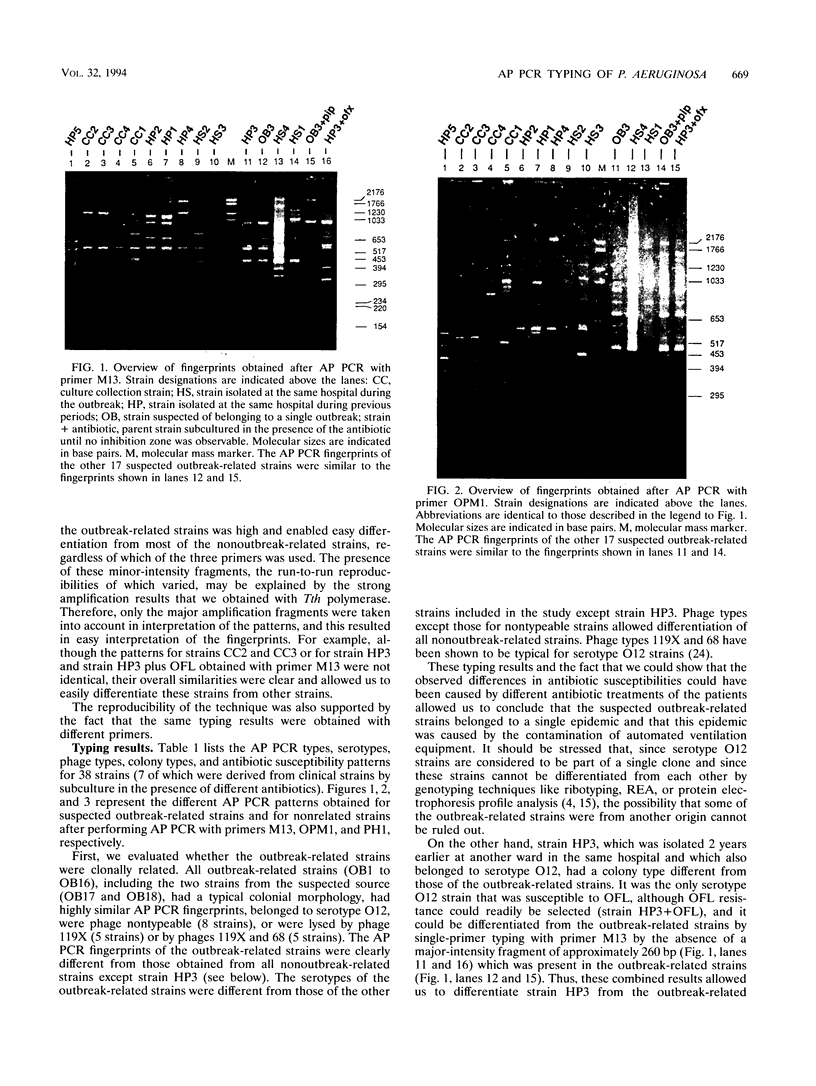
Abstract
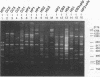
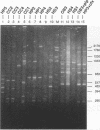
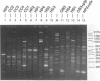
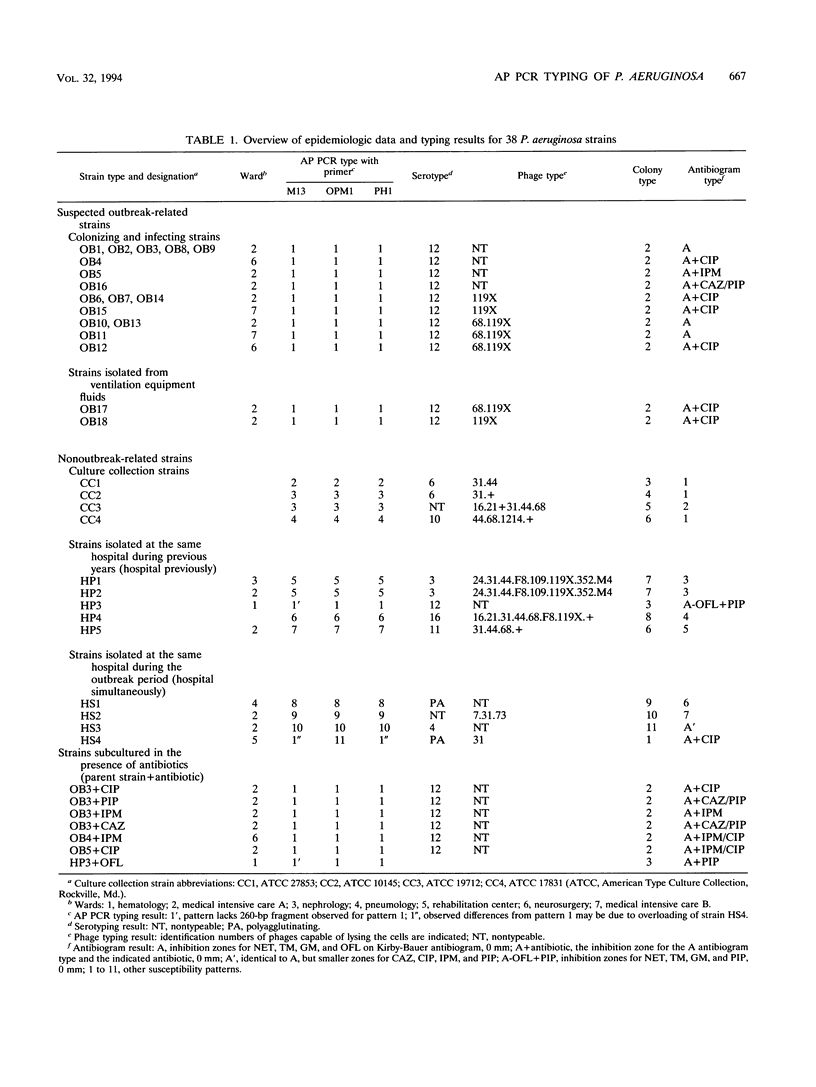
A total of 16 colonizing and infecting ofloxacin-resistant Pseudomonas aeruginosa strains and two strains isolated from ventilation equipment fluids, all with similar colonial morphologies and with minor but distinct susceptibility differences, were suspected of belonging to a single outbreak and were studied by arbitrary primer (AP) PCR. Thirteen nonrelated strains were included to evaluate the discriminatory capacity of the technique. AP PCR fingerprinting was compared with serotyping, phage typing, and antibiotic susceptibility testing. AP PCR was performed independently with three different primers. The different AP PCR typing systems yielded almost identical patterns for the epidemic strains and enabled us to differentiate most of the nonrelated strains from each other and from the outbreak strains. The combination of AP PCR typing and the phenotyping techniques that we used enabled us to conclude that an outbreak was occurring. In general, the typeability of AP PCR was greater than those of phage typing and serotyping, while the discriminatory powers of the three methods were comparable.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aufauvre-Brown A., Cohen J., Holden D. W. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J Clin Microbiol. 1992 Nov;30(11):2991–2993. doi: 10.1128/jcm.30.11.2991-2993.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc D. S., Siegrist H. H., Sahli R., Francioli P. Ribotyping of Pseudomonas aeruginosa: discriminatory power and usefulness as a tool for epidemiological studies. J Clin Microbiol. 1993 Jan;31(1):71–77. doi: 10.1128/jcm.31.1.71-77.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukadida J., de Montalembert M., Gaillard J. L., Gobin J., Grimont F., Girault D., Véron M., Berche P. Outbreak of gut colonization by Pseudomonas aeruginosa in immunocompromised children undergoing total digestive decontamination: analysis by pulsed-field electrophoresis. J Clin Microbiol. 