As part of a training set for automated image analysis, crystallization screening experiments for 269 different macromolecules were visually analyzed and a set of crystal images extracted. Outcomes and trends are analyzed.
Keywords: crystallization, image analysis, crystal images
Abstract
In the automated image analysis of crystallization experiments, representative examples of outcomes can be obtained rapidly. However, while the outcomes appear to be diverse, the number of crystalline outcomes can be small. To complement a training set from the visual observation of 147 456 crystallization outcomes, a set of crystal images was produced from 106 and 163 macromolecules under study for the North East Structural Genomics Consortium (NESG) and Structural Genomics of Pathogenic Protozoa (SGPP) groups, respectively. These crystal images have been combined with the initial training set. A description of the crystal-enriched data set and a preliminary analysis of outcomes from the data are described.
1. Introduction
A high-throughput crystallization screening laboratory housed at the Hauptman–Woodward Medical Research Institute (HWI) provides a service for the structural genomics and biological crystallography community (for a description of the method, see Luft et al., 2003 ▶). We have used data from this facility to establish a training set for automated image analysis of typical crystallization outcomes (Snell et al., 2008 ▶). This set provided representative examples of crystallization screening outcomes from a set of 96 different macromolecules and included only a small number of images (0.2%) with crystals. While our initial image-analysis developments have been successful in identifying clear and precipitate drops, developing software beyond this required a training set that included a statistically significant sampling of our ultimate target, i.e. crystals. For this reason, we have visually identified crystallization leads from 269 macromolecules encompassing samples from both the North East Structural Genomics Consortium (NESG) and Structural Genomics of Pathogenic Protozoa (SGPP) groups out of a selection of 823 targets.
In this paper, we describe the steps taken to enhance our original training set with additional crystal images and include a preliminary analysis of the crystallization results based upon these data.
2. Experimental
2.1. Samples
Our macromolecular targets included 269 macromolecules that showed crystal hits provided by the NESG and SGPP structural genomics centers, which were sent to the HWI high-throughput crystallization screening laboratory. The NESG consortium focuses on three areas: large protein-domain families, biomedical theme targets and targets nominated by the biomedical community. They provided 224 proteins for crystallization (106 of which showed at least one crystal hit). The remaining 599 eukaryotic proteins were supplied by the SGPP (163 of which showed at least one crystal hit) and included targets from major global pathogenic protozoa.
The high-throughput crystallization screening laboratory (HTS) has been described elsewhere (Luft et al., 2003 ▶). Crystallization has been carried out with three groups of cocktails: salts, polyethylene glycols (PEGs) and a group of commercial screens. The salts and PEGs groups, 1 and 2, were constructed using an incomplete factorial design (Audic et al., 1997 ▶) and are buffered with 100 mM concentrations of CAPS (pH 10.0), TAPS (pH 9.0), Tris (pH 8.0), HEPES (pH 7.5), MOPS (pH 7.0), MES (pH 6.0), sodium acetate (pH 5.0) and sodium citrate (pH 4.0). Group 1 cocktails are highly soluble salts (233 cocktails) including 36 different salts (11 cations and 14 anions) at ∼30%, ∼60% and ∼90% saturation, buffered as described. Group 2, PEG/salt (733 cocktails), includes five different molecular-weight PEGs, 20 kDa, 8 kDa, 4 kDa, 1 kDa and 400 Da, combined with 35 salts at 100 mM concentration, also buffered as described. Group 3 contains commercial screens (570 cocktails). This comprises Hampton Research Natrix, Quik, PEG/Ion, Grid (PEG 6000, Ammonium Sulfate, Sodium Chloride), Crystal Screen HT, Index and SaltRx screens. For historical reasons, the first 18 cocktails from Hampton Research Crystal Screen Cryo are distributed within groups 1 and 2. These and other occurrences of Hampton Research cryocondition cocktails serve as a control during the experimental process. The 1536 cocktails used by the HTS laboratory are reformulated each year to remove cocktails that prove problematic in formulation or that frequently produced hits that were subsequently identified as salt crystals. After 8 y, these annual changes to the cocktails are now minor; a single set of 1536 of cocktails was used throughout this study.
