Multiple crystal structures are reported of cross-linked actin dimers. Interactions that are conserved across crystal structures suggest detailed interactions that are likely to be present in F-actin filaments.
Keywords: F-actin, cross-linking, actin filaments
Abstract
The structure of actin in its monomeric form is known at high resolution, while the structure of filamentous F-actin is only understood at considerably lower resolution. Knowing precisely how the monomers of actin fit together would lead to a deeper understanding of the dynamic behavior of the actin filament. Here, a series of crystal structures of actin dimers are reported which were prepared by cross-linking in either the longitudinal or the lateral direction in the filament state. Laterally cross-linked dimers, comprised of monomers belonging to different protofilaments, are found to adopt configurations in crystals that are not related to the native structure of filamentous actin. In contrast, multiple structures of longitudinal dimers consistently reveal the same interface between monomers within a single protofilament. The reappearance of the same longitudinal interface in multiple crystal structures adds weight to arguments that the interface visualized is similar to that in actin filaments. Highly conserved atomic interactions involving residues 199–205 and 287–291 are highlighted.
1. Introduction
The protein actin self-assembles to form filamentous structures that serve critical roles in the cell. Despite intense study, our understanding of the structure of filamentous actin (F-actin) is incomplete. The structure of actin in its monomeric form is known at atomic resolution (Chik et al., 1996 ▶; Kabsch et al., 1990 ▶; Klenchin et al., 2003 ▶; McLaughlin et al., 1993 ▶; Otterbein et al., 2001 ▶; Vorobiev et al., 2003 ▶; Sablin et al., 2002 ▶), while the structure of F-actin is only understood at lower resolution. The first detailed structural models of F-actin were derived by Holmes on the basis of fiber-diffraction data extending to 8.4 Å resolution (Holmes et al., 1990 ▶). A number of subsequent models have been obtained by refinement of this model under various constraints (Holmes et al., 2003 ▶; Lorenz et al., 1993 ▶; Tirion et al., 1995 ▶). Numerous electron-microscopy investigations have provided support for the essential features of the Holmes model, as well as evidence for variations on that basic model under different conditions (Hanein et al., 1998 ▶; Milligan et al., 1990 ▶; Orlova et al., 2001 ▶; Aebi et al., 1986 ▶; Hodgkinson et al., 1997 ▶; Holmes et al., 2003 ▶). These have been at somewhat lower resolution than the original fiber-diffraction experiments, but have benefited from offsetting advantages such as the inherent presence of phase information in image reconstructions from electron micrographs. However, there remains a significant discrepancy between the levels of detail at which the actin monomer and the F-actin filament are understood. An atomic resolution view of F-actin would advance efforts to understand its complex and dynamic behavior in the cell.
The F-actin filament is composed of actin monomers arranged in two protofilaments that twist about each other. According to the terms used here, adjacent actin monomers within the same protofilament are related to each other longitudinally. Two such subunits are related to each other by a 27° rotation and a 55 Å shift along the axis of the filament, with values that vary somewhat between experiments (fiber-diffraction studies give 27.2° and 55.2 Å). The interface through which they interact is the longitudinal interface. While variable twist angles of filaments have been well documented (Egelman et al., 1982 ▶; Stokes & DeRosier, 1987 ▶; Kim, Bobkova et al., 1998 ▶; Galkin et al., 2001 ▶), in general the two protofilaments in the F-actin filament are staggered so that two adjacent monomers from different protofilaments are related to each other by a rotation of about ∼166° and by a shift along the rotation axis of about 27 Å. We refer to the relationship or the interface between two monomers related in this way as the lateral relationship or the lateral interface. Applying the lateral operation twice in succession gives the longitudinal relationship between two monomers belonging to the same protofilament.
