Abstract
Nrf2 is the key transcription factor regulating the antioxidant response. Nrf2 signaling is repressed by Keap1 at basal condition and induced by oxidative stress. Keap1 is recently identified as a Cullin 3-dependent substrate adaptor protein. A two-sites binding “hinge & latch” model vividly depicts how Keap1 can efficiently present Nrf2 as substrate for ubiquitination. Oxidative perturbation can impede Keap1-mediated Nrf2 ubiquitination but fail to disrupt Nrf2/Keap1 binding. Nrf2 per se is a redox-sensitive transcripon factor. A new Nrf2-mediated redox signaling model is proposed based on these new discoveries. Free floating Nrf2 protein functions as a redox-sensitive probe. Keap1 instead functions as a gate keeper to control the availability of Nrf2 probes and thus regulates the overall sensitivity of the redox signaling.
Keywords: Nrf2, Keap1, redox
I. Introduction: the antioxidant response
To adapt to their aerobic life style, mammalian cells have developed an elaborate yet highly efficient cytoprotective machinery. When exposed to oxidative stress, these cells can respond with a rapid and coordinated expression of a battery of gene products, including phase II detoxification enzymes/antioxidants and phase III efflux transporters [1-3]. As a result, these cells can effectively neutralize and remove excess oxidants to restore redox homeostasis. Malfunction of cellular antioxidant defense is implied in the etiology of diverse diseases, including cancer [4,5]. Gene knockout mice deficient in antioxidant response show higher susceptibility to carcinogens [6-9]. Persistent overload of reactive oxygen species (ROS) in chronic inflammation is also found correlated with an increased incidence of tumorigenesis [10-12]. Therefore, our mechanistic understanding of the regulation of the antioxidant response may eventually help us to explore new and more potent strategies of cancer prevention and therapy.
Pivotal to the antioxidant response is a transcription factor Nrf2 (NF-E2 related factor 2). Like many other transcription factors, Nrf2 signaling is regulated by compartmental segregation. Under basal conditions, Nrf2 is found mainly sequestered in the cytoplasm. When challenged by oxidative stress derived from accumulation of ROS [13-15] or reactive nitrogen species (RNS) [16,17], Nrf2 can quickly translocate into the nucleus and elicit the antioxidant response. At least four components in combine, namely Nrf2, Keap1 (Kelch-like ECH-associated protein 1), a group of small musculoaponeurotic fibrosarcoma (Maf) proteins and a cis-acting enhancer called antioxidant response element (ARE) or electrophile responsive element (EpRE), are essential for the antioxidant response [18].
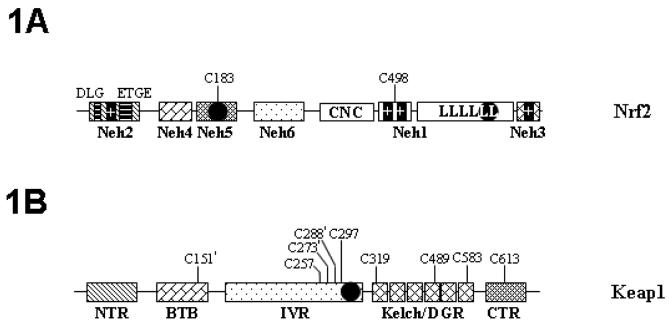
Nrf2 is a basic leucine zipper (bZIP) transcription factor featuring a Cap “n” collar (CNC) structure [19]. Nrf2 has some highly conserved domains called Nrf2-ECH homology (Neh) domains (Fig. 1A). Among them, the Neh1 domain is the CNC-bZIP domain, enabling Nrf2 to form heterodimer with the ZIP domain of small Maf proteins [1]. The Neh2 domain mediates binding with the cytosolic repressor of Nrf2, Keap1 [1]. The Neh2 domain contains a degron and involves in ubiquitin-dependent degradation [20]. The tandem of the Neh4 and Neh5 domains mediates cooperative transactivation activity of Nrf2 [21]. Recently, the Neh3 domain, which is located at the extreme end of the carboxyl terminus of Nrf2, is found to play a permissive role of Nrf2 transactivation [22]. In the absence of Nrf2 structural data, it is unknown how Neh3, together with the Neh4-Neh5 domains, exerts transactivation activity. The Neh6 domain is located in the intervening region linking the Neh5 and the Neh1 domain. The Neh6 domain may contain another degron and involve in Nrf2 degradation [23].
FIG. 1.
Schematic drawings of the molecular structure of Nrf2 and Keap1. (A) The Nrf2 molecule consists of CNC-basic leucine zipper (LLLLLL) Neh1 domain, Keap1 binding Neh2 domain (DLG and ETGE motifs are Keap1 binding motifs), a Neh3 domain, a tandem of Neh4 and Neh5 transactivation (TA) domains, and a linker Neh6 domain. The NES motifs are designated with filled circles. The bipartite NLS motif located in the basic region (+ +) is designated by double filled bars. The monopartite NLS motifs (+) located at the amino-terminus and carboxyl-terminus is designated by single filled bars. CNC, the collar “n” cap domain. The positions of putative reactive cysteines (C183 and C498) are also illustrated. (B) The Keap1 molecule consists of an amino-terminal region (NTR), a BTB region, an intervening region (IVR), a Kelch/double glycine repeat (DGR) domain and a carboxy terminal region (CTR). Keap1 also possesses a NES (filled circle) in the IVR domain. Chemically reactive cysteines are illustrated. Functionally important cysteines are labeled with asterisks.
Keap1 was first identified in a yeast two hybrid clone using the Neh2 domain as the bait [24,25]. Keap1 was found to be a potent cytosolic repressor of Nrf2. Gene knockout of Keap1 resulted in constitutively hyperactive Nrf2 signaling [26-28]. The 625 a.a. human Keap1 consists of five domains: an amino-terminal region (NTR, a.a. 1-66), a Broad complex, Tramtrack and Bric a brac domain (BTB, a.a.67-178), an intervening region (IVR, a.a. 179-321), a Kelch/double glycin repeat (DGR, a.a. 322-608) and a carboxyl terminal region (CTR, a.a. 609-625) (Fig. 1B). Keap1 mainly distributes in the cytoplasm, immobilized by binding to filamentous-actin and/or myosin VIIa of the cytoskeleton via the Kelch/DGR domain [24,29-31]. Accordingly, Keap1 is hypothesized to be the cytosolic anchor of Nrf2, sequestering Nrf2 in the cytoplasm during basal conditions. In addition, the IVR domain contains a nuclear export signal (NES) [32,33] and may dictate subcellular distribution of Keap1 [34,35]. Recently, Keap1 is also identified as a Cullin 3-dependent substrate adaptor protein for ubiquitin ligase E3 complex [36-39]. Thus, Nrf2 molecules may not only be sequestered by Keap1 but also subjected to constant degradation. Keap1 is a cysteine rich protein. Some of cysteines in Keap1 are hypothetically reactive cysteines (see below for details). Accordingly, Keap1 is hypothesized to play a role of primary redox sensor; the presence of redox stress can modify single or multiple reactive cysteine(s) in Keap1, exert a profound conformation change and consequently release Nrf2 to trigger the antioxidant response [26,40].
After Nrf2 translocates into the nucleus, Nrf2 forms heterodimer with a group of nuclear bZIP proteins called small Maf proteins [41]. The small Maf proteins, composed ofMafF, G, K, lack the transactivation domain [42]. Nrf2 heterodimerization with MafF/G/K enhances the specificity to bind to a cis-acting enhancer ARE/EpRE [43-45] located in the promoter of a battery of cytoprotective genes [42,46]. The binding of Nrf2/Maf heterodimer to ARE/EpRE subsequently initiates the transcription of these genes.
In this old model of antioxidant response, Keap1 is depicted as the primary redox sensor [26,40] and the trigger of the antioxidant response. The redox-signal encoded by Keap1 is subsequently transmitted to Nrf2 in a key process of Nrf2 release from Keap1. However, accumulating new evidence in the recent studies argues against the old model. In special, compelling evidence shows that oxidative stress fails to disrupt Keap1/Nrf2 binding. In other words, the relay of the redox signal from Keap1 to Nrf2 is unlikely to happen in vivo. How to accommodate these apparently contradictory discoveries? In this review, we will evaluate the recent progress in Nrf2/Keap1 studies. A quite different picture of redox signaling emerges from those discoveries.
II. Keap1
Keap1 as a redox sensor
Keap1 is a cysteine rich protein. Human Keap1 (hKeap1) and murine Keap1 (mKeap1) have 27 and 25 cysteine residues, respectively. In comparison, human Nrf2 (hNrf2) and mouse Nrf2 (mNrf2) only possess 6 and 7 cysteines, respectively. Keap1 thio-modification appeared more avid than Nrf2 [40]. With the assumption that Keap1 is the key regulator of Nrf2 signaling, Keap1 is prioritized as a putative candidate for a redox-sensor.
