Abstract
DNA barcodes linked to genetic features greatly facilitate screening these features in pooled formats using microarray hybridization, and new tools are needed to design large sets of barcodes to allow construction of large barcoded mammalian libraries such as shRNA libraries. Here we report a framework for designing large sets of orthogonal barcode probes. We demonstrate the utility of this framework by designing 240,000 barcode probes and testing their performance by hybridization. From the test hybridizations, we also discovered new probe design rules that significantly reduce cross-hybridization after their introduction into the framework of the algorithm. These rules should improve the performance of DNA microarray probe designs for many applications.
Keywords: hybridization, shRNA, deconvolution, library screen
ADNA barcode is a short DNA sequence that uniquely identifies a certain linked feature such as a gene or a mutation. Linking features to DNA barcodes of homogenous length and melting temperature (Tm) allows experiments to be performed on the features in a pooled format, with subsequent deconvolution by PCR followed by microarray hybridization or high throughput sequencing. DNA barcode technology greatly improves the throughput of genetic screens, making possible experiments that would otherwise be quite time-consuming or laborious. For example, DNA barcodes built into the yeast deletion collection have facilitated identification of genes whose mutants are depleted or enriched under various growth conditions or drug treatments (1–4).
For the construction of large libraries of short hairpin RNAs (5) or open-reading frames (6), it is desirable to have the libraries linked with barcodes with superior microarray hybridization characteristics. Although the DNA barcodes in the yeast deletion collection have performed well, there are only about 16,000 unique barcodes in the TAG4 set (7), which are too few for barcoding large mammalian libraries. Using random barcodes for these large libraries is less than optimal, because of the frequent off-target hybridization that occurs with random barcodes.
Numerous publications and software tools are currently available for designing DNA microarray probes (8–11). However there are no software packages or even design rules published so far specifically for DNA barcode probes. Regular probe design procedures do not fit the purpose of barcode probes very well because of one major difference in target sequence constraints. For current DNA probe design procedures, there is a fixed set of long DNA sequences (such as all yeast open-reading frames or all human RefSeq sequences) that constrain target sequences. One or more short tags (probes) are then picked that uniquely identify each target sequence and display reduced cross-hybridization to regions of other targets. In the case of barcode designs, however, the set of target sequences is not fixed. Instead, we are free to select optimal probes from the enormous space of short oligos of the same length. Also, because the probes and targets are the same sequences in the barcode case, cross-hybridization effects need to be avoided only within the probe set.
Here we present a framework for designing a large set of orthogonal DNA barcodes (DeLOB). We designed 240,000 barcodes with this procedure. From hybridization data, we found that compositions of A and C nucleotides, especially CCCC homopolymer sequences close to the 5′ end of probes, significantly affect hybridization specificity. We formulated new design rules on the basis of these observations and generated a second set of 240,000 probes. Test hybridization on these probes indicated that the introduction of new rules significantly reduced cross-hybridization. The 240,000 optimized DNA barcodes generated by our findings will be a valuable resource for constructing large libraries for genetic screening.
Results
The DeLOB Framework.
