Abstract
MicroRNAs (miRNAs) are a class of small RNAs that silence gene expression. In animal cells, miRNAs bind to the 3′ untranslated regions of specific mRNAs and inhibit their translation. The correct regulation of mRNA expression by miRNAs is believed to be important for oocyte maturation, early development and implantation. We examined the expression of 25 mRNAs involved in the microRNA processing pathway in a non human primate oocyte and embryo model. We observed that mRNAs related to miRNA splicing are downregulated during oocyte maturation while those related to miRNA processing are upregulated, indicating that there may exist a temporal difference in their activities related to transcriptional activity in germinal vesicle stage oocytes. We also observed that the vast majority of mRNAs examined were insensitive to α-amanitin at the 8-16 cell stage. The expression data did not reveal a major impact of embryo culture, and hormonal stimulation protocol affected only a small number of mRNAs, suggesting that the components of the pathway may be accumulated in the oocyte during oogenesis and resistant to exogenous insults. In comparison to published mouse array data, we observed species differences and similarities in the temporal expression patterns of some genes, suggesting that miRNA processing may be regulated differently. These data extend our understanding of the potential roles of miRNA during primate embryogenesis.
Keywords: miRNA, embryo, oocyte, gene regulation, oocyte quality, in vitro maturation, cleavage, preimplantation embryo
INTRODUCTION
MicroRNAs (miRNAs) comprise a large family of short (17-25 nucleotides), noncoding endogenous RNAs that often repress the expression of complementary messenger RNAs, thus controlling many biological processes in development, differentiation, growth, and metabolism (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001; Bartel, 2004, He and Hannon, 2004; Bagga et al., 2005). In eukaryotes, miRNAs are derived from long primary transcripts (1°miRNAs) transcribed by RNA polymerase II (Lee et al., 2004; Kim, 2005), and then cropped into the hairpin-shaped pre-miRNAs by the nuclear RNase III Drosha into 70-nt precursors (Lee et al., 2003; Reviewed by Carmel and Hannon, 2004; Tomari and Zamore, 2005) (Fig 1). Drosha/DGCR8 cleaves near the base of the stem to liberate a 60-nucleotide pre-miRNA hairpin (Denli et al., 2004; Gregory et al., 2004; Landthaler et al., 2004; Han et al., 2004, 2006). The pre-miRNAs, are exported out of the nucleus by exportin-5 (XPO5) (Yi et al., 2003; Bohnsack et al., 2004; Lund et al., 2004), and then cleaved by the cytoplasmic RNase III nuclease DICER into 22-nt miRNA duplexes (Bernstein et al., 2001; Grishok et al., 2001; Hutvágner et al., 2001; Ketting et al., 2001; Knight and Bass, 2001). One strand of the duplex usually remains as a mature miRNA (Khvorova et al., 2003; Schwarz et al., 2003; Hammond, 2005; Valencia-Sanchez et al., 2006), which is incorporated into the silencing complex RISC.
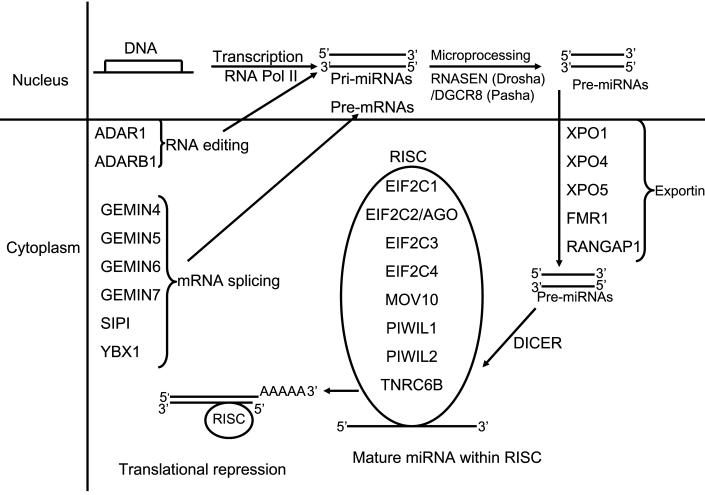
Figure 1.
Summary of miRNA processing machinery. In the nucleus miRNA are transcribed by RNA polymerase and are termed primary miRNAs (1°miRNAs). The dsRNA-specific ribonuclease RNASEN (Drosha) digests the 1°miRNAs in the nuclease to release hairpins, precursor miRNA (pre-miRNA) which are transported from the nucleus to the cytoplasm by exportins. Once in the cytoplasm, DICER cleaves the pre-miRNA approximately 19 bp from the Drosha cut site. The resulting double-stranded RNA has 1–4 nt 3′ overhangs at either end. Only one of the two strands is the mature miRNA. To control the translation of target mRNAs, the double-stranded RNA produced by DICER must strand separate, and the single-stranded mature miRNA must associate with the RISC. Post- transcriptional modifications such as splicing and editing play a part in miRNA processing.
DICER transcripts are expressed in germinal vesicle (GV) stage mouse oocytes, decline in abundance during development to the two-cell embryo stage, and remain stable during morula and blastocyst formation (Cuia et al., 2007). Mouse oocytes lacking DICER function display defects in meiotic progression and cleavage upon fertilization (Murchison et al., 2007, Tang et al., 2007). Loss of DICER function in the mouse embryo is embryonic lethal at embryonic day 7.5 (E7.5), and DICER-null embryos are deficient in stem cells(Bernstein et al., 2003).
Following fertilization of mouse eggs, the maternal-to-zygotic transition, which initiates during the one-cell stage, is clearly evident by the two-cell stage (Schultz, 1993; 1999). In macaque and human embryos, genome activation occurs at the six- to eight-cell stage (Tesarik, 1987; Tesarik et al., 1986a,b, 1988; Artley et al., 1992; Weston and Wolf, 1994; Schramm and Bavister, 1999). One outcome of this transition is that transcripts that are common to the oocyte and embryo are degraded during oocyte maturation following fertilization, and replaced with embryonic transcripts (e.g., actin) [Bachvarova et al., 1989]. In addition, a dramatic reprogramming of gene expression occurs during this transition (Latham et al., 1991; Zeng et al., 2004), and this reprogramming provides the molecular foundation for transforming the highly differentiated oocyte into the totipotent blastomeres of the early cleavage stage preimplantation embryo. More recent studies showed that loss of DICER activity leads to the enhanced expression of a large number of mRNAs that are siRNA targets fo regulation in mouse oocytes (Tam e al., 2008; Watanabe et al., 2008). The early embryonic lethal phenotype associated with DICER deficiency indicates that in mouse embryos miRNA-mediated gene silencing plays an important role in completing the maternal to zygotic transition and establishing totipotent blastomeres in the mouse. Whether this applies to primates has not been determined.
The rhesus monkey offers an ideal model system for understanding human embryogenesis. Rhesus monkey oocytes and embryos of diverse developmental potentials can be obtained through specific protocols (Schramm and Bavister, 1994). This provides an opportunity to determine whether the expression of particular genes is altered under these different circumstances, and thus may contribute to different developmental potencies. Moreover, the use of a non-human primate model permits research advances where legal and ethical restrictions inhibit the conduct of research on human embryos. For example, it is possible to employ highest quality non-human primate embryos for research, whereas a similar use of human embryos would raise significant ethical questions. However, non-human primate embryo research is quite costly, with individual oocytes and embryos representing substantial investments of resources and labor, and availability of material is limited by costs and seasonal restrictions. Because of this, the amount of molecular data that has been generated for non-human primate oocytes and embryos is lagging considerably behind other species. Thus, a system to permit molecular data to be obtained in a non-human primate species at little cost is helpful for advancing our understanding of human embryology. The Primate Embryo Gene Expression Resource (PREGER) has been established in order to permit such studies to be undertaken with little cost and effort (Zheng et al., 2004). PREGER encompasses over 200 amplified cDNA libraries representing oocytes and embryos of many different stages obtained via diverse protocols, and permits comparisons between in vivo and in vitro development, different hormonal stimulation protocols, in vivo and in vitro maturation, and different culture systems. The PREGER resource was constructed to permit investigators to obtain expression data for genes of interest in a suitable non-human primate model species, without need for direct access to a non human primate colony, and without need to expend further resources. Using the PREGER resource, we have determined the temporal patterns of expression of mRNAs encoding components of the miRNA processing machinery, and investigated whether the expression of these genes is affected by culture system or by protocols that yield oocytes and embryos of different developmental potentials. We also compared these data to published expression for the mouse in order to evaluate potential differences in the regulation, production, and function of miRNA between the two species.