Obtaining a high-resolution structure of F-actin has been an elusive challenge. X-ray crystallography provides a common route to high-resolution structures, but crystallizing a protein that forms helical assemblies presents a conundrum. On one hand, if the helical assembly is allowed to form then three-dimensional crystal growth is generally precluded. On the other hand, if polymerization is prevented then the monomeric structure may be crystallized, but critical information about the helical assembly is lost. One approach to this problem has been to crystallize small oligomers of actin, with the idea that the structures of oligomers could reveal the binding interfaces between actin monomers (Bubb et al., 2002 ▶; Dawson et al., 2003 ▶; Klenchin et al., 2005 ▶; Kudryashov et al., 2005 ▶). Defined oligomers of actin can be obtained by limited cross-linking in the filamentous state, followed by depolymerization and purification. Chemical strategies have been developed for specific cross-linking in either the longitudinal or lateral directions (Hegyi et al., 1998 ▶; Kim, Bobkova et al., 1998 ▶; Kim, Phillips et al., 1998 ▶; Knight & Offer, 1978 ▶). The first structural study of a cross-linked actin oligomer was of a trimer held together by two lateral cross-links, with polymerization blocked by a gelsolin segment 1 (GS1) bound to each protomer (Dawson et al., 2003 ▶). There were no direct cross-links between the first and third monomers, but these were intended to be longitudinally related. However, the resulting structure showed that the three subunits were not in a configuration compatible with models of native F-actin. The lateral subunit interactions, which are weaker than those between GS1 and actin, could not maintain the three monomers in an F-actin-like state.
Following work on the laterally linked trimer, the crystal structure of a longitudinally cross-linked actin dimer was reported (Kudryashov et al., 2005 ▶). In this study, the two actin monomers were observed to be related to each other in a fashion similar to the way monomers are arranged in prevailing models of the F-actin filament derived from fiber-diffraction and electron-microscopy studies. The deviations between the subunit configurations seen in the crystal structure of the dimer and those in models of the filament were in the range 5–10 Å. Arguments were presented that this was within the bounds of experimental uncertainty in fiber-diffraction and electron-microscopy studies and that the longitudinal interface visualized in the crystal structure of the actin dimer provided a faithful view of at least some of the atomic interactions between actin monomers in the native F-actin filament. This argument was supported by parallel findings reported prior to the structure of the cross-linked actin dimer, in which two actin monomers were observed in a very similar relationship in a crystal of actin bound to a formin homology 2 domain (FH2; Otomo et al., 2005 ▶). In both of those cases, the longitudinally related actin monomers were related to each other by pure translations of approximately 54 Å. No rotation was present between the actin monomers, as the two monomers were related by translational shifts within their respective crystal lattices. While the absence of the expected 28° twist between monomers presented a challenge to interpreting these structures in terms of a twisted filament, the observation of a similar interface in two distinct crystal forms lent support to the argument that the interaction is an energetically significant one. In addition, a model-building study demonstrated that it is possible to construct a model of F-actin that contains the appropriate twist between monomers while nearly maintaining the atomic interactions between monomers visualized in the crystal structures of longitudinally related actin molecules (Kudryashov et al., 2005 ▶).
Two subsequent studies have led to additional independent crystal forms of actin in which the monomers are again related by pure translation in the longitudinal direction with an interface similar to that described earlier (Allingham et al., 2005 ▶; Rizvi et al., 2006 ▶). Finally, another actin structure has been reported in which two actin monomers are present in one asymmetric unit of the crystal (Klenchin et al., 2006 ▶). In this case, the monomers were not related to each other by pure translation. A 20° twist between monomers was observed, but the axis of rotation deviates in direction from the axis of rotation in the native F-actin filament by approximately 60°. Despite this deviation, the structure provides yet another similar view of the longitudinal interface between actin monomers.
Here, we describe four additional structures of cross-linked actin dimers in new crystal forms. Consistent with previous studies by others on lateral cross-linking (Dawson et al., 2003 ▶), we find that laterally cross-linked dimers, even those with only a small toxin bound to them, do not adopt arrangements similar to those in the native actin filament. In contrast, two new crystal forms of longitudinally linked actin dimers reveal an interface that can be accommodated in F-actin and which is very similar to that which has now been observed multiple times. A comparison is provided of the available views of the longitudinal interface arising from crystal structures and filament models of actin. The data suggest a consensus regarding a set of specific atomic interactions between actin monomers in the filament.
2. Experimental procedures
2.1. Preparation of cross-linked dimers
The longitudinal dimer of rabbit skeletal actin cross-linked with N-(4-azido-2-nitrophenyl)putrescine (ANP) and labeled with TMR was prepared and purified as described previously (Kudryashov et al., 2005 ▶). The subtilisin-cleaved longitudinal dimer was prepared by incubating 6.9 mg ml−1 TMR-ANP-actin dimer with 0.014 mg ml−1 subtilisin [500:1(w:w) ratio] for 70 min at 295 K. The reaction was stopped by the addition of 0.5 mM PMSF. Under these conditions, both actin monomers in the dimer were cleaved between residues 47 and 48 as judged by the appearance of characteristic bands on an SDS gel. Despite this, electrophoresis under native conditions revealed a single band, indicating that the actin dimer remains intact.