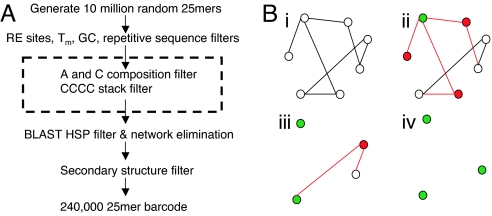
The DeLOB DNA barcode design procedure is outlined in Fig. 1A. We adopted most of the empirical rules recognized by other probe designing tools, such as unique sequences, homogeneous Tm's, and the absence of repetitive sequences and secondary structures. Special emphasis was placed on the uniqueness of probe sequence in the DeLOB procedure because cross-hybridization has to be minimized as much as possible for barcode probes. We set out to design a set of 240,000 barcode probes and generated a starting set of 10 million random 25mers as candidate probes. After excluding candidates containing restriction enzyme sites that were reserved for cloning, or those having too high or low Tm's (Tm < 58 °C or Tm > 68 °C), or those containing repetitive sequences, about 6 million candidates remained. These 6 million candidates were screened against themselves by BLAST to determine shared sequence similarity. To enforce the uniqueness of probes, we selected candidates that have the shortest BLAST high-score segment pairings (HSPs) among them. Candidates were taken as “orthogonal” if they had no shared HSPs of longer than 12 bases with each other or the set of their reverse complementary sequences. From the BLAST result, there were ≈12,000 orthogonal candidates, which were far less than the desired 240,000 probes. However, because candidates in the nonorthogonal group were nonorthogonal to only a fraction of other candidates, it was possible that a subset of candidates in the nonorthogonal group could be orthogonal to each other. We therefore designed a “network elimination algorithm” to select a subset of orthogonal candidates out of the 6 million nonorthogonal candidates.
Fig. 1.
The DeLOB DNA barcode design procedure. (A) Ten million random 25mers were generated and sequentially passed through restriction enzyme (RE) site, Tm, GC composition, and repetitive sequence filters. Candidates passing these filters were searched against themselves by BLAST and a subset of orthogonal sequences was selected on the basis of their BLAST results and the network elimination algorithm. After applying a secondary structure filter to eliminate self-folding-prone candidates, we obtained a final set of 240,000 probes. The 2 filters in the dashed box were based on rules discovered from analyzing hybridization data from first round probe design. (B) The network elimination algorithm. (i) Nonorthogonal candidate pairs were represented by a network graph. Each vertex was a candidate and each edge was a longer than 12-base match between the 2 connected candidates. (ii) One candidate was randomly chosen and placed in the orthogonal group (green). All candidates that were connected to this one were labeled in red and then eliminated from the network together with all edges incident to these red vertices. (iii) The random selection and elimination steps were repeated on the remaining network members. (iv) At the end, only orthogonal candidates were left.
A schematic illustration of the network elimination algorithm is shown in Fig. 1B. Briefly, candidates and the nonorthogonality between them were transformed into a network graph with vertices representing candidates and edges representing longer than 12-base HSPs between candidates (Fig. 1B i). One candidate was randomly picked as an orthogonal probe, and all candidates that were connected to it were eliminated from the network (Fig. 1B ii). By iterating these selection and elimination steps, we successfully separated a subset (≈400,000) of orthogonal candidates.
To increase the stringency of sequence diversity, we further excluded candidates that have more than 10 HSPs of 11 or 12 bases to other candidates in the orthogonal group. At the end, a secondary structure filter based on the UNAFold program (12) was applied to eliminate candidates that form potential intraprobe secondary structures to arrive at a final set of 240,000 probes.
Probe Hybridization Test.
To test the performance of the designed barcode probes, we performed 3 parallel microarray hybridizations. We synthesized the 240,000 oligos in 3 subpools, each containing 80,000 targets. Each subpool was labeled with Cy3 using a priming protocol that labels both strands and the mixture of all 3 pools (total) was labeled with Cy5. These 2 samples were hybridized to a microarray containing all 240,000 probes in a 1:3 ratio such that targets in each Cy3 subpool were in an equimolar ratio with their corresponding targets in the total Cy5 pool. This experimental design allows detection of intersubpool cross-hybridizations by observing the outliers of Cy3/Cy5 ratios of probes. For example, when hybridizing subpool 1 vs. total, cross-hybridization on pool 1 probes from Cy5-labeled targets of other subpools will lead to abnormally low Cy3/Cy5 ratios. In contrast, cross-hybridization on probes in pool 2 or 3 from Cy3-labeled targets of subpool 1 will cause abnormally high Cy3/Cy5 ratios for those probes.