The existence of reactive cysteines in Keap1 was first proven in mKeap1 using an irreversible alkylating agent dexamethasone 21-mesylate (Dex-mes). Tritiated Dex-mes labeled mKeap1 protein [40]. Sulforaphane (SFN), a well-known phase II inducer [47], could also label mKeap1 protein in vitro. The dithiocarbamates formed between reactive thiols of mKeap1 and the isothiocyanate group of SFN could give rise a characteristic absorption near λ=270 nm. When 0.7 μM mKeap1 was incubated with 50 μM SFN at 25°C for 90 min, an absorption at λ=270 nm was observed [40]. The kinetics of dithiocarbamate formation appeared in a SFN dose dependent manner [40]. Other phase II inducers could also modify Keap1. The reactive cysteines in Keap1 could not only be alkylated, but also be oxidized by hydroperoxide (H2O2) [48] and modified by disulfide-forming agent dipyridyl disulfide (DPDS) [40,48]. The amplitudes of thio-modification were found to correlate with the relative potency to induce antioxidant response by those compounds [40]. Keap1 contains 8 tryptophan residues, 6 of which located in the Kelch domain [48]. When Keap1 protein was treated with SFN or DPDS, substantial quenching of intrinsic tryptophan fluorescence was observed, suggesting a profound conformation change of Keap1 [48]. These data clearly show that Keap1 may function as a redox-sensor.
Great efforts were taken to elucidate the positions of reactive cysteines in Keap1. All cysteine residues in Keap1 are found highly conserved across species [24,49]. Therefore it is difficult to characterize their functional significance merely by their sequence homology. In mKeap1, 9 cysteines are flanked by one or two basic amino acid residue(s), a phenomenon that is hypothecized (local electrostatic environment model) to decrease the pKa value and increase the redox reactivity of cysteine [50]. In other words, those cysteines are hypothetically reactive cysteines. For all these 9 cysteines, two (C23, C38) are located in the NTR domain, one (C151) is located in the BTB domain, five (C241, C273, C288, C297 and C319) are located in the IVR domain and one (C613) is located in the CTR domain [26]. In contrast, it is noticeable that all 8 cysteines of the Kelch domain are not hypothetically reactive. Indeed, crystallographic data showed that they do not exist in disulfide bonds [51].
The positions of reactive cysteines in Keap1 were first characterized using the Dex-mes labeling method [26,40]. Purified and dithiothreitol-reduced mKeap1 protein was first treated with 33 folds excess of Dex-Mes at room temperature for 1 hour, followed by N-ethylmaleimide labeling of all inert cysteines. The labeled proteins were trypsinized and resolved by liquid chromatography and tandem mass spectrometry (LC-MS-MS). Four cysteines located in the IVR domain (C257, C273, C288 and C297) and one cysteine located in the CTR domain (C613) were characterized as reactive cysteines [40]. Among these five experimentally identified reactive cysteines, four are hypothetically reactive cysteines with the only exception of C257, which is flanked by an aspartic acid and a proline. Another study using a different irreversible alkylating agent biotinylated iodoacetamide (BIA) and 1-Cl-2, 4-dinitrobenzene (CDNB) further identified C151 and C319 as reactive cysteines in hKeap1 [49]. In addition, the irreversible thiol reactive modifier iodoacetyl-N-biotinylhexylenediamine (IAB) and 1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido)-butane (BMCC) as well as hydrolysable modifier SFN were used to label hKeap1 protein [52,53]. The detected sulfhydryl adducts are only partially compatible with the previous studies. C257 and C288 were found easily modified by IAB and SFN. C151 and C273 adducts were only detected occasionally. In contrast, C297 appeared to be redox-inert despite the fact that C297 is flanked by two basic residues (296RCK298). The C297 adduct was completely undetectable even at the artificially high concentration of SFN (200 μM) [52]. While the incidences of sulfhydryl modification of some hypothetically reactive cysteines are not very high, the incidences of thio-modification of some cysteines in the Kelch domain are very high. The C489 adduct was always detected when hKeap1 was treated with SFN, even at low concentration of 2 μM [52]. Like C257, C489 is flanked by an acidic glutamate (488ECY490). Likewise, another Kelch cysteine (C583) with the same context (582ECY584) also appeared reactive to SFN labeling. In contrast, another Kelch cysteine (C395) with the similar context (394DCY396) appeared not very reactive [52]. Collectively, those sulfhydryl modification data show that the local electrostatic environment model may oversimplify the prediction of redox-reactivity of a cysteine residue, and one needs to consider the complexity of three-dimensional context of a cysteine residue. Further studies are needed to address this question.
Those in vitro thio-modification studies were followed up with mutation study and loss of function assay. It was found that substitution with a serine or alanine in Cys-273 (C273S or C273A) and Cys-288 (C288S or C288A) completely abolished the basal repressive activities on Nrf2 signaling [26,34,54]. In addition, C273S mutant completely lost induced response to tert-butylhydroquinone (tBHQ) treatment. C288S in comparison, only partially lost tBHQ-induced response [34]. In contrast, mutation at C257 and C297 appeared to have no apparent functional deficit [26,34]. In an elegant in vivo reconstitution study, the function of C273A and C288A mutants were evaluated in keap1-/- nrf2-/- double knockout mouse embryonic fibroblasts (MEF) [26]. Expression of C273A or C288A mutant alone failed to repress Nrf2 signaling. However, it was interesting to observe that co-expressing C273A and C288A in equal amounts could partially restore Keap1 repression [26]. Based on these observations, it is hypothesized that C273 and C288 may form intermolecular disulfide bond and consequently causes conformation change to disrupt Nrf2 binding [26] (Fig. 3).
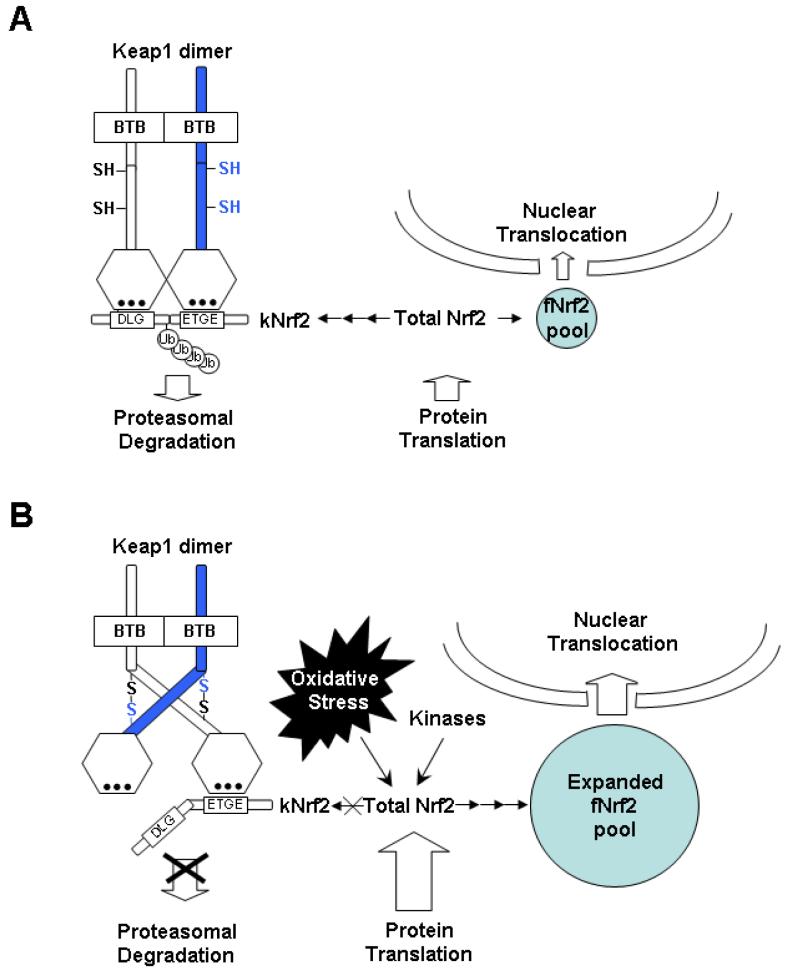
FIG. 3.
