Abstract
The identification of potential allergenic proteins is usually done by scanning a database of allergenic proteins and locating known allergens with a high sequence similarity. However, there is no universally accepted cut-off value for sequence similarity to indicate potential IgE cross-reactivity. Further, overall sequence similarity may be less important than discrete areas of similarity in proteins with homologous structure. To identify such areas, we first classified all allergens and their subdomains in the Structural Database of Allergenic Proteins (SDAP, http://fermi.utmb.edu/SDAP/) to their closest protein families as defined in Pfam, and identified conserved physicochemical property motifs characteristic of each group of sequences. Allergens populate only a small subset of all known Pfam families, as all allergenic proteins in SDAP could be grouped to only 130 (of 9318 total) Pfams, and 31 families contain more than four allergens. Conserved physicochemical property motifs for the aligned sequences of the most populated Pfam families were identified with the PCPMer program suite and catalogued in the webserver Motif-Mate (http://born.utmb.edu/motifmate/summary.php). We also determined specific motifs for allergenic members of a family that could distinguish them from non-allergenic ones. These allergen specific motifs should be most useful in database searches for potential allergens. We found that sequence motifs unique to the allergens in three families (seed storage proteins, Bet v 1, and tropomyosin) overlap with known IgE epitopes, thus providing evidence that our motif based approach can be used to assess the potential allergenicity of novel proteins.
Keywords: Allergy, Allergen classification, Cross-reactivity, Allergen motif
1. Introduction
The possibility that proteins from novel foods, drugs, or genetically modified organism may exhibit cross-reactivity with known allergens is of utmost concern to regulatory agencies, food scientists and physicians (WHO, 2003). Due to these considerations, it is important to be able to distinguish allergenic from nonallergenic proteins, and to predict potential IgE cross-reactivities (Aalberse, 2007; Breiteneder and Mills, 2006; Schein et al., 2007). Potential cross-reactive allergens often have very similar sequences (Aalberse and Stadler, 2006; Bonds et al., 2008). Thus, one of the first questions in determining potential cross-reactive foods is the degree of similarity between allergens. Allergens are referred to by names assigned by the Allergen Nomenclature Sub-Committee of the International Union of Immunological Societies (IUIS, www.allergen.org), based on the species/genus name of the source and the order they were identified (Chapman et al., 2007). This nomenclature system is independent of the biochemical and structural nature of the protein, and the names do not readily identify structural and sequence-based relationships among allergens. This means that, based on these names, one cannot easily identify the individual allergenic proteins in different organisms that could account for IgE cross-reactivity (Aalberse et al., 2001; Jenkins et al., 2005; Mirza et al., 2000; Schwietz et al., 2000).
Bioinformatics approaches and allergenic databases are now well established to identify molecular similarities of proteins as an explanation for clinically observed cross-reactivity from very different sources (Breiteneder and Mills, 2006; Brusic and Petrovsky, 2003; Furmonaviciene et al., 2005; Hileman et al., 2002; Schein et al., 2007; Thomas et al., 2005; Zorzet et al., 2004). The Structural Database of Allergenic Proteins (SDAP) (Ivanciuc et al., 2002, 2003) contains many sequence search tools that are seamlessly integrated in the design of the database. SDAP is user friendly and freely available on the Web to allergy researchers, food scientists and industrial engineers (http://fermi.utmb.edu/SDAP/). Allergy researchers can use SDAP primarily to determine food sources that might contain cross-reacting antigens. Regulators and industrial researchers can use the site tools to perform FASTA searches (Pearson, 1994) of allergenic proteins or sequence searches according to the WHO guidelines (Schein et al., 2006). FASTA searches are also helpful in clustering related allergens or suggesting the appropriate nomenclature for novel allergenic proteins. For example, cross-reactions in individuals allergic to the birch pollen allergen Bet v 1 with several fruits are a well-documented example of the pollen-food syndrome (Egger et al., 2006; Mittag et al., 2005), with symptoms ranging from local oral allergy syndrome to severe anaphylaxis. A FASTA search in SDAP quickly reveals that Bet v 1 has significant homology to the food allergens Pru av 1 from cherry (Bit score 160/Evalue 5.9e-35), Gly m 4 from soybean (Bit score 158/Evalue 3.1e-25) and Ara h 8 from peanut (Bit score 102/Evalue 4.3e-24) (Mittag et al., 2006), which could account for the cross-reactions. Pollen cross-reactivity may extend across a large number of species, and even to species from different continents (Midoro-Horiuti et al., 1999, 2003). Similar cross-reactivities among allergens with a high degree of identity have been observed for profilins, lipid transfer proteins, calcium-binding proteins, and pathogenesis-related proteins (Breiteneder and Mills, 2005b; Egger et al., 2006; Midoro-Horiuti et al., 2001; Weber, 2005). Other examples include the ficus-fruit syndrome related to the similarity of cysteine proteases in tropical fruits (Hemmer et al., 2004) or the IgE-based cross-reactivity of shrimp with other crustaceans and even non-edible arthropods such as cockroaches or dust mites due to the similarity of the muscle protein tropomyosin in these organisms (Ayuso et al., 2002a; Reese et al., 2006).
However, simple sequence similarity is not sufficient to conclusively predict IgE cross-reactivity. While short sequence elements can define an IgE epitope, short stretches of identical sequences are not long enough to predict with statistical significance cross-reactive IgE epitopes (Goodman, 2006; Ladics et al., 2006). The statistical significance can be substantially increased if the sequence is a motif that is common to many related known allergens, and is not found in related proteins that are non-allergens. Here, we define specific sequence regions with common physicochemical properties, PCP-motifs (Ivanciuc et al., 2004; Mathura et al., 2003) that may distinguish allergenic proteins.
Our work was predicated on previous studies which indicated that pollen and plant food allergens grouped to only a small number of all protein families (Breiteneder and Radauer, 2004; Jenkins et al., 2005; Radauer and Breiteneder, 2006); most of these families also contain non-allergenic proteins as well. The first step was to obtain a comprehensive assignment of all known allergens according to an existing classification scheme for protein families, Pfam (Version 22.0, http://pfam.sanger.ac.uk/)(Finn et al., 2006). These assignments have been made available on our SDAP web site. The major allergens belong to about 30 structural families, consistent with the results of others (Radauer et al., 2008). In order to discriminate the allergenic from the non-allergenic family members (Björklund et al., 2005; Brusic and Petrovsky, 2003; Furmonaviciene et al., 2005; Riaz et al., 2005; Schein et al., 2005a, 2007; Stadler and Stadler, 2003), we also determined common sequence motifs using our PCPMer program (Schein et al., 2005b,c). We show in three examples that motifs we defined as characteristic of allergens in a given Pfam coincided with previously determined IgE epitopes. The motifs thus represent a promising way to identify linear IgE epitopes that are likely to be responsible for IgE cross-reactivities. All sequence motifs for the major Pfam families with allergens can be obtained from our web server MotifMate (http://born.utmb.edu/motifmate/summary.php). The motifs can now be analyzed in screening sequence databases for potential IgE cross-reactivities (Aalberse, 2007; Hileman et al., 2002; Marti et al., 2007; Riaz et al., 2005; Saha and Raghava, 2006; Schein et al., 2007), or used in conjunction with 3D structural information on allergens to shed new light on the molecular determinants of allergenicity(Aalberse and Stadler, 2006; Breiteneder and Mills, 2006; Chapman et al., 2007; Jenkins et al., 2005; Oezguen et al., 2008).
2. Methods
2.1. Assignment of Pfam domains to all allergens
All allergen sequences from SDAP were searched in the Pfam A (Version 22.0, http://pfam.sanger.ac.uk/) (Finn et al., 2006) database for the matching family. Whenever the TrEMBL or SwissProt accession number of the allergen sequence was known, the Pfam assignment was made based on the corresponding accession number. Otherwise we performed BLAST searches to find related proteins to the SDAP allergen entry. The Pfam database has a collection of sequence alignments of related protein domains that were used to find Pfam domains for each allergen. Fragments of sequences without a significant match in Pfam where left unassigned. As a result of a direct match or individual BLAST searches, 594 out of 829 allergen protein sequences were grouped to their respective protein families and domains from Pfam A.
2.2. Generation of sequence motifs of allergens by MotifMate
MotifMate-PCP is a novel database and data mining tool developed by us to generate physicochemical property (PCP) motifs of allergens. PCP motifs were generated by our PCPMer web server (http://landau.utmb.edu:8080/WebPCPMer/). The motifs are based on the conservation of five physicochemical descriptors E1-E5 (Venkatarajan and Braun, 2001) in a multiple sequence alignment. The E1-E5 scale allows us to characterize motifs as protein regions where the side chains show conserved physicochemical properties, such as hydrophobicity, size or alpha-helical propensity, rather than strict sequence identity. We have tested the PCPMer motifs in other protein families to locate functional important regions and as meaningful fingerprints to find distantly related proteins (Mathura et al., 2003; Schein et al., 2002, 2005b,c).