MATERIALS AND METHODS
The PREGER Resource
The studies undertaken here employed the Primate Embryo Gene Expression Resource (PREGER) (www.preger.org). The resource contains a collection of reverse transcribed and polymerase chain reaction (RT-PCR)-amplified cDNA libraries corresponding to more than 200 samples of rhesus monkey oocytes and preimplantation stage embryos. The PREGER sample collection was created using a well-established method for reverse transcription (RT) and exponential cDNA amplification that maintains the quantitative representation of mRNAs by amplifying the 3' terminal regions of the entire mRNA population (Brady and Iscove, 1993; Iscove et al., 2002). After amplification, aliquots of each sample library are spotted onto filters by dot blotting as described (Quantitative Amplification and Dot Blotting, QADB). It should be noted that, because the entire mRNA population is uniformly amplified during the RT-PCR procedure, the amount of input mRNA within the range of one to four embryos does not affect the quantitative representation of sequences within the amplified cDNA population. Once the dot blots are prepared, they are hybridized to mRNA-specific probes and the hybridization results analyzed.
The QADB method has been extensively validated by previous studies in both mouse and monkey embryos, and the sensitivity and reliability of the assay documented (reviewed, Wang and Latham, 2000; Wang et al., 2001; Zheng et al., 2004). The method is applicable to small amounts of material, even single or partial embryos, provides the ability to quantify expression of a large number of mRNAs, and can provide estimates of actual mRNA abundance. These properties make the QADB method ideal for examining mRNA abundances in rhesus monkey oocyte and embryos, which are of highly limited availability and costly to obtain. The QADB method is fully quantitative and produces hybridization signals that are linear over many orders of magnitude, extending to a very low mRNA abundance (at least 500 copies/mouse embryo), and with excellent reproducibility. The method maintains quantitative representation of sequences within the cDNA population during amplification by limiting the length of the first cDNA strand, and thus minimizing selection against mRNAs of long length (Brady and Iscove, 1993). Re-amplification of cDNA through additional rounds of PCR exerts a minimal effect on results (Domachenko et al., 1997). The RT-PCR method yields highly representative cDNA libraries (Brady and Iscove, 1993; Brady et al., 1995; Cano-Gauci et al., 1993;Weaver et al., 1999; Iscove et al., 2002). The method yields expected differences in gene expression, for example revealing effects of parental chromosome origin on the preimplantation expression of the imprinted Ascl2 (Mash2), U2afi-rs1, H19, Xist, and Snrpn genes, including approximately two-fold greater levels of expression in the appropriate uniparental embryos as compared with control fertilized embryos (Latham et al., 1995; Mann et al., 1995; Rossant et al., 1998). The QADB method first revealed that the Xist RNA is first transcribed at the two-cell stage in the mouse (Latham and Rambhatla, 1995), which has been confirmed (Hartshorn et al., 2002; Zuccotti et al., 2002). Estimates of mRNA copy number in mouse embryos based on the QADB method, and the temporal and quantitative aspects of the data are consistent with data obtained by other methods, (Rambhatla et al., 1995, Wang and Latham, 2000, Wang et al., 2001). The QADB method thus has allowed us to produce a resource for gene expression study in rhesus monkey oocytes and embryos that are exceedingly limited in availability. It should be noted that, because the entire mRNA population is uniformly amplified during the RT PCR procedure, the amount of input mRNA within the range of one to four embryos does not affect the quantitative representation of sequences within the amplified cDNA population. Once the dot blots are prepared, they are hybridized to mRNA-specific probes and the hybridization results analyzed.
Oocytes and embryos
The isolation and culture of the oocytes and embryos during the construction of the PREGER sample set has been described in detail (Zheng et al., 2004). Oocytes contained in the PREGER sample set were obtained from monkeys treated with follicle stimulating hormone (FSH) only, or FSH followed by human chorionic gonadotropin (hCG), and matured either in vitro or in vivo, respectively. The sample collection also contains oocytes obtained without hormonal stimulation (non stimulated, denoted as NS). Embryos were obtained from these three categories of ooctyes, and also by natural conception (morula/blastocysts) as described (Zheng et al., 2004). Between 3 and 13 samples of one to four oocytes or embryos were obtained for each stage. The embryos included in the PREGER sample set were all high quality and healthy in appearance. Samples of eight-cell and morula-stage embryos treated with the RNA polymerase II inhibitor α-amanitin from the pronucleate stage onward in HECM9 culture were included to evaluate transcriptional dependence of mRNA expression. Details concerning the array, diversity, and origin of samples, and the sensitivity and quantitative reliability of the quantitative amplification and dot blotting method have been described previously (Zheng et al., 2004, and in other references available at the PREGER website www.preger.org). All procedures employed to obtain oocytes and embryos were conducted according to recommendations of the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act, and its amendments.
Complementary DNA Probes and Hybridization
Complementary DNA probes were obtained by PCR, which was performed in 100 μl reactions containing 4 μl of plasmid DNA product (cDNA clones obtained from Open Biosystems Huntsville, AL, USA), 10 × PCR Buffer containing 15 mM MgCl2 (Roche Diagnostics, Indianapolis, USA); 10 mM dNTPs (Roche Diagnostics); 10 μM for each of the forward and reverse primers (Table 1); and 5 U/μl TaqDNA polymerase (Roche Diagnostics). Reactions were run on a Techne PCR machine (Burlington, NJ, USA) at 94° for 5 min to denature, followed by 35 cycles of 94°C for 1 min, annealing at 55-60°C, for 1 min; 72°C for 2 min, final extension for 5 min at 72°C. PCR products were resolved on 1% agarose gels. Excised PCR products were purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. Purified PCR products were then used for probe labeling.
Table 1.
Primers employed for obtaining cDNA probes
| Gene symbol | GeneBank Accession number |
Upper primer 5′-3′ | Lower primer 5′-3- | Product size (bp) |
|---|---|---|---|---|
| ADAR | BC038227 | ACAGGACAGAGGAGGCAGAG | GACTGCAGCCATCATCACAG | 308 |
| ADARB1 | BC065545 | GACCAGTTCTCACTCACGCC | TTGAAAGCAGTCACTGGACG | 303 |
| DICER1 | BC068591 | GAGCTTAGGGCTTTGCATTG | CAGGGGATCACAGAATGCTT | 334 |
| DGCR8/PASHA | BC078147 | CTTGCTTCTGTCTTGCCTCC | TCAGACAATCAAGGCTCGC | 340 |
| EIF2C1/AGO1 | BC063275 | CATGACAAGTCTCGGAGCTG | AACTCATCAGGCATGGGAAG | 312 |
| EIF2C2/AGO2 | BC018727 | GTGGCTCACCCAAGTTCCT | TCACCACAGACCCGTATTCTT | 300 |
| EIF2C3 | BC066888 | GCCAGTTAGGAGGCTTTTCA | TGGATGCTCCTCCTCCTATG | 343 |
| EIF2C4 | BC016012 | TTGCAAGGTCTCTGGGTAAAA | ATCTTGGCTCACTGCAACCT | 303 |
| FMR1 | BC086957 | ACCATTGGAAATTTCACGTC | AAAAACATCATGACTTTGAACTGA | 302 |
| GEMIN4 | BC020062 | TGAGCAAGACCAACCCTTCT | TTGCCATGAAGTTTTGTGGA | 304 |
| GEMIN5 | BC036894 | GCCAAGATTACCAGTGTGGC | TGCCTTAGGTTGAGAGAGCC | 300 |
| GEMIN6 | BC018195 | AGGGGACCATAGAGTGAGGG | CACACTTCCTGGCCTCTCAT | 302 |
| GEMIN7 | BC007793 | TGAGCTCCCTTGGAATTTTG | CTGGTTCTTTAGCAGGAGGG | 300 |
| MOV10 | BC009312 | CTCTACCTCAGGGCCCCAC | GTTTTCTTCCTTCCCCTTCC | 305 |
| PIWIL1/HIWI | BC028581 | CTTTCAGGAGTGAGCAAGTCC | CATGTCCCAGTTCCAAAACA | 356 |
| PIWIL2/MILI | BC111751 | CCATCAGAGTTCCAGCTCCT | CAAGGCCATTTTCAGCTGTT | 318 |
| RANGAP1 | BC041396 | AGATTGTCCAGGAGCAGGAG | CACACACAGAGCACAAAGGG | 302 |
| /SCPEP1/ RISC | BC010078 | GTCTCTGGAGGCAATTTGGA | TTGACATGAAAAGGTGTTTCAGA | 355 |
| RNASEN/DROSHA | BC054003 | CCCAGATCATCATGAAGGACA | TCACCAAAGTCAAGTTTTCCG | 305 |
| SIP1/GEMIN2 | BC104968 | GGCAGCCTAACTCTGAGGAA | AGGCCATCCATTGTCTTTTG | 374 |
| TNRC6B | BC028626 | CAGAACCAGTCAGATCCCGT | TTAGTGCTGCTGCTGTTCCA | 303 |
| XPO1 | BC032847 | GTGACAGGGCTTTTCAGCTT | TTGGTCGACAAATACCCACA | 301 |
| XPO4 | BC113573 | TTGCAAAAGCACAACACAGA | CAGTGCACTTTGCAGAAAGG | 318 |
| XPO5 | BC062635 | AGCTGTAATGGAGCAAATCCC | AGGATGCCCAAAAGCTTGAT | 300 |
| YBX1 | BC015208 | CCGGCTTACCATCTCTACCA | CAGGTGCTTGCAGTTTGTTG | 376 |
Blot preparation, hybridization, and quantitative analyses were performed as described (Zheng et al., 2004). Gel-purified cDNA fragments were radiolabeled by the random primer method (Feinberg and Vogelstein, 1983). Labeled probes were hybridized to blots as described (Latham et al., 1999; Rambhatla et al., 1995). Hybridization results were visualized and quantified by storage phosphorimaging with background correction and normalization as described (Rambhatla et al., 1995). Data were expressed as the mean (± standard error of the meanSEM), counts per minute (cpm) bound value for each stage/condition of oocytes and embryos included in the analysis. Probes that yielded weak or absent hybridization signals were repeated to confirm result. The statistical significance of differences was evaluated using the t-test (P < 0.05 considered significant). Where necessary a correction for multiple testing was applied, using a significance level of P < 0.016 as indicated.