2.2. Experiment and analysis
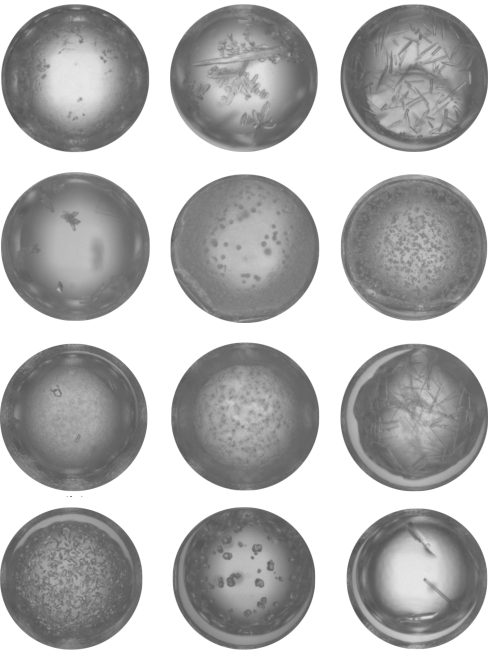
Each of the 269 macromolecular samples was submitted to the laboratory pipeline and crystallization experiments were set up in 1536-well experiment plates (Greiner BioOne, Frickenhausen, Germany) using the microbatch-under-oil method (Chayen et al., 1992 ▶). The samples from NESG were predominately supplied in Tris buffer with 5 mM DDT and 100 mM NaCl, while those from SGPP were typically in HEPES with 500 mM NaCl, 5% glycerol and 0.025% azide. The experiments were imaged over time as described in Snell et al. (2008 ▶). The software (MacroScope) used to view and classify the images was developed in-house. Images are displayed in 16 groups of 96 thumbnail images. A full-sized view of a thumbnail can be selected for closer inspection of the outcome (Snell et al., 2008 ▶). Each image, recorded at one week and four weeks from initiation (826 368 images in total), was visually classified by a single expert viewer as either a crystal, crystal and combination of other categories, e.g. precipitate, skin etc., or not a crystal. A second viewer was used to confirm the crystal classification in those images initially classified as containing crystals. Images were not classified further. Examples of crystal images from the study are shown in Fig. 1 ▶.
Figure 1.
Example images showing results classified as crystals.
3. Results
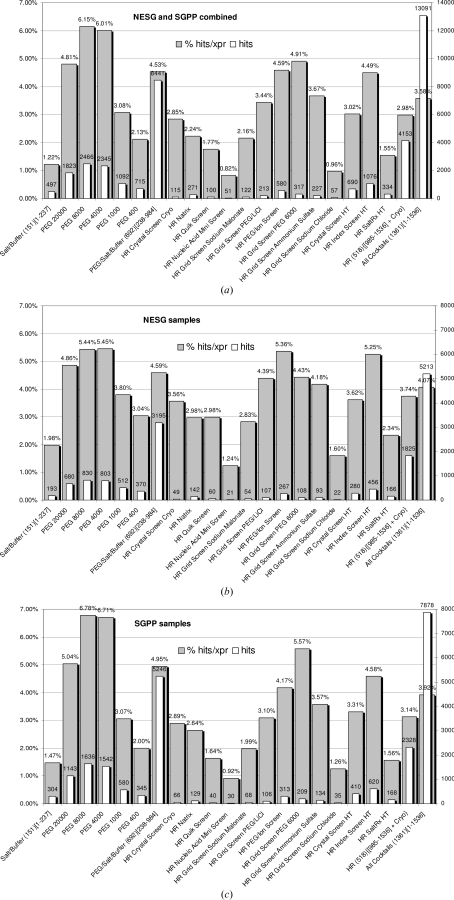
In discussing the results, it is important to remember that these represent data from crystallization screening experiments that were successful. Thus, the results show a distribution of cocktails producing initial hits for a subgroup of samples (n = 269) that crystallized. These examples do not represent a measure of general crystallization success. In Fig. 2 ▶(a), the images from both the NESG and SGPP experiments classified as showing crystals are plotted as a distribution of the different components that make up the 1536 screening cocktails. The first 233 conditions (group 1) give crystallization hits in only 1.22% of cases. The 722 PEG conditions (group 2) were somewhat more successful, with a distribution of hits observed to peak at 6.15% for PEG 8K. It is noticeable that overall the PEG conditions are quite successful crystallizing agents. Similar trends are observed when we compare the combined group 2 results (shown as PEG/Salt/Buffer) with those of the commercial screens containing PEG. The Hampton Research PEG/Ion screen and PEG 6000 Grid screens show a 4.59% and 4.91% hit rate, respectively, and the HR Index Screen, also containing PEG, shows 4.49% success.
