Two acostatin heterodimers interact together to form an αββα tetramer.
Keywords: disintegrins, acostatin, tetramer
Abstract
Disintegrins are a family of small (4–14 kDa) proteins that bind to another class of proteins, integrins. Therefore, as integrin inhibitors, they can be exploited as anticancer and antiplatelet agents. Acostatin, an αβ heterodimeric disintegrin, has been isolated from the venom of Southern copperhead (Agkistrodon contortrix contortrix). The three-dimensional structure of acostatin has been determined by macromolecular crystallography using the molecular-replacement method. The asymmetric unit of the acostatin crystals consists of two heterodimers. The structure has been refined to an R work and R free of 18.6% and 21.5%, respectively, using all data in the 20–1.7 Å resolution range. The structure of all subunits is similar and is well ordered into N-terminal and C-terminal clusters with four intramolecular disulfide bonds. The overall fold consists of short β-sheets, each of which is formed by a pair of antiparallel β-strands connected by β-turns and flexible loops of different lengths. Conformational flexibility is found in the RGD loops and in the C-terminal segment. The interaction of two N-terminal clusters via two intermolecular disulfide bridges anchors the αβ chains of the acostatin dimers. The C-terminal clusters of the heterodimer project in opposite directions and form a larger angle between them in comparison with other dimeric disintegrins. Extensive interactions are observed between two heterodimers, revealing an αββα acostatin tetramer. Further experiments are required to identify whether the αββα acostatin complex plays a functional role in vivo.
1. Introduction
Disintegrins were discovered and isolated from the venom of snakes and were given their name because of their biological function of binding to another class of protein known as integrins. Disintegrins contain a characteristic tripeptide motif, e.g. Arg-Gly-Asp (RGD), that is critical for binding integrins. Disintegrins are among the most potent known natural inhibitors of integrin function and are active at nanomolar concentrations, whereas the activity of the short linear RGD peptides is observed at the micromolar level. Disintegrins have been found to inhibit platelet aggregation, angiogenesis, metastasis and tumor growth (McLane et al., 1998 ▶; Markland, 1998 ▶). As such, disintegrins can be explored as therapeutic agents against a number of pathologies including Alzheimer’s disease, inflammation, autoimmune diseases, virus infection, asthma, osteoporosis, thrombosis and cancer (Marcinkiewicz, 2005 ▶).
Disintegrins are small disulfide-rich proteins and are isolated as soluble monomers or dimers, as well as as domains of larger membrane proteins such as the mammalian ADAM (a disintegrin and metalloproteinase) family. The monomeric forms are subdivided into short, medium and long disintegrins which contain ∼50, ∼70 and ∼84 amino-acid residues and four, six or seven disulfide bridges, respectively. Homodimeric and heterodimeric forms of disintegrins are found with ∼65 amino-acid residues per chain with four intramolecular and two intermolecular disulfide bridges. The disulfide-bond pattern is highly conserved in each group. A functional classification for disintegrins based on the tripeptide motif and their integrin-binding selectivity includes a subdivision into RGD-, MLD- and KTS-disintegrins (Marcinkiewicz, 2005 ▶).
Comparisons of the three-dimensional structures of disintegrins (Adler et al., 1991 ▶; Saudek et al., 1991 ▶; Senn & Klaus, 1993 ▶; Smith et al., 1996 ▶; Guo et al., 2001 ▶; Moreno-Murciano et al., 2003 ▶; Fujii et al., 2003 ▶; Shin et al., 2003 ▶; Bilgrami et al., 2004 ▶, 2005 ▶; Monleón et al., 2005 ▶; Janes et al., 2005 ▶; Takeda et al., 2006 ▶; Igarashi et al., 2007 ▶) revealed a remarkable similarity and a fold having an elongated overall shape. The secondary structure of disintegrins is composed of a series of short antiparallel β-sheets, with the integrin-binding motif located at the tip of one of the connecting loops. The C-terminus of each chain was found to be structurally close to the integrin-binding loop. Interactions between disintegrin subunits of dimers are mostly observed within the N-terminal residues including two disulfide bonds linking the two chains. In dimers the integrin-binding loops containing the tripeptide motif point in opposite directions. Structure–function studies of disintegrins have shown that the disulfide bonding is essential for disintegrin structural integrity and binding, whereas the RGD-flanking residues and C-terminus are relevant for integrin-binding affinity and selection (Wierzbicka-Patynowski et al., 1999 ▶; McLane et al., 2001 ▶; Yahalom et al., 2002 ▶).