Hypothetic model of Nrf2-mediated redox signaling. The Keap1 dimer functions as an ubiquitination (Ub) substrate adaptor protein. There are two pools of Nrf2 proteins, free floating Nrf2 (fNrf2) and Keap1-binding Nrf2 (kNrf2).Under homeostatic conditions (A), kNrf2 binds toa Keap1 dimer via a high affinity ETGE (hinge) motif and a low affinity DLG (hatch) motif. The two-sites binding expose the Ub-acceptor site(s) in Nrf2. Ubiquitinated Nrf2 proteins are destined to proteasomal degradation. Under homeostatic conditions, there appears to be an equilibrium between protein translation and degradation. As a result, there is only a small pool of fNrf2, contributing to basal activation. When stimulated by oxidative stress (B), the conformation change of the Keap1 dimer, probably via intermolecular disulfide bond formation, disrupts the two-sites binding. As a result, the Ub-acceptor site(s) is/are not easily accessible. The ubiquitination and the proteasomal degradation are inhibited. Keap1 is saturated by undegradated kNrf2. Protein translation however, is elevated. In combination, the pool of fNrf2 expands. fNrf2 can sense the change of redox milieu and transmit redox signals to cell nucleus via gradient nuclear translocation. In this model, redox signal detection is not relayed from Keap1 to Nrf2 via Keap1/Nrf2 dissociation. Instead, Keap1 plays a gate-keeper role; Keap1 dictates the pool size of fNrf2 and thus regulates the overall redox sensitivity.
In conformation of the BIA/IAB labeling studies, the C151 residue located in the BTB domain was also found to be important in Nrf2 repression. C151S mutation caused constitutive repression of Nrf2 signaling, both at the basal and SFN/tBHQ induced condition [34]. However, double mutation of C151S and C273S nullified repression on Nrf2 signaling, implying that C273S mutant had a dominant effect [34].
The functional significance of other chemically reactive cysteines, such as the C613 residue of the CTR, remains unclear. Since the structural data show that the amino and carboxyl terminus of the Kelch domain are located in close vicinity [51], the CTR and the IVR domain are likely located on the same side of the Kelch domain and therefore exposed to thio-modification. Further studies are needed to delineate the actual function of the C613 residue.
Thio-modification of Keap1 failed to release Nrf2 from Keap1 sequestering
After detecting sulfhydryl adducts in Keap1, the functional significance of thio-modification of Keap1 was investigated. In particular, whether thio-modification of Keap1 can trigger Nrf2 dissociation from Keap1 was examined. It was first shown that the Neh2 segment and mKeap1 could form a heterodimer under reduced conditions [40]. The Neh2/mKeap1 dimer could be disrupted by high concentrations of SFN (0.5-10 mM) as well Bis(benzylidene)acetones [40] as visualized by electrophoretic mobility shift assay (EMSA). A follow up study confirmed that the C257AC273AC288AC297A quadruple mutant of Keap1 showed attenuated binding affinity to Neh2 [26]. Based on these observations, a direct disruption model was proposed. Keap1 is hypothesized to play the role of a primary redox-sensor. Thio-modifications of Keap1 change its conformation. As a consequence, Nrf2 is released and transmits the redox signal to the nucleus. It has to be pointed out that there is an intrinsic discrepancy in these studies. Whereas 20-50 μM SFN could effectively label Keap1 [40,52], millimolar range of SFN (a 2000-folds excess of SFN to Keap1) was needed to disrupt Neh2/Keap1 binding [40]. These in vitro results appeared even more unlikely to be achieved in vivo due to the presence of excessive amount of glutathione (GSH).
Recently, the causal relationship between thio-modification of Keap1 and Nrf2 release was carefully interrogated. EMSA results showed that pre-treatment with excessive amounts of SFN (15, 35 and 80-fold) on hKeap1 failed to inhibit the formation of the Neh2/hKeap1 heterodimer [49]. In addition, the pre-formed Neh2/hKeap1 heterodimer was not disrupted by 80-folds excess SFN treatment [49]. The binding affinity between Neh2 and hKeap1 was measured using the method of isothermal calorimetry. High affinity binding between Neh2/hKeap1 (Kd=9 nM) was observed between unmodified hKeap1 and Neh2 [49] and confirmed in later experiments [55]. Pretreatment of hKeap1 with BIA or CDNB didn't change the binding affinity between thio-modified hKeap1 and Neh2 [49]. Thus, these data argue convincingly against the aforementioned direct disruption model. Recently, more experimental data were accumulated showing that substantially high amount of Nrf2/Keap1 dimers remained undisrupted in SFN and tBHQ induced antioxidant response [26,34,49,56,57]. Therefore, Keap1 and Nrf2 appear to sense and transmit redox-signals in a quite different manner.
Keap1 as a substrate adaptor protein for Cul3-dependent ubiquitination complex
Nrf2 is a short-live protein, rapidly degraded in a ubiquitin-dependent manner [23,34,58]. Recently, Keap1 is characterized as a substrate adaptor protein for ubiquitination (Ub) of Nrf2 [36-39]. As a substrate adaptor protein, Keap1 has two functional domains. The Kelch/DGR domain binds the substrate, Nrf2. The BTB domain [36-39] and probably part of the IVR domain [39] bind to the ubiquitination catalytic complex.
Ubiquitination of a substrate protein is achieved by sequential reactions catalyzed by an Ub-activating enzyme (E1), an Ub-conjugating enzyme (E2) and an Ub-ligase (E3) [59]. The Ub-E3 catalytic core consists of a scaffold Cullin protein binding to a RING box protein Rbx1 (also known as Roc1), which functions to recruit its cognate Ub-E2. To date, 7 Cullin proteins have been cloned [60]. Keap1 shows high selectivity to bind to Cullin 3 protein, weak binding to Cullin 2, and no binding activity to all other Cullins [38,39]. In comparison, other stress-transducing transcription factors show different Cullin-binding selectivity. IκB of the inflammatory response specifically binds to Cullin 1 and hypoxia inducible factor 1α (HIF-1α) specifically binds to Cullin 2. These findings suggest that the transcription factors transducing different types of stress signals are segregated in different degradation pathways [61].
The “hinge & latch” two-sites binding model
Previously it was reported that based on sequence homology and mutation analyses, there are two Keap1 binding motifs in the Neh2 domain; an ETGE motif (formulated as D/N-X-E-T/S-G-E) and a DLG motif (formulated as L-X-X-Q-D-X-D-L-G) [20,23,62]. The ETGE motif and the DLG motif can mediate a cooperative Keap1 binding. Deletion of the ETGE motif can significantly weaken Keap1 binding mediated by the DLG motif, thus the ETGE motif appears to play a permissive role for the DLG motif. Recently, isothermal calorimetry measurement also detected a two-phases binding between Keap1 and Neh2. The Keap1 binding affinity of the ETGE motif (Ka=20×107 M−1) is two orders stronger than the DLG motif (Ka=0.1×107 M−1) [55,63].
The structure of the Kelch/DGR domain of Keap1 has recently been resolved [51,64,65]. Consistent with the fact that Kelch/DGR domain consists of 6 Kelch modules (Fig. 1B), the X-ray crystallographic data depicted the Kelch/DGR domain as a hexahedron cylinder. Each Kelch module consists of 4 anti-parallel β-strands. Overall, the Kelch/DGR domain resembles a β-propeller consisting of 6 symmetric β-blades. The Neh2 binding interface is located at the bottom of this β-propeller structure, including a highly conserved arginine triad (R380, R415 and R483). These positively charged arginine residues of Keap1 may form strong electrostatic bonds with the negatively charged glutamate residues in the ETGE motif of Nrf2. When an ETGE-containing peptide was co-crystallized with the Kelch domain, the ETGE-containing peptide assumed a U-shape conformation; two short anti-parallel β-strands were linked by two overlapping type I β-turns [64,65]. This hairpin conformation of the ETGE motif may enable maximal interaction and account for high binding affinity mediated by the ETGE motif.
Since the nuclear magnetic resonance titration assay [55] suggests an optimal molecular ratio of 2:1 for Keap1/Neh2 binding, a two-sites binding model has been proposed [61]. Two Keap1 molecules form a homodimer via their BTB domain [66]. For the Neh2 domain, the high affinity binding mediated by the ETGE motif functions as a “hinge” to pin down Neh2 to Keap1. The low affinity binding mediated by the DLG motif functions as a “latch”. Linking the “hinge” and the “latch” motif is a lysine-rich central α-helix. It is very interesting to observe that in this nine-turns α-helix, 6 of 7 lysine residues are located on the same side [55]. Therefore when the “latch” is in position, it may help to set the central α-helix in an adequate orientation to expose those lysine residues, presumably as the Ub-acceptors [39]. In the Keap1 homodimer, the distances between the guanidinium nitrogens of Arg-380, Arg-415 and Arg-483 are estimated as 55, 50 and 55Å apart, whereas the central α-helix linking the DLG and ETGE motifs is estimated as 49Å [55,57]. Thus a perfect geometric match exists for two-sites binding. This “hinge & latch” model [61] therefore vividly depicts a substrate-presentation mechanism (Fig. 3A).