Murine Gene Expression Data
Expression data for murine homologues were extracted from the microarray data available in the Gene Expression Omnibus repository byeng et al (2004, which were obtained originally using the Affymetrix MOE 430A and 430B chips. The stages represented were GV stage oocytes, and embryos at the one-cell, two-cell, eight-cell, and blastocyst stages. These data were expressed as the mean (+SEM) Affymetrix array hybridization signal.
RESULTS
We examined the patterns of expression of 25 genes involved in the miRNA processing pathway. The functions of these genes are described in Table 2 and Fig. 1. Of the 25 genes examined, we detected the expression of 21 in our sample set of rhesus monkey oocytes and embryos.
Table 2.
Descriptions of miRNA processing genes functions.
| Gene | Known (or potential) function |
|---|---|
| ADAR | |
| ADARB1 | Binds to short interfering RNAs (siRNA) without editing them and suppresses siRNA-mediated RNA interference |
| DICER1 | Involved in cleaving double-stranded RNA in the RNA interference (RNAi) pathway. It produces 21 to 23 bp dsRNAs (siRNAs) which target the selective destruction of homologous RNAs |
| DGCR8 | Critical for the processing of pri-miRNA into pre-miRNA |
| EIF2C1 | |
| EIF2C2/AGO2 | |
| EIF2C3 | Provides endonuclease activity to RNA-induced silencing complexes (RISC). Cleaves siRNA/mRNA heteroduplexes bound to RISC. |
| EIF2C4 | Essential for embryonic development as well as RNA-mediated gene silencing (RNAi) |
| FMRI | RNA-binding protein. Associated with polysomes and might be involved in the transport of mRNA from the nucleus to the cytoplasm |
| GEMIN4 | |
| GEMIN5 | |
| GEMIN6 | |
| GEMIN7 | The survival of motor neuron (SMN) complex plays an essential role in spliceosomal snRNP assembly in the cytoplasm and is required for pre-mRNA splicing in the nucleus |
| SIP1/GEMIN2 | |
| MOV10 | Required to mediate microRNA-guided mRNA cleavage |
| PIWIL1 | Probable component of some RISC complex (RNA-induced silencing), which plays a role in spermatogenesis and in germ cell production, proliferation and maintenance. Inhibits endoribonuclease DICER1 activity. |
| PIWIL2 | |
| RANGAP1 | Involved in mRNA processing and transport |
| RISC/SCPEP1 | |
| RNASEN/DROSHA | Executes the initial step of microRNA (miRNA) processing in the nucleus that is cleavage of pri-miRNA to release pre- miRNA. Involved in pre-rRNA processing. Cleaves double-strand RNA and does not cleave single-strand RNA |
| TNRC6B | Required for miRNA-guided gene silencing and are present in P bodies |
| XPO | |
| XPO4 | Mediates the nuclear export of micro-RNA precursors, which form short hairpins. Also mediates the nuclear export of synthetic short hairpin |
| XPO5 | RNAs used for RNA interference, and adenovirus VA1 dsRNA. |
| YBX1 | Binds to splice sites in pre-mRNA and regulates splice site selection. Binds and stabilizes cytoplasmic mRNA. Contributes to the regulation of translation by modulating the interaction between the mRNA and eukaryotic initiation factors |
Expression of mRNAs encoding proteins involved in RNA splicing and editing during miRNA biogenesis
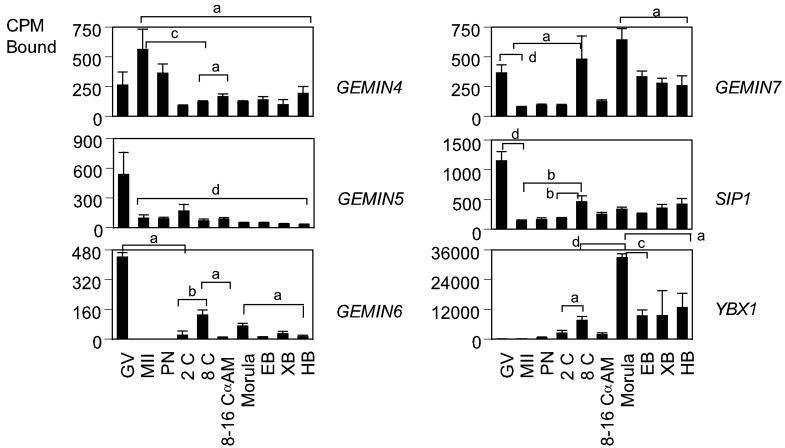
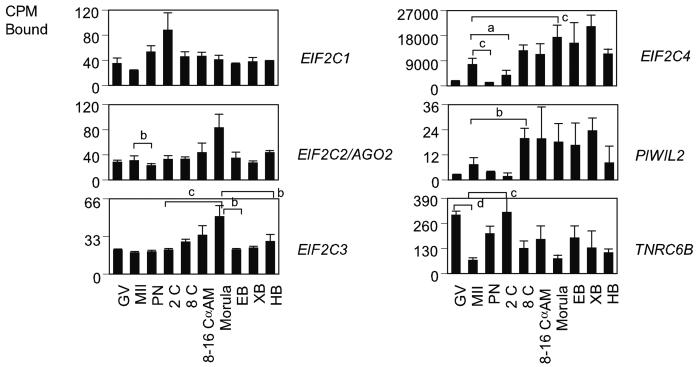
Once transcribed, the primary miRNAs are subjected to splicing and editing. Splicing is mediated by the survival of motor neuron (SMN) protein complex (Fig. 2). The SMN complex contains five proteins known as SIP1 (formerly GEMIN2; Liu et al., 1997), GEMIN3 (a DEAD-box putative RNA helicase; Charroux et al., 1999), GEMIN4 (Charroux et al., 2000), GEMIN5 (Meister et al., 2001; Gubitz et al., 2002), and GEMIN6 (Pellizzoni et al., 2002). The SMN complex plays important roles in the assembly/restructuring and function of diverse ribonucleoprotein (RNP) complexes, including spliceosomal small nuclear RNPs (snRNPs; Fischer et al., 1997; Pellizzoni et al., 1998; Meister et al., 2001), small nucleolar RNPs (snoRNPs; Jones et al., 2001; Pellizzoni et al., 2001a) heterogeneous nuclear RNPs (hnRNPs; Mourelatos et al., 2001), and transcriptosomes (Pellizzoni et al., 2001b). The YBX1 protein participates in different steps of mRNA biogenesis, including mRNA transcription, processing, and transport from the nucleus into the cytoplasm (Sommerville, 1999; Wilkinson and Shyu, 2001; Stickeler et al., 2001), binds to splice sites in pre-mRNA, and regulates splice site selection (Allemand et al., 2007). A total of six mRNAs encoding components of the SMN complex and other proteins involved in RNA splicing was analyzed (Table1, Fig. 2). The SIP1, GEMIN4, GEMIN5, GEMIN6 and GEMIN7 mRNAs were maternally expressed. SIP1, GEMIN5, and GEMIN6 mRNA were highly expressed in GV oocytes and decreased after oocyte maturation, and were then expressed throughout development. Only the GEMIN6 mRNA was significantly α-amanitin sensitive at the 8-16 cell stage. The GEMIN7 and YBX1 mRNAs increased in expression in the developing embryo, with highest signals in YBX1 mRNA and both mRNAs upregulated at the morula stage.