Here, we present the three-dimensional structure of acostatin, a heterodimeric disintegrin from the venom of the snake Agkistrodon contortrix contortrix. The gene structure encoding the β-chain precursor of acostatin is consistent with the well known pre-peptide, metalloprotease, spacer and disintegrin domains, whereas the α-chain has a short coding region encoding the disintegrin domain (Okuda et al., 2002 ▶). Acostatin purified from the venom of the Southern copperhead consists of a mature protein of 63 and 64 amino-acid residues in the α-chain and β-chain, respectively, where both chains contain the Arg-Gly-Asp (RGD) sequence motif. A predominant form of purified acostatin has been identified in which the N-terminal residue of the α-chain is a pyroglutamic acid, lacking the initial isoleucine.
2. Experimental
2.1. Crystallization and data collection
Crystals of acostatin purified from the venom of A. contortrix contortrix were grown using the hanging-drop vapor-diffusion method in which protein solution (∼16.5 mg ml−1 in 10 mM HEPES pH 7.4, 14.7 mM NaCl) was mixed with an equal volume of reservoir solution (1.8 M ammonium sulfate in 100 mM Tris buffer pH 8.5) as previously described (Moiseeva et al., 2002 ▶). Crystal characterization was performed using X-ray diffraction data collected at the SSRL (Stanford Synchrotron Radiation Laboratory, Stanford, California, USA). The final data set was collected from a flash-frozen crystal at beamline 5.0.2 of the ALS (Advanced Light Source, Berkeley, California, USA). Glycerol (12–15%) was added to the reservoir solution as a cryoprotectant. Images were processed and scaled with HKL-2000 (Otwinowski & Minor, 1997 ▶) and details of the data collection and statistics are summarized in Table 1 ▶. The crystals belonged to space group P212121 with two acostatin dimers per asymmetric unit, and diffracted to a resolution of 1.7 Å. A monoclinic crystal form of acostatin has been reported (Fujii et al., 2002 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| X-ray source | ALS BL 5.0.2 |
| Detector type | ADSC CCD Q210 |
| Crystal-to-detector distance (mm) | 180 |
| Temperature (K) | 100 |
| Oscillation range (°) | 1 |
| Exposure time (s) | 10 |
| Total angular rotation (°) | 360 |
| Wavelength (Å) | 0.9486 |
| Unique reflections | 30457 (2753) |
| Average redundancy | 13.7 (9.2) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 37.45, b = 59.81, c = 121.31 |
| Resolution (Å) | 20.0–1.7 (1.76–1.70) |
| Reflections with I > 3σ(I) (%) | 91.1 (71.2) |
| Rmerge | 0.044 (0.156) |
| VM (Å3 Da−1) | 2.5 |
| Molecules (dimers) per ASU | 2 |
| Refinement | |
| PDB code | 3c05 |
| Refinement | |
| Completeness (%) | 98.8 (90.1) |
| Resolution (Å) | 20.0–1.7 (1.745–1.701) |
| Working set of reflections (95%) | 28838 (1901) |
| Test set of reflections (5%) | 1543 (97) |