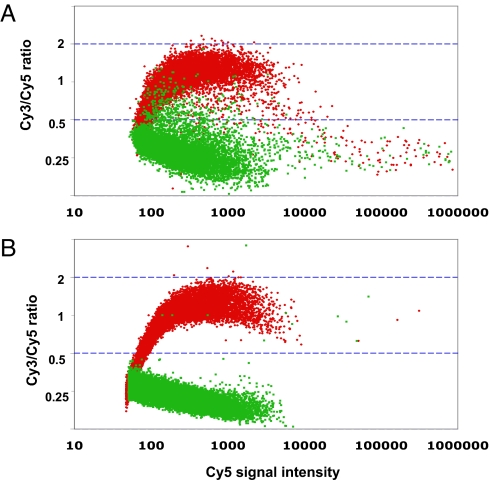
The hybridization results are summarized in Fig. 2A, where we plotted Cy3/Cy5 ratio vs. Cy5 channel signal intensity. Probes with corresponding targets in the Cy3-labeled subpool (the “present” group, in red) have an average Cy3/Cy5 ratio near 1, whereas probes that did not have corresponding targets in the Cy3-labeled subpool (the “absent” group, in green) have an average Cy3/Cy5 ratio close to 0.25. The red and green spot masses are intermixed at both extremely low and high intensities, but are more clearly separated at intermediate signals.
Fig. 2.
A representative 2-color hybridization experiment of a single subpool labeled with Cy3 vs. the entire pool labeled with Cy5. Separation of the “present group” (red, probes that have target sequences in the Cy3-labeled subpool) and “absent group” (green, probes that do not have target sequences in the Cy3-labeled subpool) in the first round design (A) and the second round of design (B). The dashed blue lines represent Cy3/Cy5 ratio of 2 and 0.5, respectively. Only 10,000 randomly sampled probes in each group were plotted for clarity.
A good probe should have 2 properties: high responsiveness and low cross-hybridization. We defined a probe as having high responsiveness if it had Cy5 channel signal within an acceptable range (signal intensity greater than 100 arbitrary fluorescent units (afu) and lower than 5,000 afu, corresponding to the 10% and 98% quantiles, respectively), and comparable Cy5 and Cy3 channel signals when its corresponding target was in the Cy3-labeled subpool (Cy3/Cy5 ratio between 0.5 and 2, i.e., the log2 ratio is within 1 unit from the center of 0, red spots between the 2 dashed blue lines in Fig. 2A). Similarly, low cross-hybridization was defined as having low Cy3 signal compared to Cy5 signal when the corresponding target was absent from the Cy3-labeled subpool (Cy3/Cy5 ratio below 0.5, green spots below the lower dashed blue line). Almost all red spots with intensity above 10,000 afu are below the lower blue line, indicating that these high signals are primarily contributed by cross-hybridization.
We found that about 84% of the probes (202,615 probes, referred to as the “good” group hereafter) passed the high-responsiveness and low cross-hybridization filters in all 3 hybridizations and were counted as acceptable probes. Of the 16% of probes performing poorly, the great majority (26,942 probes) had very low signals (signal intensity <100 afu, nonresponding or missing probes, “dim” group), 4,435 probes had very high signals (signal intensity >5,000 afu, strong cross-hybridizing probes, “bright” group), and 7,415 probes had signals in between (“medium” group).
Although intrasubpool cross-hybridization was not directly identified, its scale can be estimated to be around half of those from intersubpool cross-hybridization, as probes in the 3 subpools were randomly assigned and the 3 pools were the same size. This will correspond to about 1.8% of probes in the good group, because the intersubpool cross-hybridization rate is about 3.5% for probes of signal intensity between 100 and 5000 afu (comparing the medium group to the combined medium and good groups). But because probes having intrasubpool cross-hybridization are also very likely to have intersubpool cross-hybridization, the real number should be much lower than 1.8% in the good group after probes with intersubpool cross-hybridization have been eliminated.
Discovery of New Probe Design Rules.