Figure 2.
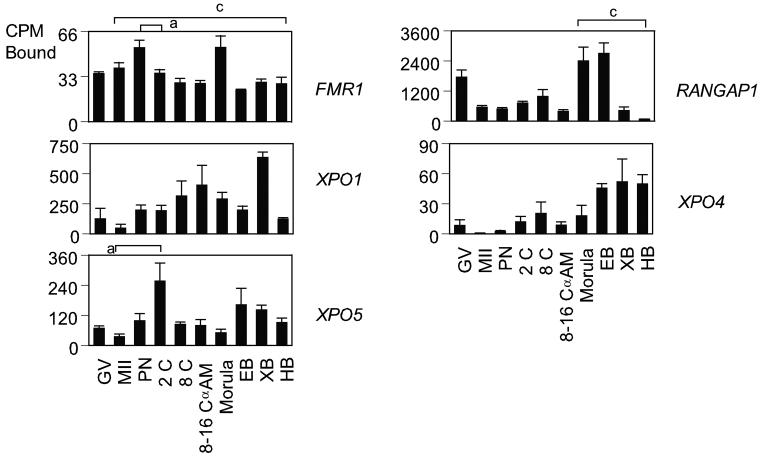
Temporal expression patterns of mRNAs encoding proteins involved in RNA splicing in rhesus monkey oocytes and embryos. Graphs show the relative levels of expression for GV and MII stage oocytes and pronucleate through hatched blastocyst stage embryos produced by in vitro fertilization of oocytes from hCG-stimulated females, and then cultured in vitro in HECM9. GV, germinal vesicle stage oocyte; MII, MII-stage oocyte; PN, pronucleate 1-cell stage embryo; 2C, 2-cell stage embryo; 8C, 8-cell stage embryo; 8–16C αAm, 8- to 16-cell stage cultured in α-amanitin; EB, early blastocyst; XB, expanded blastocyst; HB, hatched blastocyst. Expression data for the mRNAs encoding the indicated proteins are expressed as the mean CPM bound, and the standard error of mean (SEM) is indicated. Statistically significant differences in gene expression corresponding to some of the major increases or decreases in expression are denoted by the brackets (for comparisons between stages at the ends of the brackets). Letters a through d indicate P < 0.05, 0.01, 0.001, and 0.0001, respectively.
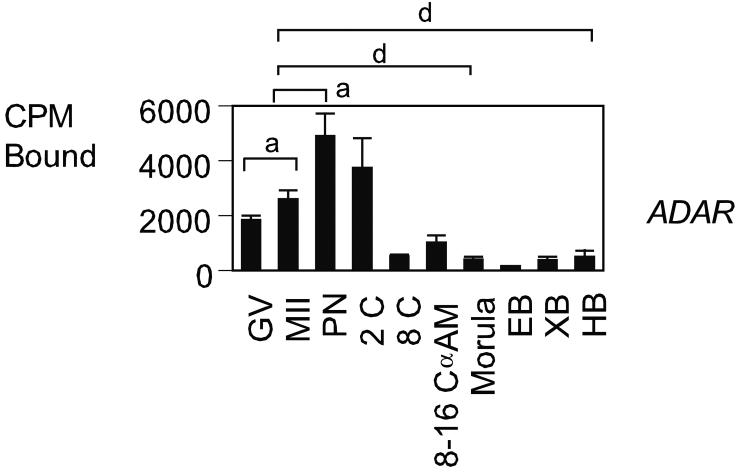
After splicing, adenosine deaminases (ADARs) edit the primary miRNAs , converting adenosine to inosine in double-stranded (ds) RNA (Bass, 2002; Knight and Bass, 2002; Tonkin and Bass, 2003; Yang et al., 2005). ADARs edit specific adenosines at crucial positions such as the glutamine/arginine site in the GluR-B transcript, which can have major consequences for the encoded protein, as inosine is translated as guanosine, leading to incorporation of a different amino acid at the edited codon. ADARs are expressed in many tissues and can bind and deaminate any dsRNA. Independent studies have shown that ADARs shuttle in and out of the nucleolus in living cells (Desterro et al., 2003; Sansam et al., 2003). Two mRNAs were analyzed in this category, ADAR1 and ADARB1 (Table 1, Fig. 3). The ADARB1 mRNA was not detected. The ADAR1 mRNA was maternally expressed and had significantly lower expression at the GV and MII oocyte stages than at the one- and two-cell stage (P < 0.05). Its expression decreased sharply thereafter, with no α-amanitin-sensitive expression evident at the 8-16 cell stage.
Figure 3.
Temporal expression patterns of mRNAs encoding proteins related to miRNA editing. Data are presented as in Fig.2.
Expression of genes involved in the RNA microprocessor complex
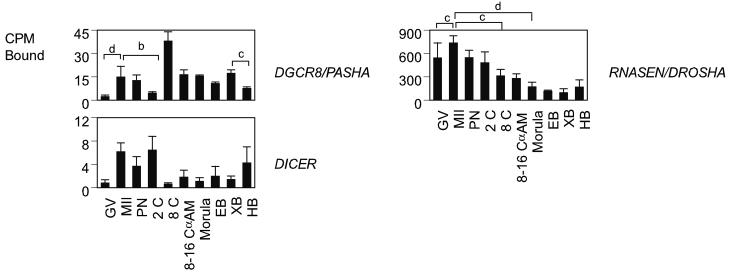
Three genes encoding components of the microprocessor complex were analyzed, RNASEN (homolog of Drosophila Drosha), DGCR8, (homolog of Drosophila Pasha) and DICER1 (Table 1, Fig. 4). RNASEN is an RNase III enzyme responsible for initiating the processing of microRNA (miRNA). It cleaves long RNA primary transcripts (1° miRNAs) to produce stem-loop structures of about 70 base pairs long, known as pre-miRNAs (Han et al., 2004). RNASEN exists as part of the microprocessor complex, which also contains the double-stranded RNA binding protein. DGCR8 is essential for RNASEN activity, and binds single-stranded fragments of the 1° miRNA that are required for proper processing. DICER1 is a member of the RNase III family of nucleases that specifically cleave double-stranded RNAs (Bernstein et al., 2001). RNASEN mRNA expression was predominantly maternal, declining in abundance during development to the 8 to 16-cell stage. Its expression was significantly increased during maturation (P < 0.001). DGCR8 mRNA expression was low at the GV stage and significantly increased during maturation (P < 0.0001). DICER1 mRNA expression appeared very low, with low hybridization signals throughout development, but hybridization data indicated an apparent increase in signal upon oocyte maturation and again at the blastocyst stage (Fig. 4). None of these three mRNA's was α-amanitin sensitive at the 8- to 16-cell, indicating a predominantly maternal ooplasmic source of expression.
Figure 4.
Temporal expression patterns of mRNAs encoding proteins involved in the RNA microprocessor complex. Data are presented as in Fig.2.
Expression of genes involved in transport of mRNA and miRNA from nucleus to cytoplasm
The traffic through the nuclear envelope is mediated by a protein family, which can be divided into exportins and importins. Binding of a molecule (a “cargo”) to exportins facilitates its export to the cytoplasm. Importins facilitate import into the nucleus. FMR1 protein is endowed with a nuclear localization and a nuclear export signal (Eberhart et al., 1996; Bardoni et al., 1997; Fridell et al., 1996), suggesting that it shuttles between nucleus and cytoplasm (Tamanini et al., 1999) and may participate in mRNA export from nucleus to cytoplasm. Exportin-5 (XPO5) mediates efficient nuclear export of short miRNA precursors (premiRNAs) and its depletion by RNA interference results in reduced miRNA levels (Lund et al., 2004). Five mRNAs involved in RNA export were analyzed, FMR1, RANGAP1, XPO1, XPO4, and XPO5 (Table 1, Fig. 5). The FMRI mRNA was distributed throughout development and had a low expression level compared to other mRNA's in the group. RANGAP1, XPO1, XPO4, and XPO5 mRNAs significantly decreased in abundance during maturation from GV-stage oocyte to the metaphase II (MII) stage (P < 0.05). The RANGAP1 mRNA was transiently elevated at the morula and early blastocyst stages. The XPO1 and XPO5 mRNAs displayed somewhat uniform patterns of expression, and the XPO4 mRNA increased gradually in expression over time. None of mRNAs in this group was α-amanitin sensitive at the 8-16 cell stage.