| Rwork (%) | 18.6 (19.2) |
| Rfree (%) | 21.5 (22.5) |
| Model | |
| No. of protein non-H atoms | 1686 |
| No. of water molecules | 303 |
| No. of sulfates | 3 |
| R.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.3 |
| Mean B factor (Å2) | 26.5 |
| Ramachandran plot: (non-Gly, non-Pro) residues in most favored regions (%) | 100.0 |
2.2. Structure solution and refinement
Initial phase estimates were derived from a molecular-replacement solution using the maximum-likelihood approach (Read, 2001 ▶) as implemented in Phaser (McCoy et al., 2007 ▶). An initial homology search model was generated using CCP4 tools (Collaborative Computational Project, Number 4, 1994 ▶) and the program CHAINSAW (Schwarzenbacher et al., 2004 ▶) with nonhomologous side chains deleted to the Cγ atom. This model was built using the trimestatin X-ray coordinates (PDB code 1j2l), truncated at the first 15 amino-acid residues and aligned with contortrostatin, another homodimeric disintegrin also purified from this snake species and thought to be the content of the crystals. The amino-acid sequences of contortrostatin and the β-chain of acostatin are identical. A clear molecular-replacement solution was found with translational Z scores of 10.20, 15.19, 19.56 and 20.81 identifying four subunits labeled A, B, C and D and oriented so as to form the characteristic intermolecular disulfide bridges. The amino-acid sequence determined from the X-ray crystallographic electron-density map and the observed weight of 13 508 Da are consistent with the presence of the predominant purified form of the heterodimeric acostatin. The molecular-replacement solution obtained was used as a starting model for automated model building using ARP/wARP (Perrakis et al., 1999 ▶) and extended to 224 of the 252 amino-acid residues contained in the Ile-lacking form of acostatin. Further model building was performed using Coot (Emsley & Cowtan, 2004 ▶). The structure was refined without noncrystallographic symmetry restraints with REFMAC5 (Murshudov et al., 1997 ▶) using the Babinet scaling option and a final overall weight of 0.45. Accessible surface area was calculated with the CCP4 program AREAIMOL using a probe radius of 1.4 Å.
3. Results and discussion
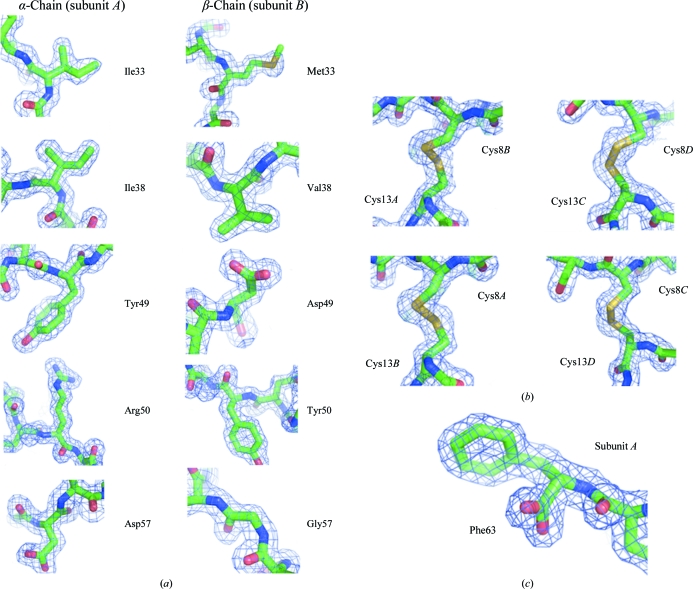
The final crystallographic model consist of 1686 protein non-H atoms from 224 amino-acid residues of two acostatin heterodimers, 293 water molecules, two sulfate ions and additional residual electron densities tentatively modeled as ten water molecules and another sulfate ion at a lower occupancy. The final refinement statistics are summarized in Table 1 ▶. The model includes amino-acid residues 5–63 for subunit A and 5–62 for subunit C of the Ile-lacking 62 amino-acid residues (2–63) of the α-chain of acostatin. The model also includes amino-acid