We generated two types of motifs: one set of motifs that represent a complete Pfam family containing allergenic proteins, i.e. these are motifs generated from the multiple sequence alignment as archived in the Pfam database, and a second set of motifs using only the allergenic proteins in a family (prepared using ClustalW). Using Perl scripts, multiple sequence alignments of all Pfam families containing allergens were downloaded to a MySQL database, PCP motifs were generated and stored in the MySQL database. Sequence alignments of only allergenic proteins in a Pfam family were manually generated with ClustalW (Chenna et al., 2003). In that phase the protein sequences were cut to the region of the known Pfam domains. In addition, the allergen proteins for each family group were submitted to a pair-wise sequence search in SDAP to eliminate almost identical proteins or protein sequences from the same allergen source. Also, protein sequences with a sequence identity of only 20% or below to the other allergens from that group were eliminated.
3. Results
3.1. Main Pfam classes for allergens
The allergens in SDAP group to only 130 of the 9318 protein families from Pfam A, and of these 31 contain multiple allergenic proteins (Table 1). A list of the allergens in the 12 Pfam families most populated with allergens is given in Table 2. The complete classification of allergens is available on our SDAP web server (http://fermi.utmb.edu/SDAP/). For each family, we determined motifs that were common to all members, and, using separate alignments of the known allergens, those motifs that were unique to allergenic proteins.
Table 1.
The most abundant Pfam A allergen families from SDAP
| No | Pfam code | Pfam domain | No allergens |
|---|---|---|---|
| 1 | PF00234 | Protease inhibitor/seed storage/LTP family | 34 |
| 2 | PF00235 | Profilin | 27 |
| 3 | PF00036 | EF hand | 23 |
| 4 | PF01357 | Pollen allergen | 19 |
| 5 | PF00188 | SCP-like extracellular protein | 19 |
| 6 | PF00407 | Pathogenesis-related protein Bet v 1 family | 16 |
| 7 | PF00261 | Tropomyosin | 16 |
| 8 | PF00190 | Cupin | 15 |
| 9 | PF00061 | Lipocalin/cytosolic fatty-acid binding protein family | 12 |
| 10 | PF03330 | Rare lipoprotein A (RlpA)-like double-psi beta-barrel | 12 |
| 11 | PF00042 | Globin | 9 |
| 12 | PF00544 | Pectate lyase | 9 |
| 13 | PF00112 | Papain family cysteine protease | 8 |
| 14 | PF00428 | 60s Acidic ribosomal protein | 8 |
| 15 | PF00082 | Subtilase family | 7 |
| 16 | PF00314 | Thaumatin family | 7 |
| 17 | PF01190 | Pollen proteins Ole e 1 family | 7 |
| 18 | PF01620 | Ribonuclease (pollen allergen) | 7 |
| 19 | PF00012 | Hsp70 protein | 6 |
| 20 | PF00578 | AhpC/TSA family | 6 |
| 21 | PF02221 | ML domain | 6 |
| 22 | PF05922 | Subtilisin N-terminal region | 6 |
| 23 | PF00089 | Trypsin | 5 |
| 24 | PF00113 | Enolase, C-terminal TIM barrel domain | 5 |
| 25 | PF00187 | Chitin recognition protein | 5 |
| 26 | PF00273 | Serum albumin family | 5 |
| 27 | PF03952 | Enolase, N-terminal domain | 5 |
| 28 | PF00151 | Lipase | 4 |
| 29 | PF00197 | Trypsin and protease inhibitor | 4 |
| 30 | PF00295 | Glycosyl hydrolases family 28 | 4 |
| 31 | PF01630 | Hyaluronidase | 4 |
Table 2.
Classification of allergens in the 12 Pfam families most populated with allergens
| Allergen | Source | Allergen | Source | Allergen | Source |
|---|---|---|---|---|---|
| PF00234: protease inhibitor/seed storage/LTP family | |||||
| Amb a 6 | Short ragweed | Ana o 3 | Cashew nut | Ara h 2 | Peanut |
| Ara h 6 | Peanut | Ber e 1 | Brazil nut | Bra j 1 | Oriental mustard |
| Bra n 1 | Rapeseed | Cor a 8 | Hazelnut | Fag e 8kD | Common buckwheat |
| Gly m 1 | soybean | Hev b 12 | Rubber (latex) | Hor v 1 | Barley |
| Hor v 21 | Barley | Jug n 1 | Black walnut | Jug r 1 | English walnut |
| Lyc e 3 | Tomato | Mal d 3 | Apple | Ory s TAI | Rice |
| Par j 1 | Parietaria judaica | Par j 2 | Parietaria judaica | Pru ar 3 | Apricot |
| Pru av 3 | Sweet cherry | Pru d 3 | European plum | Pru p 3 | Peach |
| Pyr c 3 | Pear | Ric c 1 | Castor bean | Ses i 1 | Sesame |
| Ses i 2 | Sesame | Sin a 1 | Yellow mustard | Tri a gliadin | Wheat |
| Tri a glutenin | Wheat | Tri a TAI | Wheat | Vit v 1 | Grape |
| Zea m 14 | Corn | ||||
| PF00235: profilin | |||||
| Ana c 1 | Pineapple | Api g 4 | Celery | Ara h 5 | Peanut |
| Ara t 8 | Mouse-ear cress | Bet v 2 | Birch | Cap a 2 | Bell pepper |
| Che a 2 | Lamb's-quarters | Cor a 2 | Hazelnut | Cuc m 2 | Muskmelon |
| Cyn d 12 | Bermuda grass | Dau c 4 | Carrot | Gly m 3 | Soybean |
| Hel a 2 | Sunflower | Hev b 8 | Rubber (latex) | Lit c 1 | Litchi |
| Lyc e 1 | Tomato | Mal d 4 | Apple | Mer a 1 | Mercurialis annua |
| Mus xp 1 | Banana | Ole e 2 | Olive | Par j 3 | Parietaria judaica |
| Phi p 11 | Timothy | Phl p 12 | Timothy | Pru av 4 | Sweet cherry |
| Pru p 4 | Peach | Pyr c 4 | Pear | Tri a profilin | Wheat |
| PF00036: EF hand | |||||
| Aln g 4 | Alder | Bet v 3 | Birch | Bet v 4 | Birch |
| Bos d 3 | Domestic cattle | Bra n 1 | Rapeseed | Bra n 2 | Rapeseed |
| Bra r 1 | Turnip | Che a 3 | Lamb's-quarters | Cyn d 7 | Bermuda grass |
| Cyp c 1 | Common carp | Gad c 1 | Cod | Gad m 1 | Atlantic cod |
| Hom s 4 | Human autoallergen | Jun o 4 | Prickly juniper | Ole e 3 | Olive |
| Ole e 8 | Olive | Phl p 7 | Timothy | Ran e 1 | Edible frog |
| Ran e 2 | Edible frog | Sal s 1 | Atlantic salmon | Sco j 1 | Chub mackerel |
| Syr v 3 | Lilac | The c 1 | Alaska pollock | ||
| PF01357: pollen allergen | |||||
| Ara t expansin | Mouse-ear cress | Cyn d 1 | Bermuda grass | Cyn d 15 | Bermuda grass |
| Cyn d 2 | Bermuda grass | Dac g 2 | Orchard grass | Dac g 3 | Orchard grass |
| Gly m 2 | Soybean | Hol l 1 | Velvet grass | Lol p 1 | Rye grass |
| Lol p 2 | Rye grass | Lol p 3 | Rye grass | Ory s 1 | Rice |
| Pha a 1 | Canary grass | Phl p 1 | Timothy | Phl p 2 | Timothy |
| Poa p a | Kentucky blue grass | Tri a 3 | Wheat | Tri a ps93 | Wheat |
| Zea m 1 | corn | ||||