The lateral dimer cross-linked by N,N′-p-phenylenedimaleimide (pPDM) was prepared as described previously (Knight & Offer, 1978 ▶). To purify dimers, the mixture of cross-linked actin species of different sizes was first dialyzed with three to four changes of G-actin buffer (2.0 mM Tris–HCl pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, 5 mM β-mercaptoethanol, 0.1 mM PMSF) for at least 5 d in total. Actin was then centrifuged in a TLA 60 rotor at 45 krev min−1 for 1.5 h to separate depolymerized and filamentous fractions of actin. The supernatant was applied onto a Sephacryl S-200 2.5 × 90 cm column equilibrated with G-actin buffer. To ensure better separation, the column was connected to itself, bottom to top, and run for 3 d at a flow rate that was adjusted so that the protein would go through three to four complete column volumes. Fractions containing pure pPDM cross-linked dimer were combined, concentrated, supplemented with a 1.5 molar excess of kabiramide C (KabC) and used for crystallization.
2.2. Crystallization and structure determination
All crystallizations were performed at room temperature using hanging-drop vapor diffusion. Diffraction data for one of the crystals (the laterally cross-linked pPDM dimer in space group P212121 with unit-cell parameters a = 71.1, b = 70.3, c = 75.2 Å) were collected using a Rigaku RU-200 rotating-anode X-ray generator equipped with an R-AXIS IV++ imaging-plate detector. All remaining X-ray diffraction data sets were collected at Advanced Light Source (ALS) beamline 8.2.2 equipped with an ADSC Quantum 315 CCD detector. Data were collected at 100 K.
2.2.1. Crystallization conditions for the longitudinal dimer
The drop consisted of 2.5 µl actin dimer (complexed with FH2, latrunculin A and AMPPNP) solution, 2.0 µl reservoir solution and 0.5 µl 0.1 M l-cysteine as an additive. The actin dimer complex was prepared by mixing 3 mg ml−1 actin dimer with a 1.1 molar excess of FH2. Latrunculin A was added in 1.5 molar excess over actin monomer and AMPPNP was added to a 0.001 M final concentration. The reservoir solution was prepared in a volume of 500 µl and consisted of 30% MPD, 0.1 M sodium acetate pH 4.6 and 0.02 M calcium chloride. The crystals grew as rectangular plates and belonged to space group P21, with two actin monomers in the asymmetric unit. Crystals were flash-frozen in a nitrogen-gas cryostream without the need for additional cryoprotectant. Data extending to 2.7 Å were collected at a wavelength of 1.0 Å.
2.2.2. Crystallization conditions for the subtilisin-cleaved longitudinal dimer
The drop consisted of 1 µl actin dimer (complexed with latrunculin A and ATP) solution and 1.0 µl reservoir solution. The actin dimer complex was prepared by mixing 3 mg ml−1 actin dimer with a 1.25 molar excess of latrunculin A. ATP was included at a 0.001 M final concentration. The reservoir solution was prepared in a volume of 500 µl and consisted of 30% MPD, 0.1 M sodium acetate pH 4.6 and 0.01 M calcium chloride. The crystals were isomorphous to the uncleaved longitudinal dimer described above. X-ray diffraction data were collected on the same beamline, but with noticeably stronger diffraction anisotropy. Data were deleted that fell outside the boundary of an ellipsoid in reciprocal space whose principal axes had lengths of 1/(2.7 Å), 1/(3.7 Å) and 1/(3.0 Å) along a*, b* and c*, respectively, using a procedure described previously (Strong et al., 2006 ▶).
2.2.3. Crystallization conditions for the lateral dimer
The drop consisted of 0.2 µl actin dimer (complexed with KabC and ATP) solution and 0.2 µl reservoir solution. The actin dimer complex was prepared by mixing 3 mg ml−1 actin dimer with a 1.5 molar excess of Kab C. ATP was included at 0.001 M final concentration. The reservoir solution was prepared in a volume of 100 µl and consisted of 30% PEG 4000, 0.1 M Tris–HCl pH 8.5 and 0.2 M lithium sulfate.
