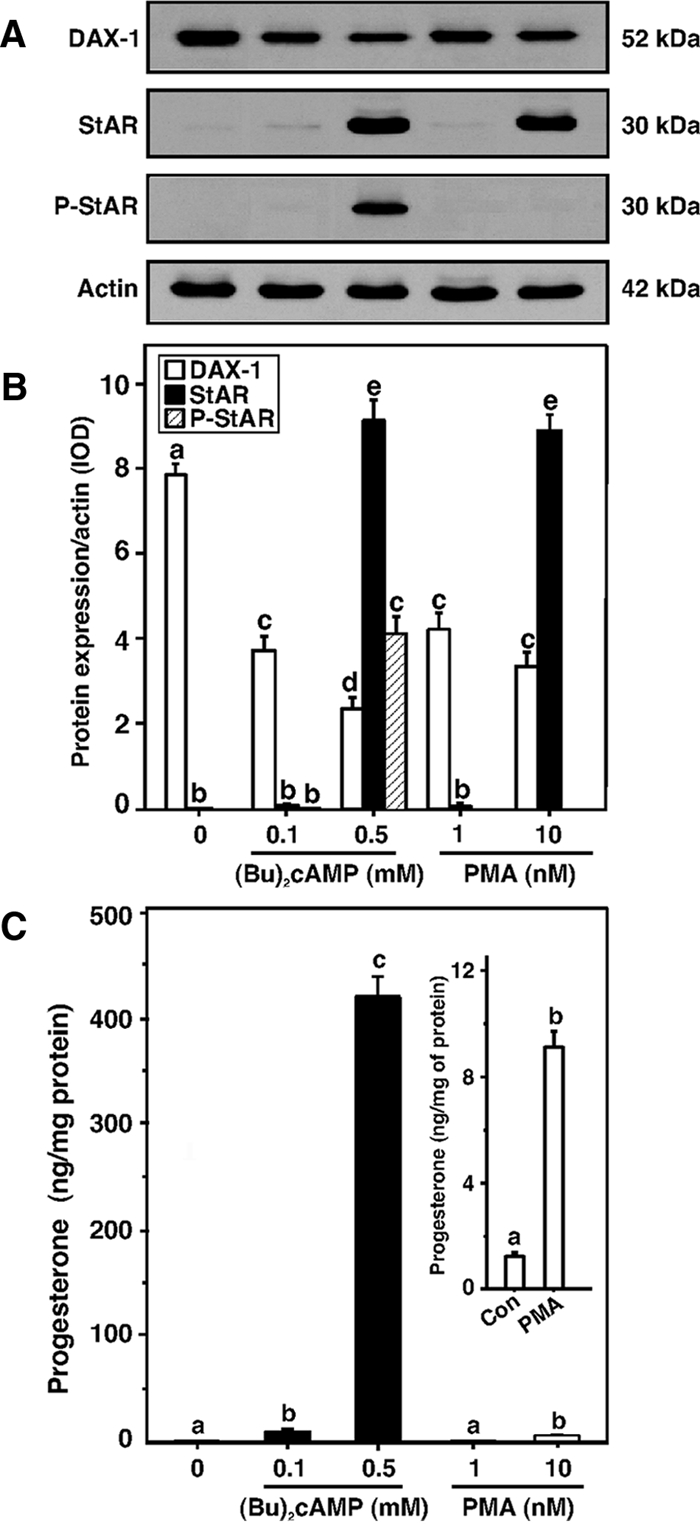
Role of PKA and PKC signaling in DAX-1, StAR, P-StAR, and progesterone levels in MA-10 cells. Cells were treated with two different concentrations of (Bu)2cAMP (0.1 and 0.5 mm) and PMA (1 and 10 nm), respectively, and subjected to preparation of whole cell lysates for Western blot analysis using 20–30 μg of protein. Representative immunoblots illustrate expression of DAX-1, StAR, and P-StAR in different treatment groups (A). Integrated OD (IOD) values of each band were quantified and compiled data (n = 4) are presented (B). C, Accumulation of progesterone in the media of the same treatment groups was determined and expressed as nanograms per milligram protein. Inset, Control (Con) and PMA (10 nm)-treated progesterone levels. Data represent the mean ± se of four independent experiments. Letters above the bars indicate that these groups differ significantly from each other, at least at P < 0.05.
The inhibition of PKA activity by H89 (0–25 μm) (17,25,30) attenuated (Bu)2cAMP (0.5 mm)-induced StAR protein and P-StAR levels, which in contrast, significantly reversed (P < 0.01) DAX-1 expression in a dose-dependent manner (Fig. 2A). H89 (25 μm) strongly decreased (≥90%) (Bu)2cAMP-stimulated StAR, P-StAR, and progesterone synthesis (not illustrated). Similarly, an inhibitor of PKC, GF-109203X (GFX; 0–25 μm) (17,25,30), decreased PMA (10 nm)-treated StAR protein levels, and P-StAR remained undetectable (Fig. 2B). GFX (5 μm) markedly diminished (P < 0.01) StAR protein expression. However, GFX used in conjunction with PMA blocked the decrease in DAX-1 protein levels. These results demonstrate the differential effects of PKA and PKC on the steroidogenic response and its inverse correlation to DAX-1 expression.
Figure 2.
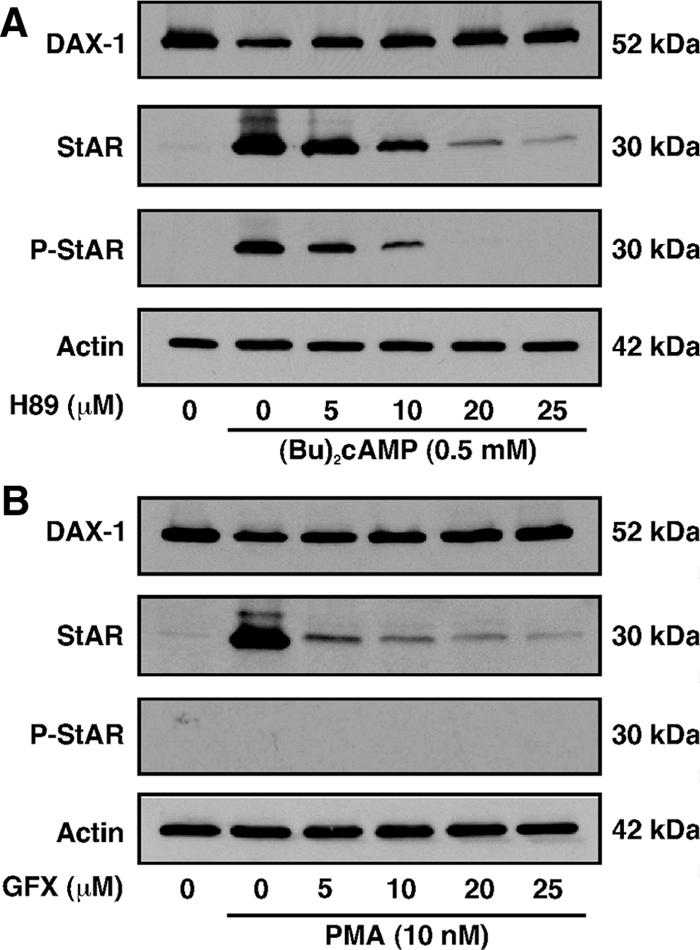
Inhibition of PKA and PKC signaling on DAX-1, StAR, and P-StAR levels in MA-10 cells. Cells were pretreated with increasing doses PKA (H89; 0–25 μm) and PKC (GFX; 0–25 μm) inhibitors for 30 min and then incubated without or with (Bu)2cAMP (0.5 mm; A) and PMA (10 nm; B) for an additional 6 h. Actin expression was assessed as a loading control (A and B). Representative immunoblots illustrate expression of DAX-1, StAR, and P-StAR using 25–30 μg of total protein. Data are representative of three independent experiments.
The results presented in Fig. 3 show the time kinetics (0–24 h) of (Bu)2cAMP- (0.5 mm) and PMA (10 nm)-mediated StAR and DAX-1 protein expression and progesterone synthesis in MA-10 cells. (Bu)2cAMP significantly increased (P < 0.05) StAR protein and progesterone levels by 1 h, elevated them maximally between 4 and 8 h, and both declined thereafter between 12 and 24 h (Fig. 3, A, C, and D). The expression of DAX-1 protein was decreased by 3 h (P < 0.05) and continued to decline with time (up to 24 h). The effects of treatment with PMA on StAR and DAX-1 levels were similar to that seen with (Bu)2cAMP (Fig 3), with PMA induction being slightly slower than that seen with the cAMP analog, significantly increasing (P < 0.05) StAR and progesterone levels by 2 and 3 h, respectively. Progesterone levels after PMA treatment for 6 h were in the neighborhood of 1–2% of those generated in response to (Bu)2cAMP. The time-response patterns of StAR and DAX-1 mRNA levels by (Bu)2cAMP and PMA were qualitatively similar (data not shown, and Refs. 29,30). These findings corroborate the results presented in Figs. 1 and 2 and reinforce the inverse relationship between DAX-1 and StAR in different steroidogenic cells (12,13,14,15,16,17).
Figure 3.