| PF00188: SCP-like extracellular protein | |||||
| Cte f 2 | Cat flea | Dol a 5 | Yellow hornet | Dol m 5 | White face hornet |
| Pol a 5 | Wasp | Pol d 5 | Mediterranean paper wasp | Pol e 5 | Paper wasp |
| Pol f 5 | Golden paper wasp | Pol g 5 | Wasp | Sol i 3 | Fire ant |
| Sol r 3 | Black fire ant | Ves f 5 | Hybrid yellowjacket | Ves g 5 | German wasp |
| Ves m 5 | Eastern yellowjacket | Ves p 5 | Western yellowjacket | Ves s 5 | Southern yellowjacket |
| Ves v 5 | Yellowjacket | Ves vi 5 | Yellowjacket | Vesp c 5 | European hornet |
| Vesp m 5 | Giant asian hornet | ||||
| PF00407: pathogenesis-related protein Bet v 1 family | |||||
| Aln g 1 | Alder | Api g 1 | Celery | Ara h 8 | Peanut |
| Bet v 1 | Birch | Car b 1 | Hornbeam | Cas s 1 | Chestnut |
| Cor a 1 | Hazelnut | Dau c 1 | Carrot | Gly m 4 | Soybean |
| Mal d 1 | Apple | Pet c PR10 | Parsley | Pha v 1 | Kidney bean |
| Pru ar 1 | Apricot | Pru av 1 | Sweet cherry | Pyr c 1 | Pear |
| Tar o RAP | Common dandelion | ||||
| PF00261: tropomyosin | |||||
| Ani s 3 | Herring worm | Cha f 1 | crab | Chi k 10 | Midge |
| Cra g 1 | Pacific oyster | Der p 10 | European house dust mite | Hal d 1 | Abalone |
| Hel as 1 | Brown garden snail | Hom a 1 | American lobster | Lep d 10 | Storage mite |
| Lep s 1 | Silverfish | Met e 1 | Greasyback shrimp | Mim n 1 | Scallop |
| Pan s 1 | Spiny lobster | Pen a 1 | Brown shrimp | Per a 7 | American cockroach |
| Per v 1 | Tropical green mussel | ||||
| PF00190: cupin | |||||
| Ana o 1 | Cashew | Ara h 1 | Peanut | Ara h 3 | Peanut |
| Ara h 4 | Peanut | Ber e 2 | Brazil nut | Cor a 11 | Hazelnut |
| Cor a 9 | Hazelnut | Fag e 1 | Common buckwheat | Gly m Bd28K | Soybean |
| Gly m conglycinin | Soybean | Gly m glycinin G1 | Soybean | Gly m glycinin G2 | Soybean |
| Jug n 2 | Black walnut | Jug r 2 | English walnut | Ses i 3 | Sesame |
| PF00061: lipocalin/cytosolic fatty-acid binding protein family | |||||
| Aca s 13 | Flour mite | Blo t 13 | Dust mite | Bos d 2 | Domestic cattle |
| Bos d 5 | Domestic cattle | Can f 1 | Dog | Can f 2 | Dog |
| Equ c 1 | Domestic horse | Fel d 4 | Cat | Lep d 13 | Storage mite |
| Mus m 1 | Mouse | Rat n 1 | Rat | Tyr p 13 | Mould mite |
| PF03330: rare lipoprotein A (RlpA)-like double-psi beta-barrel | |||||
| Ara t expansin | Mouse-ear cress | Cyn d 1 | Bermuda grass | Gly m 2 | Soybean |
| Hol l 1 | Velvet grass | Lol p 1 | Rye grass | Ory s 1 | Rice |
| Pha a 1 | Canary grass | Phl p 1 | Timothy | Poa p a | Kentucky blue grass |
| Tri a ps93 | Wheat | Zea m 1 | Corn | ||
| PF00042: globin | |||||
| Chi t 1 | Midge | Chi t 2 | Midge | Chi t 3 | Midge |
| Chi t 4 | Midge | Chi t 5 | Midge | Chi t 6 | Midge |
| Chi t 7 | Midge | Chi t 8 | Midge | Chi t 9 | Midge |
| PF00544: pectate lyase | |||||
| Amb a 1 | Short ragweed | Amb a 2 | Short ragweed | Cha o 1 | Japanese cypress |
| Cry j 1 | Japanese cedar | Cup a 1 | Arizona cypress | Cup s 1 | Common cypress |
| Jun a 1 | Texas mountain cedar | Jun o 1 | Prickly juniper | Jun v 1 | Eastern red cedar |
3.1.1. PF00234: protease inhibitor/seed storage/LTP family
This domain (InterPro IPR003612) is found in plant lipid transfer proteins, seed storage proteins, and trypsin-alpha amylase inhibitors. The domain forms a four-helical bundle in a right-handed superhelix with a folded leaf topology, which is stabilized by disulfide bonds, and which has an internal cavity. Allergens from the lipid transfer protein (LTP) family are highly resistant to both heat treatment and proteolytic digestion, and are particularly important in the Mediterranean area (Breiteneder and Mills, 2005a; Salcedo et al., 2004). Three-dimensional structures are known for three allergens from this family, namely Pru p 3 (2ALG, Fig. 1A), Hor v 1 (1JTB), Zea m 14 (1MZM). The molecular determinants of allergenicity for this family may be extracted from the known IgE epitopes, for Ara h 2 (Stanley et al., 1997), Jug r 1 (Robotham et al., 2002), Par j 1 (Asturias et al., 2003), and Par j 2 (Asturias et al., 2003). The T-cell epitopes are known only for Ara h 2 (Glaspole et al., 2005).
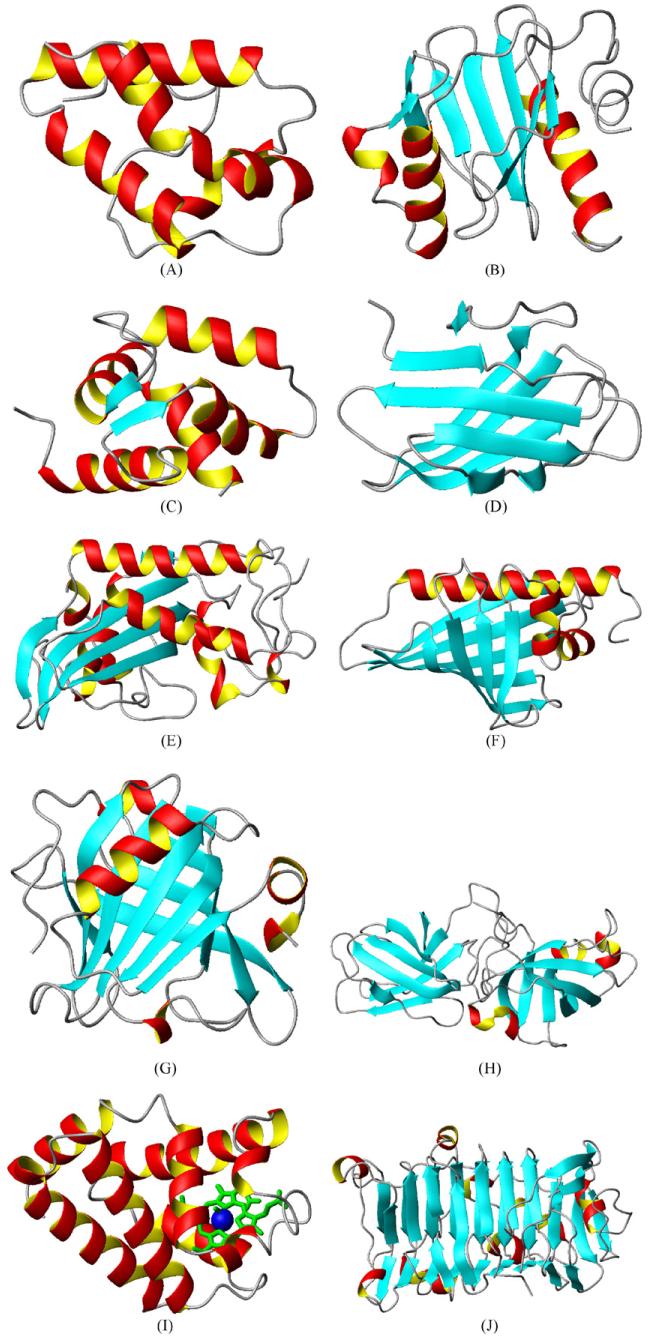
Fig. 1.
PDB structures for allergens from the most abundant Pfam families: (A) Pru p 3 (PF00234, protease inhibitor/seed storage/LTP family; 2ALG); (B) Hev b 8, (PF00235, profilin; 1G5U); (C) Bet v 4, (PF00036, EF hand; 1H4B); (D) Phl p 2, (PF01357, pollen allergen; 1WHO); (E) Ves v 5, (PF00188, SCP-like extracellular protein; 1QNX); (F) Bet v 1, (PF00407, pathogenesis-related protein Bet v 1 family; 1BV1); (G) Bos d 5, (PF00061, lipocalin/cytosolic fatty-acid binding protein family; 1GXA); (H) Phl p 1, (PF03330, rare lipoprotein A (RlpA)-like double-psi beta-barrel; 1N10); (I) Chi t 1, (PF00042, globin; 1ECO); (J) Jun a 1, (PF00544, pectate lyase; 1PXZ).
3.1.2. PF00235: profilin
Profilin (InterPro IPR002097) binds to monomeric actin in a 1:1 ratio and prevents the polymerization of actin into filaments. Three-dimensional structures for allergens in this class are available for Ara t 8 (3NUL), Bet v 2 (1CQA), and Hev b 8 (1G5U, Fig. 1B).
3.1.3. PF00036: EF hand
This family collects calcium-binding proteins (InterPro IPR002048) that contain a common domain known as the EF-hand. The EF-hand motif has a 12 residue loop flanked on both side by a twelve residue alpha-helical domain. The proteins from this class may be signaling proteins (calmodulin, troponin C) or buffering/transport proteins (calbindin D9k). PDB structures are available for Bet v 4 (1H4B, Fig. 1C), Che a 3 (1PMZ), and Phl p 7 (1K9U).
3.1.4. PF01357: pollen allergen
This family (InterPro IPR007117, Pollen allergen/expansin, C-terminal) contains expansins, proteins that mediate cell wall extension in plants. Expansins allow wall polymers to slide by breaking hydrogen bonds that keel together the wall constituents. Grass pollen allergens are the main allergens from this family (Table 2). PDB structures are available for Phl p 1 (1N10) and Phl p 2 (1WHO, Fig. 1D).