Figure 5.
Temporal expression patterns of mRNAs encoding proteins involved in the transport of mRNA and miRNA from nucleus to cytoplasm. Data are presented as in Fig.2.
Expression of mRNAs encoding components of the RISC complex
ARGONAUTE family proteins are highly conserved and have been implicated in RNAi function and related phenomena in several organisms. In addition to roles in RNAi-like mechanisms, ARGONAUTE proteins influence development in C. elegans by the two small temporal RNAs (stRNAs) lin-4 and let-7 (Lee et al., 1993; Reinhart et al., 2000), and at least a subset are involved in stem cell fate determination (Brower-Toland, 2007; Carmel et al., 2002; Lin, 2007). They contain two common domains named PAZ and PIWI domains (Cerutti et al., 2000; Song et al., 2003). The PAZ domain proteins include AGO proteins, also known as EIF2Cs in mammals (Lingel et al., 2004), and PIWI domain proteins include PIWIL proteins. Both domains are components of RNA-induced silencing complex (RISC) [Tahbaz et al., 2004]. The expression of 6 mRNAs encoding ARGONAUTE family proteins was examined (Table1, Fig. 6). Three PAZ domain mRNAs EIF2C1, EIF2C2, and EIF2C3 were expressed at nearly constant levels throughout development. The EIF2C4 mRNA had a predominantly embryonic expression pattern and yielded the strongest hybridization signal for the group. The EIF2C3 mRNA produced weak hybridization signals in oocytes and embryos. None of the four PAZ domain mRNAs analyzed was α-amanitin sensitive at the 8-16 cell stage. Two PIWI domain mRNAs were analyzed, PIWIL1 and PIWIL2. PIWI is essential for maintaining germline stem cells in adult Drosophila ovaries and testes (Cox et al., 1998). The PIWIL2 mRNA displayed an overall low expression level and was detectable in oocytes, with increased expression from the eight-cell stage onward. Its pattern was similar to that of EIF2C4 mRNA. The PIWIL1 mRNA was not detected.
Figure 6.
Temporal expression patterns of mRNAs related to components of the RISC complex. Data are presented as in Fig. 2.
The TNRC6 family proteins are required for miRNA-guided gene silencing and are present in P bodies in C. elegans and D. melanogaster (Ding et al., 2005; Rehwinkel et al., 2005). TNRC6B and MOV10 are required for miRNA-guided mRNA cleavage in vivo and specifically associate with EIF2C proteins (Meister et al., 2005). The TNRC6B mRNA significantly decreased in abundance during maturation from the GV stage oocyte to the MII stage P < 0.0001 (Fig. 6), and was then expressed throughout development. The TNRC6B mRNA was not α-amanitin sensitive at 8-16 cell stage. The MOV10 mRNA was not detected.
Analysis of Transcription Factor Binding Sites
To explore whether the 25 genes examined share possible aspects of gene regulation, we applied the oPOSSUM analysis software (http://www.cisreg.ca/cgibin/oPOSSUM/opossum) (Ho-Sui et al., 2005) to the list of genes. This software combines methods for detecting transcription factor binding sites (TFBS) documented in the JASPAR database, statistical methods for identifying over-represented TFBS amongst a group of genes, and a database of DNA regions that are conserved amongst moderately divergent organisms (i.e., phylogenetic footprinting). A combination of a Z score > 10 and Fisher P value < 0.01 provides minimal likelihood of false positive results. This analysis revealed one transcription factor-binding site (IRF2) that is significantly over-represented (found in TNR6CB, RANGAP1, FMR1, GEMIN7) (Table 3). One other transcription factor-binding site (STAT1) yielded borderline significance scores (found in TNR6CB, RANGAP1, FMR1, GEMIN7, XPO5, MOV10, ADARB1). The failure to detect expression of four of the miRNA pathway-associated mRNAs (ADARB1, FMR1, MOV10, PIWIL1) was reminiscent of the low or undetected expression of these mRNAs in the mouse (Zeng et al., 2004). We therefore also sought to determine whether these four genes might share any aspect of transcriptional regulation. This analysis revealed two transcription factor-binding sites with marginal significance for over-representation, PAX6 (found in FMR1, MOV10) and STAT1 (found in ADARB1, FMR1, MOV10). Further studies will be needed to ascertain whether these transcription factors indeed contribute to the regulation of this complex pathway in oocytes and embryos.
Table 3.
Over-Represented Transcription Factor Binding Site Analysis
| Transcription Factor | No. Target Genes | Z-score | Fisher P |
|---|---|---|---|
| Total gene set | |||
| IRF2 | 4 | 13.5 | 0.00396 |
| STAT1 | 7 | 6.93 | 0.00427 |
| Non-expressed genes | |||
| PAX6 | 2 | 10.2 | 0.0251 |
| STAT1 | 3 | 8.94 | 0.00105 |
Effects of hormonal stimulation protocol and embryo culture on mRNA expression
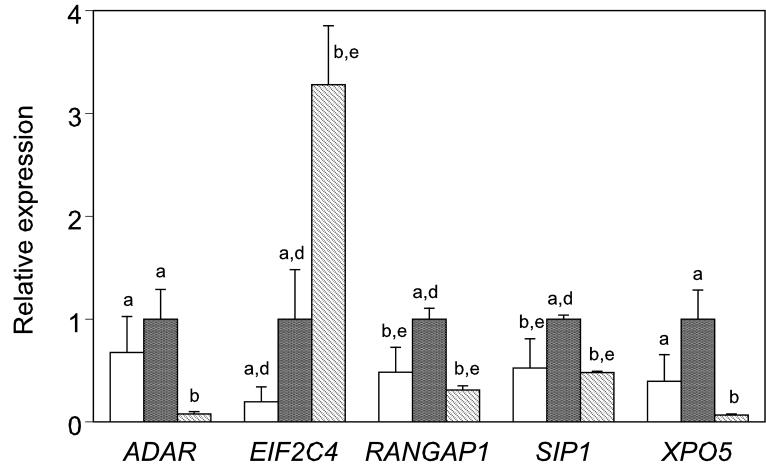
We next wished to determine whether the expression of mRNAs related to the miRNA function pathway are affected by the hormonal stimulation and in vitro maturation conditions employed to produce the oocytes prior to in vitro fertilization. The PREGER sample set contains oocytes of high quality resulting from in vivo maturation (hCG oocytes), intermediate quality resulting from FSH stimulation in vivo followed by in vitro maturation (FSH oocytes), and low quality resulting from in vitro maturation of oocytes from non-stimulated ovaries (NS oocytes) (Schramm and Bavister, 1994), along with cultured embryos derived from these three kinds of oocytes following in vitro fertilization. The resource also contains embryos flushed directly from the reproductive tract and lysed, which can be used to evaluate possible effects of embryo culture. We find that embryo culture had no significant effect on the expression of the mRNAs under study, however hormonal stimulation protocol and oocyte quality affected a small number of transcripts. In previous studies we observed that many maternal mRNAs are precociously eliminated by the two-cell stage with embryos from either FSH or NS oocytes (Zheng et al., 2005) as compared to embryos from FSH+hCG cycles. Of the 21 mRNAs for which expression was detected, 13 displayed values of 50 cpm bound or greater, and were selected for examining effects of hormonal stimulation protocol and IVM on expression. Of these 13, four (ADAR, RANGAP1,SIP1, XPO5) displayed significantly reduced (p < 0.05) expression, two of which (RANGAP1, SIP1) passed the more stringent level of p < 0.016, and were likewise reduced in embryos from FSH oocytes (Figure 7). One mRNA (EIF2C4) was elevated in NS embryos.
Figure 7.
Effect of hormonal stimulation in two-cell stage embryos produced by in vitro fertilization. Stimulation protocols: white bar, FSH, females given FSH injection only, fully grown oocytes from large antral follicles matured in vitro; black bar, hCG, females injected with FSH and hCG, fully grown oocytes from large antral follicles matured in vivo; stippled bar, NS, females received no stimulation, fully grown oocytes from small antral follicles matured in vitro. Genes are listed alphabetically. Values for average expression and S.E.M. are normalized to the average hCG value (assigned a value of 1.0) for each mRNA. Different letters (a-c) denote statistically significant differences within stage P < 0.05; d-f denotes P < 0.016.