Figure 2.
Frequency of crystals as a function of the crystallization cocktail for (a) all the samples (NESG and SGPP combined) and as a function of the individual groups: (b) NESG and (c) SGPP. Shaded bars indicate the crystal observations as a percentage of the cocktails sampled in the group, while unshaded bars give the actual number of crystals in the group. The results are broken down into groups of cocktails and subsets of the individual groups.
Overall, where crystals occur, the average percentage of conditions that show hits in the 1536-condition screen is 3.58%, or ∼55 hits per sample. As seen in Fig. 1 ▶, these can be crystals that visually appear to require little optimization or those that may require significant effort to optimize.
The same data are broken down into individual groups from NESG and SGPP in Figs. 2 ▶(b) and 2 ▶(c). Each of these structural genomics groups focuses on different targets. The two groups used different techniques to clone, express, purify and formulate their targets; however, the trends in crystallization behavior are strikingly similar. Because of the dissimilarity between the NESG and SGPP samples, we conclude that the overall trends seen in Fig. 2 ▶(a) can be regarded as a representative sample of the distribution of successful crystallization lead conditions for a general population of soluble biological macromolecules where crystallization is possible.
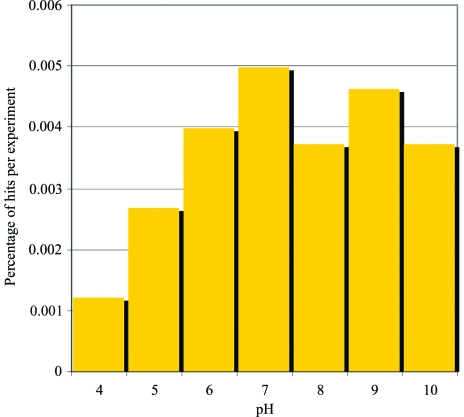
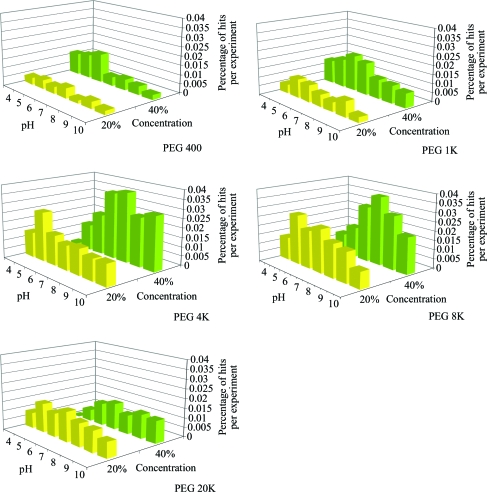
The distribution of hits occurring in the highly soluble salt cocktails (group 1) is shown in Fig. 3 ▶. There is an increase from lower to higher pH values, but no abrupt drop in crystallization success with the highest pH cocktails. In Fig. 4 ▶, the crystal hits from the PEG screen cocktails (group 2) are shown. The number of crystal hits peaks in PEG 4K and 8K, with a reduced number of hits beyond the range of these two molecular weights. It would seem that the choice of PEG molecular weights sampled in group 2 is a valid choice. There is a slightly increased hit rate for the 40% concentration versus the 20% concentration: 54% versus 46%, respectively. There also appears to be a slight preference for neutral pH in the 4K and 8K cases, especially for 40% concentration.
Figure 3.
Distribution of crystal hits in group 1, highly soluble salts, as a function of pH.
Figure 4.
The distribution of crystal results for various molecular-weight PEGs at 20% and 40% concentration as a function of pH.