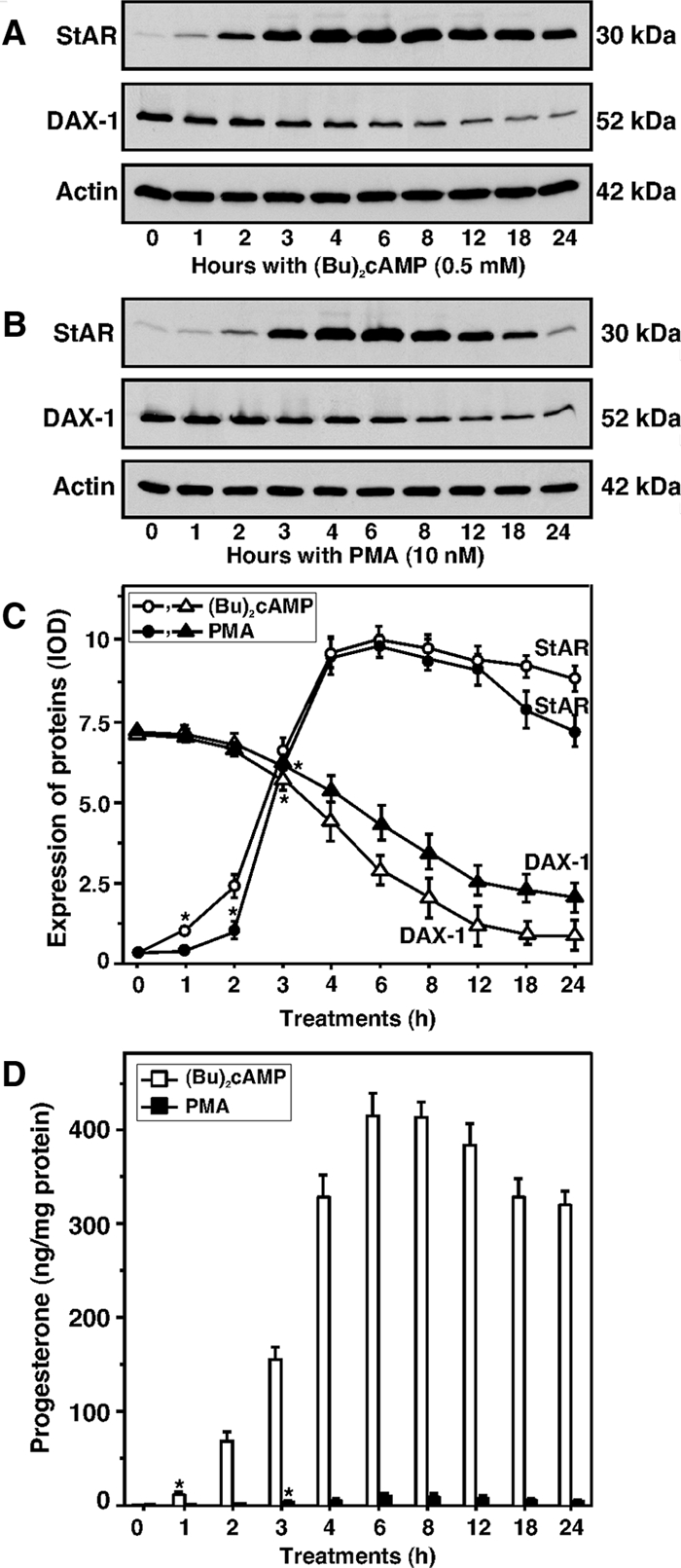
Time-course expression patterns of StAR and DAX-1 proteins and progesterone synthesis in response to (Bu)2cAMP (0.5 mm) and PMA (10 nm). MA-10 cells were treated for 0–24 h at the indicated doses of (Bu)2cAMP (A) and PMA (B), and 22–25 μg of total protein were used for Western blot analyses. Representative immunoblots illustrate the expression of StAR and DAX-1 in different treatment groups. Integrated OD (IOD) values for each band were quantified and compiled data from three independent experiments for both proteins are presented (C). Actin expression was assessed as a loading control. Accumulation of progesterone in the media of the same treatment groups was determined and expressed as nanograms per milligram protein, which represent the mean ± se of three independent experiments (D). Asterisks indicate the first time point at which values were significantly different (P < 0.05) from controls.
Repression of DAX-1 by PKA and PKC requires transcription and de novo protein synthesis
To determine the nature of DAX-1 suppression by PKA and PKC signaling, MA-10 cells were treated with (Bu)2cAMP (0.5 mm) and PMA (10 nm) for 6 h, and transcription and translation were subsequently inhibited by coincubation with actinomycin D (Act; 5 μm) and cycloheximide (Chx; 5 μm), respectively. The results presented in Fig. 4 (A and B) demonstrate that (Bu)2cAMP and PMA decreased (P < 0.01) the expression of DAX-1 mRNA, responses that were essentially reversed by either Act or Chx. Act and Chx alone had no significant effects on DAX-1 expression. Notably, the decreases in DAX-1 mRNA were found to be associated with increases in progesterone levels in the media (Fig. 4B). These data indicate that attenuation of DAX-1 in response to PKA and PKC requires transcription and translation of a regulatory factor(s) involved in DAX-1 gene transcription or degradation of DAX-1 mRNA in MA-10 mouse Leydig tumor cells.
Figure 4.
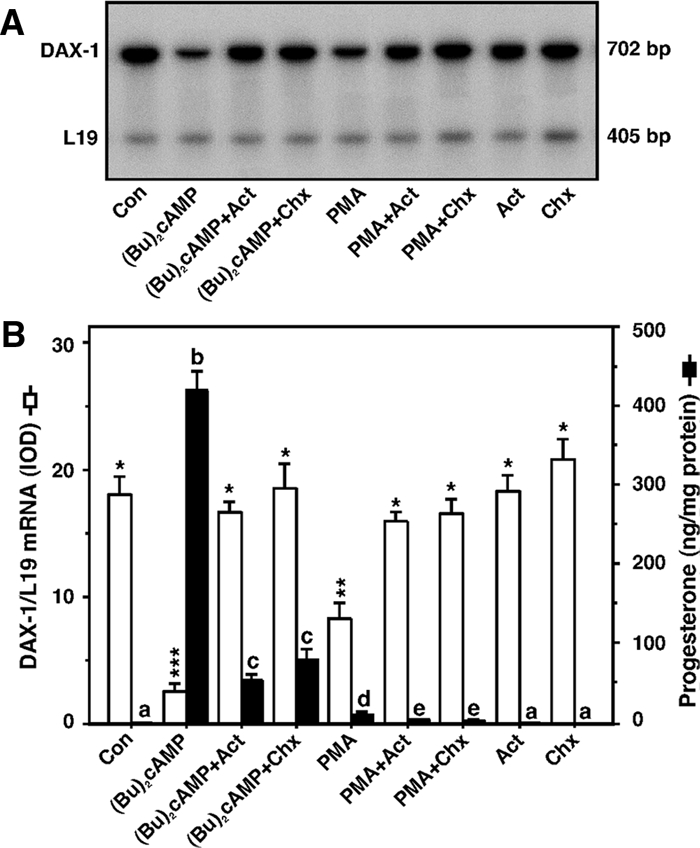
Inhibition of DAX-1 expression by PKA and PKC signaling requires transcription and de novo protein synthesis. MA-10 cells were treated without (Con) or with (Bu)2cAMP (0.5 mm) and PMA (10 nm) in the absence or presence of Act (5 μm) and Chx (5 μm) for 6 h and subjected to DAX-1 mRNA expression by RT-PCR analysis (A). A representative autoradiogram illustrates the expression of DAX-1 and L19 in different treatment groups using 2 μg of total RNA. Integrated OD (IOD) values of each band were quantified and normalized to the corresponding L19 bands (B). Accumulation of progesterone in the media was determined and expressed as nanograms per milligram protein (B). Results represent the mean ± se of three independent experiments. Note the different scales on the graph. Letters above the bars indicate that these groups differ significantly from each other at least at P < 0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. control DAX-1 mRNA.
Interaction of DAX-1 with the StAR proximal promoter represses StAR gene expression
Because the repression of DAX-1 is associated with an elevation of StAR expression and steroid synthesis, the mechanism of its regulation by PKA and PKC signaling is of considerable interest. We have previously demonstrated that the proximal −151/−1-bp region of the mouse StAR promoter, which contains the recognition motifs for several trans-regulatory factors, is critical in transcription of the StAR gene (29,47,53,56). ChIP analyses revealed that DAX-1 associates with the proximal StAR promoter (−170/−1 bp) and that both (Bu)2cAMP (0–1.0 mm, Fig. 5A) and PMA (0–20 nm, Fig. 5B) dose-dependently suppressed protein-DNA interactions. No association of DAX-1 with the distal StAR promoter (−3596/−3428 bp) was observed (Fig. 5C), nor was any DAX-1 binding seen with DNA immunoprecipitated with IgG. As a positive control, we examined the association of P-CREB with the proximal StAR promoter (Fig. 5D), and P-CREB association was found to increase in response to (Bu)2cAMP (53). These results demonstrate the specificity of the DAX-1-DNA interaction with the proximal StAR promoter and that the PKA and PKC signaling diminish this interaction.
Figure 5.
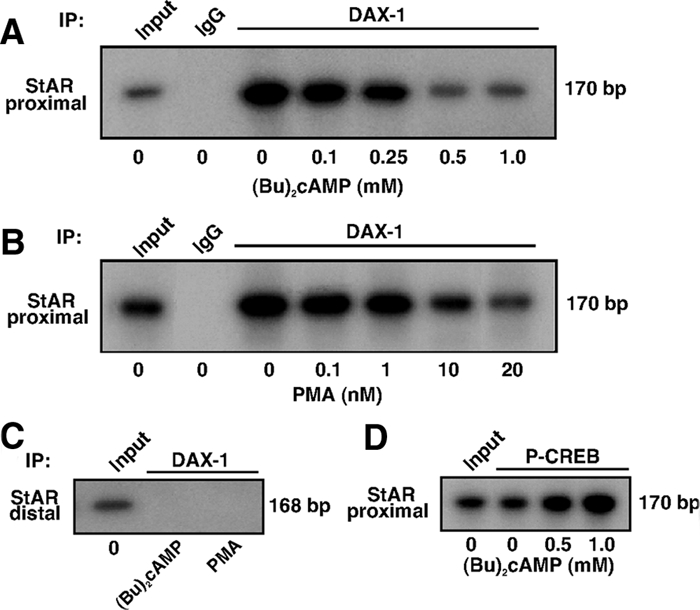
Effects of PKA and PKC on the association of DAX-1 with the StAR promoter. MA-10 cells were treated without or with increasing doses of (Bu)2cAMP (0–1.0 mm, A) and PMA (0–20 nm, B) for 6 h, and ChIP assays were carried out as described in Materials and Methods. Cross-linked sheared chromatin obtained from (Bu)2cAMP and PMA-treated cells was immunoprecipitated (IP) with either IgG or anti-DAX-1 and anti-P-CREB (D) Abs. Recovered chromatin was subjected to PCR analysis using primers (−170/−1 bp, A, B, and D) encompassing the DAX-1 binding site or primers (3596/−3428 bp, C) approximately 3500 bp upstream of the DAX-1 region. Representative autoradiograms illustrate the association of DAX-1 and P-CREB with the StAR promoter in response to (Bu)2cAMP and PMA. Data shown are representative of three independent experiments.