1991 Sep;29(9):2068–2071. doi: 10.1128/jcm.29.9.2068-2071.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattard F., Gaudin O. G., Pozzetto B., Ros A., Mbida A. D. Genotypic homogeneity of nosocomial Pseudomonas aeruginosa O12 strains demonstrated by analysis of protein profiles, DNA fingerprints and rRNA gene restriction patterns. Eur J Clin Microbiol Infect Dis. 1993 Jan;12(1):57–61. doi: 10.1007/BF01997061. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- HABS I. Untersuchungen über die O-Antigene von Pseudomonas aeruginosa. Z Hyg Infektionskr. 1957;144(3):218–228. [PubMed] [Google Scholar]
- Lehmann P. F., Lin D., Lasker B. A. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J Clin Microbiol. 1992 Dec;30(12):3249–3254. doi: 10.1128/jcm.30.12.3249-3254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier S. I., Wernars K. Typing of Listeria strains by random amplification of polymorphic DNA. Res Microbiol. 1992 Jun;143(5):499–505. doi: 10.1016/0923-2508(92)90096-7. [DOI] [PubMed] [Google Scholar]
- Myers L. E., Silva S. V., Procunier J. D., Little P. B. Genomic fingerprinting of "Haemophilus somnus" isolates by using a random-amplified polymorphic DNA assay. J Clin Microbiol. 1993 Mar;31(3):512–517. doi: 10.1128/jcm.31.3.512-517.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard C., Brousseau R., Mouton C. Application of polymerase chain reaction with arbitrary primer (AP-PCR) to strain identification of Porphyromonas (Bacteroides) gingivalis. FEMS Microbiol Lett. 1992 Aug 15;74(2-3):163–168. doi: 10.1111/j.1574-6968.1992.tb05360.x. [DOI] [PubMed] [Google Scholar]
- Ogle J. W., Janda J. M., Woods D. E., Vasil M. L. Characterization and use of a DNA probe as an epidemiological marker for Pseudomonas aeruginosa. J Infect Dis. 1987 Jan;155(1):119–126. doi: 10.1093/infdis/155.1.119. [DOI] [PubMed] [Google Scholar]
- Pitt T. L. Epidemiological typing of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 1988 Apr;7(2):238–247. doi: 10.1007/BF01963095. [DOI] [PubMed] [Google Scholar]