residues 4–62 for subunit B and 4–59 for subunit D of the 64 amino-acid residues of the β-chain of acostatin. Electron densities are connected for all backbone atoms at the 1σ level except for residues Arg43D–Gly44D and the tentatively assigned Lys61C–His62C C-terminal residues. Residual electron densities are visible and could potentially be explained on the basis of disorder in the amino-terminal and carboxy-terminal residues and potential alternative conformations including the side chains of Met33B, Lys14C and Glu35D. The model has been refined to crystallographic R work and R free values of 18.6% and 21.5%, respectively, using all data in the 20.0–1.7 Å resolution range, with root-mean-square deviations (r.m.s.d.s) in bond lengths and bond angles of 0.013 Å and 1.3°, respectively. The geometry of the model was analyzed with MOLPROBITY (Davis et al., 2007 ▶) and showed 100% of the residues to be in the core region of the Ramachandran plot. Additional stereochemistry was analyzed using Coot and was found to be in agreement with expected values. One outlier is found in the rotamer conformation of Cys13 from all subunits. Fig. 1 ▶ shows representative electron-density fit including Cys13 and a carboxy-terminal group at residue Phe63 from the α-type subunit A.
Figure 1.
Electron-density fit of the model showing (a) observed differences in the amino-acid sequence of the α- and β-chains of acostatin represented by subunits A and B, respectively, (b) all Cys13 residues identified as rotamer outliers and (c) the carboxyl group of the C-terminal residue Phe63 of subunit A. This figure was prepared using PyMOL (DeLano, 2002 ▶).
3.1. Acostatin subunit structures
The overall fold of all acostatin subunits (A, B, C, D) is similar and is depicted for the heterodimer AB in Fig. 2 ▶(a). Each subunit structure can be divided into two distinct clusters: an amino-terminal cluster (up to residue 19) and a carboxy-terminal cluster (residue 20 and beyond). In subunits A, B and D, the structure contains three β-sheets each formed by a pair of antiparallel β-strands consisting of residues 8–9 with 14–15, residues 27–28 with 31–32 and residues 38–40 with 49–50; in subunit C only the latter two β-sheets are found. The β-strands are connected by β-turns and flexible loops of different lengths consisting of 4–10 residues. The typical intra-chain disulfide bridges found in the disintegrin family are also observed in the acostatin structure. For all subunits, the distances calculated between the S atoms of the pairs of Cys residues 7–30, 21–27, 26–51 and 39–58 are all within expected disulfide-bond distances. The high content of disulfide bridges in these polypeptides is likely to contribute to the formation of a stable and well defined three-dimensional structure.
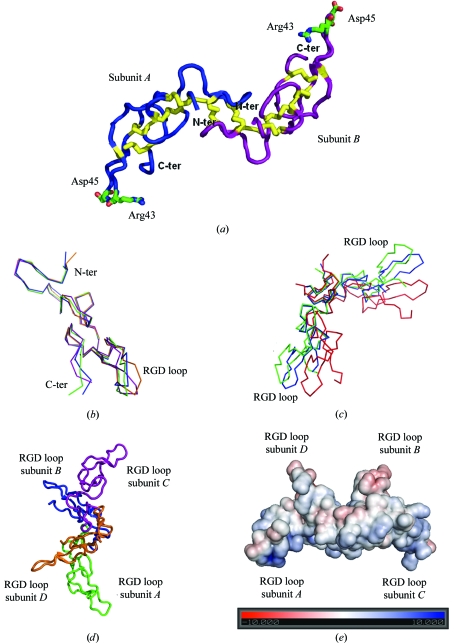
Figure 2.