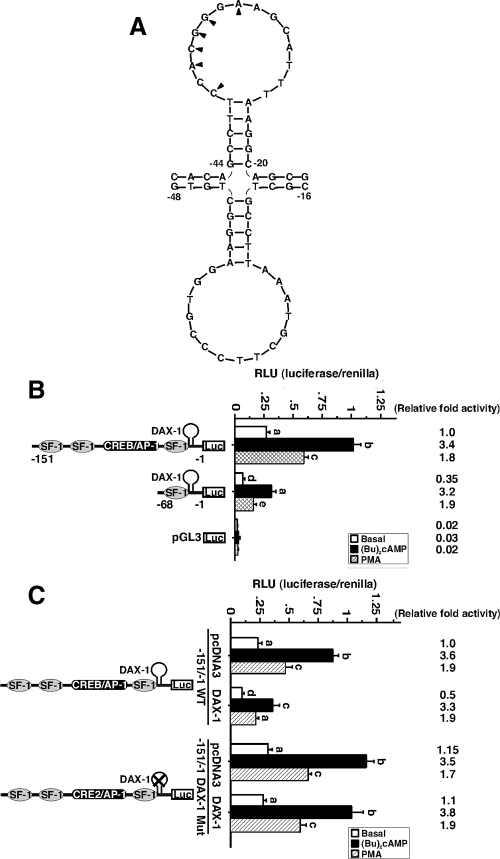
Closer analysis of the proximal region in the mouse StAR promoter predicted a DNA hairpin structure (Fig. 6A) at position −44/−20 bp (containing a stem of five nucleotides and a loop of 15 nucleotides; evaluated using Mfold software, (57)], and thus, the role of this hairpin on DAX-1 binding was analyzed. MA-10 cells transfected with the −151/−1 StAR reporter segment demonstrated 3.4 ± 0.5- and 1.8 ± 0.3-fold increases in luciferase activity by (Bu)2cAMP (0.5 mm) and PMA (10 nm), respectively, over untreated cells (Fig. 6B). Deletion of the 5′-flanking region to −68/−1 bp (which still contains the hairpin loop), diminished (P < 0.05) basal and consequently (Bu)2cAMP and PMA mediated StAR reporter activity without affecting fold responsiveness, when compared with the −151 construct. Overexpression of DAX-1, in the presence of the −151/−1 bp reporter, decreased basal promoter activity approximately 50% but did not affect (Bu)2cAMP- and PMA-mediated fold activity (Fig. 6C). The −151/−1 bp construct was chosen based on higher reporter responses in comparison with the −68/−1-bp StAR segment. The introduction of mutations in the hairpin loop resulted in a StAR promoter that was no longer inhibited by DAX-1 but rather was modestly increased (∼15%) in basal and (Bu)2cAMP-/PMA-mediated StAR gene expression (Fig. 6C). These findings point to the functional importance of the −44/−20 bp motif in recruiting DAX-1.
Figure 6.
Identification of a DNA hairpin structure in the mouse StAR promoter and the effect of DAX-1 on PKA- and PKC-mediated StAR promoter activity. A, Presence of a putative DAX-1 binding motif in the mouse StAR gene. This motif is located between −44 and −20 bp and is composed of five nucleotides stem and 15 nucleotides loop. B, MA-10 cells were transfected with different StAR reporter plasmids (−151/−1 and −68/−1 bp) in the presence of 20 ng of pRL-SV40 vector (renilla luciferase for determining transfection efficiency). Schematic representations of different StAR reporters (−151 and −68 bp) are shown (bottom panel). pGL3 basic (pGL3) was used as a control. Cells were also transfected with empty vector (pcDNA3) or DAX-1 expression plasmid, within the context of the −151/−1 wild-type and mutant StAR reporter segments as indicated, in the presence of pRL-SV40 (C). After 36 h of transfection, cells were incubated for a further 6 h in the absence (basal) or presence of either (Bu)2cAMP (0.5 mm) or PMA (10 nm), and luciferase activity in the cell lysates was determined and expressed as relative light units (RLU, luciferase/renilla). Relative fold activity is illustrated above bar diagrams. Data represent the mean ± se of four independent experiments. Letters above the bars indicate that these groups differ significantly from each other at least at P < 0.05.
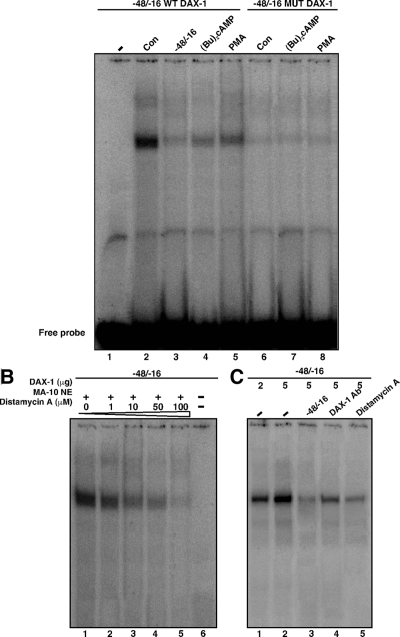
To obtain more insight into these mechanisms, the ability of the predicted hairpin structure to bind DAX-1 was studied using EMSA. As illustrated in Fig. 7A, a 32P-labeled probe (−48/−16 bp) encompassing the hairpin loop binds to a protein in MA-10 NE, and this DNA-protein complex was nearly abolished with cold competitor (Fig. 7A; compare lanes 2 and 3). NE obtained from (Bu)2cAMP- (lane 4) and PMA (Fig. 7A, lane 5)-treated MA-10 cells contained a significantly diminished (P < 0.01) DNA-protein complex. Importantly, protein-DNA binding was reduced when the probe was mutated such that the loop would be disrupted (Fig. 7A, lanes 6–8), suggesting the importance of this loop in DAX-1 binding. Figure 7B (lanes 1–5) demonstrated that the binding of MA-10 NE to the −48/−16 bp was inhibited with the minor groove binder, distamycin A, in a concentration-dependent manner (0–100 μm), suggesting this compound affects DNA conformation (6,58). The hairpin loop was also able to bind in vitro-translated DAX-1 (Fig. 7C, lanes 1–5), and the resulting protein-DNA complex was further enhanced with an increasing amount of DAX-1 (Fig. 7C; compare lanes 1 and 2). The binding of in vitro-translated DAX-1 to the −48/−16 was also strongly inhibited by DAX-1 Ab and distamycin A (Fig. 7C; compare lanes 2, 4, and 5). These data demonstrate that DAX-1 specifically binds to the DNA hairpin structure and represses StAR gene transcription.
Figure 7.
The binding of the hairpin region in the StAR promoter to endogenous DAX-1 in MA-10 NE and in vitro-translated DAX-1 using EMSAs. MA-10 NE (12–15 μg) obtained from control (Con, lanes 2, 3, and 6), 0.5 mm (Bu)2cAMP (lanes 4 and 7), and 10 nm PMA (lanes 5 and 8)-treated cells were incubated with the 32P-labeled wild-type (WT) and Mut DAX-1 probes (−48/−16 bp) encompassing the hairpin loop (A) as described in Materials and Methods. The specificity of DAX-1 binding (−48/−16 bp) in untreated MA-10 NE (lanes 1–6) was also assessed with increasing amounts (1–100 μm) of distamycin A (B). Binding of in vitro-translated DAX-1 protein (lanes 1–5) was evaluated with the −48/−16 probe (C). Protein-DNA binding was augmented with an increasing amount of DAX-1 (lanes 1–5). Binding was challenged with cold competitor (lane 3), DAX-1 Ab (lane 4), and distamycin A (lane 5). Cold competitor (−48/−16 bp) was used at 100-fold molar excess. Migration of free probes is shown in A. These experiments were repeated three times, and representative phosphor images from each group are illustrated.
Silencing of DAX-1 increases StAR expression and steroidogenesis
Given the complex interplay between PKA/PKC signaling and DAX-1 function, the suppressive effects of DAX-1 on StAR expression and progesterone synthesis were studied by silencing DAX-1 in MA-10 cells. Cells transfected with 100 nm of either of two DAX-1-specific siRNAs (see Materials and Methods) diminished DAX-1 expression (not illustrated). However, cells transfected with 50 nm of each siRNA (100 nm total) drastically decreased (80–90%) DAX-1 protein and DAX-1 mRNA levels (Fig. 8). As shown in Western (Fig. 8A) and RT-PCR (Fig. 8B) analyses, (Bu)2cAMP (0.5 mm) and PMA (10 nm) treatments decreased (P < 0.01) DAX-1 expression in cells transfected with a negative control siRNA. Interestingly, the silencing of DAX-1 augmented basal and (Bu)2cAMP/PMA mediated StAR expression and progesterone synthesis between 1.6- and 3.8-fold (Fig. 8C), demonstrating further that DAX-1 decreases the steroidogenic response.
Figure 8.
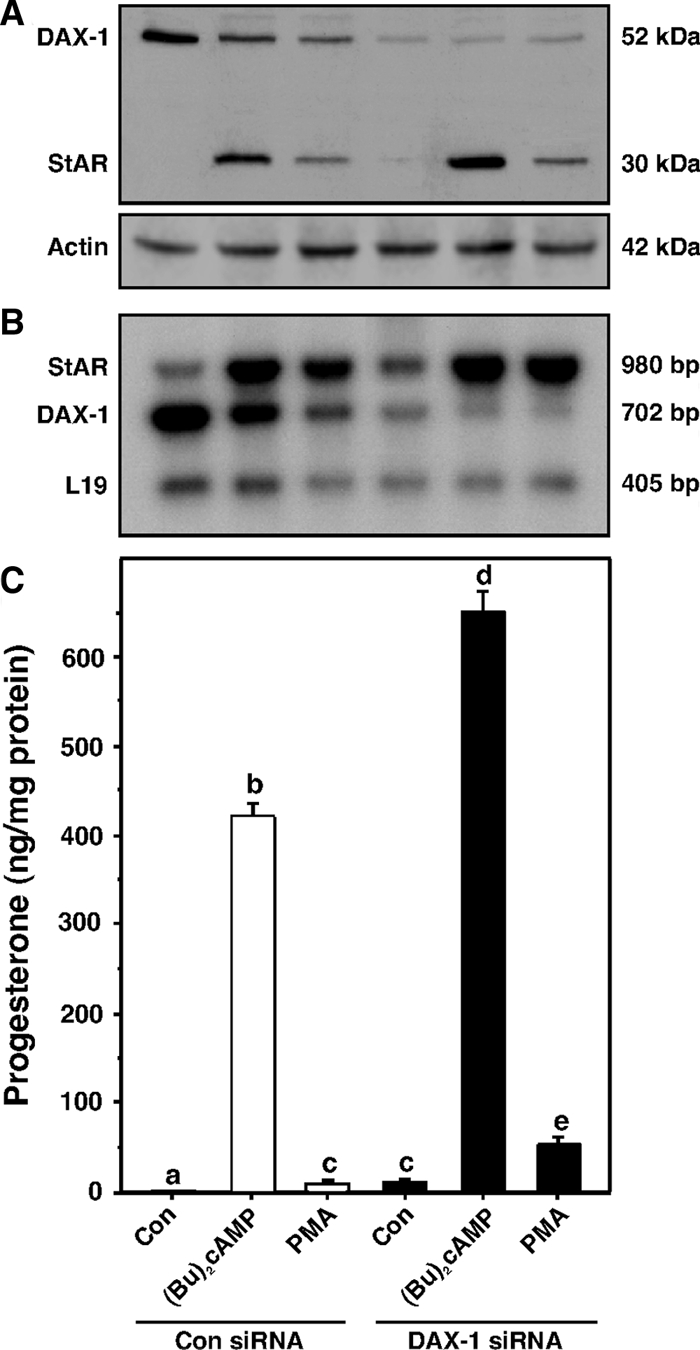
Silencing of DAX-1 and its relevance to StAR expression and progesterone synthesis. MA-10 cells were transfected with either a negative control siRNA at 100 nm (Con siRNA) or a mixture of two DAX-1-specific siRNAs at 50 nm (100 nm total, DAX-1 siRNA), as described in Materials and Methods. After 36 h of transfection, cells were treated without (Con) or with (Bu)2cAMP (0.5 mm) and PMA (10 nm) for an additional 6 h and subjected to Western (DAX-1, StAR, and actin) and RT-PCR (StAR, DAX-1, and L19) analyses. Representative immunoblots and autoradiograms illustrate the expression of DAX-1, StAR, and actin (A) and StAR, DAX-1, and L19 (B) in different treatment groups, respectively. Actin and L19 were used as controls in Western and RT-PCR analyses, respectively. Similar results were obtained from three different experiments. C, Accumulation of progesterone in the media was determined and expressed as nanograms per milligram protein, and the data shown represent the mean ± se of three independent experiments. Letters above the bars indicate that these groups differ significantly from each other, at least at P < 0.05.
