Abstract
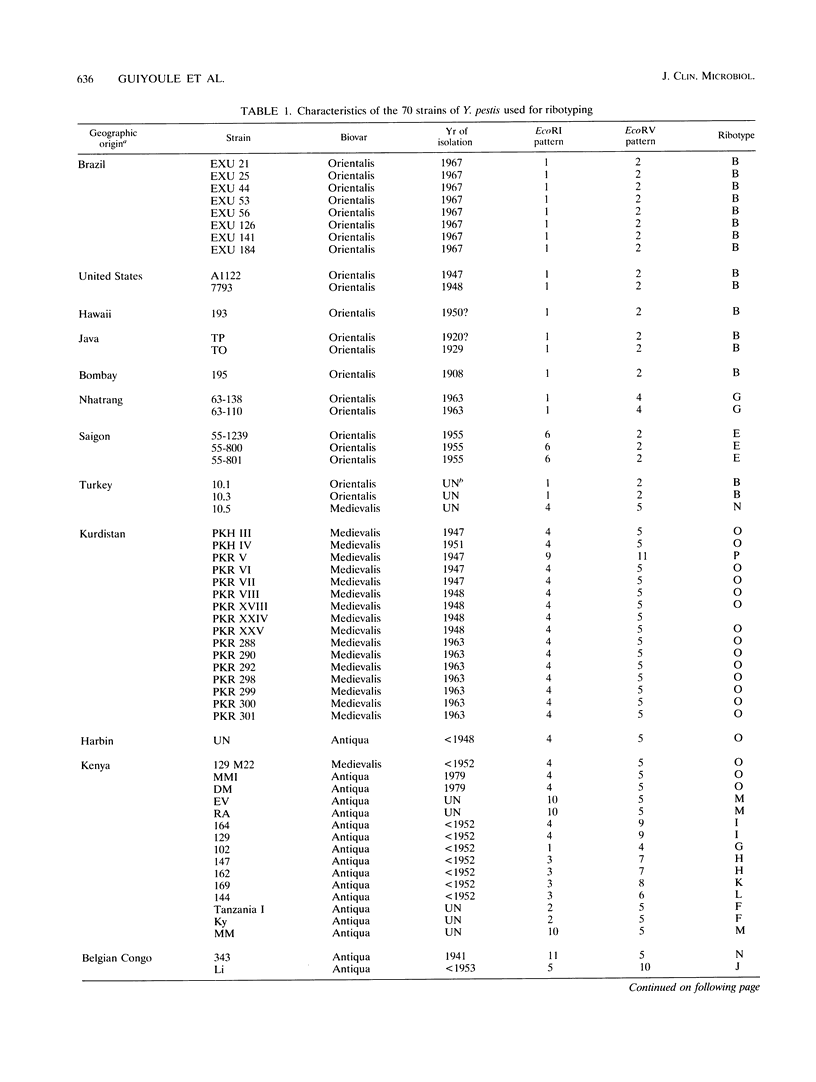
Yersinia pestis is the causative agent of plague, a disease which has caused the deaths of millions of people and which persists now in endemic foci. The rRNA gene restriction patterns (i.e., ribotypes) of 70 strains of Y. pestis, isolated on the five continents over a period of 72 years, were determined by hybridization with a 16S-23S rRNA probe from Escherichia coli. The combination of the EcoRI and EcoRV patterns resulted in the elucidation of 16 ribotypes. Two of them (B and O) characterized 65.7% of the strains studied, while the 14 other ribotypes were found in no more than three strains each. A relationship was established between biovars and ribotypes: strains of biovar Orientalis were of ribotypes A to G, those of biovar Antiqua were of ribotypes F to O, and those of biovar Medievalis were of ribotypes O and P. Great heterogeneity in rRNA restriction patterns was found among strains isolated in Africa; this heterogeneity was less pronounced among Asian isolates and was completely absent from the American strains. Pulsed-field gel electrophoresis was performed on the DNAs of some strains, but it appeared that different colonies from the same strain displayed different pulsed-field gel electrophoresis patterns and therefore that this technique was not suitable for comparison of Y. pestis isolates. In contrast, the ribotypes of individual colonies within a given strain were stable and were not modified after five passages in vivo. A clear correlation between the history of the three plague pandemics and the ribotypes of the strains could be established.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen J. K., Saunders N. A. Epidemiological typing of Yersinia enterocolitica by analysis of restriction fragment length polymorphisms with a cloned ribosomal RNA gene. J Med Microbiol. 1990 Jul;32(3):179–187. doi: 10.1099/00222615-32-3-179. [DOI] [PubMed] [Google Scholar]
- Blumberg H. M., Kiehlbauch J. A., Wachsmuth I. K. Molecular epidemiology of Yersinia enterocolitica O:3 infections: use of chromosomal DNA restriction fragment length polymorphisms of rRNA genes. J Clin Microbiol. 1991 Nov;29(11):2368–2374. doi: 10.1128/jcm.29.11.2368-2374.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel E., Guiyoule A., Mercereau-Puijalon O., Mollaret H. H. Chromosomal marker for the 'high pathogenicity' phenotype in Yersinia. Contrib Microbiol Immunol. 1991;12:192–197. [PubMed] [Google Scholar]