If there are any probe characteristics that are specifically associated with performance of probes, it should be possible to form new design rules on the basis of these characteristics to improve future probe design. Therefore, we compared BLAST scores, Tm's, nucleotide compositions, and repetitive nucleotide stack compositions among the 4 groups identified as dim, medium, good, and bright.
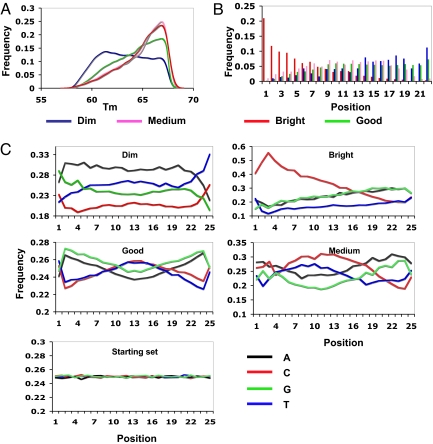
We did not find a significant difference between the groups on probe BLAST scores, probably because the BLAST scores were already very homogeneous after the probes were selected from a total of 10 million candidates. There were, however, differences in the distributions of Tm's between probe groups (Fig. 3A). Probes in the bright and medium groups were strongly biased toward having high Tm's (higher than 65 °C), whereas the dim group was biased toward having low Tm's (lower than 62 °C). However, this statistical observation is not very helpful in forming new probe designing rules because there were also many good probes having Tm's in these ranges.
Fig. 3.
Analysis of probe composition and activity. (A) Distribution of Tm's in the 4-probe groups. (B) Distribution of CCCC motifs along probe lengths in the 4 groups. In the bright group, CCCCs were highly biased toward the very 5′ end, whereas in other groups, CCCCs were depleted from the very 5′ end of probes. (C) Nucleotide compositions at each of the 25 bases on probes in the 4 groups and the starting set of 10 million candidates 25mers. Dim probes had high A and low C compositions along the probe except for the 2 ends. Bright probes had extremely skewed C composition at the 5′ half of probes. The starting set had equal compositions for the 4 nucleotides at all 25 positions.
We postulated that difference in signal intensities between groups might be caused by differences in overall GC content of probes. The G + C contents in the 4 groups were indeed in the expected order, with the bright and dim groups having the highest and lowest G + C contents, respectively (Table 1). However, the differences were rather small to account for the disparity in their hybridization properties. Instead, the most striking differences were in C and A nucleotide compositions. For the good group, each of the 4 nucleotides comprised roughly 25% of the total. In the dim group, there was a markedly higher percentage of A nucleotides (29.4%) and low C (20.9%) while both G and T remained at ≈25%. In contrast, the bright group had both A and T around 25%, but with extremely high C (34.4%) and low G (16.5%). The low G was likely a compensation effect because we set the G + C to be around 50% when designing the probes. From this analysis, we concluded that high A and low C nucleotide composition is associated with low hybridization signals, and high C nucleotide composition is associated with high hybridization signals.
Table 1.
Comparison of nucleotide compositions among 4 groups of probes having different hybridization behavior: Single nucleotide compositions
| Probes | G + C % | A% | C% | G% | T% |
|---|---|---|---|---|---|
| Good | 49.2 | 25.0 | 24.7 | 24.5 | 25.8 |
| Bright | 50.9 | 24.5 | 34.4 | 16.5 | 24.6 |
| Medium | 50.4 | 26.6 | 26.6 | 23.8 | 22.9 |
| Dim | 46.4 | 29.4 | 20.9 | 25.6 | 24.2 |
To test whether different nucleotide compositions at varying positions within probes will affect their hybridization behavior, we compared the nucleotide compositions at each of the 25 probe positions between the 4 groups. All 4 nucleotides in the good group stay around the designed 25% level across the probe length, but show an interesting “twisting” pattern (Fig. 3C). This pattern did not exist in the starting set of 10 million probes (Fig. 3C), so it must be the result of passing through serial filters in the DeLOB procedure. The dim group had continuous high A (around 30%) and low C (around 20%) except on the ends of the probes. Again, the bright group showed the most striking pattern for distribution of C: all of the first 12 nucleotides had very high C composition (higher than 30%), reaching a maximum of 55% at position 3.