Role of Nur77 and SF-1 on StAR gene expression and their consequences with DAX-1
To better understand the involvement of DAX-1 in the transcriptional repression of the StAR gene, the roles of Nur77 and SF-1 with respect to PKA and PKC signaling were studied. Earlier studies have demonstrated that SF-1 can bind to the −46/−40 bp of the StAR promoter (this sequence lies within the hairpin) (37,48,59), and this motif is also a target for Nur77 (46). Thus, it was of interest whether the hairpin structure could recognize Nur77 and SF-1. It can be seen in Fig. 9A that the binding of MA-10 NE to the −48/−16 bp region could be markedly inhibited with unlabeled probe (Fig. 9A, lane 3) and that a DAX-1 Ab (Fig. 9A, lane 4) also strongly decreased complex formation. Additionally, protein-DNA binding was attenuated (P < 0.05) by the presence of unlabeled consensus NBRE sequence (Fig. 9A; compare lanes 2 and 5). The binding was also competed with an unlabeled SF-1/2 consensus binding site sequence (SF-1/2 BS; −48/−35 bp, Fig. 9A; compare lanes 2 and 6) that has previously been demonstrated to bind SF-1 in MA-10 NE (48). More specifically, the binding of MA-10 NE with the −48/−35 bp sequence (SF-1/2 site) was shown to be effectively inhibited by its consensus sequence or by a SF-1 Ab. In the present study, partial competition of the protein-DNA complex by unlabeled SF-1 and Nur77 binding sequences suggests that DAX-1 predominantly binds to the −48/−16-bp region. CREB Ab that recognizes the CRE family proteins had no effect on the protein-DNA complex, indicating CREB does not bind to this region and confirms previous findings (supplemental Fig. 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org) (29). Importantly, the protein-DNA complex could not be competed by unlabeled probe bearing mutations localized to the hairpin loop but that left the SF-1/Nur77 binding element intact (Fig. 9A, lane 7). These results indicate that the DNA hairpin structure can bind both Nur77 and SF-1 and that mutation of the predicted loop appears to decrease the binding of these factors within this region.
Figure 9.
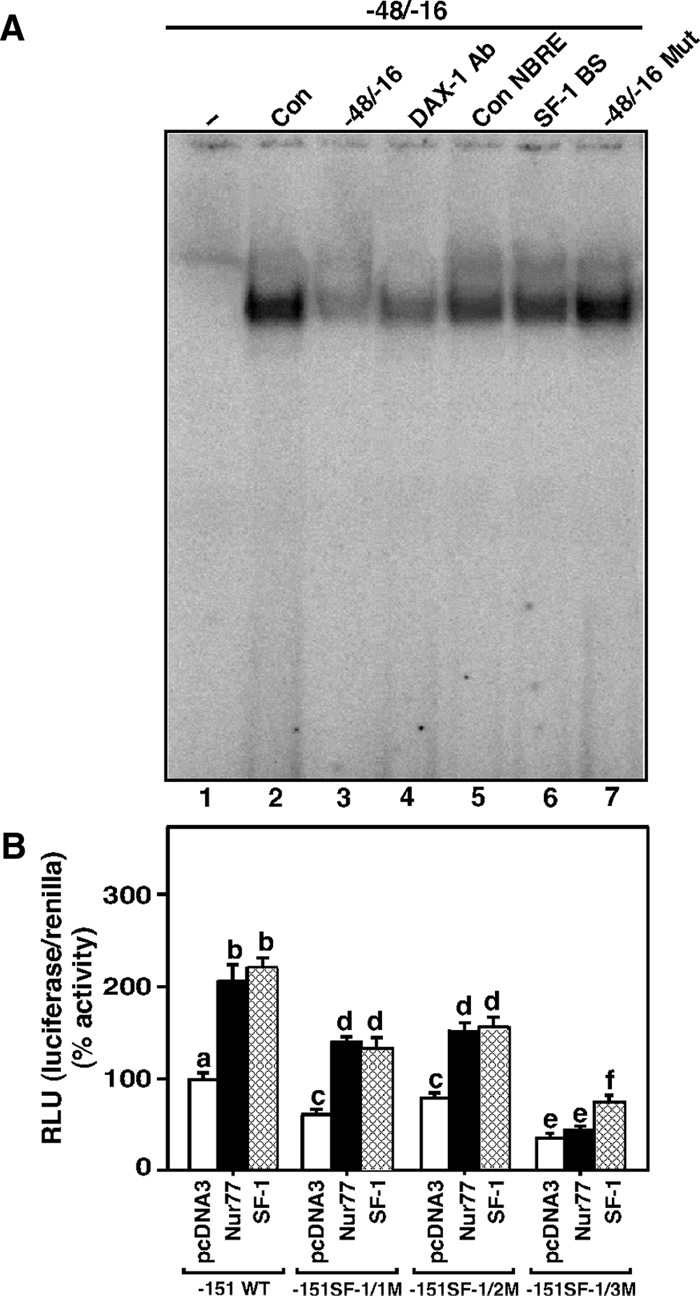
Assessment of Nur77 and SF-1 binding to the DAX-1 hairpin structure and roles of these factors in StAR promoter responsiveness. A, MA-10 NE (15 μg) was used to determine protein binding to the 32P-labeled DAX-1 (−48/−16 bp) motif (lanes 2–7). Binding of untreated MA-10 NE (Con, lane 2) to the labeled probe (−48/−16) was challenged with its unlabeled (−48/−16 bp) sequence (lane 3), DAX-1 Ab (lane 4), consensus NBRE (Con NBRE, lane 5), a confirmed SF-1 BS (lane 6), and with a mutant DAX-1 sequence (−48/−16 Mut, lane 7). MA-10 cells were transfected with empty vector (pcDNA3), Nur77, and SF-1 expression plasmids, within the context of one of the −151/−1 StAR reporter segments: either wild-type (−151 WT) or a reporter containing mutations in one of the SF-1 binding sites (SF-1/1M, SF-1/2M, SF-1/3M), in the presence of 15–20 ng of pRL-SV40 (B). After 48 h of transfection, cells were collected and luciferase activity in the cell lysates determined by [relative light units (RLU), luciferase/renilla] and presented in terms of percent activity. Data represent the mean ± se of four independent experiments. Letters above the bars indicate that these groups differ significantly from each other, at least at P < 0.05.
The influence of Nur77 and SF-1 was further investigated by determining StAR promoter responsiveness within the context of the −151/−1 StAR reporter segment, which is known to contain three SF-1 recognition motifs (46,48). MA-10 cells transfected with either the Nur77 or SF-1 expression plasmids, in the presence of the −151/−1 StAR reporter segment, demonstrated approximately 2-fold increases in StAR reporter activity when compared with controls (Fig. 9B). Specific mutations targeting each of the SF-1 recognition motifs decreased (P < 0.05) basal as well as Nur77 and SF-1 induced StAR promoter activity with the order of importance being SF-1/3>SF-1/1>SF-1/2. Notably, transactivation of the StAR gene by Nur77 was lacking when the SF-1/3 mutation was used. These data demonstrate that both Nur77 and SF-1 can target three SF-1 binding sites present within the −151/−1 bp region of the StAR promoter and positively influence transcription of the StAR gene (46,48).
In additional studies, MA-10 cells transfected with the Nur77 expression plasmid in the presence of the −151/−1 StAR reporter segment increased (Bu)2cAMP- (1.9 ± 0.4-fold) and PMA (1.6 ± 0.2-fold)-mediated StAR reporter activity relative to the responses seen in mock-transfected cells (Fig. 10A). However, cells coexpressing Nur77 and DAX-1 demonstrated decreased (P < 0.05) basal and (Bu)2cAMP-/PMA-mediated StAR reporter activity when compared with Nur77-induced responses. SF-1 expression also augmented both basal and (Bu)2cAMP-/PMA-stimulated StAR promoter responsiveness, and the magnitude of the responses over mock-transfected cells were 2.9 ± 0.7- and 2.1 ± 0.4-fold by (Bu)2cAMP and PMA, respectively. SF-1 mediated StAR gene transcription was also diminished in cells coexpressing SF-1 and DAX-1 (Fig. 10A). These data demonstrate that DAX-1 represses transactivation potential of Nur77 and SF-1 in regulating StAR gene transcription.
Figure 10.