DISCUSSION
The correct regulation of mRNA expression by miRNAs is believed to be important for oocyte maturation and early development (Murchison et al., 2007; Tang et al., 2007; Bernstein et al., 2003). We provide here the first report of expression of genes that comprise this important pathway in a non human primate oocyte and embryo model species. Several striking features of the data can be noted. First, we observe that mRNAs related to miRNA splicing (GEMIN5, GEMIN6, GEMIN7, SIP1) are downregulated during oocyte maturation most likely via maternal mRNA degradation. Conversely one of the mRNAs related to miRNA processing (DGCR8) undergoes an apparent upregulation during maturation, most likely the result of increased mRNA polyadenylation. DICER also appeared to increase in hybridization signal during maturation, but the signal obtained was overall quite low. There may exist a temporal difference in maximal activities of these two portions of the pathway, splicing and processing, related, respectively, to transcriptional activity in the GV stage oocyte, and cytoplasmic processes upon maturation and after fertilization related to elimination of maternal mRNAs. This is further supported by the strongly maternal pattern of RNASEN mRNA expression. The YBX1 and GEMIN7 mRNAs were elevated at the morula stage, possibly related to a need for mRNA splicing at the time of embryonic genome activation. The ADAR1 mRNA was most highly expressed as a maternal transcript, indicating a potentially important role for this protein in the transcriptionally active GV stage and early embryo.
The second striking result is that the vast majority of the mRNAs examined displayed insensitivity to α-amanitin at the 8-16 cell stage. Only GEMIN6 was α-amanitin sensitive, YBX1 and GEMIN7 mRNAs displayed pronounced increases in expression at the morula stage, and RANGAP1 and XPO4 mRNAs displayed a clear increase in expression after the morula stage. This obviously does not exclude active transcription after the morula stage, however, in contrast to many other mRNAs examined in earlier studies (Zheng et al.,2005, Mtango and Latham, 2007), this group of mRNAs appears notable for persistent contribution of maternally inherited transcripts throughout early and mid cleavage development. It is tempting to speculate that if correct regulation by miRNAs is important for early development, then one aspect of high oocyte quality could be a sufficient supply and persistence of maternal transcripts encoding these proteins.
Third, we do not observe a major impact of either embryo culture or hormonal stimulation/oocyte maturation protocol on the expression of the mRNAs studied here. This suggests that the components of the pathway may be accumulated in the oocyte early during oogenesis and be relatively refractory to exogenous insults. This may reflect the dynamic nature of oocyte-somatic cell interactions leading to an ever-changing ooplasmic state during oogenesis, with a significant role played by this pathway in preparing the oocyte for successful maturation, as strikingly demonstrated by the deficiency in meiotic progression of DICER deficient mouse oocytes (Murchison et al., 2007).
While we do not observe a major impact of either embryo culture or hormonal stimulation and oocyte maturation protocols on the expression of these mRNAs, we do observe that the RANGAP1 mRNA is diminished in abundance in two-cell embryos from both FSH and NS oocytes, which have diminished developmental potential relative to hCG oocytes (Schramm and Bavister, 1994). RANGAP1 is a key regulator of bidirectional transport of proteins and ribonucleoproteins across the nuclear pore complex, and is also a component of the spindle (Bamba et al., 2002). Thus, in addition to its role in miRNA production, RANGAP1 regulates a myriad of processes at the level of nuclear-cytoplasmic exchange. Loss of the ubiquitin E2 ligase UBC9 results in a failure of RANGAP1 accumulation at the nuclear pore complex, and defects in blastocyst expansion and inner cell mass development leading to early postimplantation developmental arrest (DeGregori et al., 1994; Nacerddine et al., 2005). It is also interesting to note that in Drosophila, the Segregation Distorter locus encodes a mutant RanGap protein that mislocalizes to the nucleus, disrupting nuclear RAN pathway signaling, and leading to lethality, and that overexpression of RANGAP1 can also lead to segregation distortion via excessive localization to the nucleus (Merrill et al., 1999; Kusano et al., 2002). These observations indicate that the correct level of expression of RANGAP1 is essential for normal development. Deficient expression of RANGAP1 mRNA, therefore, could contribute to reduced developmental potential in embryos derived from FSH and NS oocytes. Whereas in vivo matured oocytes (hCG treated females) can support approximately 30% blastocyst formation, FSH oocytes are much less able (4%) to do so, and NS oocytes do not support blastocyst formation following standard IVM and embryo culture (Schramm and Bavister, 1994).
Available data indicate that a lack of hormonal stimulation likely compromises correct transcriptional regulation during oogenesis, as GV stage oocytes from NS females do not display the expected nucleolar rimming, but FSH oocytes do undergo rimming and display enhanced ability to undergo IVM and to support embryogenesis (Schramm et al., 1993, Schramm and Bavister, 1994). Cumulus enclosed oocytes display superior characteristics after IVM as compared to denuded oocytes (Schramm and Bavister, 1994). Moreover, whilst NS oocyte culture with exogenous gonadotropins leads to enhanced meiotic maturation, activation, and early cleavage, but not enhanced developmental potential thereafter (Schramm and Bavister, 1995), co-culture of NS oocytes in vitro with granulosa cells from FSH-stimulated females leads to enhanced development (Schramm and Bavister, 1996). This is not seen with granulosa cells from NS females (Schramm and Bavister, 1996). In vitro matured rhesus monkey oocytes also display deficiencies in embryonic transcriptional activation (Schramm et al., 2003). Last, we reported previously that many maternal mRNAs may be precociously diminished in oocytes from NS females, leading to deficient expression after fertilization (Zheng et al., 2005). Collectively, these data indicate that hormonal stimulation operating in concert with signals from somatic cells in the follicle enhances both nuclear and cytoplasmic processes that later favor both meiosis and embryogenesis, and that interruption of these signals can disrupt the normal course of events during the period immediately preceding maturation and during meiotic progression, as well as during embryogenesis (Schramm and Bavister, 1999). Oocytes matured in vitro from FSH stimulated females display moderately reduced developmental potential, whilst those from NS females display severely reduced developmental potential, indicates that the beneficial effects of hormonal stimulation and possibly oocyte-somatic cell interactions may continue even beyond the time when oocytes are typically removed for IVM. The identification of genes such as RANGAP1 that display deficient expression in embryos from both FSH and NS females should help us to understand the normal course of events that establish high developmental potential, and the maternal components of the oocyte that may be affected when oocyte-somatic cell signaling is disrupted.