When examining the probe sequences of the bright group, we found that many probes had a pattern of 4 consecutive Cs (CCCC stacks) in them. As we already excluded candidates containing 5 or longer single nucleotide repeats in the designing procedure, 4-nucleotide repeats were the longest in the orthogonal set. To see whether quadruplet stacks were associated with probe behavior, we compared the compositions of AAAA, CCCC, GGGG, and TTTT stacks in the 4 groups (Table 2). Similar to what we observed in single nucleotide compositions, the dim group had CCCC stacks significantly depleted and AAAA stacks significantly enriched, whereas the bright group had CCCC extremely enriched and GGGG depleted. Interestingly, the good group had both CCCC and AAAA significantly depleted suggesting that both AAAA and CCCC should be avoided in designing probes.
Table 2.
Abundance of N4 compositions among probe classes
| Total | Dim | Medium | Bright | Good | |
|---|---|---|---|---|---|
| Probes | 241399 | 26942 | 7415 | 4426 | 202615 |
| CCCC | 11448 | 490 | 1358 | 2712 | 6888 |
| (P = 7.1 × 10−113) | (P = 0) | (P = 0) | (P = 7 × 10−178) | ||
| AAAA | 13042 | 2545 | 522 | 248 | 9727 |
| (P = 1.9 × 10−189) | (P = 4.5 × 10−10) | (P = 0.55) | (P = 4 × 10−33) | ||
| GGGG | 11503 | 1636 | 370 | 32 | 9465 |
| (P = 7.4 × 10−24) | (P = 0.36) | (P = 1.6 × 10−36) | (P = 0.05) | ||
| TTTT | 12978 | 1401 | 357 | 234 | 10986 |
| (P = 0.20) | (P = 0.03) | (P = 0.79) | (P = 0.36) |
To examine whether there is a position effect of quadruplet stacks along a probe, we checked the locations of stacks in the 4 probe groups. There was no significant difference in distributions of AAAA, GGGG, and TTTT stacks along the probe between groups (data not shown). Interestingly, we again observed opposing patterns of CCCC distribution between the bright and dim groups (Fig. 3B). In the bright group, CCCC stacks were predominantly located at the very 5′ of probes, whereas in the dim group, they were more enriched at the very 3′ of probes. The good group also had CCCC stacks depleted at their 5′ ends. Collectively, these observations suggest that CCCC stacks in the 5′ half of probes are correlated with strong cross-hybridization.
On the basis of these nucleotide composition analyses, we derived 2 new probe design rules: (i) to improve probe responsiveness, the nucleotide composition of A in a probe should be limited to below 28%, and AAAA stacks should be avoided in probe sequences; (ii) to reduce cross-hybridization effects but still maintain reasonable probe response, the C nucleotide composition of probes should be limited to between 22 and 28%, and CCCC stack or 4 nonconsecutive Cs in any 6 consecutive nucleotides in the first 12 positions of a probe should be avoided.
Second Round Probe Design and Hybridization Test.
We designed a second set of 240,000 probes after incorporating the 2 new rules into the DeLOB. Before the candidates were screened against themselves by BLAST, they were first screened against the good probes that were recovered from the first round of design to eliminate candidates that were not orthogonal to the original good probes. This was done so that the barcodes from both batches could later be combined into a single large pool without compromising hybridization performance.