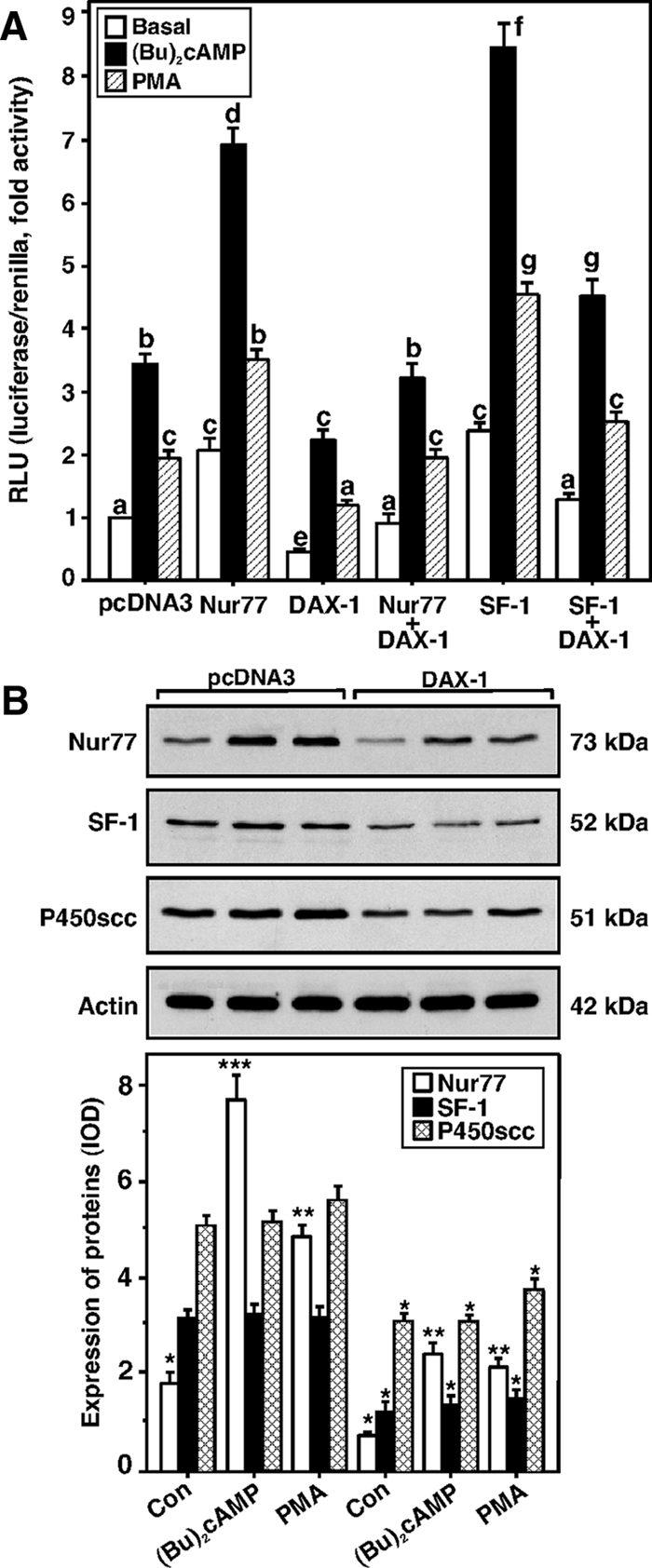
The effects of the expression of Nur77, SF-1, and DAX-1, independently or in combination on StAR promoter activity; the effects of DAX-1, (Bu)2cAMP, and PMA on Nur77, SF-1; and P450scc expression in MA-10 cells. A, Using the −151/−1-bp StAR reporter segment, cells were transfected with empty vector (pcDNA3), Nur77, SF-1, DAX-1, or a combination thereof as indicated in the presence of 15–20 ng of pRL-SV40. After 36 h of transfection, cells were incubated for an additional 6 h in the absence (basal) or presence of (Bu)2cAMP (0.5 mm) and PMA (10 nm), and luciferase activity in the cell lysates was determined [relative light units (RLU), luciferase/renilla] and expressed in terms of fold activity. Data represent the mean ± se of three to five independent experiments. Cells were also transfected with either empty vector (pcDNA3) or DAX-1 expression plasmids and subjected to immunoblotting for Nur77, SF-1, and P450scc expression using 20–25 μg of total cellular protein (B). Cells were treated with (Bu)2cAMP (0.5 mm) and PMA (10 nm) for 6 h, and the expression of Nur77, SF-1, and P450scc proteins was determined by immunoblotting. Actin expression demonstrates equal loading. Representative immunoblots illustrate the expression of Nur77, SF-1, and P450scc proteins in different treatment groups. Integrated OD (IOD) values of each band were quantified and compiled data from three independent experiments are presented (lower panel). Letters above the bars indicate that these groups differ significantly from each other, at least at P < 0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001 represent significant differences in comparison with respective controls.
DAX-1 overexpression in MA-10 cells was also capable of decreasing (P < 0.05) Nur77 and SF-1 protein levels between 45 and 70% when compared with their respective mock-transfected controls (Fig. 10B). Similarly, DAX-1 decreased expression of the P450scc protein expression by about 25% (Fig. 10B and supplemental Fig. 2) (10,14). Cells treated with (Bu)2cAMP (0.5 mm) and PMA (10 nm) demonstrated 4.2 ± 1.3- and 2.9 ± 0.7-fold increases in Nur77 protein expression over untreated cells, respectively, and these levels were decreased (P < 0.05) after DAX-1 expression. Conversely, neither (Bu)2cAMP nor PMA was able to alter SF-1 protein levels, suggesting these analogs do not affect SF-1 protein synthesis (60,61). Whereas (Bu)2cAMP had no effect on P450scc protein levels, PMA slightly elevated P450scc expression (Fig. 10B). These findings demonstrate that the suppression of StAR gene expression by DAX-1 occurs, at least in part, due to a transrepression mechanism initiated as a consequence of DAX-1 simultaneously inhibiting the expression of both Nur77 and SF-1 in MA-10 mouse Leydig tumor cells.
Discussion
Regulation of the StAR protein and steroid biosynthesis in steroidogenic tissues has been demonstrated to be mediated through multiple signaling pathways and transcription factors that generate both positive and negative effects on the steroidogenic response (6,12,21,27,56,62,63,64,65). Whereas many well-described signals up-regulate the steroidogenic response, the roles of inhibitory events in StAR expression and steroidogenesis are not as well understood. The nuclear receptor DAX-1 is endowed with transcriptional repressor activity that is invariably abolished in individuals harboring DAX-1 mutations, suggesting that the impairment of its transrepressor function is linked to the pathogenesis of adrenal hypoplasia congenita (4,7). DAX-1 is an atypical nuclear receptor that lacks a zinc-finger DNA binding motif; instead, it harbors an amino-terminal Gly- and Ala-rich repetitive region that serves as a DNA binding domain (6,8,31,66). Nevertheless, studies have demonstrated that DAX-1 does not necessarily require binding to DNA to exhibit its repressive effects but can inhibit the transcription of several genes through discrete mechanisms (31,33,34,35,67).
The present findings outline the events by which the activation of the PKA [by (Bu)2cAMP] and PKC (by PMA) signaling pathways can down-regulate DAX-1 and concomitantly up-regulate StAR expression in a time-dependent fashion. The down-regulation of DAX-1 by PKA and PKC signaling requires de novo transcription and translation of an unknown gene or set of genes, suggesting that the degradation of DAX-1 is dependent on the synthesis of a protein induced by either of these pathways. Consequently, DAX-1 serves as a potent repressor of steroidogenesis, the effects of which are relieved by activation of either PKA or PKC. It should be noted that in MA-10 cells, the steroid production elicited by PKC alone is compromised by the obligatory need for PKA to phosphorylate and activate StAR (31,33,34,35,67). As such, the effects of DAX-1 are more clearly seen at the level of StAR protein expression rather than at the level of steroid biosynthesis. Interestingly, the repression of DAX-1 by (Bu)2cAMP and PMA was found to be essentially reversed by the pathway specific inhibitors H89 and GFX, respectively, which, in contrast, attenuated StAR expression and steroid synthesis. Therefore, it is conceivable that a balance between the repressor function of DAX-1 and the inducer functions of PKA and PKC may allow for a finer-tuning of the steroidogenic machinery in mouse Leydig cells. One possible source of contention in evaluating these data, however, is that the selected doses of H89 and GFX used (5–25 μm) here to inhibit the PKA and PKC pathways have also been shown to affect several other kinases, albeit in different systems, that are under the influence of these same signaling pathways (68,69,70,71). Studies have documented that 5- to 10-μm concentrations of H89 and GFX can be ineffective in inhibiting the PKA and PKC pathways (72,73,74); however, these inhibitors have been used at 40–50 μm in different cellular models (73,75,76,77), suggesting that differing cellular levels of PKA and PKC, and other variations can give rise to the need to use varying amounts of these inhibitors to achieve similar effects. Whereas the results from this (the present data, and Refs. 17,25,26,30,78) and other (79,80,81) laboratories have demonstrated that relevant concentrations (10–50 μm) of H89 and GFX are required for the inhibition of PKA and PKC in Leydig cells, it is not currently known whether these inhibitors affect other kinases in MA-10 mouse Leydig tumor cells.
An intriguing aspect of the present findings is the identification of a novel DNA hairpin structure at position −44/−20 bp in the mouse StAR promoter that appears to be responsible for the recruitment of DAX-1. The functional relevance of this hairpin was verified by three independent approaches demonstrating that DAX-1 binding to this site effectively represses steroid synthesis by impairing transcription of the StAR gene. First, in vivo ChIP data showed a clear association of DAX-1 with the StAR proximal promoter, and both (Bu)2cAMP and PMA progressively diminished this interaction. Second, reporter gene analyses suggested that DAX-1 binds to a putative hairpin loop in the StAR promoter and suppresses StAR gene expression, whereas disruption of the loop abrogated the inhibitory function of DAX-1. Third, EMSAs demonstrated that the binding of a protein present in MA-10 NE to the −48/−16 bp site mirrored the complex formation seen using in vitro-translated DAX-1 and that this DNA-protein complex was specifically affected by cold competitors, a disruption of the hairpin loop, by distamycin A or a DAX-1 Ab. Inhibition of protein-DNA binding by distamycin A indicated that DAX-1 interacts with the minor groove of the DNA helix, a characteristic of high-mobility group box proteins, and suggests that this drug alters the secondary structure to which DAX-1 appears to bind (6,58). Previous work demonstrated that DAX-1 binding to the DNA hairpin requires an intact stem loop and that the binding was lacking in the absence of a loop. Furthermore, the binding of DAX-1 to DNA stem loops appears to be stronger with higher stem lengths than with short stems and is also dependent on the nucleotide compositions in the loop (6,66). In this study we demonstrated that a predicted hairpin structure at −44/−20 containing five paired stem nucleotides and 15 loop nucleotides binds DAX-1 and that the disruption of the hairpin loop blocks the ability of DAX-1 to bind and prevents the repression that DAX-1 imparts on the promoter. However, we cannot exclude the possibility that mutations may affect DAX-1 binding through a mechanism independent of the disruption of the hairpin loop. Whereas these results are in agreement with earlier studies demonstrating that DAX-1 binds to a DNA hairpin at −61/−27 bp in the human StAR promoter and decreases StAR expression and steroidogenesis (6,66), more conclusive demonstration of a hairpin loop structure in the mouse StAR promoter will require additional investigation. Two stable hairpins that bind DAX-1 have already been shown to be present in the mouse DAX-1 promoter (6). In contrast, suppression of the HDL receptor gene transcription by DAX-1 does not require DAX-1 binding to a DNA hairpin (35). It has been reported that the ligand-binding domain of DAX-1 can bind to mRNAs that in turn modulate the activity of the DAX-1 amino-terminal repeats (82). Therefore, it appears unlikely that a single mechanism is used by DAX-1 as it exerts its transcriptional repression on the different enzymes and genes involved in steroidogenesis.