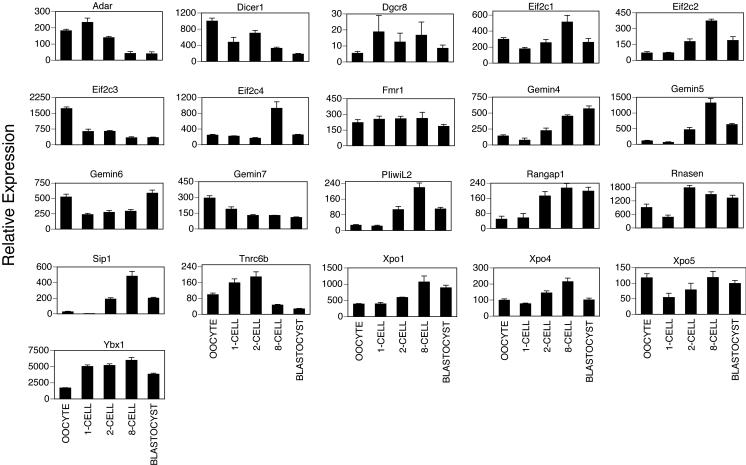
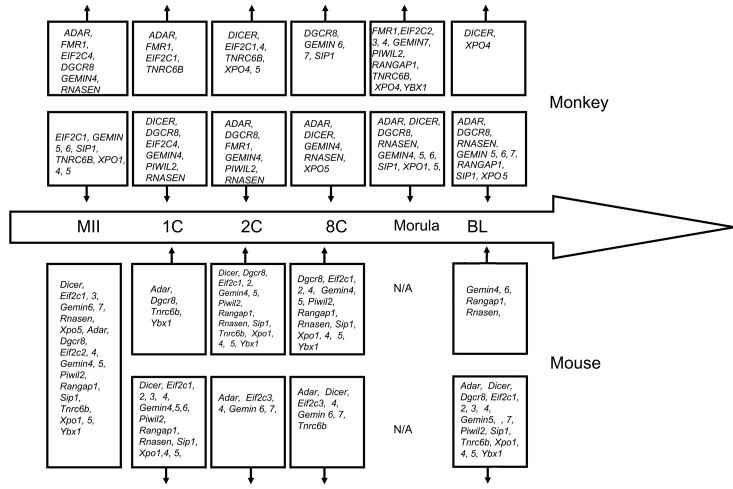
The availability of array data for gene expression in mouse embryos (Zeng et al., 2004; Fig. 8) provides the opportunity to evaluate the similarity and difference between the patterns of gene expression in rhesus monkey and a rodent model organism (Fig. 9). We observe some similarities between the two species for miRNA processing genes. Both species display predominantly maternal expression of ADAR1. Similar constitutive patterns are seen for DGCR8, FMR1, XPO5, andEIF2C1. In both species, the XPO4 and PIWIL2 mRNAs are upregulated over time. However, differences are also apparent. The GEMIN family of mRNAs displays quite different patterns of expression. The RANGAP1 mRNA increases in abundance in both species around the time of transcriptional activation, but thereafter this mRNA declines in abundance to a greater degree in the monkey. Some mRNAs display more pronounced differences in expression pattern. The Dicer mRNA is more strongly expressed as a maternal transcript in mouse oocytes compared to mouse embryos. The Tnrc6b mRNA shows a comparatively larger decrease in expression in the later cleavage stages in the mouse than in the monkey. These observations indicate that the activity of some parts of this pathway likely follow a similar pattern between the two species, however differences in temporal patterns of expression of DICER, GEMIN, TNRC6B, RANGAP1 and RNASEN mRNAs indicate that some processes may be regulated differently between the two species. Additional studies will be needed to ascertain whether the differences in DICER and RNASEN mRNA expression patterns between the two species reflect different temporal requirements for these gene products during oogenesis and embryogenesis, perhaps associated with different temporal kinetics of the maternal to embryonic transition and elimination of maternally inherited mRNAs. The mRNA expression data provided here will provide a useful foundation of information for designing detailed functional studies of miRNA production in rhesus monkey oocytes and embryos. Further detailed studies of the relevant proteins and their contributions to development will further advance the field. Moreover, the emerging ability to examine miRNA expression in early stage embryos (Tang et al., 2006) will provide the opportunity to compare between species the conservation of roles for individual miRNAs in oocytes and early embryos. Gene targeting studies in the mouse have revealed that a number of the genes examined here (Eif2c2, Dgcr8, Dicer Rangap1, Sip1) play essential roles in early embryo development (Morita et al., 2007; Wang et al., 2007; Bernestein et al., 2003; Deng and Lin., 2002; Carmel et al., 2007; Watanabe et al., 2008). Sip1 (a.k.a. Gemin2) homozygous and heterozygous mutant and and wild type preimplantation embryos showed no morphological alterations (Jablonka et al., 2002). Dgcr8 knockouts display abnormal development by embryonic day 6.5 (Wang et al., 2007). Eif2c2 (a.k.a. Ago2), Eif2c3 (a.k.a. Ago3), and Dicer mRNAs are detected by RT-PCR throughout oocyte growth and Eif2c4 (a.k.a. Ago4) and Mili mRNAs at early stages of oocytes growth. Deficiency for Eif2c2 mRNA leads to an elevation of many DICER target mRNAs (Watanabe et al., 2008) indicating the importance of AGO2 in regulating the maternal mRNA population. Further detailed studies of the expression and post-translational control of these proteins, and other functional studies in the rhesus monkey should provide additional insight into how the non human primate embryo correctly regulates this important class of molecules, the role of this pathway during embryogenesis, and how this pathway may be disrupted in response either to endogenous or exogenous factors.
Figure 8.
Expression patterns of microRNA processing genes in mouse oocytes and embryos. Expression data for murine homologues were extracted from the microarray data deposited in the Gene Expression Omnibus repository by Zeng et al (2004), which were obtained originally using the Affymetrix MOE 430A and 430B chip. The stages represented were GV stage oocytes, and embryos at the one-cell, two-cell, 8-cell, and blastocyst stages. These data were expressed as the mean (±SEM) Affymetrix array hybridization signal.
Figure 9.
Developmental transitions in expression of miRNA pathway genes in rhesus monkey and mouse (from array data of Zeng et al., 2004) oocytes and embryos. Genes initially expressed at MII stage in mouse are indicated. The stages at which individual genes undergo increases or decreases in mRNA expression are indicated by upward and downward pointing vertical arrows, respectively.
ACKNOWLEDGEMENTS
We thank Bela Patel, Malgorzata McMenamin, Judy Procknow and Ann Marie Paprocki for their technical assistance. We also thank R. Dee Schramm for his contribution to the development of the PREGER resource. This work was supported by grants from the National Institutes of Health, RR15253 from the National Centers for Research Resources (NCRR), HD43092 from the National Institute for Child Health and Human Development (NICHD), and CA95569 from the National Cancer Institute.
REFERENCES
- Allemand E, Hastings ML, Murray MV, Myers MP, Krainer AR. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid binding protein YB-1. Nat Struct Mol Biol. 2007;7:630–638. doi: 10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- Artley JK, Braude PR, Johnson MH. Gene activity and cleavage arrest in human pre-embryos. Hum Reprod. 1992;7:1014–1021. doi: 10.1093/oxfordjournals.humrep.a137761. [DOI] [PubMed] [Google Scholar]
- Bachvarova R, Cohen EM, De Leon V, Tokunaga K, Sakiyama S, Paynton BV. Amounts and modulation of actin mRNAs in mouse oocytes and embryos. Development. 1989;106:561–565. doi: 10.1242/dev.106.3.561. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bamba C, Bobinnec Y, Fukuda M, Nishida E. The GTPase Ran regulates chromosome positioning and nuclear envelope assembly in vivo. Curr Biol. 2002;12:503–507. doi: 10.1016/s0960-9822(02)00741-8. [DOI] [PubMed] [Google Scholar]
- Bardoni B, Sittler A, Shen Y, Mandel JL. Analysis of domains affecting intracellular localization of the FMRP protein. Neurobiol Dis. 1997;4:329–336. doi: 10.1006/nbdi.1997.0142. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Girlish D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G, Billia F, Knox J, Hoang T, Kirsch IR, Voura EB, Hawley RG, Cumming R, Buchwald M, Siminovitch K. Analysis of gene expression in a complex differentiation hierarchy by global amplification of cDNA from single cells. Curr Biol. 1995;5:909–22. doi: 10.1016/S0960-9822(95)00181-3. [DOI] [PubMed] [Google Scholar]
- Brady G, Iscove NN. Construction of cDNA libraries from single cells. Meth Enzymol. 1993;225:611–623. doi: 10.1016/0076-6879(93)25039-5. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SCR, Lin H. Drosophila PIWI associates with chromatin and interacts directly with Hp1a. Genes and Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gauci DF, Lualdi JC, Ouellette AJ, Brady G, Iscove NN, Buick RN. In vitro cDNA amplification from individual intestinal crypts: a novel approach to the study of differential gene expression along the crypt-villus axis. Exp Cell Res. 1993;20:344–9. doi: 10.1006/excr.1993.1255. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuia X, Shena X, Kim N. Dicer1 expression in preimplantation mouse embryos: Involvement of Oct3/4 transcription at the blastocyst stage. Biochemical and Biophysical Research Communications. 2007;352:231–236. doi: 10.1016/j.bbrc.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;32:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Russ A, von Melchner H, Rayburn H, Priyaranjan P, Jenkins NA, Copeland NG, Ruley HE. A murine homolog of the yeast RNA1 gene is required for postimplantation development. Genes Dev. 1994;8:265–76. doi: 10.1101/gad.8.3.265. [DOI] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Domachenko A, Latham KE, Hatton KS. Expression of myc-family, myc-interacting, and myc target genes during preimplantation mouse development. Mol Reprod Dev. 1997;47:57–65. doi: 10.1002/(SICI)1098-2795(199705)47:1<57::AID-MRD8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, Warren ST. The Fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B, Addendum A. A technique for radiolabeling DNA restriction fragmetns to high specific activity. Anal Bioch. 1983;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Benson RE, Hua J, Bogerd HP, Cullen BR. A nuclear role for the Fragile X mental retardation protein. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5: A novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- Hammond S. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Letters. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hartshorn C, Rice JE, Wangh LJ. Developmentally-regulated changes of Xist RNA levels in single preimplantation mouse embryos, as revealed by quantitative real-time PCR. Mol Reprod Dev. 2002;61:425–36. doi: 10.1002/mrd.10037. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- Jablonka S, Holtmann B, Meisterdagger G, Bandilladagger M, Rossoll W, Fischerdagger U, Sendtner M. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. PNAS. 2002;99:10126–10131. doi: 10.1073/pnas.152318699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J Biol Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- Kusano A, Staber C, Ganetzky B. Segregation distortion induced by wild-type RanGAP in Drosophila. Proc Natl Acad Sci U S A. 2002;99:6866–6670. doi: 10.1073/pnas.102165099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Latham KE, De La Casa E, Schultz R. Analysis of mRNA expression during preimplantation development. In: Tuan RS, Lo CW, editors. Methods in Molecular Biology: Developmental Biology Protocols. II. 1999. pp. 315–331. [DOI] [PubMed] [Google Scholar]