(a) Overall structure of the acostatin heterodimer represented by a Cα tracing of subunits A (in blue) and B (in magenta) with disulfide bridges in yellow and the side chains of the RGD binding loops. (b) Superimposition of the Cα tracing of the acostatin ABCD subunits (A subunit in green, B in blue, C in purple and D in orange). (c) Superimposition of the Cα tracing of acostatin AB (green) and CD (blue) dimers on the dimer from E. carinatus (red). (d) Overall structure of the tetrameric arrangement of all acostatin subunits represented by a Cα tracing. (e) The electrostatic surface of the acostatin tetramer on the scale ±10 kT/e. The color map is from red (negative electrostatic potential) to blue (positive electrostatic potential). The figure was prepared in PyMOL (DeLano, 2002 ▶) and the electrostatic potential was calculated using APBS (Baker et al., 2001 ▶).
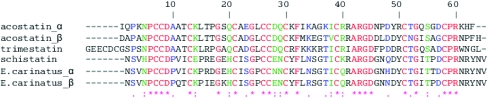
Comparison of the structure of the α- and β-chains of acostatin does not reveal major structural changes (Fig. 2 ▶ b). The r.m.s.d. for the superimposition of the Cα atoms (residues 5–59) of the α-chains (A/C) and the β-chains (B/D) are 0.88 and 1.02 Å, respectively. In comparison, superimposition of mixed chain types gives r.m.s.d.s of 1.03 Å (A/B), 1.04 Å (A/D), 1.12 Å (C/D) and 1.57 Å (B/C). For all overlays, the deviations are mainly located in the region of residues 38–50, designated as the Arg-Gly-Asp-containing (RGD) loop, with the largest deviation of 4.3 Å at Asp45 when comparing subunits B and C. We also observed that the C-terminal residues 60–62 visible in subunits A, B and C and located adjacent to the RGD loops are found in different orientations. Most of the observed differences can be accounted for by crystal contacts. The comparison of the acostatin fold with the previously determined disintegrin structures of the monomeric trimestatin, the schistatin homodimer and the heterodimer from Echis carinatus does not indicate any major structural rearrangements, as expected from their homologous sequences (Fig. 3 ▶). The calculated r.m.s.d. of 1.2–1.5 Å in the superimposition of acostatin with other disintegrin structures is comparable to the overlay of the different chain types of acostatin. Additional conformational differences are also observed in the N-terminal residues.
Figure 3.
Sequence alignment of acostatin with trimestatin, schistatin and the E. carinatus heterodimer.
3.2. The αβ acostatin dimer
Specific interactions are found between the α- and β-chains in both the AB and CD dimers. The N-terminal clusters of each pair of subunits are responsible for dimer formation (Fig. 2 ▶ a). In both heterodimers, we observed that the distances calculated between the S atoms of Cys residue 8 in one chain and Cys residue 13 in the other chain are all within expected disulfide-bond distances. This pattern of disulfide bridges is identical to the pattern of intermolecular disulfide bonds observed in the homodimer of schistatin (Bilgrami et al., 2004 ▶) and the heterodimer from the E. carinatus disintegrin (Bilgrami et al., 2005 ▶). These two intermolecular disulfide bridges per heterodimer certainly contribute to the stability and rigidity of the dimer. In addition, two hydrogen-bond distances are observed in the heterodimer AB between the side-chain N atoms of Asn5 and the carbonyl O atoms of Ala10. In heterodimer CD the side chains of Asn5 adopt a different rotamer conformation and the carbonyl O atoms of Ala10 and the N atoms of Cys7 are found to interact with the same water molecules. Overall, 8.3% and 9.7% of the accessible surface area of the subunits is buried in the formation of the AB and CD dimers, respectively.