2.2.4. Structure solution and refinement
The crystal structures were determined by molecular replacement using the program Phaser (McCoy et al., 2005 ▶), with a single protomer of rabbit actin (PDB code 2a5x) serving as the search model, and subsequently refined. Model building was performed using the graphics program Coot (Emsley & Cowtan, 2004 ▶). To benefit from the use of TLS refinement (Winn et al., 2001 ▶), the models were refined with the program REFMAC5 (Murshudov et al., 1997 ▶). The geometric quality of the models was assessed with the following structure-validation tools: ERRAT (Colovos & Yeates, 1993 ▶), PROCHECK (Laskowski et al., 1993 ▶) and WHAT IF (Vriend & Sander, 1991 ▶). Protein structures were illustrated using the program PyMOL (DeLano, 2002 ▶). The structural superimpositions illustrated in Figs. 1, 2 and 4 were computed with SSM (Krissinel & Henrick, 2004 ▶) within the Coot interface.
2.3. Structure comparisons
Atomic coordinates from distinct actin crystal structures were superimposed using the program LSQMAN (Kleywegt & Jones, 1994 ▶) using a 2 Å threshold for the overlap of equivalent atoms. The optimal rotations between sets of equivalent atoms in the actin dimer interface were determined as described by Kabsch (1976 ▶). For comparisons of the different instances of the longitudinal interface, the following 35 residues from the top region of the lower actin monomer were used for superposition: 35–38, 66–69, 187, 190, 193–209 and 242–249. The 28 residues used from the bottom region of the upper monomer were 149, 166–167, 171, 282–283, 285–296, 314 and 321–328.
Coordinate sets were analyzed using principal component analysis as first described by Diamond (1974 ▶). Briefly, coordinate sets were first shifted to have their centers of mass at the origin. Each coordinate set of n atoms was then treated as a single column vector of length 3n. Vectors from each of the m coordinate sets were used to construct a 3n × m matrix A. The eigenvalues and eigenvectors were then extracted from the 3n × 3n semi-positive definite matrix AA T. The vector for each coordinate set was then projected onto the first two eigenvectors, with the results shown in Fig. 2(c). In the calculations performed here, n was 28 and m was 9. The top two eigenvectors accounted for 74% of the variance between the nine coordinate sets.
To evaluate conserved interactions, the closest approach between residue i in the lower monomer and residue j in the upper monomer was calculated for each structure of the longitudinal interface visualized. The values were calculated as the shortest distance between the centers of any two atoms belonging to the residues in question. The calculations were restricted to the list of residues in the interface noted above. Contact maps were prepared to illustrate the pattern of contacts between residues on the two sides of the interface. These were colored by dividing the range of distances into 11 bins, with ten cutoff values from 5.4 to 12 Å; the ten cutoff values were spaced equally in terms of their squared values in order to better spread the observed distances into separate bins. To tabulate close contacts, pairs of residues were identified whose closest approach was less than 4.5 Å and the degree of conservation of these contacts across the various instances of the longitudinal interface was evaluated.
3. Results
3.1. Formation of cross-linked dimers
Two different kinds of actin dimers were prepared: longitudinally linked and laterally linked. Following previously reported methods, the longitudinal construct was prepared by cross-linking Gln41 from the bottom monomer to Cys374 in the upper monomer using a heterobifunctional photo-activated reagent, ANP [N-(4-azido-2-nitrophenyl)putrescine; Hegyi et al., 1998 ▶; Kim, Bobkova et al., 1998 ▶; Kim, Phillips et al., 1998 ▶]. This dimer has been studied extensively and its atomic structure has already been reported in one crystal form (Kudryashov et al., 2005 ▶). The relationship between monomers in the dimer is compatible with that found in F-actin, as suggested by our modeling studies (Kudryashov et al., 2005 ▶). Furthermore, in solution these dimers can reassemble into filaments which exhibit similar kinetic and equilibrium parameters for the S1 myosin interaction with actin, including the K m and V max values of the actomyosin ATPase (Kim et al., 2002 ▶).