3.1.5. PF00188: SCP-like extracellular protein
This family (InterPro IPR001283, Allergen V5/Tpx-1 related) includes venom antigen 5 from wasps (Dol a 5 from the yellow hornet Dolichovespula arenaria, Dol m 5 from the white face hornet Dolichovespula maculata, Pol a 5 from the paper wasp Polistes annularies, Pol d 5 from the Mediterranean paper wasp Polistes dominulus, Pol e 5 from the paper wasp Polistes exclamans, Pol f 5 from the paper wasp Polistes fuscatus, Pol g 5 from the paper wasp Polistes gallicus, Ves f 5 from the downy yellowjacket Vespula flavopilosa, Ves g 5 from the German yellowjacket Vespula germanica, Ves m 5 from the Eastern yellow jacket Vespula maculifrons, Ves p 5 from the Western yellowjacket Vespula pennsylvanica, Ves s 5 from the Southern yellowjacket Vespula squamosa, Ves v 5 from the common yellowjacket Vespula vulgaris, Ves vi 5 from the wasp Vespula vidua, Vesp c 5 from the European hornet Vespa crabo, Vesp m 5 from the giant asian hornet Vespa mandarina) and venom antigen 3 (Sol i 3 from the fire ant Solenopsis invicta and Sol r 3 from the black imported fire ant Solenopsis richteri), which both are potent allergens that mediate allergic reactions to insect stings of the Hymenoptera family. The structure (1QNX, Fig. 1E) and T-cell epitopes of Ves v 5(Bohle et al., 2005) are known.
3.1.6. PF00407: pathogenesis-related protein Bet v 1 family
The most important allergen from this class (InterPro IPR000916) is the major white birch (Betula verrucosa) pollen antigen. Bet v 1, which is the main cause of type I allergies observed in spring. The Bet v 1 allergens are formed by six anti-parallel betastrands and three alpha-helices. Four of the beta-strands dominate the global fold, and two of the helices form a C-terminal amphipathic helical motif. The family contains pathogenesis-related (PR-10) allergens (Midoro-Horiuti et al., 2001), such as Aln g 1, Api g 1, Ara h 8, Bet v 1, Cor a 1, Dau c 1, Gly m 4, Mal d 1, Pru ar 1, Pru av 1, and Pyr c 1. PDB structures are reported for Api g 1 (2BK0), Bet v 1 (1BV1, Fig. 1F), and Pru av 1 (1E09). The conformational IgE epitopes of Bet v 1 were identified (Mirza et al., 2000).
3.1.7. PF00261: tropomyosin
Tropomyosins (InterPro IPR000533) are alpha-helical proteins that form a coiled-coil structure of two parallel helices containing two sets of seven alternating actin-binding sites. In striated muscles, tropomyosin regulates the muscle contraction by mediating the interactions between the troponin complex and actin. Allergies to crustaceans, such as shrimp, crab, crawfish and lobster, are mainly induced by tropomyosin (Reese et al., 1999). IgE epitopes are known for the shrimp allergens Pen a 1 (Ayuso et al., 2002a) and Pen i 1 (Shanti et al., 1993). The high conservation of tropomyosin sequences among invertebrates explains why the cross-reactivity of allergens from shellfish and mollusks are often cross-reactive (Chu et al., 2000; Jeong et al., 2006). However, vertebrate tropomyosins are not known to be allergenic.
3.1.8. PF00190: cupin
The cupin family (InterPro IPR006045) contains the conserved barrel domain of the cupin superfamily (cupa is the Latin term for a small barrel), and is comprised of 11S and 7S plant seed storage proteins. The IgE epitopes for five members of this family are reported in the literature: Ara h 1 (Shin et al., 1998), Ara h 3 (Rabjohn et al., 1999), Fag e 1 (Yoshioka et al., 2004), Gly m glycinin G1 (Beardslee et al., 2000) and Gly m glycinin G2 (Helm et al., 2000).
3.1.9. PF00061: lipocalin/cytosolic fatty-acid binding protein family
Lipocalins (InterPro IPR000566) are proteins that transport small hydrophobic molecules, such as lipids, retinoids, and steroids. The fold is an eight-strand anti-parallel beta-barrel enclosing the binding site. The structures of several allergens from this family are known: Bos d 2 (1BJ7), Bos d 5 (1GXA, Fig. 1G), Equ c 1 (1EW3), Mus m 1 (1MUP) and Rat n 1 (2A2U).
3.1.10. PF03330: rare lipoprotein A (RlpA)-like double-psi beta-barrel
The rare lipoprotein A (RlpA) fold (InterPro IPR005132) is found in bacterial and eukaryotic lipoproteins, and represents a double-psi beta-barrel fold. This domain may be found in the N-terminal part of several pollen allergens. The 3D structure of only one allergen, Phl p 1 (1N10, Fig. 1H), is known.
3.1.11. PF00042: globin
Globins (InterPro IPR000971) are heme-containing proteins involved in binding and/or transporting oxygen. Hemoglobin is a protein that in vertebrates transports oxygen from lungs to other tissues, containing two alpha and two beta chains with the characteristic three-dimensional globin fold. Monomeric and dimeric hemoglobins have been identified as major allergenic components in insects. The antigenic determinants of this family from Chironomus thummi thummi (midge) have been characterized as regions with dominant polar amino acids and high flexibility (Baur et al., 1986). The global fold of the monomeric allergen Chit 1 is shown in Fig. 1I (PDB code 1ECO).
3.1.12. PF00544: pectate lyase
Pectate lyase (InterPro IPR002022) is an enzyme involved in the cleavage of pectate, which occurs during the maceration and rotting of plant tissue. This family contains several major pollen allergens, such as those from short ragweed (Ambrosia artemisiifolia), Amb a 1 and Amb a 2. The most common pollen allergen in Japan is Cry j 1, a glycoprotein from the Japanese cedar (Cryptomeria japonica). Other cedar allergens are Jun a 1 (Juniperus ashei, mountain cedar), Jun o 1 (Juniperus oxycedrus, prickly juniper), Jun v 1 (Juniperus virginiana, eastern red cedar). Pollen from several cypress species contains allergens homologous with pectate lyase, namely Cup a 1 (Cupressus arizonica, cypress), Cup s 1 (Cupressus sempervirens, common cypress), Cha o 1 (Chamaecyparis obtuse, Japanese cypress). The IgE epitopes are known for Cry j 2 (Tamura et al., 2003) and Jun a 1 (Midoro-Horiuti et al., 2003, 2006), and the T-cell epitopes were identified for Cha o 1 (Sone et al., 2005), Cry j 1 (Sone et al., 1998), and Cry j 2 (Sone et al., 1998). The structure of one allergen for this family has been deposited in PDB: Jun a 1 (1PXZ, Fig. 1J) (Czerwinski et al., 2005). All allergens from this family have similar sequences and there are significant cross-reactivities to food allergens (Bonds et al., 2008). Schwietz et al. studied the in vivo and in vitro cross-reactivity between pollen extracts of mountain cedar and 11 other Cupressaceae species, one Taxodiaceae species (Japanese cedar), one Pinaceae species, and an angiosperm, and found that the 12 Cupressaceae and the Japanese cedar are cross-reactive (Schwietz et al., 2000).
3.2. Sequence motifs characteristic of allergens may correlate with cross-reactivity
Proteins in the same Pfam class are homologous, are expected to share a similar 3D-structure, and often have common biochemical functions (Finn et al., 2006). High overall sequence similarity is a good indicator of cross-reactivity (Aalberse, 2007). However, as antibodies bind to surface patches of folded proteins, cross-reactivity may be better indicated by matching specific areas of the protein structure rather than just the global fold. To differentiate local sequence areas of known allergens, we first generated sequence motifs that are characteristic for the complete family of all those Pfam classes that contain allergens. These “Full-Pfam motifs” can be used to classify novel proteins, and to determine whether it belongs to a Pfam with many allergenic members. In addition, “Allrg-Pfam” motifs were defined that were derived from alignments of only the allergens within each protein family. This procedure allows us to distinguish allergen specific motifs from those that are common to all proteins in the family. All Full-Pfam sequence motifs are publicly available on our MotifMate web server (http://born.utmb.edu/motifmate/summary.php).
We next compared the motifs that were specific for the allergens with known IgE epitopes, to see if there was a correlation that could account for clinically significant cross-reactivities between allergens. Three major Pfam families were chosen: the seed storage proteins (a subset of PF00234), the pathogenesis-related protein Bet v 1 family (PF00407) and tropomyosin (PF00261). The motifs common to the allergen members of each family were compared to known IgE epitopes (Table 3). Motifs of Full-Pfam and Allrg-Pfam in equivalent positions in the Pfam domains are listed on the same line and referred to with the number in column 1. For the seed storage proteins, there are five motifs in Allrg-Pfam (numbered 1, 3-6) and four Full-Pfam motifs (2, 3, 4, 7). Motifs 1, 5 and 6 are unique to the allergens (Allrg-Pfam). A novel protein that contained some or all of the Full-Pfam motifs would probably be a member of this Pfam. If there were a significant match to motifs 1, 5 or 6 that characterized the allergenic proteins, it would be also flagged as potentially allergenic. The only representative of this family for which IgE epitopes have been determined is the walnut allergen Jug r 1. The epitope QGLRGEEMEEMV (Robotham et al., 2002) partially overlaps with motif 6 that is characteristic of allergens (bold letters in Table 3). This suggests that this common sequence could play a role in observed clinical cross-reactivities among allergens of this protein family (Comstock et al., 2004; Goetz et al., 2005; Robotham et al., 2005).