In essence, this “hinge & latch” model can discern the difference between a one-site “hinge” binding and a two-sites “hinge & latch” binding. The one-site binding is a mere physical binding between Keap1 and Neh2. Only the two-sites binding assumes the substrate presentation function. Indeed, the deletion of a segment (a.a. 17-32) containing the DLG domain could stabilize this Nrf2 mutant [23]. Unlike the one-site binding, the two-sites binding is relatively unstable, dictated by the low-affinity “latch” binding. The low affinity “latch” binding may be more vulnerable to the conformation change of Keap1 dimer as a result of oxidative perturbation (Fig. 3B). The switch from the two-sites binding to the one-site binding can explain the observed facts that the inhibition of Nrf2 degradation is not at the expense of Keap1/Nrf2 binding.
The “hinge & latch” model can also explain how thio-modification disrupts the substrate presentation condition and consequently cause an impedance of Nrf2 ubiquitination. Recently, Keap1 has been characterized as a Zn2+-binding metalloprotein [48]. The metal-coordination function may explain how under reducing conditions Keap1 maintains the geometric constraint needed for a two-sites binding. Keap1 is able to bind zinc under reducing conditions. Keap1 was found to have similar Zn2+-binding affinity as zinc finger proteins [48]. Stoichiometry measurements show that one Keap1 molecule binds 1 zinc ion. Mutation at C273 and C288 significantly attenuates zinc-binding affinity of Keap1 [48]. Thus C273 and C288 are hypothesized to function as ligands in zinc coordination [48]. In other words, the active thiolates of C273 and C288 are preserved in zinc-binding and poised for redox sensing. Treatment of oxidants can displace zinc and modify C273 and C288 residues [48]. When C273 and C288 are modified, a possible intermolecular disulfide bond formation [26] may profoundly change the conformation of the Keap1 dimer [48]. As a result, the distance of arginine triads between two Kelch domains changes, becoming incompatible with a two-sites binding (Fig. 3B). This conformation change however, may be not sufficient to disrupt physical binding between Nrf2 and Keap1 mediated by high affinity binding of the ETGE motif. In other words, thio-modification only negates Keap1-mediated Nrf2 ubiquitination but does not disrupt Nrf2/Keap1 binding.
The thio-modification may simultaneously have a profound effect on Keap1 self-ubiquitination. Intense Keap1 degradation in parallel with attenuated Nrf2 ubiquitination was reported in tBHQ induced antioxidant response [67,68]. However, unlike Nrf2 degradation, the Keap1 self-ubiquitination triggered a 26S proteasome-independent degradation [67].
Alternatively, it is possible that Keap1 homodimerization is vulnerable to oxidative perturbation. Previously, it was reported that the treatments of pro-oxidant 100 μM PDTC (pyrrolidinedithiocarbamate) and phenethyl isothiocyanate (PEITC) could disrupt Keap1 homodimer [66]. Mutation of Ser-104 in the BTB domain of Keap1 was found to be able to disrupt Keap1 homodimerization [66]. Unfortunately, this experiment could not be duplicated using high concentration of SFN (90 μM) or Ser-104 mutation [57] and thus remains controversial. Dissociation of Keap1 homodimer but not the Keap1/Nrf2 heterodimer may have functional implications. The Keap1/Nrf2 heterodimer may shuttle to the nucleus together [33,35] to evade cytosolic proteasomal degradation. Mutation of the NES motif of Keap1 appeared to be able to arrest Keap1/Nrf2 in the nucleus [35]. One confusing question is how Keap1 escapes from cytoskeletal anchoring [29]. In addition, it is unknown whether the NES motif of Keap1 can be disabled under physiological conditions. However, the nuclear translocation of Keap1/Nrf2 heterodimer may have functional significance. Recently, a BTB-bZIP transcription repressor protein Bach2 was found to be recruited into the promyelocytic leukemia (PML) nuclear body via the BTB domain [69]. Since Nrf2 lacks the BTB domain, it is fascinating to speculate that the formation of Keap1/Nrf2 heterodimer may help to recruit Nrf2 into the PML nuclear body. Further studies are needed to address these questions.
III. Nrf2
Multiple NES/NLS motifs determine the subcellular distribution of Nrf2
Recently, mechanisms governing nuclear importing and exporting of Nrf2 have been elucidated. In human Nrf2 protein (in a 589 amino acids frame), one bipartite nuclear localization signal (NLS) was identified in the basic region [70,71] called bNLS. In addition, a monopartite NLS was characterized at the amino-terminus (NLSN) and another monopartite NLS was identified at the carboxyl-terminus (NLSC) of human and murine Nrf2 [72]. One NES motif was characterized in the ZIP dimerization domain (537L-K-K-Q-L-S-T-L-Y-L546) [70,71] called NESzip. The NESzip motif conforms to the canonical leucine-rich NES [73,74]. Canonical leucine-rich NES motif can be formulated as Φ1-(X)2-3-Φ2-(X)2-3-Φ3-X-Φ4, where 4 positions of Φ need to be hydrophobic amino acid residues and X can be any amino acid [73,74]. Recently, another NES (175L-L-S-I-P-E-L-Q-C-L-N-I186) was characterized in the Neh5 transactivation (TA) domain [35,75] called NESTA. These NLS and NES motifs differ in driving forces. The bNLS motif appears equal in driving force to the NESTA motif and stronger than the NESzip motif [75]. In full-length wild type Nrf2, the combined nuclear exporting activities of NESTA and NESzip appeared to be able to counter balance the combined nuclear importing activities mediated by the bNLS, NLSN and NLSC motifs. When an enhanced green fluorescence protein (EGFP) tagged Nrf2 was expressed in HeLa cells, a heterologous distribution pattern of EGFP-Nrf2 was observed. Nearly 60% of the cells showed whole cell distribution, 30% of the cells showed nuclear distribution and 12% of the cells showed cytosolic distribution [75]. Mutation of a single key leucine residue in the NES motif, for example, L544 in the NESzip motif and L184 in the NESTA motif, was sufficient to nullify their nuclear exporting activity [71,75]. Mutation of either the L184 residue or the L544 residue in full length Nrf2 was sufficient to convert the heterologous distribution of EGFP-Nrf2 into a predominantly nuclear distribution pattern [75]. In other words, both the NESTA and NESzip motifs are indispensable to counter balance the combined nuclear importing activity mediated by the bNLS, NLSN and NLSC motifs.
Overlapping localization of NES/NLS motifs
One salient feature of the NES/NLS motifs of Nrf2 is that they co-localize with other functional motifs (Fig. 1A). The bNLS is located in the basic region, a positively charged domain involved in DNA binding. The NESzip motif co-localizes with the ZIP dimerization domain [71]. It is well known that Nrf2 cannot form homodimer [76]. Nrf2 has to form heterodimer with small Maf proteins to bind to ARE [41]. The ZIP dimerization domain forms a parallel coiled coil [77] that consists of 4-6 heptads formulated as (abcdefg)4-6. The position “a” and “d” need to be hydrophobic residues. In the process of dimerization, the “a” and “d” residue in one monomer interact with the complementary “d” and “a” residue in the opposite monomer, respectively [76]. The interaction forms a hydrophobic core essential for dimer stability [78]. In this NESzip motif, the Φ1 (L537) and Φ3 (L544) residues locate at the “d” position in the fifth and sixth heptad of ZIP domain, respectively. The Φ2 (L541) residue locates at the “a” position in the sixth heptad. In other words, this NESzip motif occupies three key positions in the dimerization domain. The overlap between the NESzip motif and the ZIP motif implies that when Nrf2 forms heterodimer via leucine zipper with its obligatory binding partner small Maf proteins, the NESzip motif can be simultaneously masked. Indeed, we have found that MafG/K can enhance nuclear retention of Nrf2 (Li, W. Yu, S.W., Kong, A.-N.T., unpublished data). Recently a tyrosine-560 (Y560) was identified as a phosphorylation site for tyrosine kinase Fyn [79]. In addition, glycogen synthase kinase 3β (GSK-3β) was characterized as an upstream kinase of Fyn [80]. Indeed, activation of GSK-3β could promote Nrf2 nuclear exporting and attenuate phase II gene induction [80,81]. Interestingly, this Y560 is a neighbor of a legitimate MAPK site (S561 as in 559PYSP562). This S561 residue could be phosphorylated by ERK1/2 and p38 (Yu, S., Li, W., Kong, A.N., unpublished data). The juxtaposition of Y560 and S561 implies that MAPK pathway and GSK-3β/Fyn pathway may converge on Nrf2. GSK-3β/Fyn pathway and MAPK pathways may crosstalk and regulate Nrf2 signaling in a cooperative way. Further studies are needed to elucidate whether the vicinity of these two phosphorylation sites can influence Nrf2 nuclear export mediated by NESzip.