In the lateral dimer, Lys191 is linked to Cys374 (presumably from the monomer belonging to the opposite protofilament) using a different homobifunctional reagent, pPDM (N,N′-p-phenylenedimaleimide; Kim et al., 2002 ▶; Knight & Offer, 1980 ▶). The ability of purified covalently cross-linked actin oligomers to nucleate actin assembly has been described (Lal et al., 1984 ▶), showing that in solution such dimers and oligomers can adopt state(s) compatible with F-actin structure. Crystallization experiments were undertaken for both types of dimers under a variety of conditions and treatments.
3.2. Crystal structures of the lateral dimer
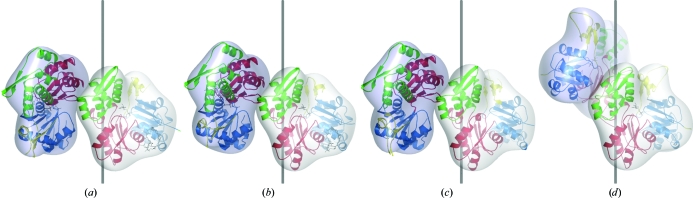
The lateral dimer crystallized in two different crystal forms (Table 1 ▶). The first crystal form included ATP, Ca2+ and kabiramide C (KabC). KabC is an actin-specific macrolide toxin which inhibits fibril growth in the longitudinal direction by binding and blocking one surface of the longitudinal interface (a patch between subdomains 1 and 3). Diffraction data were collected with 98.5% completeness to a resolution of 2.2 Å. The structure was solved using molecular replacement and refined to R work and R free values of 0.165 and 0.227, respectively (Table 2 ▶). However, the pPDM cross-link was not visible in difference electron-density maps. In the crystal, the two actin monomers that are inferred by their proximity to be cross-linked to each other are related by a 21 screw axis in the crystal. The arrangement of the two monomers deviates by approximately 90° from the configuration expected for a lateral F-actin dimer (Fig. 1 ▶). It was determined that there were no significant differences between this structure and a previously deposited entry in an isomorphous crystal form (PDB code 1qz5; Klenchin et al., 2003 ▶), so these coordinates were not deposited in the Protein Data Bank.
Table 1. Crystal structures containing laterally cross-linked dimers.
| Reference | This work | This work† | Dawson et al. (2003 ▶) |
|---|---|---|---|
| PDB code | 2q36 | N/A | 1mdu |
| Crystallization components | KabC, ATP, Ca2+ | KabC, ATP, Ca2+ | ATP, Ca2+ |
| Cross-link | pPDM | pPDM | pPDM |
| Space group | P212121 | P212121 | P21 |
| Resolution (Å) | 2.5 | 2.2 | 2.2 |
| Unit-cell parameters | |||
| a (Å) | 40.5 | 71.1 | 67.2 |
| b (Å) | 74.2 | 70.3 | 75.9 |
| c (Å) | 144.4 | 75.2 | 96.7 |
| β (°) | 91.8 | ||
| Actin monomers per ASU | 1 | 1 | 2 |
Table 2. X-ray data-collection and refinement statistics.
Values in parentheses are for the last shell.
| Data-collection statistics | ||||
| Cross-link | Longitudinal by ANP | Longitudinal by ANP | Lateral by pPDM | Lateral by pPDM |
| Subtilisin cleavage | No | Yes | No | No |
| Additional components | Latrunculin, AMPPNP, Ca2+, TMR† | Latrunculin, TMR, ATP, Ca2+ | KabC, ATP, Ca2+ | KabC, ATP, Ca2+ |
| Space group | P21 | P21 | P212121 | P212121 |
| Resolution (Å) | 2.7 (2.80–2.70) | 2.7 (2.80–2.70) | 2.4 (2.49–2.40) | 2.2 (2.28–2.20) |
| Unit-cell parameters | ||||
| a (Å) | 108.1 | 108.2 | 40.5 | 71.1 |
| b (Å) | 71.8 | 71.9 | 74.2 | 70.3 |
| c (Å) | 54.8 | 54.7 | 144.4 | 75.2 |
| β (°) | 104.7 | 104.0 | ||
| Monomers per ASU | 2 | 2 | 1 | 1 |
| No. of images collected | 181 | 180 | 225 | 203 |
| Oscillation range per image (°) | 1.0 | 1.0 | 1.0 | 1.0 |
| Exposure time (s) | 4 | 5 | 4 | 300 |
| Measured reflections | 81583 | 56566 | 139584 | 155978 |
| Unique reflections | 23524 | 19970 | 17711 | 19510 |
| Completeness (%) | 99.5 (97.1) | 87.5 (55.8) | 99.9 (99.8) | 98.5 (96.8) |
| Rmerge‡ | 0.067 (0.315) | 0.140 (0.336) | 0.085 (0.500) | 0.093 (0.420) |
| I/σ(I) | 13.6 (2.1) | 5.0 (1.0§) | 17.5 (4.0) | 17.8 (4.6) |
| Refinement statistics | ||||
| Rwork | 0.228 | 0.249 | 0.175 | 0.165 |
| Rfree | 0.277 | 0.288 | 0.223 | 0.227 |
| No. of reflections used for Rfree | 1147 | 764 | 800 | 1009 |
| R.m.s.d. bond lengths (Å) | 0.009 | 0.006 | 0.008 | 0.012 |
| R.m.s.d. bond angles (°) | 1.46 | 1.04 | 1.53 | 2.12 |
| PDB code | 2q1n | 2q31 | 2q36 | Isomorphous to 1qz5 |
FH2 was also included but was not observed in electron-density map.