- Latham KE, Rambhatla L. Expression of X-linked genes in androgenetic, gynogenetic, and normal mouse preimplantation embryos. Dev Genet. 1995;17:212–22. doi: 10.1002/dvg.1020170306. [DOI] [PubMed] [Google Scholar]
- Latham KE, Rambhatla L, Hayashizaki Y, Chapman VM. Stage-specific induction and regulation by genomic imprinting of the mouse U2afbp-rs gene during preimplantation development. Dev Biol. 1995;168:670–676. doi: 10.1006/dbio.1995.1111. [DOI] [PubMed] [Google Scholar]
- Latham KE, Solter D, Schultz RM. Activation of a two-cell stage-specific gene following transfer of heterologous nuclei into enucleated mouse embryos. Mol Reprod Dev. 1991;30:182–186. doi: 10.1002/mrd.1080300303. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. piRNAs in the Germ Line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain. Nature Structural & Molecular Biology. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Mann M, Latham KE, Varmuza S. Identification of genes showing altered expression in preimplantation and early postimplantation parthenogenetic embryos. Dev Genet. 1995;17:223–32. doi: 10.1002/dvg.1020170307. [DOI] [PubMed] [Google Scholar]
- Meister G, Bühler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- Morita S, Horiia T, Kimuraa M, Gotoc Y, Ochiyad T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtango NR, Latham KE. Differential Expression of Cell Cycle Genes in Rhesus Monkey Oocytes and Embryos of Different Developmental Potentials. Biol Reprod. 2008;78:254–66. doi: 10.1095/biolreprod.107.064931. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–79. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr Biol. 2001a;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol. 2001b;152:75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J Biol Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- Rambhatla L, Patel B, Dhanasekaran N, Latham KE. Analysis of G protein a subunit mRNA abundance in preimplantation mouse embryos using a rapid, quantitative RT-PCR approach. Mol Reprod Dev. 1995;41:314–324. doi: 10.1002/mrd.1080410306. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rossant J, Guillemot F, Tanaka M, Latham K, Gertenstein M, Nagy A. Mash2 is expressed in oogenesis and preimplantation development but is not required for blastocyst formation. Mech Dev. 1998;73:183–91. doi: 10.1016/s0925-4773(98)00051-3. [DOI] [PubMed] [Google Scholar]
- Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci U S A. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm RD, Bavister BD. Follicle-stimulating hormone priming of rhesus monkeys enhances meiotic and developmental competence of oocytes matured in vitro. Biol Reprod. 1994;51:904–12. doi: 10.1095/biolreprod51.5.904. [DOI] [PubMed] [Google Scholar]
- Schramm RD, Bavister BD. Effects of granulosa cells and gonadotrophins on meiotic and developmental competence of oocytes in vitro in non-stimulated rhesus monkeys. Hum Reprod. 1995;10:887–895. doi: 10.1093/oxfordjournals.humrep.a136056. [DOI] [PubMed] [Google Scholar]
- Schramm RD, Bavister BD. Granulosa cells from follicle stimulating hormone-primed monkeys enhance the development competence of in-vitro-matured oocytes from non-stimulated rhesus monkeys. Hum Reprod. 1996;11:1698–1702. doi: 10.1093/oxfordjournals.humrep.a019472. [DOI] [PubMed] [Google Scholar]
- Schramm RD, Bavister BD. A macaque model for studying mechanisms controlling oocyte development and maturation in human and non-human primates. Hum Reprod. 1999;14:2544–2555. doi: 10.1093/humrep/14.10.2544. [DOI] [PubMed] [Google Scholar]
- Schramm RD, Paprocki AM, VandeVoort CA. Causes of developmental failure of in-vitro matured rhesus monkey oocytes: impairments in embryonic genome activation. Hum Reprod. 2003;18:826–833. doi: 10.1093/humrep/deg144. [DOI] [PubMed] [Google Scholar]
- Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15:531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- Schultz RM. The regulation and reprogramming of gene expression in the preimplantation embryo. Adv Dev Biochem. 1999;5:127–162. [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sommerville J. Activities of cold-shock domain proteins in translation control. Bioessays. 1999;21:319–325. doi: 10.1002/(SICI)1521-1878(199904)21:4<319::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz N, Kolb FA, Zhang F, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO reports. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Hajkova P, Barton SC, O'Carroll D, Lee C, Lao K, Surani MA. 220-plex microRNA expression profile of a single cell. Nat Protoc. 2006;1:1154–1159. doi: 10.1038/nprot.2006.161. [DOI] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanini F, Bontekoe C, Bakker CE, van Unen L, Anar B, Willemsen R, Yoshida M, Galjaard H, Oostra BA, Hoogeveen AT. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum Mol Genet. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- Tesarik J, Kopecny V, Plachot M, Mandelbaum J. Activation of nucleolar and extranucleolar RNA synthesis and changes in the ribosomal content of human embryos developing in vitro. J Reprod Fertil. 1986a;78:463–470. doi: 10.1530/jrf.0.0780463. [DOI] [PubMed] [Google Scholar]
- Tesarik J, Kopecny V, Plachot M, Mandelbaum J, DaLage C, Flechon JE. Nucleologenesis in the human embryo developing in vitro: ultrastructural and autoradiographic analysis. Dev Biol. 1986b;115:193–203. doi: 10.1016/0012-1606(86)90240-x. [DOI] [PubMed] [Google Scholar]
- Tesarik J, Kopecny V, Plachot M, Mandelbaum J. High-resolution autoradiographic localization of DNA-containing sites and RNA synthesis in developing nucleoli of human preimplantation embryos: a new concept of embryonic nucleologenesis. Development. 1987;101:777–791. doi: 10.1242/dev.101.4.777. [DOI] [PubMed] [Google Scholar]
- Tesarik J, Kopecny V, Plachot M, Mandelbaum J. Early morphological signs of embryonic genome expression in human preimplantation development as revealed by quantitative electron microscopy. Dev Biol. 1988;128:15–20. doi: 10.1016/0012-1606(88)90261-8. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chung YG, DeVries WN, Struwe M, Latham KE. Role of protein synthesis in the development of a transcriptionally permissive state in one-cell stage mouse embryos. Biol Reprod. 2001;65:748–754. doi: 10.1095/biolreprod65.3.748. [DOI] [PubMed] [Google Scholar]
- Wang Q, Latham KE. Translation of maternal mRNAs encoding transcription factors during genome activation in early mouse embryos. Biol Reprod. 2000;62:969–978. doi: 10.1095/biolreprod62.4.969. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, i Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani A, Sakaki Y, i Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Weaver DL, Núñez C, Brunet C, Bostock V, Brady G. Single-cell RT-PCR cDNA subtraction. Methods Mol Biol. 1999;97:601–9. doi: 10.1385/1-59259-270-8:601. [DOI] [PubMed] [Google Scholar]
- Weston AM, Wolf DP. Timing of the maternal to embryonic transition in rhesus monkey embryos. Biol.Reprod. 1994;50 Abstract. [Google Scholar]
- Wilkinson MF, Shyu AB. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays. 2001;23:775–787. doi: 10.1002/bies.1113. [DOI] [PubMed] [Google Scholar]
- Yang W, Wang Q, Howell KL, Lee JT, Cho DS, Murray JM, Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation development. Developmental Biology. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zheng P, Patel B, McMenamin M, Reddy SE, Paprocki AM, Schramm RD, Latham KE. The primate embryo gene expression resource: a novel resource to facilitate rapid analysis of gene expression patterns in non-human primate oocytes and preimplantation stage embryos. Biol Reprod. 2004;70:1411–1418. doi: 10.1095/biolreprod.103.023788. [DOI] [PubMed] [Google Scholar]
- Zheng P, Patel B, McMenamin M, Moran E, Paprocki AM, Kihara M, Schramm RD, Latham KE. Effects of follicle size and oocyte maturation conditions on maternal messenger RNA regulation and gene expression in rhesus monkey oocytes and embryos. Biol Reprod. 2005;72:890–897. doi: 10.1095/biolreprod.104.035881. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Boiani M, Ponce R, Guizzardi S, Scandroglio R, Garagna S, Redi CA. Mouse Xist expression begins at zygotic genome activation and is timed by a zygotic clock. Mol Reprod Dev. 2002;61:14–20. doi: 10.1002/mrd.1126. [DOI] [PubMed] [Google Scholar]