Table 3.
AutoMotifs for allergens and entire Pfam families for seed storage proteins, Bet v 1-related family, and tropomyosin
| No | Allergens only |
Entire Pfam family |
||
|---|---|---|---|---|
| E | Motif | E | Motif | |
| PF00234: protease inhibitor/seed storage/LTP family (subgroup B)-seed sequence: Jug r 1 | ||||
| 1 | 1.7 | 1 CQYYLR 6 | ||
| 2 | 0.5 | 10 RSGGYDED 17 | ||
| 3 | 1.8 | 26 CCQQLS 31 | 0.9 | 26 CCQQLSQI 33 |
| 4 | 2.0 | 37 CQCEGLR 43 | 0.5 | 37 CQCEGL 42 |
| 5 | 1.7 | 49 QQQQ 52 | ||
| 6 | 1.8 | 59 EMEEMVQSA 67 | ||
| 7 | 1.2 | 67 ARDLPKEC 74 | ||
| PF00407: pathogenesis-related protein Bet v 1 family-seed sequence: Bet v 1 | ||||
| 1 | 2.0 | 6 ETETTSVIPA 15 | ||
| 2 | 1.3 | 15 AARLFKA 21 | ||
| 3 | 2.0 | 31 PKVAP 35 | 1.2 | 25 DGDNLFPKVAP 35 |
| 4 | 2.0 | 42 ENIEGNGGPGTIK 54 | 1.8 | 46 GNGGPG 51 |
| 5 | 1.8 | 69 DRVDEVD 75 | 1.5 | 68 KDRVDEVD 75 |
| 6 | 1.7 | 81 YNYSVIEGGPI 91 | ||
| 7 | 2.0 | 110 GGSILK 115 | 1.5 | 110 GGSILK 115 |
| 8 | 2.0 | 120 YHTKG 124 | 1.0 | 120 YHTKGD 125 |
| 9 | 0.7 | 129 KAEQVKASK 137 | ||
| 10 | 2.0 | 145 RAVESYLLAH 154 | 1.2 | 145 RAVESYLLAH 154 |
| PF00261: tropomyosin-seed sequence: Pen a 1 | ||||
| 1 | 2.0 | 7 ENDLD 11 | ||
| 2 | 1.8 | 14 QESL 17 | ||
| 3 | 2.0 | 20 ANIQ 23 | 0.8 | 20 ANIQLV 25 |
| 4 | 2.0 | 33 NAEGEVA 39 | ||
| 5 | 1.0 | 39 AALNRR 44 | ||
| 6 | 2.0 | 47 LLEEDLERSEER 58 | 1.0 | 54 RSEER 58 |
| 7 | 2.0 | 65 KLAEASQAADESERMRKVLE 84 | 1.4 | 62 ATTKLAEASQAADE 75 |
| 8 | 1.5 | 79 MRKVLENR 86 | ||
| 9 | 2.0 | 90 DEERMDALENQLKEAR 105 | 1.5 | 90 DEERM 94 |
| 10 | 1.6 | 98 ENQLKEA 104 | ||
| 11 | 2.0 | 108 AEEADRKYDEVARKLAMVEADLERAEERAE 137 | 1.5 | 108 AEEADRKYDEVA 119 |
| 12 | 1.2 | 130 ERAEERAETGE 140 | ||
| 13 | 2.0 | 145 ELEEELRVVGNNLKSLEVSEEKANQRE 171 | 1.1 | 147 EEELR 151 |
| 14 | 1.0 | 155 NNLKS 159 | ||
| 15 | 1.1 | 166 KANQREEAYK 175 | ||
| 16 | 2.0 | 174 YKEQIKTL 181 | ||
| 17 | 2.0 | 184 KLKAAEARA 192 | 1.4 | 186 KAAEARAEFAE 196 |
| 18 | 2.0 | 195 AERSVQKLQKEVDRLEDELVNEKEKYK 221 | 1.2 | 201 KLQKEVDRLE 210 |
| 19 | 2.0 | 225 DELD 228 | ||
Similarly, unique Allrg-Pfam motifs 1, 3-8 and 10 characterize allergens in the Bet v 1 family (Table 3). Here again, a conformational IgE epitope, 42ENIEGNGGPGT52 70R 72D 76H 86I 97K (Mirza et al., 2000) correlates with sequences within these Allrg-Pfam motifs. The entire linear part of the IgE epitope is found in the Allrg-Pfam motif 4, and the individual residues 70R, 72D and 86I are in motifs 4 and 5. The cross-reactivites observed between allergens from this family (Aalberse et al., 2001; Kazemi-Shirazi et al., 2000; Wensing et al., 2002) may be explained by the conservation of this physico-chemical profile for the Bet v1 IgE epitope across all these allergens. As in the first example, the experimentally documented IgE epitope sequence correlates better with motifs derived from the known allergens than for those that characterize the whole Pfam class.
Numerous studies have related the similar structures of members of the tropomyosin family to clinically significant cross-reactions (Ayuso et al., 2002b; Chu et al., 2000; Fernandes et al., 2003; Jeong et al., 2006; Wild and Lehrer, 2005; Zhang et al., 2006). We previously demonstrated that tropomyosin allergens are difficult to discriminate from non-allergenic tropomyosins with the current web servers for allergenicity prediction (Schein et al., 2007). In this work, we found that Allrg-Pfam and Full-Pfam motifs showed distinctions between the two groups. MotifMate identified 19 common motifs in the highly conserved sequences of tropomyosins. Five of these, 1, 2, 4, 16 and 19, are characteristic of the allergenic family members. We then mapped the sequences of nine linear IgE motifs that were identified for the shrimp tropomyosin allergen, Pen a 1 (Ayuso et al., 2002a). While areas of the epitopes are found in motifs common to all tropomyosins, the sequences for the most part correlate with the Allrg-Pfam motifs that are specific for the allergenic tropomyosins. In particular, the Allrg-Pfam motifs 1 and 19 match well to epitope sequences. These three examples all indicate that distinguishing local areas of conserved physicochemical properties that are common to allergenic members of the same Pfam can be useful in predicting determinants of IgE reactivity, and potential cross-reactivity.
4. Discussion
One major goal of our SDAP database is to provide a rapid way for researchers to identify common features of allergenic proteins, as a basis for identifying proteins that could be expected to cause cross-reactions in patients. The sequence comparison tools in SDAP can be used for that purpose. However, grouping all the allergens in SDAP according to protein families within Pfam (Tables 1 and 2) now makes this determination even faster, and more accurate as distinct domains of the allergens are assigned to different Pfam. Further, this grouping allowed us to define a series of known 3D protein structures (Fig. 1) to characterize the folds of the majority of allergens. We also derived new sequence specific motifs of proteins in those protein families with a large number of allergens and demonstrated that we also can generate specific motifs that can distinguish them from homologous but non-allergenic proteins in the same protein family (Table 3). Finally, we could show that specific motifs did indeed correlate with allergenicity, as they corresponded to experimentally determined linear IgE epitopes for three different examples.
However, the conserved motifs that are characteristic only for allergens from a Pfam family are not restricted to the set of IgE epitopes. These motifs may be buried, in which case they represent structurally important residues. Alternatively, the group of residues could give the necessary conformational flexibility to an antigenic site, thus distinguishing them from the rest of the family. The results reported in Table 3 demonstrate that allergens within a Pfam family have distinct conserved regions as compared to the entire Pfam family.
Our data correlate well with previous attempts to group allergenic proteins according to common sequences, structures (Aalberse, 2000), and functional classes (Breiteneder and Ebner, 2000; Ebner et al., 2001; Midoro-Horiuti et al., 2001). Our finding demonstrate novel applications of allergen classifications (Breiteneder and Mills, 2005b; Breiteneder and Mills, 2006; Jenkins et al., 2005; Radauer and Breiteneder, 2006; Radauer et al., 2008), and allowed us to also analyze finer details of the sequences that correlate with allergenicity. As most of the known allergens can be grouped to only 31 Pfam, this indicates that bioinformatics approaches should be useful for predicting allergenicity for novel proteins. The MotifMate approach outlined here indicates further that sequence fingerprints of allergens and non-allergens within each Pfam family could provide a useful tool to predict cross-reactivity of allergens with similar sequences.
These findings represent a considerable advance over the original decision tree for combining computational and experimental tests to determine whether a protein is a potential allergen (WHO, 2000, 2001, 2003). There, cross-reactivity was predicted based on overall sequence similarity of 35% of sequence identity in a window of 80 residues, from FASTA alignments, or on identical regions, as short as six to eight residues, in the protein sequences. While the FAO/WHO procedure is available in SDAP (Schein et al., 2006, 2007), our results and those of others (e.g. Hileman et al., 2002) indicated that too many non-allergenic proteins are detected by the suggested thresholds. We suggest that these guidelines be modified, to use more sophisticated comparisons to the known allergen sequences, and particularly allergen specific motifs such as those we define here.