- Pitt T. L., Livermore D. M., Pitcher D., Vatopoulos A. C., Legakis N. J. Multiresistant serotype O 12 Pseudomonas aeruginosa: evidence for a common strain in Europe. Epidemiol Infect. 1989 Dec;103(3):565–576. doi: 10.1017/s095026880003096x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C. L., Yeo C. C., Tay L. Genome fingerprinting by pulsed-field gel electrophoresis and ribotyping to differentiate Pseudomonas aeruginosa serotype O11 strains. Eur J Clin Microbiol Infect Dis. 1992 Sep;11(9):817–822. doi: 10.1007/BF01960881. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Jincharadze A. G., Prosnyak M. I., Ivanov P. L., Limborska S. A. M13 phage DNA as a universal marker for DNA fingerprinting of animals, plants and microorganisms. FEBS Lett. 1988 Jun 20;233(2):388–392. doi: 10.1016/0014-5793(88)80467-8. [DOI] [PubMed] [Google Scholar]
- Samadpour M., Moseley S. L., Lory S. Biotinylated DNA probes for exotoxin A and pilin genes in the differentiation of Pseudomonas aeruginosa strains. J Clin Microbiol. 1988 Nov;26(11):2319–2323. doi: 10.1128/jcm.26.11.2319-2323.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speert D. P., Campbell M. E., Farmer S. W., Volpel K., Joffe A. M., Paranchych W. Use of a pilin gene probe to study molecular epidemiology of Pseudomonas aeruginosa. J Clin Microbiol. 1989 Nov;27(11):2589–2593. doi: 10.1128/jcm.27.11.2589-2593.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struelens M. J., Deplano A., Godard C., Maes N., Serruys E. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J Clin Microbiol. 1992 Oct;30(10):2599–2605. doi: 10.1128/jcm.30.10.2599-2605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassart G., Georges M., Monsieur R., Brocas H., Lequarre A. S., Christophe D. A sequence in M13 phage detects hypervariable minisatellites in human and animal DNA. Science. 1987 Feb 6;235(4789):683–684. doi: 10.1126/science.2880398. [DOI] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Pretzman C., Postic D., Saint Girons I., Baranton G., McClelland M. Genomic fingerprinting by arbitrarily primed polymerase chain reaction resolves Borrelia burgdorferi into three distinct phyletic groups. Int J Syst Bacteriol. 1992 Jul;42(3):370–377. doi: 10.1099/00207713-42-3-370. [DOI] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]