We performed the same hybridization test for the second batch of probes as was performed on the first batch. The results are summarized in Fig. 2B, which shows 2 major differences when compared to Fig. 2A. First, there is a cleaner separation of the present group (in red) from the absent group (in green) at signal intensity above 100 afu, although the average Cy3/Cy5 ratios of the 2 groups are still around 1 and 0.25, respectively. Second, the number of spots with an intensity >5000 afu was decreased more than 7-fold, and the long tail of intermixed red and green spots at intensity >10,000 afu disappeared. These hybridization results suggest that introduction of the new design rules significantly reduces cross-hybridization. At the same time, the percentage of good probes increased from 84% to 87% with the same high responsiveness and low cross-hybridization filter applied on the first batch data. This improvement is not as striking mainly because there are more nonresponding probes in the second round (31,627 compared to 26,942 in the first round) even though we normalized the 2 batches of hybridization data to have the same median.
We combined the good probes from the 2 rounds of design and eliminated probes with the lowest signal intensities to obtain a desired final set of 240,000 probes that can be used as orthogonal DNA barcodes in future experiments. Probe sequences and implementation of the network elimination algorithm are available from our lab Web site (http://elledgelab.bwh.harvard.edu/Barcode).
Discussion
DNA barcodes should have homogenous Tm's, high sensitivity, and specificity in hybridization to correctly deconvolute pool compositions. On the basis of empirical observations and theoretical calculations, the currently accepted DNA probe design rules include that probes should have roughly equal Tm's, low sequence similarities, and lack of secondary structures (11). However, for reasons that are not well understood, there are often exceptional probes that have very low responsiveness or high cross-hybridization, despite having been designed according to the commonly accepted rules.
We applied the currently known rules of microarray probe design to generate a set of 240,000 orthogonal 25mers that can be used as DNA barcodes. We sought to minimize cross-hybridization among probes by reducing sequence similarities as much as possible. In the well-validated 20mer barcodes in the yeast deletion collection (4), the longest contiguous matches were 9 bases, which was 45% of the probe length. It was also reported that cross-hybridization significantly dropped when the longest match was shorter than 40% of probe length for probes of 50 to 70 bases (13, 14). We therefore estimated that in 25mers, less than 50% of contiguous sequence match (12 bases or shorter) might be a reasonable cutoff for probe sequence similarities. When we define orthogonality as having stretches of no longer than 12 bases of contiguous matches to any other probes, it is very difficult to design libraries as large as 240,000 orthogonal probes directly based on BLAST results, as the great majority of candidates had some nonorthogonal matches in the candidate set. However, we noticed that in the nonorthogonal candidate network, many of these disqualified probes were not directly connected, allowing us to remove some “connecting” candidates to filter out a set of orthogonal candidates. We therefore implemented a network elimination algorithm for selecting orthogonal probes. Because the number of edges incident to vertices were quite homogeneous, the numbers of finally selected orthogonal probes did not vary greatly, regardless of how we randomly chose candidates as orthogonal. This algorithm can generate multiple sets of probes that are orthogonal inside each set, but not between sets. By reusing candidates in the nonorthogonal group, we had a larger set of orthogonal candidates upon which to apply additional constraints to arrive at a desired number of probes. The 240,000 barcode probes ultimately generated in this fashion will be a valuable resource for constructing large-scale libraries. It should be noted that this set of 240,000 orthogonal barcodes could be expanded to 480,000 barcodes with their reverse complementary sequences if a single-stranded hybridization sample, such as a sample made of directional RNAs, were used as probe instead of a double-stranded sample. Furthermore, using a single-stranded sample should reduce cross-hybridization for the 240,000 set by 50%.
It was surprising that it was not the overall G + C composition of probes but C alone that was contributing most to cross-hybridization. This unexpected finding reflects the fact that some fundamentals of DNA hybridization are still not well understood regardless of its wide application (15). Similarly it was only A but not T composition that was associated with low hybridization signal. Although some of the low signals may be the result of missing targets, the strong association of high A and low C compositions with the dim group suggests that probes in this category indeed hybridize poorly. These observations also clearly suggest that nucleotides A and T, or C and G are not equal in determining probe behavior. We speculate that these different behaviors may be caused by different probe structures, and molecular dynamics simulations of DNA molecules on glass surfaces (16) might provide hints to solve this puzzle.