R
merge = 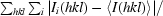
 .
.
Statistics reported before ellipsoidal truncation of anisotropic data.
Figure 1.
Crystal structures of laterally cross-linked actin dimers showing subunit rearrangements that are not consistent with the arrangements of actin monomers in the F-actin filament. (a) A structure of the lateral dimer complexed with KabC determined in space group P212121 (see Table 1 ▶). (b) A second structure of the lateral dimer complexed with KabC, also in space group P212121 but with different unit-cell parameters to (a). It was found to be isomorphous to a previously reported structure of monomeric actin (PDB code 1qz5; Klenchin et al., 2003 ▶). (c) A third lateral interaction (also cross-linked by pPDM) reported in an actin trimer (Dawson et al., 2003 ▶). (d) A lateral pair of actin monomers from the Holmes model of the F-actin filament (Holmes et al., 1990 ▶) shown for reference. Two actin monomers are shown, each colored according to the actin subdomains (subdomains 1, 2, 3 and 4 are colored blue, yellow, red and green, respectively).
The second laterally cross-linked crystal form also contained KabC, Ca2+ and ATP. Diffraction data were collected with 99.9% completeness to 2.5 Å resolution. The structure was solved using molecular replacement and refined to an R work of 0.175 and an R free of 0.223 (Table 2 ▶). The pPDM cross-link was again not visible in difference electron-density maps, but the two actin monomers presumably cross-linked in this crystal were likewise related by a 21 screw axis and were oriented in nearly the same fashion as in the crystal form described above (Fig. 1 ▶). Furthermore, the arrangement observed in the two crystal forms reported here was found to be similar to that first observed by Dawson and coworkers for the laterally linked trimer bound to GS1 (Dawson et al., 2003 ▶). Although this arrangement has now been observed in three different crystal forms of actin and thus may reflect a possible mode of actin–actin interaction, it does not resemble the native filamentous form of actin. Neither is it similar to the ribbon-like arrangement described for actin molecules in complex with profilin (Schutt et al., 1993 ▶). These laterally linked actin structures are therefore not discussed further in the present study.
3.3. Crystal structures of the longitudinal dimer
Two crystals obtained for the longitudinally linked dimer were distinct from the crystal form reported previously (Kudryashov et al., 2005 ▶). The first crystal obtained was grown in the presence of formin homology 2 domain (FH2). It was reasoned that the addition of FH2 might stabilize a pair of longitudinally linked dimers in the crystal, analogous to its role in the cell as a nucleation factor for the growth of actin filaments. A nonhydrolyzable ATP analog, adenylyl-imidodiphosphate (ANPPNP), was also added, along with latrunculin, which inhibits polymerization in the longitudinal direction by tethering subdomains 2 and 4 within a single protomer. The crystals belong to space group P21 with unit-cell parameters a = 108.1, b = 71.8, c = 54.8 Å, β = 104.7° (Table 2 ▶). The structure was solved using molecular replacement and refined at a resolution of 2.7 Å with final R work and R free values of 0.228 and 0.277, respectively (Table 2 ▶). The FH2 domain was evidently excluded from the crystal, as it did not appear in the electron-density map. Although the ANP cross-link was similarly invisible in difference electron-density maps, the arrangement of the two monomers in the asymmetric unit suggests that they are cross-linked not to each other but to monomers related by a 54.7 Å unit-cell translation. This crystal form therefore contains two independent linear arrays of actin molecules. These two longitudinally linked arrays of molecules in the crystal are nearly identical to each other in structure.