The NESTA motif is located in the Neh5 transactivation domain [75]. Previously it was reported that the Neh4 and Neh5 domains bind with a transcription co-activator CREB binding protein (CBP) [21,82,83] and silencing mediator for retinoid and thyroid hormone receptors (SMRT) [84]. The functional significance of overlapping localization of the NESTA motif with the Neh5 domain remains unclear.
Nrf2 as a redox-sensor
The identification of multiple NES/NLS motifs in Nrf2 naturally raised a question whether these NES/NLS motifs are redox-sensitive. There is a cysteine-498 embedded in the bNLS motif (Fig. 1A). Serine substitution of this cysteins residue (C498S) changed the DNA binding affinity but not the subcellular distribution of Nrf2 [85]. Therefore, the bNLS motif per se may have no impact on redox-sensitive distribution of Nrf2. The redox-sensitivity of the NLSN and NLSC motif has not been examined. Since there is no cysteine residue embedded in these two NLS motifs, they may be redox-insensitive. The NESzip motif was found redox-insensitive [71]. The redox-insensitivity of NESzip may be explained by the absence of cysteinein the NESzip motif [71].
The NESTA motif was found to be redox-sensitive. An EGFP tagged Nrf2 segment (a.a.162-295) containing the NESTA motif (EGFP-NESTA) exhibited a cytosolic distribution that could be nullified by a variety of oxidants including SFN, tBHQ and H2O2 [75]. It is very interesting to observe that EGFP-NESTA exhibits a graded nuclear translocation in response to SFN treatment. The kinetics of SFN-induced EGFP-NESTA translocation can be described by two parameters. One is the maximal accumulation (Amax) of EGFP-NESTA in the nucleus. The other parameter is the half time to achieve maximal translocation (t1/2). The EGFP-NESTA showed a dose-dependent nuclear translocation in response to SFN treatments. In the four tested concentrations (5, 12.5, 25 and 50 μM), the Amax values of EGFP-NESTA appeared to be positively correlated with the SFN concentrations, i.e. the stronger the oxidative signal, more EGFP-NESTA proteins translocate into the nucleus. The t1/2 values were inversely correlated with SFN concentrations, suggesting that the stronger the oxidative signal, the faster Nrf2 influx [75]. The graded response of the NESTA motif implies that the NESTA motif can not only sense the presence of oxidative signals, but also precisely transmit the “intensity” of oxidative signals to the nucleus and up-regulate gene transcription accordingly, particularly at physiologically achievable conditions [86].
There is a C183 residue embedded in this NESTA motif (Fig. 1A), bearing a resemblance to the reported redox-sensitive NES motifs of yeast AP-1 like transcription factor (YAP-1) [87] and mammalian bZIP protein Bach2 [88]. The C183A mutation remarkably slowed down EGFP-NESTA nuclear translocation kinetics and attenuated the ARE-luciferase reporter gene activity [75]. Mutation of C183A in the full length Nrf2 also significantly attenuated the redox-sensitivity of Nrf2 [75]. Based on these observations, it is hypothesized that the redox-sensitivity of the NESTA motif is mediated by the C183 residue.
The C183 residue is flanked by a glutamine and a leucine (182QCL184). In other words, the C183 residue is not a hypothetically reactive cysteine [50]. As previously described in the Keap1 section, the local electrostatic environment model may fail to predict precisely whether a cysteine residue is a reactive cysteine, as disproved by many experimental evidences. The fact that the reactive cysteine (619LCS621) embedded in the YAP-1 redox-sensitive NES is also not a hypothetically reactive cysteine [87] provides further corroboration. In addition, when we labeled hNrf2 proteins with BIA, we detected modified C183 adducts using the MS-MS analysis (Li, W., Liu, T., Li, H., Kong, A.-N.T., unpublished data). Thus, it is probable that C183 is a reactive cysteine.
It is possible that direct sulfhydryl modification of the C183 residue may inhibit the access and binding of nuclear exportin chromosome region maintenance 1 (CRM1) to the NESTA motif and consequently abrogate cytosolic distribution of EGFP-NESTA. Similar results have been reported in the YAP-1 protein [87]. Alternatively, intra-molecular disulfide bond formation may also disable the NESTA activity [89]. What kind of specific thio-modification of the C183 residue requires further study.
Based on these observations, we propose a new Nrf2 signaling model [75]. The Nrf2 molecule possesses multiple NES/NLS motifs, and their relative driving forces are represented by the size and direction of the arrows (Fig. 2). Under basal conditions (Fig. 2A), the combined nuclear exporting forces of NESTA and NESzip can counteract the combined nuclear importing force of the bNLS, NLSN and NLSC motifs. As a dynamic balance, Nrf2 exhibits a predominantly whole cell distribution. The residual nuclear Nrf2 may account for the basal or constitutive Nrf2 activities. The observation of a small percentage of cells exhibiting nuclear and cytosolic distribution of Nrf2 [75] may reflect the hyper- and hypo-oxidative condition of individual cells, respectively. When challenged with oxidative stress (Fig. 2B), the redox-sensitive NESTA is disabled but the redox-insensitive NESzip remains functional [71] and the bNLS, NLSN and NLSC motifs may remain functionally uninterrupted [85]. Since the driving force of the NESzip motif is weaker than the bipartite bNLS motif, the nuclear importing force mediated by the bNLS, NLSN and NLSC motifs prevails and triggers Nrf2 nuclear translocation (Fig. 2B). In this model, Nrf2 per se is redox-sensitive. Nrf2 consists of a constitutively active NESzip bNLS-NLSN-NLSC tandem and a conditional NESTA motif. The NESTA motif functions as a redox-sensitive switch that can be turned on/off by oxidative signals and determine the subcellular localization of Nrf2. This model may not only account for the repression and activation of Nrf2 signaling but may also account for the inactivation of Nrf2 signaling. Accumulating evidence shows that Nrf2 activation can consequently elevate the expression and enzymatic activities of γGCS/GST as well as the GSH level in cells [90,91]. Considering the reversible nature of sulfhydryl modification by SFN, the elevated GSH levels may favor a restoration of the NESTA activity and trigger Nrf2 nuclear export. In addition, oxidative stress can elicit nuclear translocation of thioredoxin [92], which may subsequently reduce Nrf2 in the nucleus. Further studies are needed to examine this possibility.
FIG. 2.
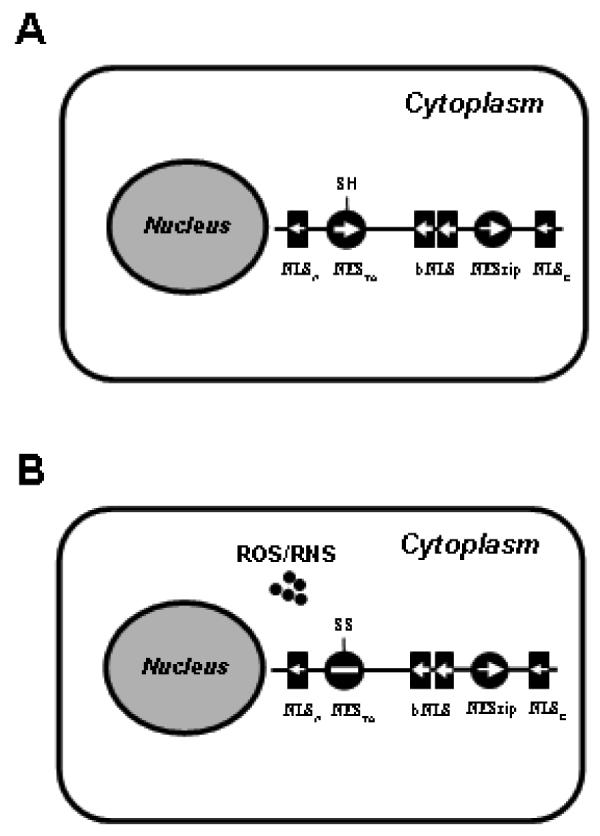
Nrf2 functions as a redox probe. The identified NES and NLS motifs of Nrf2 are symbolized by filled circles and boxes, respectively. Their driving forces are designated by the direction and size of the arrows. During the unstimulated condition (A), two NES motifs can counter balance the combined driving force of the bNLS, NLSN and NLSC motifs and sequester Nrf2 in the cytoplasm. When challenged by oxidative stress (B), the reactive cysteine in the NESTA can detect the presence of reactive oxygen species (ROS) or reactive nitrogen species (RNS) and disable the NESTA (plain line). As a result, the driving force of the bNLS, NLSN and NLSC motif prevails and causes Nrf2 nuclear translocation.