Another interesting finding in this study is the involvement of Nur77 in StAR expression and the observation that DAX-1 negatively modulates Nur77-induced transactivation of the StAR gene. The overexpression of Nur77 enhanced (Bu)2cAMP- and PMA-mediated StAR gene expression, and these increases were diminished by DAX-1. Like StAR, the expression of Nur77 is increased by both (Bu)2cAMP and PMA and down-regulated by DAX-1, suggesting that the induction of Nur77 and the attenuation of DAX-1 are crucial for steroidogenesis in Leydig cells. Previous studies have shown that DAX-1 interacts with Nur77 and inhibits its transcriptional effects by competing with Nur77 binding to the steroid receptor coactivator-1 (83). Nur77 has been reported to trans-activate the genes for 17α-hydroxylase, 21-hydroxylase, and 20α-hydroxysteroid dehydrogenase using cell-specific signal transduction pathways and/or combinations of cofactors, i.e. steroid receptor coactivator, CREB binding protein (CBP), p300/CBP-associated factor, and vitamin D receptor-associated protein-205 (43,44,84). Studies have demonstrated that the induction of CREB by (Bu)2cAMP is correlated with its increased presence on chromatin and the recruitment of the coregulator CBP/p300 to the StAR promoter (53,64,85). Hence, it is likely that the PKA- and PKC-mediated induction of Nur77 results in the recruitment of CBP/p300 to function in regulating StAR transcription. This hypothesis agrees with earlier studies demonstrating that (Bu)2cAMP and human chorionic gonadotropin can induce Nur77 expression in Leydig and adrenal cell lines (42). Notably, in K28 mouse Leydig cells, Nur77 expression correlates with increased steroid synthesis but not StAR expression (42). In contrast, we observed that the increased levels of Nurr77 that occur in conjunction with the activation of either PKA or PKC were tightly associated with increased StAR gene expression, an observation that is consistent with findings that were published during the preparation of this manuscript (46). This seeming contradiction could be due to differences in cell signaling specificity, experimental conditions, and/or the presence of unknown factor(s) involved in StAR expression. Our current data suggest that Nur77 is capable of binding to three SF-1 binding sites at −138/−133 bp (SF-1/1), −46/−40 bp (SF-1/2), and −102/−96 bp (SF-1/3) within the −151/−1 bp region of the mouse StAR promoter and that the mutation of these SF-1 binding sites represses Nur77 mediated transactivation of the StAR gene (46). Previously, it has been demonstrated that the −46/−40 bp region (SF-1/2) of the mouse StAR promoter binds SF-1 and that protein-DNA binding was markedly decreased with its unlabeled sequence or SF-1 Ab (48). Consequently, we predict that the DAX-1 stem loop containing the SF-1/2 motif can bind to both Nur77 and SF-1 because their binding was competed for with consensus NBRE and SF-1 binding (−46/−40 bp) sequences, respectively, but was not affected after the disruption of the hairpin loop. Further studies to elucidate these mechanisms are currently underway and will lead to a better understanding of the involvement of Nur77 in regulating StAR gene transcription.
The physiological roles and tissue distribution of DAX-1 are strikingly similar to those of the nuclear receptor SF-1, and this factor is known to play pivotal roles in the cAMP-regulated expression of several steroidogenic enzymes and genes, including StAR (33,36,37,56,60). In the present study, DAX-1 was demonstrated to inhibit SF-1-induced transactivation of the StAR gene by PKA and PKC, an observation in agreement with the repression of SF-1 mRNA and steroidogenesis by DAX-1 seen in previous studies (24,32). However, neither (Bu)2cAMP nor PMA was able to alter SF-1 protein levels, suggesting that these agents influence posttranslational modification of SF-1. Indeed, forskolin has been shown to enhance the phosphorylation of SF-1 leading to an increase in SF-1 binding to the StAR proximal promoter and a concomitant increase in StAR gene transcription (60). In agreement with our previous findings (48), we now provide evidence that the −48/−16 promoter region encompassing the DNA hairpin binds to SF-1 and that the disruption of the hairpin loop prevents formation of the protein-DNA complex. Furthermore, (Bu)2cAMP and PMA were able to partially rescue the transactivation potentials of Nur77 and SF-1 in DAX-1-mediated repression of the StAR gene (3). DAX-1 has been shown to repress both Müllerian-inhibiting substance and the HDL receptor gene transcription by affecting the synergism between SF-1 and GATA-4 (34) and SF-1 and sterol regulatory element-binding protein-1a (35), respectively. Whether DAX-1 binding to the DNA hairpin affecting the neighboring GATA-4 binding site (−66/−61 bp) (86) requires further investigation. Nevertheless, it is tempting to speculate that combinatorial effects of other factors, which interact/cooperate with SF-1 (for example, GATA-4, CCAAT/enhancer-binding protein-β, CREB/cAMP response element modulator, specificity protein-1) in transcriptional regulation of the StAR gene, may also be negatively influenced by DAX-1.
The expression of DAX-1 strongly suppresses StAR transcription, demonstrating one of the important and effective means by which DAX-1 inhibits steroid biosynthesis, and these events are influenced by Nur77 and SF-1. In this study we were able to corroborate previous findings (10,14), demonstrating that in MA-10 cells overexpressing DAX-1 the expression of P450scc is inhibited. DAX-1-deficient male mice up-regulate aromatase expression, an event associated with Leydig cell hyperplasia and degeneration of germinal epithelium, leading to infertility (11). DAX-1 is expressed in Sertoli cells, regulated developmentally and is involved in the development of spermatogenic cells (3,40). Therefore, in addition to the physiological importance of DAX-1 in the development and function of the adrenal glands, it also appears to play key roles in controlling testicular function.
In summary, these findings provide insight into the mechanisms by which DAX-1 represses StAR expression and steroidogenesis and extend our understanding of the interplay between PKA/PKC signaling and the repressional events governing the StAR promoter in MA-10 mouse Leydig tumor cells. DAX-1 binds to a DNA hairpin structure in the mouse StAR promoter and effectively suppresses steroid synthesis by inhibiting transcription of the StAR gene. Furthermore, the down-regulation of DAX-1 increases the expression of Nur77 and SF-1, two transcription factors that can activate the StAR promoter. Thus, removing the potent repression elicited by DAX-1 is a key event in regulating StAR-mediated steroidogenesis.
Supplementary Material
Acknowledgments
The authors thank Dr. P. Sassone-Corsi (University of Louis Pasteur, Strasbourg, France), Dr. S. R. King (Baylor College of Medicine, Houston, TX), Dr. W. L. Miller (University of California, San Francisco, San Francisco, CA), and Dr. K. Morohashi (National Institute for Basic Biology, Okazaki, Japan) for the generous gifts of antibodies to DAX-1, P-StAR, total StAR, and SF-1, respectively. We also thank Dr. H.-S. Choi (Chonnam National University, Kwangju, Republic of Korea) and Dr. K. L. Parker (University of Texas Southwestern Medical Center, Dallas, TX) for Nur77 and SF-1 expression plasmids, respectively.
Footnotes
This work was supported by National Institutes of Health Grant HD-17481 and funds from the Robert A. Welch Foundation Grant B1-0028.
Disclosure Statement: The authors have nothing to disclose.
First Published Online September 11, 2008
Abbreviations: Ab, Antibody; Act, actinomycin D; (Bu)2cAMP, dibutyryl cAMP; CBP, CREB binding protein; ChIP, chromatin immunoprecipitation; Chx, cycloheximide; CREB, cAMP response element-binding protein; DAX-1, dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene 1; GFX, GF-109203X; HDL, high-density lipoprotein; Mut, mutated; NBRE, nerve growth factor-induced clone B response element; NE, nuclear extract; P-CREB, phosphorylation of CREB; PKA, protein kinase A; PKC, protein kinase C; PMA, phorbol 12-myristate 13-acetate; P450scc, P450 cholesterol side-chain cleavage enzyme; P-StAR, phosphorylation of StAR; SF-1, steroidogenic factor-1, SF-1 BS, SF-1 binding site; siRNA, small interfering RNA; StAR, steroidogenic acute regulatory protein.