In Table 1 ▶, the macromolecules that produced crystal hits are summarized as a function of the group 1 (highly soluble salts) and group 2 (PEGs) components of the HWI cocktail component of the screen. From the group 1 and group 2 components of the screen, crystals resulted for 254 of the 269 samples (94.4%), with the remaining crystal hits occurring in the commercial screens. The incomplete factorial design of group 1 and group 2 is highly successful in capturing the majority of crystallization leads. Of the 254 hits, 115 macromolecules had hits in group 1 and 247 had hits in group 2. A total of seven of the hits in group 1 were unique to that group; no hits occurred elsewhere in the screen. For group 2, 139 of these hits were unique. As a percentage of the total macromolecules that crystallized in groups 1 and 2, 45.3% occurred in group 1 and 97.2% in group 2. Of these, 2.8% were unique to group 1 and 54.7% were unique to group 2. When the number of cocktails in both group 1 and group 2 are taken into account, group 1 has a 49.4% success rate and group 2 has a 33.9% success rate. However, group 2 is substantially better in producing unique hits: 19.1% compared with 3.0% for group 1.
Table 1. The number of cocktails that produced crystals as a function of the cocktail grouping.
Note that multiple hits within the same cocktail group for a single macromolecule are only counted as one hit in this analysis.
| No. of cocktails producing crystals | ||||
|---|---|---|---|---|
| Group 1, highly soluble salts | Group 2, PEGs | Unique to group 1 | Unique to group 2 | |
| No. of conditions | 233 | 733 | 233 | 733 |
| No. of macromolecules | 115 | 247 | 7 | 139 |
| Hits in HWI cocktails (%) | 45.3 | 97.2 | 2.8 | 54.7 |
| Percentage of No. of cocktails (%) | 49.4 | 33.7 | 3.0 | 19.0 |
| Percentage overall (%) | 42.8 | 91.8 | 2.6 | 51.7 |
In Table 2 ▶, the group 2 data are broken down as a function of the PEG molecular weight. The numbers of cocktail conditions for each PEG are comparable and the peak performance seems to occur for PEG 4K and 8K, in agreement with Fig. 4 ▶. There are a very small number of unique hits for the different molecular-weight PEGs. Of the small number of unique hits, PEG 4K and PEG 8K provided 14 and ten unique hits, respectively. PEG 400 had two unique hits, while PEG 1K and 20K both had only a single unique hit. This leads us to conclude that where a crystal hit occurs in one PEG condition, it is also likely to be seen in others. This is illustrated in Table 3 ▶, where the number of macromolecules giving crystals is tabulated against how many PEG conditions produced crystals. For 28 macromolecules a single PEG condition produced crystals, with the remaining 226 being produced by two or more different molecular-weight PEG conditions. For 73 of the 254 macromolecules all five different molecular-weight PEGs produced crystal hits.
Table 2. The number of cocktails that produced crystals from group 2 (the PEG group), separated by PEG molecular weight.
Again, multiple hits within the same cocktail group for a single macromolecule are only counted as one hit in this analysis.
| No. of cocktails giving crystals | |||||
|---|---|---|---|---|---|
| PEG 20K | PEG 8K | PEG 4K | PEG 1K | PEG 400 | |
| No. of conditions | 145 | 153 | 148 | 145 | 142 |
| No. of macromolecules | 180 | 207 | 209 | 141 | 111 |
| Hits in HWI cocktails (%) | 70.9 | 81.5 | 82.3 | 55.5 | 43.7 |
| Percentage overall (%) | 66.9 | 77.0 | 77.7 | 52.4 | 41.3 |
| No. unique | 1 | 14 | 10 | 1 | 2 |
| Percentage unique in HWI cocktails (%) | 0.8 | 5.5 | 3.9 | 0.4 | 0.8 |
Table 3. The number of macromolecules that had a crystal hit in a unique PEG for the group of PEG 20K through to PEG 400, in multiples of those PEGs.
| No. of cocktails giving crystals | |||||
|---|---|---|---|---|---|
| 1 PEG only | 2 PEGs | 3 PEGs | 4 PEGs | 5 PEGs | |
| Macromolecules | 28 | 32 | 65 | 49 | 73 |
The data presented in Tables 1 ▶, 2 ▶ and 3 ▶ are broken down into groups of cocktails where multiple hits within a cocktail group for an individual sample are counted as a single hit for that cocktail group. In Table 4 ▶, the performance of the cocktail groups in terms of the total number of hits within each cocktail group for these 269 proteins is presented. Overall, 55 of the 1536 conditions resulted in crystallization hits on average (for these 269 macromolecules where crystal hits occurred). Multiple hits were seen in both groups 1 and 2.