Unlike the previous Keap1 model, this new model hypothesizes a Keap1-independent Nrf2 signaling. Indeed, knocking down endogenous Keap1 activity with siRNA failed to change tBHQ-induced nuclear translocation of Nrf2 [75]. Definitive proof may require replicating EGFP-NESTA translocation experiments in keap1 null MEFs. Although Nrf2 appears to be self sufficient to transduce redox signals, this new Keap1-independent model does not exclude Keap1's involvement in redox signaling. Keap1 may provide an additional regulation controlling the Nrf2 availability, especially via the constant ubiquitination and proteasomal degradation. Therefore, this new model favors the debate that Keap1 function is alternative but not exclusive in regulating Nrf2 signaling [93].
IV. A new redox-signaling model of the antioxidant response
In a brief summary, accumulating evidence shows that Keap1 mediated Nrf2 ubiquitination has high redox-sensitivity. In contrast, Keap1/Nrf2 dissociation has very low redox-sensitivity. The low redox sensitivity of Keap1/Nrf2 dissociation renders the “relay” step in the old model very unlikely. In addition, Nrf2 nuclear translocation has high redox-sensivity.
To reconcile these observations, we propose a new Keap1/Nrf2 redox-signaling model (Fig. 3). In cells, there may exist two pools of Nrf2 proteins; one is free floating Nrf2 (fNrf2) and the other is Keap1-bound Nrf2 (kNrf2), which is destined for ubiquitination and proteasomal degradation. Under homeostatic conditions, there is constitutive synthesis of new Nrf2 protein and persistent ubiquitin-dependent degradation of Nrf2. An equilibrium is maintained between Nrf2 synthesis and degradation. As a consequence, only a small fNrf2 pool exists, sufficient for basal or constitutive activity (Fig. 3A). When cells are exposed to oxidative stress, the redox-sensitive Nrf2 ubiquitination is impeded. Consequently, Nrf2-binding capacity of Keap1 is saturated and even diminished due to Keap1 self-ubiquitination [67,68]. However, Nrf2 translation is enhanced [94]. As a consequence, the fNrf2 pool size is enlarged (Fig. 3B). Since the fNrf2 proteins have high redox-sensitivity and high mobility [75], they are poised to assume surveillance of the fluctuation of intracellular redox conditions and transmit redox-signals into cell nucleus. In other words, the fNrf2 proteins are ready-to-react redox probes, whereas kNrf2 proteins are redox-inert. The observation of a graded Nrf2 nuclear translocation implies that the relative abundance of fNrf2 may determine the actual amplitude of an antioxidant response. If cells have more fNrf2, the same dosage of ROS/RNS may elicit more Nrf2 influx into the nucleus and consequently induce a stronger antioxidant response. In this new model, the redox-signals are encoded and transduced into the nucleus by fNrf2. The overall redox-sensitivity of the cell is determined by the pool size of fNrf2, which is regulated by Keap1 in a redox-sensitive manner. In other words, Keap1 functions as a redox-sensitive gate keeper. The key difference between the old and new model is that the redox-signals are not relayed from Keap1 to Nrf2. Both Nrf2 and Keap1 have high sensitivity for redox signals.
This new model may also explain the acute effect elicited by phase II inducers. The treatment of low dosage of phase II inducers can impede Keap1-mediated ubiquitination of Nrf2 and expand the fNrf2 pool. Thus, the phase II inducers exert a “priming” effect to tune up the overall redox sensitivity of the cell. Subsequently, when the cell confronts oxidative stress, the cell can readily trigger amore effective antioxidant response.
The relative expression ratio of Keap1/Nrf2 may be critical for Nrf2 signaling. In addition, many other factors may also be involved in the regulation of the sensitivity of this redox-signaling process. For example, the initial substrate recognition step of Nrf2 ubiquitination is mediated by the ETGE “hinge” motif. Recently, a member of phosphoglycerate mutase family, called PGAM5, was found to bind Keap1 [95]. This PGAM5 molecule possesses an N-X-E-S-G-E motif but no DLG motif. Therefore, PGAM5 may function as a competitive antagonist of Nrf2. The high abundance of PGAM5 in cells [95] may have a profound impact on the pool size of fNrf2. Recently, a cancer and Parkinson's disease associated protein called DJ-1/PARK7 was found to be able to stabilize Nrf2 via preventing Keap1 binding [96]. For a long time, a wealth of evidence has been accumulated showing that many dietary compounds can induce Nrf2 signaling via activating diverse phosphorylation pathways, including MAPK [68,83,97-104], PKC [105-108], PI3K [109], PERK [110], CK2 [111] and GSK3 β/Fyn [79-81] pathways. Unfortunately, the actual phosphorylation sites in Nrf2 or Keap1 are mainly unknown. However, at least one phosphorylation site has been elucidated. PKC phosphorylation of the Serine-40 (S40) residue of Nrf2 was reported to facilitate Keap1/Nrf2 dissociation and Nrf2 nuclear translocation [105,106]. The position of S40 is located between the DLG motif and the ETGE motif. It is unknown whether phosphorylation at S40 can disrupt “the hinge and latch” two sites binding. An artificial phosphorylation at the threonine residue of the D-X-E-T-G-E motif indeed disrupts Neh2/Keap1 binding [64]. Furthermore, a CAND1 molecule was found to be importantto regulate Keap1/Cul3 binding and recycling of the Cul3 complex [112]. Studies are needed to determine whether the expression and activities of those molecules are redox-sensitive. There are observations that oxidized n-3 fatty acids can react with Keap1 and induce Keap1/Cul3 dissociation [54,113].
V. Parallel channels of redox signaling?
Nrf2 has two homologues, Nrf1 and Nrf3. Nrf2 shares high sequence homology and probably similar origin with Nrf1, including the amino-terminus [114]. In contrast, Nrf2 shares poor homology with Nrf3, especially in the amino-terminus [115]. Until recently, it was unclear why three different Nrf isofomers exist with apparently functional redundancy.
Recently, two independent studies elegantly elucidate that Nrf1 and Nrf2 are actually located in different subcellular compartments and employ different activation mechanisms [116,117]. Like Nrf2, Nrf1 also possesses a functionally intact Neh2 domain, including the ETGE “hinge” motif and the DLG “latch” domain, and is capable of binding with Keap1 [116,117]. However, in its extensive amino-terminus, Nrf1 possesses an extra transmembrane domain, which is absent in Nrf2 [116,117]. Therefore, unlike Nrf2 proteins, which exhibit a Keap1-dependent uniform distribution in the cytoplasm, Nrf1 proteins are found mainly tethered to the membrane of endoplasmic reticulum (ER) in a Keap1-independent manner [116,117]. Fluorescent microscopic studies confirmed that Nrf1 was indeed co-localized with ER-specific protein calreticulin [116]. Differential centrifugation studies also confirmed that Nrf1 proteins were mainly detected in ER membrane P100 fraction [116]. Consistent withits ER localization, ER stressor tunicamycin could effectively mobilize Nrf1 into the nucleus [116]. A careful examination showed that the size of the nuclear Nrf1 (110 kD) was smaller than ER-bound Nrf1 (120 kD), suggesting that the nuclear Nrf1 is a cleavage product of the ER-bound Nrf1 [116].
These observations suggest that Nrf1 and Nrf2 may encode ER stress signals in different manners. Nrf1 can directly detect the ER stress in the lumen of ER. Nrf2 can also sense various ER stress signals elicited by the treatments of tunicamycin (glycolysation inhibitor), thapthiagargen (calcium-releasing inhibitor) and glucose deprivation [118]. However, ER stress signals encoded by Nrf2 is relayed to Nrf2 via an ER-membrane spanning PKR-related ER kinase (PERK) [110,118]. Therefore, it is an interesting question whether the ER stress signals encoded by Nrf1 are the same as those encoded by Nrf2. Considering the fact that the ER lumen is more oxidized than the cytoplasm [119], the redox-sensitivity of Nrf1 may be different than that of Nrf2. Nrf1 does possess a NESTA motif like Nrf2, however the NESTA in Nrf1 lacks the embedded reactive cysteine residue [75] suggesting that the NESTA motif in Nrf1 may not be redox-sensitive.
Up until now, the induced ER stress responses have mainly been examined in wild type cells. Future examination of the ER stress response in nrf1-/- or nrf2-/- MEFs may help to dissect the specificity of ER stress signaling mediated by Nrf2 and Nrf1, respectively. Furthermore, it is unknown whether the induced gene expression profiles elicited by Nrf1 and Nrf2 are the same. One microarray study examining tunicamycin induced gene expression in wild type and nrf2-/- mice has been published [120]. Comparison of those data with microarray data collected from nrf1-/- mice may help to discover whether Nrf1-driven genes are the same or different from Nrf2-driven genes. Recently, it was reported that small Maf homodimers exhibited different sequence-specific binding affinity compared to small Maf/CNC heterodimer [43]. It is tempting to speculate that different composite small Maf/CNC dimers, such as MafG/Nrf2 and MafG/Nrf1 dimers, may bind to different cis-acting enhancers, thus eliciting transcription of discrete subsets of cytoprotective genes. If it can be proven that different sets of cyto-protective genes are induced, then cytoplasmic and ER stress signals may be sorted through different channels.