Others have also suggested that motif-based methods could identify allergenic proteins more specifically. The MEME protein motifs (Mari, 2005; Saha and Raghava, 2006; Stadler and Stadler, 2003), for example, have average lengths of 50 residues (Marti et al., 2007), and are thus not as specific as the physicochemical properties motifs we were able to extract. Our motifs correspond better to the length expected for epitopes. The MotifMate motifs can be used to filter large genomic databases directly, either before or after a preliminary classification to eliminate all sequences that do not belong to Pfam families in SDAP. In this way, a large number of sequences will be eliminated from the first step, without time-consuming computations.
The advantages of a motif-based approach are clear from the examples presented above. Our MotifMate comparisons discriminated between allergenic and non-allergenic tropomyosins, a difficult task as allergenic tropomysosins, from mite (Der p 10) and shrimp (Pen a 1), are highly similar (80.28%, 228/284, E score 7.2e-73) to the mammalian homologs that are not allergenic (Schein et al., 2007). Of four programs tested for their ability to distinguish four allergenic tropomysosins from four non-allergens, only WebAllergen (Riaz et al., 2005) found that while all eight proteins have five wavelet allergenic motifs (Li et al., 2004) in common, the allergenic tropomyosins have several additional wavelet motifs that may distinguish them. Both Allermatch (Fiers et al., 2004), which applies the FAO/WHO allergenicity guidelines (WHO, 2000, 2001, 2003) and AlgPred (a support vector machines classifier) (Saha and Raghava, 2006) found all eight tropomyosins to be allergens, while MEME motifs (Stadler and Stadler, 2003) predicted all eight to be non-allergens.
We should at this point note that PCPMer motifs, Allrg-Pfam and Full-Pfam, are numerical vectors based on the E1-E5 physico-chemical properties. They have been translated, for convenience, into representative amino acid sequences in Table 3. They can be used, in combination with other SDAP tools, to compare the physicochemical properties of these motifs to those of novel proteins.
5. Conclusions
The identification of potential allergenic proteins is usually done by global sequence similarity searches. Tools to do overall similarity searching are now incorporated in SDAP. The classification of allergens into Pfam domains reveals the structural relationship between various allergens, thus providing a basis for identifying allergenic determinants. Our results show that allergens can be represented by a small fraction of possible protein families and folds. Out of the 9318 protein families from Pfam, only 130 families are currently listed for all allergens in SDAP, and 31 families contain more than four allergens. The most populated Pfam families are protease inhibitor/seed storage/lipid transfer protein, profilin, EF hand, group I pollen allergens, SCP-like extracellular protein, pathogenesis-related protein Bet v 1 family, tropomyosin, and cupins. Details for the Pfam classification of all allergens can be accessed from the SDAP web site (http://fermi.utmb.edu/SDAP/). The sequence motifs characteristic for Pfam classes are available via our web server MotifMate (http://born.utmb.edu/MotifMate/). Those motifs represent sequence-based fingerprints that characterize the major Pfam families with allergens. In addition we also showed, for three important Pfam classes that contain many allergens, how specific motifs correspond to known IgE epitopes. These allergen-specific motifs are the basis of an original method to predict the potential allergenicity of novel proteins and clinical cross-reactivity.
Acknowledgements
This work was supported by a contract from the U.S. Food and Drug Administration (HHSF223200710011I), and grants from the National Institute of Health (R01 AI 064913), and the U.S. Environmental Protection Agency under a STAR Research Assistance Agreement (No. RD 833137). The article has not been formally reviewed by the EPA, and the views expressed in this document are solely those of the authors.
References
- Aalberse RC. Structural biology of allergens. Journal of Allergy and Clinical Immunology. 2000;106:228–238. doi: 10.1067/mai.2000.108434. [DOI] [PubMed] [Google Scholar]
- Aalberse RC. Assessment of allergen cross-reactivity. Clinical and Molecular Allergy. 2007;5:2. doi: 10.1186/1476-7961-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aalberse RC, Akkerdaas J, van Ree R. Cross-reactivity of IgE antibodies to allergens. Allergy. 2001;56:478–490. doi: 10.1034/j.1398-9995.2001.056006478.x. [DOI] [PubMed] [Google Scholar]
- Aalberse RC, Stadler BM. In silico predictability of allergenicity: from amino acid sequence via 3D structure to allergenicity. Molecular Nutrition & Food Research. 2006;50:625–627. doi: 10.1002/mnfr.200500270. [DOI] [PubMed] [Google Scholar]
- Asturias JA, Gomez-Bayon N, Eseverri JL, Martinez A. Par j 1 and Par j 2, the major allergens from Parietaria judaica pollen, have similar immunoglobulin E epitopes. Clinical and Experimental Allergy. 2003;33:518–524. doi: 10.1046/j.1365-2222.2003.01631.x. [DOI] [PubMed] [Google Scholar]
- Ayuso R, Lehrer SB, Reese G. Identification of continuous, allergenic regions of the major shrimp allergen Pen a 1 (tropomyosin) International Archives of Allergy and Immunology. 2002a;127:27–37. doi: 10.1159/000048166. [DOI] [PubMed] [Google Scholar]
- Ayuso R, Reese G, Leong-Kee S, Plante M, Lehrer SB. Molecular basis of arthropod cross-reactivity: IgE-binding cross-reactive epitopes of shrimp, house dust mite and cockroach tropomyosins. International Archives of Allergy and Immunology. 2002b;129:38–48. doi: 10.1159/000065172. [DOI] [PubMed] [Google Scholar]
- Baur X, Aschauer H, Mazur G, Dewair M, Prelicz H, Steigemann W. Structure, antigenic determinants of some clinically important insect allergens: chironomid hemoglobins. Science. 1986;233:351–354. doi: 10.1126/science.2425431. [DOI] [PubMed] [Google Scholar]
- Beardslee TA, Zeece MG, Sarath G, Markwell JP. Soybean glycinin G1 acidic chain shares IgE epitopes with peanut allergen Ara h 3. International Archives of Allergy and Immunology. 2000;123:299–307. doi: 10.1159/000053642. [DOI] [PubMed] [Google Scholar]
- Björklund ÅK, Soeria-Atmadja D, Zorzet A, Hammerling U, Gustafsson MG. Supervised identification of allergen-representative peptides for in silico detection of potentially allergenic proteins. Bioinformatics. 2005;21:39–50. doi: 10.1093/bioinformatics/bth477. [DOI] [PubMed] [Google Scholar]
- Bohle B, Zwolfer B, Fischer GF, Seppala U, Kinaciyan T, Bolwig C, Spangfort MD, Ebner C. Characterization of the human T cell response to antigen 5 from Vespula vulgaris (Ves v 5) Clinical and Experimental Allergy. 2005;35:367–373. doi: 10.1111/j.1365-2222.2005.02180.x. [DOI] [PubMed] [Google Scholar]
- Bonds RS, Midoro-Horiuti T, Goldblum R. A structural basis for food allergy: the role of cross-reactivity. Current Opinion in Allergy and Clinical Immunology. 2008;8:82–86. doi: 10.1097/ACI.0b013e3282f4177e. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Ebner C. Molecular and biochemical classification of plant-derived food allergens. Journal of Allergy and Clinical Immunology. 2000;106:27–36. doi: 10.1067/mai.2000.106929. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Mills ENC. Nonspecific lipid-transfer proteins in plant foods and pollens: an important allergen class. Current Opinion in Allergy and Clinical Immunology. 2005a;5:275–279. doi: 10.1097/01.all.0000168794.35571.a5. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Mills ENC. Plant food allergens-structural and functional aspects of allergenicity. Biotechnology Advances. 2005b;23:395–399. doi: 10.1016/j.biotechadv.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Mills ENC. Structural bioinformatic approaches to understand cross-reactivity. Molecular Nutrition & Food Research. 2006;50:628–632. doi: 10.1002/mnfr.200500274. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Radauer C. A classification of plant food allergens. Journal of Allergy and Clinical Immunology. 2004;113:821–830. doi: 10.1016/j.jaci.2004.01.779. [DOI] [PubMed] [Google Scholar]
- Brusic V, Petrovsky N. Bioinformatics for characterisation of allergens, allergenicity and allergic crossreactivity. Trends in Immunology. 2003;24:225–228. doi: 10.1016/s1471-4906(03)00075-9. [DOI] [PubMed] [Google Scholar]
- Chapman MD, Pomés A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. Journal of Allergy and Clinical Immunology. 2007;119:414–420. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu KH, Wong SH, Leung PSC. Tropomyosin is the major mollusc allergen: reverse transcriptase polymerase chain reaction, expression and IgE reactivity. Marine Biotechnology. 2000;2:499–509. doi: 10.1007/s101260000035. [DOI] [PubMed] [Google Scholar]