References
- Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, Guo W, Lalli E, Moser C, Walker AP, McCabe ER, Meitinger T, Monaco AP, Sassone-Corsi P, Camerino G An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372:635–641 [DOI] [PubMed] [Google Scholar]
- Muscatelli F, Strom TM, Walker AP, Zanaria E, Recan D, Meindl A, Bardoni B, Guioli S, Zehetner G, Rabl W, Schwarz HP, Kaplan J-C, Camerino G, Meitinger T, Monaco AP 1994 Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 372:672–676 [DOI] [PubMed] [Google Scholar]
- Tamai KT, Monaco L, Alastalo TP, Lalli E, Parvinen M, Sassone-Corsi P 1996 Hormonal and developmental regulation of DAX-1 expression in Sertoli cells. Mol Endocrinol 10:1561–1569 [DOI] [PubMed] [Google Scholar]
- Reutens AT, Achermann JC, Ito M, Gu WX, Habiby RL, Donohoue PA, Pang S, Hindmarsh PC, Jameson JL 1999 Clinical and functional effects of mutations in the DAX-1 gene in patients with adrenal hypoplasia congenita. J Clin Endocrinol Metab 84:504–511 [DOI] [PubMed] [Google Scholar]
- Meeks JJ, Weiss J, Jameson JL 2003 Dax1 is required for testis determination. Nat Genet 34:32–33 [DOI] [PubMed] [Google Scholar]
- Zazopoulos E, Lalli E, Stocco DM, Sassone-Corsi P 1997 DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature 390:311–315 [DOI] [PubMed] [Google Scholar]
- Lalli E, Bardoni B, Zazopoulos E, Wurtz JM, Strom TM, Moras D, Sassone-Corsi P 1997 A transcriptional silencing domain in DAX-1 whose mutation causes adrenal hypoplasia congenita. Mol Endocrinol 11:1950–1960 [DOI] [PubMed] [Google Scholar]
- Crawford PA, Dorn C, Sadovsky Y, Milbrandt J 1998 Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol Cell Biol 18:2949–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, McCabe ER 2005 DAX1 origin, function, and novel role. Mol Genet Metab 86:70–83 [DOI] [PubMed] [Google Scholar]
- Lalli E, Melner MH, Stocco DM, Sassone-Corsi P 1998 DAX-1 blocks steroid production at multiple levels. Endocrinology 139:4237–4243 [DOI] [PubMed] [Google Scholar]
- Wang ZJ, Jeffs B, Ito M, Achermann JC, Yu RN, Hales DB, Jameson JL 2001 Aromatase (Cyp19) expression is up-regulated by targeted disruption of Dax1. Proc Natl Acad Sci USA 98:7988–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman H, Murigande C, Nadakal A, Capponi AM 2002 Repression of DAX-1 and induction of SF-1 expression. Two mechanisms contributing to the activation of aldosterone biosynthesis in adrenal glomerulosa cells. J Biol Chem 277:41259–41267 [DOI] [PubMed] [Google Scholar]
- Tajima K, Dantes A, Yao Z, Sorokina K, Kotsuji F, Seger R, Amsterdam A 2003 Down-regulation of steroidogenic response to gonadotropins in human and rat preovulatory granulosa cells involves mitogen-activated protein kinase activation and modulation of DAX-1 and steroidogenic factor-1. J Clin Endocrinol Metab 88:2288–2299 [DOI] [PubMed] [Google Scholar]
- Jo Y, Stocco DM 2004 Regulation of steroidogenesis and steroidogenic acute regulatory protein in R2C cells by DAX-1 (dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene-1). Endocrinology 145:5629–5637 [DOI] [PubMed] [Google Scholar]
- Manna PR, Chandrala SP, Jo Y, Stocco DM 2006 cAMP-independent signaling regulates steroidogenesis in mouse Leydig cells in the absence of StAR phosphorylation. J Mol Endocrinol 37:81–95 [DOI] [PubMed] [Google Scholar]
- Yu CC, Li PH 2006 In vivo inhibition of steroidogenic acute regulatory protein expression by dexamethasone parallels induction of the negative transcription factor DAX-1. Endocrine 30:313–323 [DOI] [PubMed] [Google Scholar]
- Manna PR, Jo Y, Stocco DM 2007 Regulation of Leydig cell steroidogenesis by extracellular signal-regulated kinase 1/2: role of protein kinase A and protein kinase C signaling. J Endocrinol 193:53–63 [DOI] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM 1994 The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR). J Biol Chem 269:28314–28322 [PubMed] [Google Scholar]
- Lin D, Sugawara T, Strauss III JF, Clark BJ, Stocco DM, Saenger P, Rogol A, Miller WL 1995 Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science 267:1828–1831 [DOI] [PubMed] [Google Scholar]
- Stocco DM, Clark BJ 1996 Regulation of the acute production of steroids in steroidogenic cells. Endocr Rev 17:221–244 [DOI] [PubMed] [Google Scholar]
- Manna PR, Stocco DM 2005 Regulation of the steroidogenic acute regulatory protein expression: functional and physiological consequences. Curr Drug Targets Immune Endocr Metab Disord 5:93–108 [DOI] [PubMed] [Google Scholar]
- Miller WL 2007 StAR search—what we know about how the steroidogenic acute regulatory protein mediates mitochondrial cholesterol import. Mol Endocrinol 21:589–601 [DOI] [PubMed] [Google Scholar]
- Clark BJ, Combs R, Hales KH, Hales DB, Stocco DM 1997 Inhibition of transcription affects synthesis of steroidogenic acute regulatory protein and steroidogenesis in MA-10 mouse Leydig tumor cells. Endocrinology 138:4893–4901 [DOI] [PubMed] [Google Scholar]
- Manna PR, Pakarinen P, El-Hefnawy T, Huhtaniemi IT 1999 Functional assessment of the calcium messenger system in cultured mouse Leydig tumor cells: regulation of human chorionic gonadotropin-induced expression of the steroidogenic acute regulatory protein. Endocrinology 140:1739–1751 [DOI] [PubMed] [Google Scholar]
- Jo Y, King SR, Khan SA, Stocco DM 2005 Involvement of protein kinase C and cyclic adenosine 3′,5′-monophosphate-dependent kinase in steroidogenic acute regulatory protein expression and steroid biosynthesis in Leydig cells. Biol Reprod 73:244–255 [DOI] [PubMed] [Google Scholar]
- Manna PR, Kero J, Tena-Sempere M, Pakarinen P, Stocco DM, Huhtaniemi IT 2001 Assessment of mechanisms of thyroid hormone action in mouse Leydig cells: regulation of the steroidogenic acute regulatory protein, steroidogenesis, and luteinizing hormone receptor function. Endocrinology 142:319–331 [DOI] [PubMed] [Google Scholar]
- Stocco DM, Wang X, Jo Y, Manna PR 2005 Multiple signaling pathways regulating steroidogenesis and steroidogenic acute regulatory protein expression: more complicated than we thought. Mol Endocrinol 19:2647–2659 [DOI] [PubMed] [Google Scholar]
- Arakane F, King SR, Du Y, Kallen CB, Walsh LP, Watari H, Stocco DM, Strauss III JF 1997 Phosphorylation of steroidogenic acute regulatory protein (StAR) modulates its steroidogenic activity. J Biol Chem 272:32656–32662 [DOI] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Eubank DW, Clark BJ, Lalli E, Sassone-Corsi P, Zeleznik AJ, Stocco DM 2002 Regulation of steroidogenesis and the steroidogenic acute regulatory protein by a member of the cAMP response-element binding protein family. Mol Endocrinol 16:184–199 [DOI] [PubMed] [Google Scholar]
- Manna PR, Chandrala SP, King SR, Jo Y, Counis R, Huhtaniemi IT, Stocco DM 2006 Molecular mechanisms of insulin-like growth factor-I mediated regulation of the steroidogenic acute regulatory protein in mouse Leydig cells. Mol Endocrinol 20:362–378 [DOI] [PubMed] [Google Scholar]
- Ito M, Yu R, Jameson JL 1997 DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol 17:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna PR, Tena-Sempere M, Huhtaniemi IT 1999 Molecular mechanisms of thyroid hormone-stimulated steroidogenesis in mouse Leydig tumor cells. Involvement of the steroidogenic acute regulatory (StAR) protein. J Biol Chem 274:5909–5918 [DOI] [PubMed] [Google Scholar]
- Hanley NA, Rainey WE, Wilson DI, Ball SG, Parker KL 2001 Expression profiles of SF-1, DAX1, and CYP17 in the human fetal adrenal gland: potential interactions in gene regulation. Mol Endocrinol 15:57–68 [DOI] [PubMed] [Google Scholar]
- Tremblay JJ, Viger RS 2001 Nuclear receptor Dax-1 represses the transcriptional cooperation between GATA-4 and SF-1 in Sertoli cells. Biol Reprod 64:1191–1199 [DOI] [PubMed] [Google Scholar]
- Lopez D, Shea-Eaton W, Sanchez MD, McLean MP 2001 DAX-1 represses the high-density lipoprotein receptor through interaction with positive regulators sterol regulatory element-binding protein-1a and steroidogenic factor-1. Endocrinology 142:5097–5106 [DOI] [PubMed] [Google Scholar]
- Sugawara T, Kiriakidou M, McAllister JM, Holt JA, Arakane F, Strauss III JF 1997 Regulation of expression of the steroidogenic acute regulatory protein (StAR) gene: a central role for steroidogenic factor 1. Steroids 62:5–9 [DOI] [PubMed] [Google Scholar]
- Wooton-Kee CR, Clark BJ 2000 Steroidogenic factor-1 influences protein-deoxyribonucleic acid interactions within the cyclic adenosine 3,5-monophosphate-responsive regions of the murine steroidogenic acute regulatory protein gene. Endocrinology 141:1345–1355 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Takeda Y, Shikayama T, Mukai T, Hisano S, Morohashi KI 2001 Comparative localization of Dax-1 and Ad4BP/SF-1 during development of the hypothalamic-pituitary-gonadal axis suggests their closely related and distinct functions. Dev Dyn 220:363–376 [DOI] [PubMed] [Google Scholar]
- Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, De Rooij DG, Themmen AP, Behringer RR, Parker KL 2004 Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol 18:1610–1619 [DOI] [PubMed] [Google Scholar]
- Kojima Y, Sasaki S, Hayashi Y, Umemoto Y, Morohashi K, Kohri K 2006 Role of transcription factors Ad4bp/SF-1 and DAX-1 in steroidogenesis and spermatogenesis in human testicular development and idiopathic azoospermia. Int J Urol 13:785–793 [DOI] [PubMed] [Google Scholar]
- Murphy EP, Conneely OM 1997 Neuroendocrine regulation of the hypothalamic pituitary adrenal axis by the nurr1/nur77 subfamily of nuclear receptors. Mol Endocrinol 11:39–47 [DOI] [PubMed] [Google Scholar]
- Song KH, Park JI, Lee MO, Soh J, Lee K, Choi HS 2001 LH induces orphan nuclear receptor Nur77 gene expression in testicular Leydig cells. Endocrinology 142:5116–5123 [DOI] [PubMed] [Google Scholar]
- Kovalovsky D, Refojo D, Liberman AC, Hochbaum D, Pereda MP, Coso OA, Stalla GK, Holsboer F, Arzt E 2002 Activation and induction of NUR77/NURR1 in corticotrophs by CRH/cAMP: involvement of calcium, protein kinase A, and MAPK pathways. Mol Endocrinol 16:1638–1651 [DOI] [PubMed] [Google Scholar]
- Stocco CO, Lau LF, Gibori G 2002 A calcium/calmodulin-dependent activation of ERK1/2 mediates JunD phosphorylation and induction of nur77 and 20α-hsd genes by prostaglandin F2α in ovarian cells. J Biol Chem 277:3293–3302 [DOI] [PubMed] [Google Scholar]