Our observation that unusual compositions of nucleotide A and C abundance and CCCC stacks affects probe sensitivity and specificity is consistent with previous analyses on Affymetrix and Nimblegen arrays. In analyzing Affymetrix mismatch (MM) probes of high outlier signal intensities, Wang et al. (17) observed high C and low A compositions at the 5′ half of these probes, which is very similar to what we observed in this study. This is also consistent with what Wei et al. found on Nimblegen microarrays that protruding ends contributed more to signal intensity than tethered ends (18). In a reexamination of the representative MM probes listed in Wang et al.'s report (17), we found that all of the high-intensity MM probes had CCCC in their sequences (data not shown). In another study, Wu et al. analyzed concordance of Affymetrix probes by comparing signal correlations between neighboring probes (19). They observed the strongest cross-hybridization effect on probes containing GGGG stacks, which did not show cross-hybridization in our study. However, they also found that probes containing CCCC also tend to result in increased cross-hybridization. On the basis of these data, it appears that cross-hybridization to probes containing a large number of Cs or having CCCC stacks is a common phenomenon in both Agilent and Affymetrix chips. Our second round hybridization test showed that cross-hybridization was significantly reduced after eliminating CCCC stacks and lowering C compositions at the 5′ half of probes. This rule thus should be adopted in designing any DNA microarray probes to reduce cross-hybridization.
Materials and Methods
The DeLOB Protocol.
Ten million 25mer oligo DNA sequences were generated as candidates with the “makenucseq” program in the EMBOSS package (20). These DNA sequences were sequentially fed into a restriction enzyme filter which exclude sequences containing restrictive enzyme sites that are reserved for library cloning (EcoR1, XhoI, BglII, MluI, AvrII, FseI, and MfeI), a Tm filter based on the “nearest neighbor model” (21) to exclude sequences of Tm below 58 °C or above 68 °C, a GC composition filter to exclude sequences of GC below 40% or above 60%, and a repetitive sequence filter to exclude sequences containing repetitive tracts (5 or longer single nucleotide repeats or 4 or longer double nucleotides repeats). Candidates that passed all these filters were compared to each other for sequence similarity using the BLAST program with the “−F” option turned off. We defined 2 candidates to be orthogonal to each other if they do not have stretches longer than 12 bases of HSPs between them. On the basis of BLAST results, candidates were divided into 2 groups: those with no HSPs of 13 bases or longer to any other candidate (orthogonal probes I), and those with longer than 12 bases HSPs to at least 1 of other candidates (nonorthogonal probes). For the latter group, we applied a “network elimination” algorithm (see below) to obtain a subset of candidates that were orthogonal to each other (orthogonal probes II), and combine with orthogonal probes I. These orthogonal probes were then fed into a secondary structure filter, which was based on the “hybrid-ss” program in the UNAFold package (12) to exclude probes that form intraprobe secondary structures (self-folding energy < −2 kJ/mol at 50 °C).
The Network Elimination Algorithm.
We first constructed a network from all nonorthogonal candidates. Each vertex in the network represented a candidate and an edge represented the existence of a longer than 12-base HSP between the 2 connected candidates. We randomly chose 1 candidate and placed it in the inclusion group (orthogonal probes II). Candidates that were connected to this one were placed into the exclusion group. We then eliminated all candidates in the exclusion group from the network, together with all edges incident to these candidates. This selection-and-elimination procedure was then repeated on the remaining network till all candidates were put into either of the 2 groups. Candidates in the inclusion group were orthogonal to each other.
Microarray Hybridization.