- Comstock SS, McGranahan G, Peterson WR, Teuber SS. Extensive in vitro cross-reactivity to seed storage proteins is present among walnut (Juglans) cultivars and species. Clinical and Experimental Allergy. 2004;34:1583–1590. doi: 10.1111/j.1365-2222.2004.02049.x. [DOI] [PubMed] [Google Scholar]
- Czerwinski EW, Midoro-Horiuti T, White MA, Brooks EG, Goldblum RM. Crystal structure of Jun a 1, the major cedar pollen allergen from Juniperus ashei, reveals a parallel beta-helical core. Journal of Biological Chemistry. 2005;280:3740–3746. doi: 10.1074/jbc.M409655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner C, Hoffmann-Sommergruber K, Breiteneder H. Plant food allergens homologous to pathogenesis-related proteins. Allergy. 2001;56:43–44. doi: 10.1034/j.1398-9995.2001.00913.x. [DOI] [PubMed] [Google Scholar]
- Egger M, Mutschlechner S, Wopfner N, Gadermaier G, Briza P, Ferreira F. Pollen-food syndromes associated with weed pollinosis: an update from the molecular point of view. Allergy. 2006;61:461–476. doi: 10.1111/j.1398-9995.2006.00994.x. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Reshef A, Patton L, Ayuso R, Reese G, Lehrer SB. Immunoglobulin E antibody reactivity to the major shrimp allergen, tropomyosin, in unexposed Orthodox Jews. Clinical and Experimental Allergy. 2003;33:956–961. doi: 10.1046/j.1365-2222.2003.01722.x. [DOI] [PubMed] [Google Scholar]
- Fiers M, Kleter GA, Nijland H, Peijnenburg A, Nap JP, van Ham R. Aller-match (TM), a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines. Bmc Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer ELL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Research. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furmonaviciene R, Sutton BJ, Glaser F, Laughton CA, Jones N, Sewell HF, Shakib F. An attempt to define allergen-specific molecular surface features: a bioinformatic approach. Bioinformatics. 2005;21:4201–4204. doi: 10.1093/bioinformatics/bti700. [DOI] [PubMed] [Google Scholar]
- Glaspole IN, de Leon MP, Rolland JM, O'Hehir RE. Characterization of the T-cell epitopes of a major peanut allergen, Ara h 2. Allergy. 2005;60:35–40. doi: 10.1111/j.1398-9995.2004.00608.x. [DOI] [PubMed] [Google Scholar]
- Goetz DW, Whisman BA, Goetz AD. Cross-reactivity among edible nuts: double immunodiffusion, crossed immunoelectrophoresis and human specific IgE serologic surveys. Annals of Allergy Asthma & Immunology. 2005;95:45–52. doi: 10.1016/S1081-1206(10)61187-8. [DOI] [PubMed] [Google Scholar]
- Goodman RE. Practical and predictive bioinformatics methods for the identification of potentially cross-reactive protein matches. Molecular Nutrition & Food Research. 2006;50:655–660. doi: 10.1002/mnfr.200500277. [DOI] [PubMed] [Google Scholar]
- Helm RM, Cockrell G, Connaughton C, Sampson HA, Bannon GA, Beilinson V, Nielsen NC, Burks AW. A soybean G2 glycinin allergen. 2. Epitope mapping and three-dimensional modeling. International Archives of Allergy and Immunology. 2000;123:213–219. doi: 10.1159/000024446. [DOI] [PubMed] [Google Scholar]
- Hemmer W, Focke M, Götz M, Jarisch R. Sensitization to Ficus benjamina: relationship to natural rubber latex allergy and identification of foods implicated in the ficus-fruit syndrome. Clinical and Experimental Allergy. 2004;34:1251–1258. doi: 10.1111/j.1365-2222.2004.02046.x. [DOI] [PubMed] [Google Scholar]
- Hileman RE, Silvanovich A, Goodman RE, Rice EA, Holleschak G, Astwood JD, Hefle SL. Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. International Archives of Allergy and Immunology. 2002;128:280–291. doi: 10.1159/000063861. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O, Oezguen N, Mathura VS, Schein CH, Xu Y, Braun W. Using property based sequence motifs and 3D modeling to determine structure and functional regions of proteins. Curr Med Chem. 2004;11:583–593. doi: 10.2174/0929867043455819. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O, Schein CH, Braun W. Data mining of sequences and 3D structures of allergenic proteins. Bioinformatics. 2002;18:1358–1364. doi: 10.1093/bioinformatics/18.10.1358. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O, Schein CH, Braun W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Research. 2003;31:359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins JA, Griffiths-Jones S, Shewry PR, Breiteneder H, Mills ENC. Structural relatedness of plant food allergens with specific reference to cross-reactive allergens: an in silico analysis. Journal of Allergy and Clinical Immunology. 2005;115:163–170. doi: 10.1016/j.jaci.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Jeong KY, Hong CS, Yong TS. Allergenic tropomyosins and their cross-reactivities. Protein and Peptide Letters. 2006;13:835–845. doi: 10.2174/092986606777841244. [DOI] [PubMed] [Google Scholar]
- Kazemi-Shirazi L, Pauli G, Purohit A, Spitzauer S, Froschl R, Hoffmann-Sommergruber K, Breiteneder H, Scheiner O, Kraft D, Valenta R. Quantitative IgE inhibition experiments with purified recombinant allergens indicate pollen-derived allergens as the sensitizing agents responsible for many forms of plant food allergy. Journal of Allergy and Clinical Immunology. 2000;105:116–125. doi: 10.1016/s0091-6749(00)90186-6. [DOI] [PubMed] [Google Scholar]
- Ladics GS, Bardina L, Cressman RF, Mattsson JL, Sampson HA. Lack of cross-reactivity between the Bacillus thuringiensis derived protein Cry1F in maize grain and dust mite Der p7 protein with human sera positive for Der p7-IgE. Regulatory Toxicology and Pharmacology. 2006;44:136–143. doi: 10.1016/j.yrtph.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Li KB, Issac P, Krishnan A. Predicting allergenic proteins using wavelet transform. Bioinformatics. 2004;20:2572–2578. doi: 10.1093/bioinformatics/bth286. [DOI] [PubMed] [Google Scholar]
- Mari A. Importance of databases in experimental and clinical allergology. International Archives of Allergy and Immunology. 2005;138:88–96. doi: 10.1159/000087848. [DOI] [PubMed] [Google Scholar]
- Marti P, Truffer R, Stadler MB, Keller-Gautschi E, Crameri R, Mari A, Schmid-Grendelmeier P, Miescher SM, Stadler BM, Vogel M. Allergen motifs and the prediction of allergenicity. Immunology Letters. 2007;109:47–55. doi: 10.1016/j.imlet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Mathura VS, Schein CH, Braun W. Identifying property based sequence motifs in protein families and superfamilies: application to DNase-1 related endonucleases. Bioinformatics. 2003;19:1381–1390. doi: 10.1093/bioinformatics/btg164. [DOI] [PubMed] [Google Scholar]
- Midoro-Horiuti T, Brooks EG, Goldblum RM. Pathogenesis-related proteins of plants as allergens. Annals of Allergy Asthma & Immunology. 2001;87:261–271. doi: 10.1016/S1081-1206(10)62238-7. [DOI] [PubMed] [Google Scholar]
- Midoro-Horiuti T, Goldblum RM, Kurosky A, Wood TG, Schein CH, Brooks EG. Molecular cloning of the mountain cedar (Juniperus ashei) pollen major allergen, Jun a 1. Journal of Allergy and Clinical Immunology. 1999;104:613–617. doi: 10.1016/s0091-6749(99)70332-5. [DOI] [PubMed] [Google Scholar]
- Midoro-Horiuti T, Mathura V, Schein CH, Braun W, Yu SN, Watanabe M, Lee JC, Brooks EG, Goldblum RM. Major linear IgE epitopes of mountain cedar pollen allergen Jun a 1 map to the pectate lyase catalytic site. Molecular Immunology. 2003;40:555–562. doi: 10.1016/s0161-5890(03)00168-8. [DOI] [PubMed] [Google Scholar]
- Midoro-Horiuti T, Schein CH, Mathura V, Braun W, Czerwinski EW, Togawa A, Kondo Y, Oka T, Watanabe M, Goldblum RM. Structural basis for epitope sharing between group 1 allergens of cedar pollen. Molecular Immunology. 2006;43:509–518. doi: 10.1016/j.molimm.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, Gajhede M. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. Journal of Immunology. 2000;165:331–338. doi: 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- Mittag D, Batori V, Neudecker P, Wiche R, Friis EP, Ballmer-Weber BK, Vieths S, Roggen EL. A novel approach for investigation of specific and cross-reactive IgE epitopes on Bet v 1 and homologous food allergens in individual patients. Molecular Immunology. 2006;43:268–278. doi: 10.1016/j.molimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Mittag D, Vieths S, Vogel L, Wagner-Loew D, Starke A, Hunziker P, Becker WM, Ballmer-Weber BK. Birch pollen-related food allergy to legumes: identification and characterization of the Bet v 1 homologue in mungbean (Vigna radiata), Vig r 1. Clinical and Experimental Allergy. 2005;35:1049–1055. doi: 10.1111/j.1365-2222.2005.02309.x. [DOI] [PubMed] [Google Scholar]
- Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, Braun W. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Molecular Immunology. 2008;45:3740–3747. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. Using the FASTA program to search protein and DNA sequence databases. Methods in Molecular Biology. 1994;25:365–389. doi: 10.1385/0-89603-276-0:365. [DOI] [PubMed] [Google Scholar]
- Rabjohn P, Helm EM, Stanley JS, West CM, Sampson HA, Burks AW, Bannon GA. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. Journal of Clinical Investigation. 1999;103:535–542. doi: 10.1172/JCI5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C, Breiteneder H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. Journal of Allergy and Clinical Immunology. 2006;117:141–147. doi: 10.1016/j.jaci.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. Journal of Allergy and Clinical Immunology. 2008;121:847–52 e7. doi: 10.1016/j.jaci.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Reese G, Ayuso R, Lehrer SB. Tropomyosin: an invertebrate pan-allergen. International Archives of Allergy and Immunology. 1999;119:247–258. doi: 10.1159/000024201. [DOI] [PubMed] [Google Scholar]
- Reese G, Schicktanz S, Lauer I, Randow S, Lüttkopf D, Vogel L, Lehrer SB, Vieths S. Structural, immunological and functional properties of natural recombinant Pen a 1, the major allergen of Brown Shrimp, Penaeus aztecus. Clinical and Experimental Allergy. 2006;36:517–524. doi: 10.1111/j.1365-2222.2006.02454.x. [DOI] [PubMed] [Google Scholar]
- Riaz T, Hor HL, Krishnan A, Tang F, Li KB. WebAllergen: a web server for predicting allergenic proteins. Bioinformatics. 2005;21:2570–2571. doi: 10.1093/bioinformatics/bti356. [DOI] [PubMed] [Google Scholar]
- Robotham JM, Teuber SS, Sathe SK, Roux KH. Linear IgE epitope mapping of the English walnut (Juglans regia) major food allergen, Jug r 1. Journal of Allergy and Clinical Immunology. 2002;109:143–149. doi: 10.1067/mai.2002.120558. [DOI] [PubMed] [Google Scholar]
- Robotham JM, Wang F, Seamon V, Teuber SS, Sathe SK, Sampson HA, Beyer K, Seavy M, Roux KH. Ana o 3, an important cashew nut (Anacardium occidentale L.) allergen of the 2S albumin family. Journal of Allergy and Clinical Immunology. 2005;115:1284–1290. doi: 10.1016/j.jaci.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Saha S, Raghava GPS. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Research. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo G, Sanchez-Monge R, Diaz-Perales A, Garcia-Casado G, Barber D. Plant non-specific lipid transfer proteins as food and pollen allergens. Clinical and Experimental Allergy. 2004;34:1336–1341. doi: 10.1111/j.1365-2222.2004.02018.x. [DOI] [PubMed] [Google Scholar]
- Schein CH, Ivanciuc O, Braun W. Common physical-chemical properties correlate with similar structure of the IgE epitopes of peanut allergens. Journal of Agricultural and Food Chemistry. 2005a;53:8752–8759. doi: 10.1021/jf051148a. [DOI] [PubMed] [Google Scholar]
- Schein CH, Ivanciuc O, Braun W. Structural database of allergenic proteins (SDAP) In: Maleki SJ, Burks AW, Helm RM, editors. Food Allergy. ASM Press; Washington, DC: 2006. pp. 257–283. [Google Scholar]
- Schein CH, Ivanciuc O, Braun W. Bioinformatics approaches to classifying allergens and predicting cross-reactivity. Immunology and Allergy Clinics of North America. 2007;27:1–27. doi: 10.1016/j.iac.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Özgün N, Izumi T, Braun W. Total sequence decomposition distinguishes functional modules, „molegos” in apurinic/apyrimidinic endonucleases. BMC Bioinformatics. 2002;3:37. doi: 10.1186/1471-2105-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Zhou B, Braun W. Stereophysicochemical variability plots highlight conserved antigenic areas in Flaviviruses. Virology Journal. 2005b;2:40. doi: 10.1186/1743-422X-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Zhou B, Oezguen N, Mathura VS, Braun W. Molego-based definition of the architecture and specificity of metal-binding sites. Proteins: Structure, Function, and Bioinformatics. 2005c;58:200–210. doi: 10.1002/prot.20253. [DOI] [PubMed] [Google Scholar]
- Schwietz LA, Goetz DW, Whisman BA, Reid MJ. Cross-reactivity among conifer pollens. Annals of Allergy Asthma & Immunology. 2000;84:87–93. doi: 10.1016/S1081-1206(10)62746-9. [DOI] [PubMed] [Google Scholar]
- Shanti KN, Martin BM, Nagpal S, Metcalfe DD, Rao PVS. Identification of tropomyosin as the major shrimp allergen and characterization of its IgE-binding epitopes. Journal of Immunology. 1993;151:5354–5363. [PubMed] [Google Scholar]
- Shin DS, Compadre CM, Maleki SJ, Kopper RA, Sampson H, Huang SK, Burks AW, Bannon GA. Biochemical and structural analysis of the IgE binding sites on Ara h1, an abundant and highly allergenic peanut protein. Journal of Biological Chemistry. 1998;273:13753–13759. doi: 10.1074/jbc.273.22.13753. [DOI] [PubMed] [Google Scholar]
- Sone T, Dairiki K, Morikubo K, Shimizu K, Tsunoo H, Mori T, Kino K. Identification of human T cell epitopes in Japanese cypress pollen allergen, Cha o 1, elucidates the intrinsic mechanism of cross-allergenicity between Cha o 1 and Cry j 1, the major allergen of Japanese cedar pollen, at the T cell level. Clinical and Experimental Allergy. 2005;35:664–671. doi: 10.1111/j.1365-2222.2005.02221.x. [DOI] [PubMed] [Google Scholar]
- Sone T, Morikubo K, Miyahara M, Komiyama N, Shimizu K, Tsunoo H, Kino K. T cell epitopes in Japanese cedar (Cryptomeria japonica) pollen allergens: choice of major T cell epitopes in Cry j 1 and Cry j 2 toward design of the peptide-based immunotherapeutics for the management of Japanese cedar pollinosis. Journal of Immunology. 1998;161:448–457. [PubMed] [Google Scholar]
- Stadler MB, Stadler BM. Allergenicity prediction by protein sequence. Faseb Journal. 2003;17:1141–1143. doi: 10.1096/fj.02-1052fje. [DOI] [PubMed] [Google Scholar]
- Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, Helm RM, West CM, Bannon GA. Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Archives of Biochemistry and Biophysics. 1997;342:244–253. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Kawaguchi J, Serizawa N, Hirahara K, Shiraishi A, Nigi H, Taniguchi Y, Toda M, Inouye S, Takemori T, Sakaguchi M. Analysis of sequential immunoglobulin E-binding epitope of Japanese cedar pollen allergen (Cry j 2) in humans, monkeys and mice. Clinical and Experimental Allergy. 2003;33:211–217. doi: 10.1046/j.1365-2222.2003.01579.x. [DOI] [PubMed] [Google Scholar]
- Thomas K, Bannon G, Hefle S, Herouet C, Holsapple M, Ladics G, MacIntosh S, Privalle L. In silico methods for evaluating human allergenicity to novel proteins. International Bioinformatics Workshop Meeting Report, February 23-24, 2005. Toxicological Sciences. 2005;88:307–310. doi: 10.1093/toxsci/kfi277. [DOI] [PubMed] [Google Scholar]
- Venkatarajan MS, Braun W. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical-chemical properties. Journal of Molecular Modeling. 2001;7:445–453. [Google Scholar]
- Weber RW. Cross-reactivity of pollen allergens: recommendations for immunotherapy vaccines. Current Opinion in Allergy and Clinical Immunology. 2005;5:563–569. doi: 10.1097/01.all.0000191240.28255.ab. [DOI] [PubMed] [Google Scholar]
- Wensing M, Akkerdaas JH, van Leeuwen A, Stapel SO, Bruijnzeel-Koomen C, Aalberse RC, Bast B, Knulst AC, van Ree R. IgE to Bet v 1 and profilin: cross-reactivity patterns and clinical relevance. Journal of Allergy and Clinical Immunology. 2002;110:435–442. doi: 10.1067/mai.2002.126380. [DOI] [PubMed] [Google Scholar]
- WHO . Report of a Joint FAO/WHO Expert Consultation. World Health Organization; Geneva: 2000. Safety aspects of genetically modified foods of plant origin. [Google Scholar]
- WHO . Report of a Joint FAO/WHO Expert Consultation. World Health Organization; Geneva: 2001. Evaluation of allergenicity of genetically modified foods. [Google Scholar]
- WHO . Codex ad hoc inter-governmental task force on foods derived from biotechnology. World Health Organization; Yokohama: 2003. Joint FAO/WHO food Standards Programme.http://www.codexalimentarius.net/ [Google Scholar]
- Wild LG, Lehrer SB. Fish and shellfish allergy. Current Allergy and Asthma Reports. 2005;5:74–79. doi: 10.1007/s11882-005-0059-z. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Ohmoto T, Urisu A, Mine Y, Adachi T. Expression and epitope analysis of the major allergenic protein Fag e 1 from buckwheat. Journal of Plant Physiology. 2004;161:761–767. doi: 10.1016/j.jplph.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Matsuo H, Morita E. Cross-reactivity among shrimp, crab and scallops in a patient with a seafood allergy. Journal of Dermatology. 2006;33:174–177. doi: 10.1111/j.1346-8138.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- Zorzet A, Gustafsson M, Hammerling U. Prediction of food protein allergenicity: a bioinformatic learning systems approach. In Silico Biology. 2004;2:525–534. [PubMed] [Google Scholar]