Table 4. Distribution of the hits per macromolecule as a function of the cocktail.
| Cocktail groups | ||||||
|---|---|---|---|---|---|---|
| Salts | PEG 20K | PEG 8K | PEG 4K | PEG 1K | PEG 400 | |
| No. of conditions | 233 | 145 | 153 | 148 | 145 | 142 |
| Average No. of hits per macromolecule | 3.0 | 11.0 | 14.9 | 14.2 | 6.6 | 4.3 |
| Standard deviation | 3.1 | 7.8 | 9.2 | 9.0 | 5.9 | 4.2 |
| Maximum No. of hits | 22 | 32 | 39 | 35 | 26 | 19 |
| Minimum No. of hits | 1 | 1 | 1 | 1 | 1 | 1 |
For any particular macromolecule, where a hit occurs in multiple components or subcomponents of those groups, these hits are not necessarily chemically related other than having the same PEG or a salt. To understand the data in a statistically meaningful manner, a far larger sample of images will need to be classified. Similarly, the results are grouped into a binary distribution, i.e. crystal (or crystal with other category) and no crystal. A result that is classified as not a crystal could be a precipitate, clear or some other outcome. Classifying the results more completely will produce a better understanding of the performance of the cocktails.
4. Discussion and concluding remarks
This study relied on two viewers classifying images, with the first viewer initially classifying the complete set of images as containing a crystal or not. The second viewer then looked at these, confirming the classification, to give 17 895 examples of crystals. Three viewers were used for each image in our parallel 96-protein study (Snell et al., 2008 ▶). In this case, all the viewers had to agree for a unanimous classification, i.e. a drop that was classified by two viewers as containing just a crystal but that was classified by a third as a crystal and something else was not regarded as unanimously classified. In the case of the work presented here, more relaxed criteria were used for crystal classification, i.e. a crystal was any image classified as having a crystal present. With the larger image sample in this study, having the second viewer confirm those images initially classified as crystals ensured that those images could be reliably used for the training set without imposing an undue workload for multiple viewers to examine every image considered.
In looking at where crystals result in the screen (Fig. 2 ▶), certain favored regions can be seen. PEG 4K and 8K produced more hits than the other PEGs within group 2. The Hampton Research screens containing PEG, i.e. the PEG 6000 Grid Screen, PEG/LiCl Screen and Index Screen, also show this trend. The poor performers are the Nucleic Acid Mini Screen and the Sodium Chloride Grid Screen. The samples are not nucleic acids so the results in the former are not surprising. The Sodium Chloride Grid Screen is a fine screen around a narrow range of conditions, so its low success rate is also not surprising. The grid screens are incorporated within the crystallization screening cocktails for a dual purpose: (i) to relate the other results to commonly sampled conditions and (ii) to determine the effect of small chemical shifts on the sample being studied. The result with ammonium sulfate (3.67% success) is significantly different from that with sodium chloride (0.96% success). In sodium chloride, protein interactions show very little salt-dependence up to very high salt concentrations, whereas in ammonium sulfate proteins show a sharp decrease in the second virial coefficient with increasing salt concentration beyond a certain threshold (Dumetz et al., 2007 ▶). The second virial coefficient has been used as a measure of how ‘attractive’ macromolecules are to each other. Values that have been associated with a crystallization slot with values between −1 × 10−4 and −8 × 10−4 mol ml g−2 (George & Wilson, 1994 ▶) indicate protein–protein interactions that are slightly to moderately attractive (Wilson, 2003 ▶). The results here lend some evidence to the observations of a decrease in the second virial coefficient associated with crystallization. Another interesting observation is the performance of the Sodium Malonate Grid Screen. This grid screen was developed from an observation by McPherson (2001 ▶) that sodium malonate was almost twice as successful as sodium acetate, sodium tartrate, sodium formate and ammonium sulfate in crystallizing 23 different macromolecules tested. In our data sodium malonate is successful compared with sodium chloride, but still significantly less successful than ammonium sulfate. The data also illustrate the coverage provided by groups 1 and 2 of the cocktail screens in comparison to the commercial screens sampled. Groups 1 and 2 have an overall hit rate of 4.53%, compared with the Hampton Research cocktails, which average 2.98%. This should not be taken as an indication that the commercial screens are inferior. The sampling scheme used for the commercial screens is generally much sparser, sampling a wider region of chemical space, and they were designed for vapor-diffusion crystallization methods with lower precipitant concentrations than the group 1 and 2 cocktails, which were designed specifically for the batch method.