- Carniel E., Mercereau-Puijalon O., Bonnefoy S. The gene coding for the 190,000-dalton iron-regulated protein of Yersinia species is present only in the highly pathogenic strains. Infect Immun. 1989 Apr;57(4):1211–1217. doi: 10.1128/iai.57.4.1211-1217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEVIGNAT R. Variétés de l'espèce Pasteurella pestis; nouvelle hypothèse. Bull World Health Organ. 1951;4(2):247–263. [PMC free article] [PubMed] [Google Scholar]
- Fetherston J. D., Schuetze P., Perry R. D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992 Sep;6(18):2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- Grimont F., Chevrier D., Grimont P. A., Lefevre M., Guesdon J. L. Acetylaminofluorene-labelled ribosomal RNA for use in molecular epidemiology and taxonomy. Res Microbiol. 1989 Sep;140(7):447–454. doi: 10.1016/0923-2508(89)90065-x. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Iteman I., Baril C., Saint Girons I., Carniel E. Pulse field electrophoresis of the chromosome of the pathogenic yersiniae. Contrib Microbiol Immunol. 1991;12:198–202. [PubMed] [Google Scholar]
- Iteman I., Guiyoule A., de Almeida A. M., Guilvout I., Baranton G., Carniel E. Relationship between loss of pigmentation and deletion of the chromosomal iron-regulated irp2 gene in Yersinia pestis: evidence for separate but related events. Infect Immun. 1993 Jun;61(6):2717–2722. doi: 10.1128/iai.61.6.2717-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARIMI Y. CONSERVATION NATURELLE DE LA PESTE DANS LE SOL. Bull Soc Pathol Exot Filiales. 1963 Nov-Dec;56:1183–1186. [PubMed] [Google Scholar]
- Koblavi S., Grimont F., Grimont P. A. Clonal diversity of Vibrio cholerae O1 evidenced by rRNA gene restriction patterns. Res Microbiol. 1990 Jul-Aug;141(6):645–657. doi: 10.1016/0923-2508(90)90059-y. [DOI] [PubMed] [Google Scholar]
- Lucier T. S., Brubaker R. R. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J Bacteriol. 1992 Apr;174(7):2078–2086. doi: 10.1128/jb.174.7.2078-2086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Pasquier N., Picard B., Heeralal S., Krishnamoorthy R., Goullet P. Correlation between ribosomal DNA polymorphism and electrophoretic enzyme polymorphism in Yersinia. J Gen Microbiol. 1990 Aug;136(8):1655–1666. doi: 10.1099/00221287-136-8-1655. [DOI] [PubMed] [Google Scholar]
- Schaffer H. E., Sederoff R. R. Improved estimation of DNA fragment lengths from Agarose gels. Anal Biochem. 1981 Jul 15;115(1):113–122. doi: 10.1016/0003-2697(81)90533-9. [DOI] [PubMed] [Google Scholar]