The second crystal of the longitudinally linked dimer was obtained following subtilisin cleavage between actin residues Met47 and Gly48 of the flexible subdomain 2. ATP and latrunculin were included in the crystallization. The hope here was that the cleaved segments of subdomain 2 would be less constrained by other factors (allosteric changes owing to latrunculin, lattice forces etc.) from forming contacts with residues in subdomain 1 above. The resulting crystals were nearly isomorphous to that described above (space group P21, unit-cell parameters a = 108.2, b = 71.9, c = 54.7 Å, β = 104.0°). However, the crystals containing the cleaved dimer were distinguished by stronger anisotropy, diffracting to 2.3 Å along the a* direction but to only 3.7 Å along b*. Diffraction data were collected to 2.7 Å resolution with 87.5% completeness. The structure was refined to an R work of 0.249 and an R free of 0.288 (Table 2 ▶). The structures of both the cleaved and uncleaved longitudinal dimer were very similar. Both contain bound nucleoside triphosphate and latrunculin. Residues 38–65 (corresponding to most of subdomain 2) were also disordered in both.
Residues that comprise the longitudinal interface include residues 199–205 from subdomain 4 (in the lower protomer) and residues 287–291 from subdomain 3 (in the upper protomer). The detailed atomic interactions visualized are discussed subsequently. Subdomain 2 from the lower monomer is also believed to make important interactions with the monomer above. However, a large region of subdomain 2 (residues 38–65) is missing from the present structural model owing to crystal disorder; subdomain 2 has been found to be disordered to varying degrees in previous structures. As a result, the interface visualized here can only account for part of the native interface.
3.4. Comparison of the longitudinal interface observed in multiple structures
A number of comparisons were made of the interface between longitudinally related actin monomers visualized in various crystals and filament models. Eight different coordinate sets were prepared for the longitudinal actin dimer. One of these coordinate sets was obtained from the new crystal form of the cross-linked actin dimer described above. One coordinate set was obtained from the crystal structure of the cross-linked actin dimer reported previously (Kudryashov et al., 2005 ▶); this crystal is isomorphous with one obtained from a noncross-linked actin mutant in complex with either ATP or ADP (Rould et al., 2006 ▶). Four other coordinate sets were obtained from structures in which actin monomers were not cross-linked to each other but were nonetheless seen to form a similar interface between longitudinally related monomers in the crystal (Allingham et al., 2005 ▶; Klenchin et al., 2006 ▶; Otomo et al., 2005 ▶; Rizvi et al., 2006 ▶; Table 3 ▶). These six models are derived from six independent crystal forms whose unit-cell parameters and crystal contacts (apart from those representing the longitudinal interface) are distinct. Finally, two coordinate sets were obtained from models of the F-actin filament. The first of these was the canonical Holmes model (Holmes et al., 1990 ▶). The other was a model of the F-actin filament constructed by applying the known helical parameters of F-actin while attempting to maintain the atomic interactions visualized in the first crystal structure of the longitudinally cross-linked actin dimer as described previously (Kudryashov et al., 2005 ▶). The overall similarities and differences between the models are shown in Fig. 2 ▶(a).
Table 3. Crystal structures with longitudinal interfaces.