VI. Perspective and future directions
Two common features are unanimously observed in various antioxidant response induced by different compounds; i.e. increased amount of steady state Nrf2 and Nrf2 nuclear translocation. However, mechanistic explanations for the repression and activation of Nrf2 signaling differ drastically. Great progress has been made in understanding Keap1/Nrf2 interplay and the regulation of the antioxidant response. However, there remain many intriguing issues. With the rapid progress of technologies, such asthe protein crystallography, LC-MS-MS analysis, genetic manipulation (transgene mice or gene knockdown using siRNA or shRNA) and DNA/RNA microarray among others, many enigmatic issues may be clarified in the near future.
Enormously fruitful is the combination of the structural study with the functional study, as illustrated by the proposition of the “hinge & latch” model. In future, the crystallographic resolution of the entire molecule of Keap1 combined with mutation and artificial thio-modification, may enable us to visualize whether zinc-coordination can stabilize Keap1/Nrf2 binding; whether the formation of intermolecular disulfide bond between C273 and C288 can change the distance of the arginine triads between two different Kelch domains; and what is the mechanism that switches Nrf2-ubiquitination to Keap1 self-ubiquitination. Resolution of the crystal structure of Nrf2 may enable us to visualize whether the formation of disulfide bond or other types of thio-modification can change the conformation and function of Nrf2.
Another promising direction is the application of LC-MS-MS in the detection of reactive cysteines in Keap1 and Nrf2 when treated with different structural inducers. Thus far, a major methodological drawback is the difficulty in analyzing hydrolysable adducts. With the progress in the optimization of sample handling, we may soon be able to standardize the analysis protocol, making feasible of high throughput analyses. Progress in this area may enable us to find out whether different inducers result in distinct thio-modification maps in Nrf2 and Keap1.
Collectively, progress in these areas may not only enable us to gain deeper mechanistic understanding of Nrf2 signaling, it may also help us to design and optimize drugs with higher potency for chemoprevention of cancer, aging, inflammation and neurodegenerative diseases [64,121].
Acknowledgement
We thank Drs. Tong Liu and Hong Li for the assistance in mass spectrometry analysis of sulfhydryl adducts of hNrf2 proteins. We thank Drs. John Mieyal, Lesa Beamer, Karen Liby, Walter Watson for constructive discussion.
Supported by NIH R01 CA94828 to A.-N.T. K.
References
- 1.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Klaassen CD, Slitt AL. Regulation of hepatic transporters by xenobiotic receptors. Curr Drug Metab. 2005;6(4):309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- 3.Mandlekar S, Hong JL, Kong AN. Modulation of metabolic enzymes by dietary phytochemicals: a review of mechanisms underlying beneficial versus unfavorable effects. Curr Drug Metab. 2006;7(6):661–675. doi: 10.2174/138920006778017795. [DOI] [PubMed] [Google Scholar]
- 4.Lee JM, Li J, Johnson DA, et al. Nrf2, a multi-organ protector? Faseb J. 2005;19(9):1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 5.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38(4):769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 6.Yates MS, Kwak MK, Egner PA, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66(4):2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 7.Xu C, Huang MT, Shen G, et al. Inhibition of 7,12-Dimethylbenz(a)anthracene-Induced Skin Tumorigenesis in C57BL/6 Mice by Sulforaphane Is Mediated by Nuclear Factor E2-Related Factor 2. Cancer Res. 2006;66(16):8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 8.Iida K, Itoh K, Kumagai Y, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64(18):6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 9.Ramos-Gomez M, Dolan PM, Itoh K, Yamamoto M, Kensler TW. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24(3):461–467. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Sanchez G, Giuliani A. Cellular redox status regulates hypoxia inducible factor-1 activity. Role in tumour development. J Exp Clin Cancer Res. 2007;26(1):39–50. [PubMed] [Google Scholar]
- 11.Bubici C, Papa S, Dean K, Franzoso G. Mutual cross-talk between reactive oxygen species and nuclear factor-kappa B: molecular basis and biological significance. Oncogene. 2006;25(51):6731–6748. doi: 10.1038/sj.onc.1209936. [DOI] [PubMed] [Google Scholar]
- 12.Ushio-Fukai M, Alexander RW. Reactive oxygen species as mediators of angiogenesis signaling: role of NAD(P)H oxidase. Mol Cell Biochem. 2004;264(1-2):85–97. doi: 10.1023/b:mcbi.0000044378.09409.b5. [DOI] [PubMed] [Google Scholar]
- 13.Osburn WO, Wakabayashi N, Misra V, et al. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-mediated formation of superoxide anion. Arch Biochem Biophys. 2006;454(1):7–15. doi: 10.1016/j.abb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson LA, Gemin A, Espiritu R, Singh G. ets-1 is transcriptionally up-regulated by H2O2 via an antioxidant response element. Faseb J. 2005;19(14):2085–2087. doi: 10.1096/fj.05-4401fje. [DOI] [PubMed] [Google Scholar]
- 15.Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 2004;36(10):1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 16.Dhakshinamoorthy S, Porter AG. Nitric oxide-induced transcriptional up-regulation of protective genes by Nrf2 via the antioxidant response element counteracts apoptosis of neuroblastoma cells. The Journal of biological chemistry. 2004;279(19):20096–20107. doi: 10.1074/jbc.M312492200. [DOI] [PubMed] [Google Scholar]
- 17.Park EY, Kim SG. NO signaling in ARE-mediated gene expression. Methods Enzymol. 2005;396:341–349. doi: 10.1016/S0076-6879(05)96028-X. [DOI] [PubMed] [Google Scholar]
- 18.Dinkova-Kostova AT, Holtzclaw WD, Kensler TW. The role of Keap1 in cellular protective responses. Chem Res Toxicol. 2005;18(12):1779–1791. doi: 10.1021/tx050217c. [DOI] [PubMed] [Google Scholar]
- 19.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh Y, Iida K, Kang MI, et al. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Archives of biochemistry and biophysics. 2005;433(2):342–350. doi: 10.1016/j.abb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6(10):857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 22.Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. 2005;25(24):10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. The Journal of biological chemistry. 2004;279(30):31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 24.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhakshinamoorthy S, Jaiswal AK. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2001;20(29):3906–3917. doi: 10.1038/sj.onc.1204506. [DOI] [PubMed] [Google Scholar]
- 26.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(7):2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339(1):79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 28.Wakabayashi N, Itoh K, Wakabayashi J, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35(3):238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 29.Kang MI, Kobayashi A, Wakabayashi N, Kim SG, Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(7):2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velichkova M, Guttman J, Warren C, et al. A human homologue of Drosophila kelch associates with myosin-VIIa in specialized adhesion junctions. Cell Motil Cytoskeleton. 2002;51(3):147–164. doi: 10.1002/cm.10025. [DOI] [PubMed] [Google Scholar]
- 31.Kang KW, Lee SJ, Park JW, Kim SG. Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol Pharmacol. 2002;62(5):1001–1010. doi: 10.1124/mol.62.5.1001. [DOI] [PubMed] [Google Scholar]
- 32.Karapetian RN, Evstafieva AG, Abaeva IS, et al. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol Cell Biol. 2005;25(3):1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol. 2005;25(11):4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23(22):8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the nrf2-mediated antioxidant response by escorting nuclear export of nrf2. Mol Cell Biol. 2007;27(18):6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25(1):162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi A, Kang MI, Okawa H, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24(24):10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol. 1995;15(8):4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motohashi H, O'Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294(1-2):1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto T, Kyo M, Kamiya T, et al. Predictive base substitution rules that determine the binding and transcriptional specificity of Maf recognition elements. Genes Cells. 2006;11(6):575–591. doi: 10.1111/j.1365-2443.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- 44.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. The Journal of biological chemistry. 1991;266(18):11632–11639. [PubMed] [Google Scholar]
- 45.Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(16):6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 47.Talalay P, Fahey JW, Holtzclaw WD, Prestera T, Zhang Y. Chemoprotection against cancer by phase 2 enzyme induction. Toxicol Lett. 1995;82-83:173–179. doi: 10.1016/0378-4274(95)03553-2. [DOI] [PubMed] [Google Scholar]
- 48.Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44(18):6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- 49.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(29):10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder GH, Cennerazzo MJ, Karalis AJ, Field D. Electrostatic influence of local cysteine environments on disulfide exchange kinetics. Biochemistry. 1981;20(23):6509–6519. doi: 10.1021/bi00526a001. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. The Journal of biological chemistry. 2004;279(52):54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 52.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18(12):1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 53.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. The Journal of biological chemistry. 2005;280(36):31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 54.Levonen AL, Landar A, Ramachandran A, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378(Pt 2):373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26(8):2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi A, Kang MI, Watai Y, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26(1):221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. The Journal of biological chemistry. 2006;281(34):24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 58.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. The Journal of biological chemistry. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 59.Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8(3):499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 60.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 61.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387(10-11):1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi M, Itoh K, Suzuki T, et al. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7(8):807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 63.Tong KI, Padmanabhan B, Kobayashi A, et al. Different Electrostatic Potentials Define ETGE and DLG Motifs as Hinge and Latch in Oxidative Stress Response. Mol Cell Biol. 2007 doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. Embo J. 2006;25(15):3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Padmanabhan B, Tong KI, Ohta T, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21(5):689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 66.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. The Journal of biological chemistry. 2002;277(39):36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 67.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. The Journal of biological chemistry. 2005;280(34):30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 68.Yuan X, Xu C, Pan Z, et al. Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog. 2006;45(11):841–850. doi: 10.1002/mc.20234. [DOI] [PubMed] [Google Scholar]