The overall fold of the acostatin AB and CD dimers is essentially similar. Superimposition of the dimers gives a calculated r.m.s.d. on Cα atoms of 1.82 Å. We observed that dimerization through the N-terminal domains takes place such that the C-terminal domains are facing away from each other. The C-terminal domains in the heterodimeric acostatin are widely separated from each other: the distances between the tips of the C-terminal domains at the Cα atoms of Asp45 are 69.5 and 69.8 Å for the AB and CD dimers, respectively. With such an orientation of the N- and C-terminal regions, it is not surprising to find that the slight differences between the heterodimers are located in this RGD-containing segment. Comparison of the acostatin dimers with previously reported dimeric disintegrins reveals a major overall difference. We observed that the dimerization through the N-terminal clusters generated a different hinge region between the C-terminal domains (Fig. 2 ▶ c), with a larger angle in the acostatin dimers. This larger angular hinge moves the tips of the C-terminal domains in acostatin further apart. In comparison, the calculated distances between the Cα atoms of Asp45 in schistatin and the E. carinatus heterodimeric disintegrin are 57.7 and 59.1 Å, respectively.
3.3. An αββα acostatin tetramer
We observed considerable interactions between the acostatin AB and CD heterodimers, as shown in Figs. 2 ▶(d) and 2 ▶(e). A substantial part (8.2%) of the accessible surface area of the subunits, mostly spread over the N- and C-terminal clusters of the β chains (B and D), is buried in this heterodimer–heterodimer interaction. We identified two regions of hydrophobic interaction involving residue Leu15 in subunit B with Phe32 and Ile54 in subunit D and vice versa. Two hydrogen bonds are also formed, with the carbonyl O atom of Ser19 interacting with the side-chain N atom of Gln20. Two additional hydrogen bonds are formed between subunits B and C; one involved the side-chain carboxyl group of Glu35 (subunit B) and the N atom of Leu15 (subunit C) and the other is made between the carbonyl O atom of Asn52 (subunit B) and the side-chain amino group of Lys14 (subunit C). These residues adopt a different conformation in the A/D subunits and the interactions found between them are mediated through a network of water molecules. The surface complementarities of the AB and CD dimers suggest the possibility of a tetrameric form of acostatin that is best represented by an αββα acostatin tetramer. In the tetrameric form the RGD loops are all pointing in different and almost orthogonal directions. The distance between the Cα atoms of Asp45 of the β-chains is 40.9 Å and that between the α-chains is 80.6 Å. The equivalent distances measured between the AD and BC subunits are 40.5 and 38.7 Å, respectively. This tetrameric arrangement is new among known disintegrin structures but could be an artifact of crystallization. Further experiments are required to identify whether this αββα acostatin complex plays a functional role in vivo.
Supplementary Material
PDB reference: acostatin, 3c05, r3c05sf
Acknowledgments
We thank the SSRL, the ALS and their staff for providing access to the beamline facility. This work was supported in part by funds provided to MA by the Department of Energy under contract No. DEAC02-98CH10866, the National Institutes of Health/National Institute of General Medical Sciences under agreement Y1 GM-0080-03 and the Brookhaven National Laboratory/Laboratory Directed Research and Development Program. RB acknowledges partial support from the National Science Foundation (grant CHE-98-16294) as well as from the Zumberge Research Innovation Fund of the University of Southern California.