Target sequences were synthesized on Agilent arrays in 3 individual subpools, each containing 80,000 targets. The oligos were designed such that 3 25mer target sequences were concatenated by EcoRI and XhoI sites for future cloning purpose and flanked by PCR primer sites at the 5′ and 3′ ends. These subpools were cleaved from the arrays by Agilent and PCR amplified. Targets in each subpool were PCR amplified using PCR primers with T7 sites and labeled with Cy3 using a T7 primer. An equal proportion mixture of the 3 subpools (the total) was labeled with Cy5. No restriction enzyme digestion of oligos was applied at any step. Then each subpool was hybridized vs. the total in a 1:3 ratio by amount of DNA onto a microarray that contains the designed 240,000 probes. Microarray hybridization and feature extraction were performed following the standard Agilent protocol.
Hybridization Data Analysis and New Probe-Designing Rule Discovery.
Intensity data were median normalized on both Cy5 and Cy3 channels to have an arbitrary median of 200. Specifically, while the median value for the Cy5 channel was computed from all probes, the median value for the Cy3 channel was calculated from probes that had their corresponding targets in the subpool. Probes that had a Cy3/Cy5 ratio greater than 0.5 when the corresponding targets were not in the subpool hybridized to the array were considered as having significant cross-hybridization. These cross-hybridizing probes were further divided into 3 groups on the basis of their signal intensity: bright probes with intensities greater than 5000 afu, dim probes with intensities below 100 afu, and medium probes with intensities between 100 and 5000 afu.
Various sequence characteristics of probes in the noncross-hybridization group and the 3 cross-hybridization groups were compared. These characteristics include distributions of Tm's, BLAST scores, overall nucleotide compositions, and nucleotide compositions at each of the 25 positions of probes. We also counted the occurrence of AAAA, CCCC, GGGG, and TTTT repeats in probes of the 4 groups and assessed statistical significance of enrichment or depletion of the 4 repeats in each group by the χ2 test. Positions of the nucleotide quadruplet distribution along probes were also compared between groups.
Acknowledgments.
We thank the Research Information Technology Group at Harvard Medical School for providing access to its computation facility and M. Li for technical assistance. This work is supported by Department of Defense Breast Cancer Innovator Awards (to S.J.E. and G.J.H.). G.J.H. and S.J.E. are Investigators with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 2.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 3.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker DD, et al. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 5.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 6.Rual JF, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierce SE, et al. A unique and universal molecular barcode array. Nat Methods. 2006;3:601–603. doi: 10.1038/nmeth905. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen HB, Wernersson R, Knudsen S. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 2003;31:3491–3496. doi: 10.1093/nar/gkg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 11.Hu G, et al. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics. 2007;8:350. doi: 10.1186/1471-2105-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 13.He Z, et al. Empirical establishment of oligonucleotide probe design criteria. Appl Environ Microbiol. 2005;71:3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kane MD, et al. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozhitkov AE, Tautz D, Noble PA. Oligonucleotide microarrays: widely applied–poorly understood. Brief Funct Genomic Proteomic. 2007;6:141–148. doi: 10.1093/bfgp/elm014. [DOI] [PubMed] [Google Scholar]
- 16.Wong KY, Pettitt BM. Orientation of DNA on a surface from simulation. Biopolymers. 2004;73:570–578. doi: 10.1002/bip.20004. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Characterization of mismatch and high-signal intensity probes associated with Affymetrix genechips. Bioinformatics. 2007;23:2088–2095. doi: 10.1093/bioinformatics/btm306. [DOI] [PubMed] [Google Scholar]
- 18.Wei H, et al. A study of the relationships between oligonucleotide properties and hybridization signal intensities from NimbleGen microarray datasets. Nucleic Acids Res. 2008;36:2926–2938. doi: 10.1093/nar/gkn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, et al. Short oligonucleotide probes containing G-stacks display abnormal binding affinity on Affymetrix microarrays. Bioinformatics. 2007;23:2566–2572. doi: 10.1093/bioinformatics/btm271. [DOI] [PubMed] [Google Scholar]
- 20.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 21.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]