- 69.Tashiro S, Muto A, Tanimoto K, et al. Repression of PML nuclear body-associated transcription by oxidative stress-activated Bach2. Mol Cell Biol. 2004;24(8):3473–3484. doi: 10.1128/MCB.24.8.3473-3484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. The Journal of biological chemistry. 2005;280(32):29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 71.Li W, Jain MR, Chen C, et al. Nrf2 Possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J Biol Chem. 2005;280(31):28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 72.Theodore M, Kawai Y, Yang J, et al. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. The Journal of biological chemistry. 2008;283(14):8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82(3):475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 74.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82(3):463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 75.Li W, Yu SW, Kong AN. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. The Journal of biological chemistry. 2006;281(37):27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 76.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22(18):6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McLachlan AD, Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975;98(2):293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 78.Thompson KS, Vinson CR, Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32(21):5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 79.Jain AK, Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. The Journal of biological chemistry. 2006;281(17):12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 80.Jain AK, Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. The Journal of biological chemistry. 2007;282(22):16502–16510. doi: 10.1074/jbc.M611336200. [DOI] [PubMed] [Google Scholar]
- 81.Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. The Journal of biological chemistry. 2006;281(21):14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 82.Zhu M, Fahl WE. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem Biophys Res Commun. 2001;289(1):212–219. doi: 10.1006/bbrc.2001.5944. [DOI] [PubMed] [Google Scholar]
- 83.Shen G, Hebbar V, Nair S, et al. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. The Journal of biological chemistry. 2004;279(22):23052–23060. doi: 10.1074/jbc.M401368200. [DOI] [PubMed] [Google Scholar]
- 84.Ki SH, Cho IJ, Choi DW, Kim SG. Glucocorticoid receptor (GR)-associated SMRT binding to C/EBPbeta TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression. Mol Cell Biol. 2005;25(10):4150–4165. doi: 10.1128/MCB.25.10.4150-4165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bloom D, Dhakshinamoorthy S, Jaiswal AK. Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2002;21(14):2191–2200. doi: 10.1038/sj.onc.1205288. [DOI] [PubMed] [Google Scholar]
- 86.Hu R, Hebbar V, Kim BR, et al. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther. 2004;310(1):263–271. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- 87.Yan C, Lee LH, Davis LI. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. Embo J. 1998;17(24):7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoshino H, Kobayashi A, Yoshida M, et al. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem. 2000;275(20):15370–15376. doi: 10.1074/jbc.275.20.15370. [DOI] [PubMed] [Google Scholar]
- 89.Kuge S, Arita M, Murayama A, et al. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol Cell Biol. 2001;21(18):6139–6150. doi: 10.1128/MCB.21.18.6139-6150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee TD, Yang H, Whang J, Lu SC. Cloning and characterization of the human glutathione synthetase 5′-flanking region. Biochem J. 2005;390(Pt 2):521–528. doi: 10.1042/BJ20050439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 92.Hansen JM, Watson WH, Jones DP. Compartmentation of Nrf-2 redox control: regulation of cytoplasmic activation by glutathione and DNA binding by thioredoxin-1. Toxicol Sci. 2004;82(1):308–317. doi: 10.1093/toxsci/kfh231. [DOI] [PubMed] [Google Scholar]
- 93.Magin TM. A keaper and a striker maintain epidermal homeostasis. Nat Genet. 2003;35(3):202–204. doi: 10.1038/ng1103-202. [DOI] [PubMed] [Google Scholar]
- 94.Purdom-Dickinson S, Sheveleva E, Sun H, Chen QM. Translational Control of Nrf2 Protein in Activation of Antioxidant Response by Oxidants. Mol Pharmacol. 2007 doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 95.Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. The Journal of biological chemistry. 2006;281(49):37893–37903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- 96.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer-and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu R, Chen C, Mo YY, et al. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. The Journal of biological chemistry. 2000;275(51):39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- 98.Yu R, Lei W, Mandlekar S, et al. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. The Journal of biological chemistry. 1999;274(39):27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 99.Keum YS, Yu S, Chang PP, et al. Mechanism of Action of Sulforaphane: Inhibition of p38 Mitogen-Activated Protein Kinase Isoforms Contributing to the Induction of Antioxidant Response Element-Mediated Heme Oxygenase-1 in Human Hepatoma HepG2 Cells. Cancer Res. 2006;66(17):8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 100.Xu C, Yuan X, Pan Z, et al. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther. 2006;5(8):1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- 101.Chen C, Pung D, Leong V, et al. Induction of detoxifying enzymes by garlic organosulfur compounds through transcription factor Nrf2: effect of chemical structure and stress signals. Free Radic Biol Med. 2004;37(10):1578–1590. doi: 10.1016/j.freeradbiomed.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 102.Papaiahgari S, Kleeberger SR, Cho HY, Kalvakolanu DV, Reddy SP. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. The Journal of biological chemistry. 2004;279(40):42302–42312. doi: 10.1074/jbc.M408275200. [DOI] [PubMed] [Google Scholar]
- 103.Keum YS, Han YH, Liew C, et al. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm Res. 2006;23(11):2586–2594. doi: 10.1007/s11095-006-9094-2. [DOI] [PubMed] [Google Scholar]
- 104.Yu R, Mandlekar S, Lei W, Fahl WE, Tan TH, Kong AN. p38 mitogen-activated protein kinase negatively regulates the induction of phase II drug-metabolizing enzymes that detoxify carcinogens. The Journal of biological chemistry. 2000;275(4):2322–2327. doi: 10.1074/jbc.275.4.2322. [DOI] [PubMed] [Google Scholar]
- 105.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. The Journal of biological chemistry. 2002;277(45):42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 106.Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. The Journal of biological chemistry. 2003;278(45):44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 107.Numazawa S, Ishikawa M, Yoshida A, Tanaka S, Yoshida T. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am J Physiol Cell Physiol. 2003;285(2):C334–342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- 108.Cho MK, Kim WD, Ki SH, et al. Role of Galpha12 and Galpha13 as novel switches for the activity of Nrf2, a key antioxidative transcription factor. Mol Cell Biol. 2007;27(17):6195–6208. doi: 10.1128/MCB.02065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee JM, Hanson JM, Chu WA, Johnson JA. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. The Journal of biological chemistry. 2001;276(23):20011–20016. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]
- 110.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23(20):7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pi J, Bai Y, Reece JM, et al. Molecular mechanism of human Nrf2 activation and degradation: role of sequential phosphorylation by protein kinase CK2. Free Radic Biol Med. 2007;42(12):1797–1806. doi: 10.1016/j.freeradbiomed.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26(4):1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao L, Wang J, Sekhar KR, et al. Novel N-3 fatty acid oxidation products activate Nrf2 by destabilizing the association between keap1 and cullin3. The Journal of biological chemistry. 2006 doi: 10.1074/jbc.M607622200. [DOI] [PubMed] [Google Scholar]
- 114.Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci U S A. 1993;90(23):11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kobayashi A, Ito E, Toki T, et al. Molecular cloning and functional characterization of a new Cap‘n’ collar family transcription factor Nrf3. J Biol Chem. 1999;274(10):6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 116.Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. The Journal of biological chemistry. 2006;281(28):19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 117.Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J. 2006;399(3):373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. The Journal of biological chemistry. 2004;279(19):20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 119.Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992;356(6366):260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- 120.Nair S, Xu C, Shen G, et al. Toxicogenomics of endoplasmic reticulum stress inducer tunicamycin in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Toxicol Lett. 2007;168(1):21–39. doi: 10.1016/j.toxlet.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen C, Kong AN. Dietary cancer-chemopreventive compounds: from signaling and gene expression to pharmacological effects. Trends Pharmacol Sci. 2005;26(6):318–326. doi: 10.1016/j.tips.2005.04.004. [DOI] [PubMed] [Google Scholar]