References
- Adler, M., Lazarus, R. A., Dennis, M. S. & Wagner, G. (1991). Science, 253, 445–448. [DOI] [PubMed]
- Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001). Proc. Natl Acad. Sci. USA, 98, 10037–10041. [DOI] [PMC free article] [PubMed]
- Bilgrami, S., Tomar, S., Yadav, S., Kaur, P., Kumar, J., Jabeen, T., Sharma, S. & Singh, T. P. (2004). J. Mol. Biol.341, 829–837. [DOI] [PubMed]
- Bilgrami, S., Yadav, S., Kaur, P., Sharma, S., Perbandt, M., Betzel, C. & Singh, T. P. (2005). Biochemistry, 44, 11058–11066. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S. & Richardson, D. C. (2007). Nucleic Acids Res.35, W375–W383. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, California, USA. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fujii, Y, Okuda, D., Fujimoto, Z., Morita, T. & Mizuno, H. (2002). Acta Cryst. D58, 145–147. [DOI] [PubMed]
- Fujii, Y., Okuda, D., Horii, K., Morita, T. & Mizuno, H. (2003). J. Mol. Biol.332, 1115–1122. [DOI] [PubMed]
- Guo, R. T., Chou, L. J., Chen, Y. C., Chen, C. Y., Pari, K., Jen, C. J., Lo, S. J., Huang, S. L., Lee, C. Y., Chang, T. W. & Chaung, W. J. (2001). Proteins, 43, 499–508. [DOI] [PubMed]
- Igarashi, T., Araki, S., Mori, H. & Takeda, S. (2007). FEBS Lett.581, 2416–2422. [DOI] [PubMed]
- Janes, P. W., Saha, N., Barton, W. A., Kolev, M. V., Wimmer-Kleikamp, S. H., Nievergall, E., Blobel, C. P., Himanen, J.-P., Lackmann, M. & Nikolov, D. B. (2005). Cell, 123, 291–304. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- McLane, M. A., Kuchar, M. A., Brando, C., Santoli, D., Paquette-Straub, C. A. & Miele, M. E. (2001). Haemostasis, 31, 177–182. [DOI] [PubMed]
- McLane, A., Marcinkiewicz, C., Vijay-Kumar, S., Wierzbicka-Patynowski, I. & Niewiarowski, S. (1998). Proc. Soc. Exp. Biol. Med.219, 109–119. [DOI] [PubMed]
- Marcinkiewicz, C. (2005). Curr. Pharm. Des.11, 815–827. [DOI] [PubMed]
- Markland, F. S. (1998). Toxicon, 36, 1749–1800. [DOI] [PubMed]
- Moiseeva, N., Swenson, S. D., Markland, F. S. Jr & Bau, R. (2002). Acta Cryst. D58, 2122–2124. [DOI] [PubMed]
- Monleón, D., Esteve, V., Kovacs, H., Calvete, J. J. & Celda, B. (2005). Biochem. J.387, 57–66. [DOI] [PMC free article] [PubMed]
- Moreno-Murciano, M. P., Monleón, D., Marcinkiewicz, C., Calvete, J. J. & Celda, B. (2003). J. Mol. Biol.329, 135–145. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Okuda, D., Koike, H. & Morita, T. (2002). Biochemistry, 41, 14248–14254. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Saudek, V., Atkinson, R. A. & Pelton, J. T. (1991). Biochemistry, 30, 7369–7372. [DOI] [PubMed]
- Schwarzenbacher, R., Godzik, A., Grzechnik, S. K. & Jaroszewski, L. (2004). Acta Cryst. D60, 1229–1236. [DOI] [PubMed]
- Senn, H. & Klaus, W. (1993). J. Mol. Biol.232, 907–925. [DOI] [PubMed]
- Shin, J., Hong, S. Y., Chung, K., Kang, I., Jang, Y., Kim, D. S. & Lee, W. (2003). Biochemistry, 42, 14408–14415. [DOI] [PubMed]
- Smith, K. J., Jaseja, M., Lu, X., Williams, J. A., Hyde, E. I. & Trayer, I. P. (1996). Int. J. Pept. Protein Res.48, 220–228. [DOI] [PubMed]
- Takeda, S., Igarashi, T., Mori, H. & Araki, S. (2006). EMBO J.25, 2388–2396. [DOI] [PMC free article] [PubMed]
- Wierzbicka-Patynowski, I., Niewiarowski, S., Marcinkiewicz, C., Calvete, J. J., Marcinkiewicz, M. M. & McLane, M. A. (1999). J. Biol. Chem.274, 37809–37814. [DOI] [PubMed]
- Yahalom, D., Wittelsberger, A., Mierke, D. F., Rosenblatt, M., Alexander, J. M. & Chorev, M. (2002). Biochemistry, 41, 8321–8331. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: acostatin, 3c05, r3c05sf