The group 1 cocktails produce very few unique hits that are not seen in other cocktails, while group 2 is a far better performer: 3.0% versus 19.1%, respectively, when normalized to account for the smaller number of cocktails in the group 1 screen. The number of unique hits within the group 2 cocktails is small, suggesting that group 2 is somewhat oversampled within the screen or that PEGs are especially effective crystallizing agents. The large number of multiple hits for each macromolecule is especially interesting. 269 macromolecules are represented of 823 that were sampled. It is unlikely, given the number of multiple hits, that the samples that did not show any leads would have produced leads if finer sampling of the same chemical crystallization space had taken place. Significantly different steps in orthogonal regions of chemical space may be more likely to lead to successful crystallization of these samples. The NESG macromolecules were more successful in producing crystal hits (∼47%) than the SGPP samples (∼27%). The success rates seen for the general biomedical community samples that come through the HTS laboratory, based on a study of 96 representative samples, is ∼51% (Snell et al., 2008 ▶). While the different success rates have yet to be analyzed in detail, an important factor may be that the NESG samples are prokaryotic while the SGPP samples are eukaryotic.
The data are based on the analysis of crystal hits only. Where no crystals were produced, the images were not categorized further, i.e. we do not know if the experiment precipitated or if it was still clear from the initial analysis. Without further analysis of this, we can only draw simple conclusions from these data. As noted in our companion paper (Snell et al., 2008 ▶), the HWI high-throughput crystallization laboratory is a unique resource. Since its inception, every macromolecule that has come through the laboratory has been screened and the results have been imaged and stored together with biochemical information using the same protocols. To date, over 10 000 macromolecular samples have been screened by the laboratory, generating over 90 million images of crystallization experiments in progress. Even with a simple analysis of crystal hits from a subset of these images, we have produced useful information on the performance of screens used for crystallization. The development of automated image analysis, aided by the training set we have established, combined with the biochemical data and incomplete factorial approach used from the outset will provide a unique insight into general crystallization behavior and trends for biological macromolecules.
Acknowledgments
The John R. Oishei Foundation, the Cummings Foundation and NIH grants U54 GM074899-01, U54 GM07495-04 and GM-64655 are acknowledged for financial support. NESG and SGPP are acknowledged for their valuable collaborations, generating the targets used in this analysis. The anonymous referees are acknowledged for helpful comments.
References
- Audic, S., Lopez, F., Claverie, J. M., Poirot, O. & Abergel, C. (1997). Proteins, 29, 252–257. [DOI] [PubMed]
- Chayen, N. E., Shaw Stewart, P. D. & Blow, D. M. (1992). J. Cryst. Growth, 122, 176–180.
- Dumetz, A. C., Snellinger-O’Brien, A. M., Kaler, E. W. & Lenhoff, A. M. (2007). Protein Sci.16, 1867–1877. [DOI] [PMC free article] [PubMed]
- George, A. & Wilson, W. W. (1994). Acta Cryst. D50, 361–365. [DOI] [PubMed]
- Luft, J. R., Collins, R. J., Fehrman, N. A., Lauricella, A. M., Veatch, C. K. & DeTitta, G. T. (2003). J. Struct. Biol.142, 170–179. [DOI] [PubMed]
- McPherson, A. (2001). Protein Sci.10, 418–422. [DOI] [PMC free article] [PubMed]
- Snell, E. H. et al. (2008). Acta Cryst. D64, 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W. W. (2003). J. Struct. Biol.142, 56–65. [DOI] [PubMed]