- Martin LJ, Tremblay JJ 2005 The human 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase type 2 promoter is a novel target for the immediate early orphan nuclear receptor Nur77 in steroidogenic cells. Endocrinology 146:861–869 [DOI] [PubMed] [Google Scholar]
- Martin LJ, Boucher N, Brousseau C, Tremblay JJ 2008 The orphan nuclear receptor Nur77 regulates hormone-induced StAR transcription in Leydig cells through a cooperation with CaMKI. Mol Endocrinol 22:2021–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna PR, Eubank DW, Stocco DM 2004 Assessment of the role of activator protein-1 on transcription of the mouse steroidogenic acute regulatory protein gene. Mol Endocrinol 18:558–573 [DOI] [PubMed] [Google Scholar]
- Manna PR, Eubank DW, Lalli E, Sassone-Corsi P, Stocco DM 2003 Transcriptional regulation of the mouse steroidogenic acute regulatory protein gene by the cAMP response-element binding protein and steroidogenic factor 1. J Mol Endocrinol 30:381–397 [DOI] [PubMed] [Google Scholar]
- Ascoli M 1981 Characterization of several clonal lines of cultured Leydig tumor cells: gonadotropin receptors and steroidogenic responses. Endocrinology 108:88–95 [DOI] [PubMed] [Google Scholar]
- Dyson MT, Jones JK, Kowalewski MP, Manna PR, Alonso M, Gottesman ME, Stocco DM 2008 Mitochondrial A-kinase anchoring protein 121 binds type II protein kinase A and enhances steroidogenic acute regulatory protein-mediated steroidogenesis in MA-10 mouse Leydig tumor cells. Biol Reprod 78:267–277 [DOI] [PubMed] [Google Scholar]
- Bose HS, Whittal RM, Baldwin MA, Miller WL 1999 The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule. Proc Natl Acad Sci USA 96:7250–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Zanger UM, Honda S, Hara M, Waterman MR, Omura T 1993 Activation of CYP11A and CYP11B gene promoters by the steroidogenic cell-specific transcription factor, Ad4BP. Mol Endocrinol 7:1196–1204 [DOI] [PubMed] [Google Scholar]
- Manna PR, Stocco DM 2007 Crosstalk of CREB and Fos/Jun on a single cis-element: transcriptional repression of the steroidogenic acute regulatory protein gene. J Mol Endocrinol 39:261–277 [DOI] [PubMed] [Google Scholar]
- Caron KM, Ikeda Y, Soo SC, Stocco DM, Parker KL, Clark BJ 1997 Characterization of the promoter region of the mouse gene encoding the steroidogenic acute regulatory protein. Mol Endocrinol 11:138–147 [DOI] [PubMed] [Google Scholar]
- Gruber F, Hufnagl P, Hofer-Warbinek R, Schmid JA, Breuss JM, Huber-Beckmann R, Lucerna M, Papac N, Harant H, Lindley I, de Martin R, Binder BR 2003 Direct binding of Nur77/NAK-1 to the plasminogen activator inhibitor 1 (PAI-1) promoter regulates TNFα-induced PAI-1 expression. Blood 101:3042–3048 [DOI] [PubMed] [Google Scholar]
- Manna PR, Wang XJ, Stocco DM 2003 Involvement of multiple transcription factors in the regulation of steroidogenic acute regulatory protein gene expression. Steroids 68:1125–1134 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003 Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraku Y, Oikawa S, Kawanishi S 2002 Distamycin A, a minor groove binder, changes enediyne-induced DNA cleavage sites and enhances apoptosis. Nucleic Acids Res Suppl 95–96 [DOI] [PubMed] [Google Scholar]
- Sugawara T, Kiriakidou M, McAllister JM, Kallen CB, Strauss III JF 1997 Multiple steroidogenic factor 1 binding elements in the human steroidogenic acute regulatory protein gene 5′-flanking region are required for maximal promoter activity and cyclic AMP responsiveness. Biochemistry 36:7249–7255 [DOI] [PubMed] [Google Scholar]
- Gyles SL, Burns CJ, Whitehouse BJ, Sugden D, Marsh PJ, Persaud SJ, Jones PM 2001 ERKs regulate cyclic AMP-induced steroid synthesis through transcription of the steroidogenic acute regulatory (StAR) gene. J Biol Chem 276:34888–34895 [DOI] [PubMed] [Google Scholar]
- Desclozeaux M, Krylova IN, Horn F, Fletterick RJ, Ingraham HA 2002 Phosphorylation and intramolecular stabilization of the ligand binding domain in the nuclear receptor steroidogenic factor 1. Mol Cell Biol 22:7193–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhoff TW, McLean MP 1999 Repression of the rat steroidogenic acute regulatory (StAR) protein gene by PGF2α is modulated by the negative transcription factor DAX-1. Endocrine 10:83–91 [DOI] [PubMed] [Google Scholar]
- Christenson LK, Strauss III JF 2001 Steroidogenic acute regulatory protein: an update on its regulation and mechanism of action. Arch Med Res 32:576–586 [DOI] [PubMed] [Google Scholar]
- Hiroi H, Christenson LK, Chang L, Sammel MD, Berger SL, Strauss III JF 2004 Temporal and spatial changes in transcription factor binding and histone modifications at the steroidogenic acute regulatory protein (StAR) locus associated with stAR transcription. Mol Endocrinol 18:791–806 [DOI] [PubMed] [Google Scholar]
- Silverman E, Yivgi-Ohana N, Sher N, Bell M, Eimerl S, Orly J 2006 Transcriptional activation of the steroidogenic acute regulatory protein (StAR) gene: GATA-4 and CCAAT/enhancer-binding protein β confer synergistic responsiveness in hormone-treated rat granulosa and HEK293 cell models. Mol Cell Endocrinol 252:92–101 [DOI] [PubMed] [Google Scholar]
- Lalli E, Sassone-Corsi P 2003 DAX-1, an unusual orphan receptor at the crossroads of steroidogenic function and sexual differentiation. Mol Endocrinol 17:1445–1453 [DOI] [PubMed] [Google Scholar]
- Altincicek B, Tenbaum SP, Dressel U, Thormeyer D, Renkawitz R, Baniahmad A 2000 Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J Biol Chem 275:7662–7667 [DOI] [PubMed] [Google Scholar]
- Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H 1990 Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem 265:5267–5272 [PubMed] [Google Scholar]
- Alessi DR 1997 The protein kinase C inhibitors Ro 318220 and GF 109203X are equally potent inhibitors of MAPKAP kinase-1β (Rsk-2) and p70 S6 kinase. FEBS Lett 402:121–123 [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P 2000 Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 351:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts NA, Marber MS, Avkiran M 2004 Specificity of action of bisindolylmaleimide protein kinase C inhibitors: do they inhibit the 70 kDa ribosomal S6 kinase in cardiac myocytes? Biochem Pharmacol 68:1923–1928 [DOI] [PubMed] [Google Scholar]
- Kleppisch T, Nelson MT 1995 Adenosine activates ATP-sensitive potassium channels in arterial myocytes via A2 receptors and cAMP-dependent protein kinase. Proc Natl Acad Sci USA 92:12441–12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode HP, Moormann B, Dabew R, Goke B 1999 Glucagon-like peptide 1 elevates cytosolic calcium in pancreatic β-cells independently of protein kinase A. Endocrinology 140:3919–3927 [DOI] [PubMed] [Google Scholar]
- Boterman M, Elzinga CR, Wagemakers D, Eppens PB, Zaagsma J, Meurs H 2005 Potentiation of β-adrenoceptor function in bovine tracheal smooth muscle by inhibition of protein kinase C. Eur J Pharmacol 516:85–92 [DOI] [PubMed] [Google Scholar]
- von Lindern M, Parren-van Amelsvoort M, van Dijk T, Deiner E, van den Akker E, van Emst-de Vries S, Willems P, Beug H, Lowenberg B 2000 Protein kinase Cα controls erythropoietin receptor signaling. J Biol Chem 275:34719–34727 [DOI] [PubMed] [Google Scholar]
- Thodeti CK, Nielsen CK, Paruchuri S, Larsson C, Sjolander A 2001 The epsilon isoform of protein kinase C is involved in regulation of the LTD(4)-induced calcium signal in human intestinal epithelial cells. Exp Cell Res 262:95–103 [DOI] [PubMed] [Google Scholar]
- Kwon G, Pappan KL, Marshall CA, Schaffer JE, McDaniel ML 2004 cAMP dose-dependently prevents palmitate-induced apoptosis by both protein kinase A- and cAMP-guanine nucleotide exchange factor-dependent pathways in β-cells. J Biol Chem 279:8938–8945 [DOI] [PubMed] [Google Scholar]
- Rao RM, Jo Y, Leers-Sucheta S, Bose HS, Miller WL, Azhar S, Stocco DM 2003 Differential regulation of steroid hormone biosynthesis in R2C and MA-10 Leydig tumor cells: role of SR-B1-mediated selective cholesteryl ester transport. Biol Reprod Endocr Res 68:114–121 [DOI] [PubMed] [Google Scholar]
- Laurich VM, Trbovich AM, O'Neill FH, Houk CP, Sluss PM, Payne AH, Donahoe PK, Teixeira J 2002 Mullerian inhibiting substance blocks the protein kinase A-induced expression of cytochrome p450 17α-hydroxylase/C(17–20) lyase mRNA in a mouse Leydig cell line independent of cAMP responsive element binding protein phosphorylation. Endocrinology 143:3351–3360 [DOI] [PubMed] [Google Scholar]
- Millena AC, Reddy SC, Bowling GH, Khan SA 2004 Autocrine regulation of steroidogenic function of Leydig cells by transforming growth factor-α. Mol Cell Endocrinol 224:29–39 [DOI] [PubMed] [Google Scholar]
- Chen YC, Huang YL, Huang BM 2005 Cordyceps sinensis mycelium activates PKA and PKC signal pathways to stimulate steroidogenesis in MA-10 mouse Leydig tumor cells. Int J Biochem Cell Biol 37:214–223 [DOI] [PubMed] [Google Scholar]
- Lalli E, Ohe K, Hindelang C, Sassone-Corsi P 2000 Orphan receptor DAX-1 is a shuttling RNA binding protein associated with polyribosomes via mRNA. Mol Cell Biol 20:4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KH, Park YY, Park KC, Hong CY, Park JH, Shong M, Lee K, Choi HS 2004 The atypical orphan nuclear receptor DAX-1 interacts with orphan nuclear receptor Nur77 and represses its transactivation. Mol Endocrinol 18:1929–1940 [DOI] [PubMed] [Google Scholar]
- Wansa KD, Harris JM, Muscat GE 2002 The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J Biol Chem 277:33001–330011 [DOI] [PubMed] [Google Scholar]
- Clem BF, Hudson EA, Clark BJ 2005 Cyclic adenosine 3′,5′-monophosphate (cAMP) enhances cAMP-responsive element binding (CREB) protein phosphorylation and phospho-CREB interaction with the mouse steroidogenic acute regulatory protein gene promoter. Endocrinology 146:1348–1356 [DOI] [PubMed] [Google Scholar]
- Silverman E, Eimerl S, Orly J 1999 CCAAT enhancer-binding protein β and GATA-4 binding regions within the promoter of the steroidogenic acute regulatory protein (StAR) gene are required for transcription in rat ovarian cells. J Biol Chem 274:17987–17996 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.