| Label† | A | — | B | C | D | E | F |
|---|---|---|---|---|---|---|---|
| Reference | This work | This work | Kudryashov et al. (2005 ▶) | Otomo et al. (2005 ▶) | Allingham et al. (2005 ▶) | Rizvi et al. (2006 ▶) | Klenchin et al. (2006 ▶) |
| PDB code | 2q1n | 2q31 | 2a5x | 1y64 | 2aso | 2fxu | 2hmp |
| Crystal information | |||||||
| Ligands and additives | Latrunculin, AMPPNP, Ca2+ (FH2 not observed) | Subtilisin-cleaved actin, latrunculin AMPPNP, Ca2+ | Latrunculin, AMPPNP, Ca2+ | ATP, Ca2+,, FH2 | Sphinxolide, ATP, Ca2+, | Bistramide A, ATP, Ca2+, | Protease ECP32-cleaved actin, ATP, Sr2+ |
| Cross-link | Longitudinal by ANP | Longitudinal by ANP | Longitudinal by ANP | None | None | None | None |
| Space group | P21 | P21 | C2 | C2 | C2 | C2 | P212121 |
| Resolution (Å) | 2.7 | 2.7 | 2.5 | 3.05 | 1.7 | 1.35 | 1.9 |
| Unit-cell parameters | |||||||
| a (Å) | 108.1 | 108.2 | 207.3 | 232.0 | 58.2 | 60.1 | 63.9 |
| b (Å) | 71.8 | 71.9 | 54.4 | 56.2 | 55.0 | 56.5 | 198.1 |
| c (Å) | 54.8 | 54.7 | 36.2 | 100.9 | 103.5 | 101.6 | 69.6 |
| β (°) | 104.7 | 104.0 | 98.6 | 107.7 | 92.4 | 94.6 | |
| Actin monomers per ASU | 2 | 2 | 1 | 1 | 1 | 1 | 2 |
| Other crystallization components | 30% MPD, 0.1 M sodium acetate pH 4.6 | 30% MPD, 0.1 M sodium acetate pH 4.6 | 35% MPD, 0.1 M sodium acetate pH 4.7 | 40 mM Tris–HCl pH 7.6 | 12% PEG 5000 methyl ether, 0.1 M sodium/MES/acetate pH 5.5, 1 mM TCEP | 24% PEG 15K, 1 M MES pH 6.0, 1 mM TCEP | 10% ethylene glycol, 15% dimethyl PEG 5000, 50 mM triethanolamine, 10 mM spermidine pH 7.75 |
Coordinate sets are labeled for subsequent comparisons.
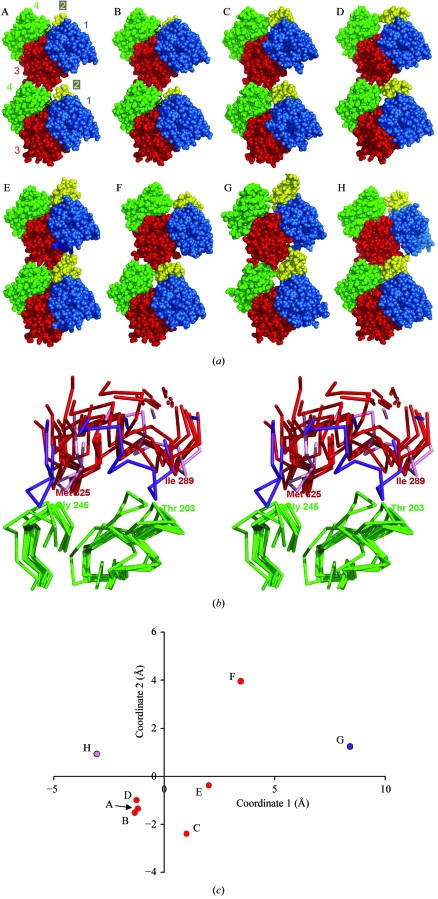
Figure 2.
New crystal structure of the longitudinally cross-linked actin dimer and a comparison to previously reported arrangements of actin molecules. (a) Eight models showing similar arrangements of longitudinally related actin monomers. The actin subdomains are colored as in Fig. 1 ▶. The models are labeled A–H as in Tables 3 ▶ and 4 ▶. Where present, the bound nucleotide is shown in black. (b) Stereoview showing the protein backbones of the eight models of the longitudinal interface. The lower monomers have been superimposed. Subdomain 4 of the lower monomer is shown in green. Subdomain 3 of the upper monomer is shown in red for the six crystal structures and in purple and pink for the two filament models G and H, respectively. (c) The degree of variation between the different instances of the longitudinal actin interface, based on a principal component analysis. The atomic positions analyzed are those of the atoms in the bottom region of the upper monomer after the lower monomers had been superimposed. The plotted positions of the various models give an indication of the r.m.s. deviation between them (see Table 4 ▶).
To make comparisons possible between different instances of the longitudinal interface, the bottom actin monomers from all the models were first superimposed on each other. Whatever transformations were applied to the bottom monomers in order to overlap them, corresponding transformations were applied to the top monomers. Following this strategy, differences between different longitudinal interfaces are manifested by differences between the coordinates of the top monomers derived from the eight different models. In order to avoid potential problems arising from flexibility and structural deviations of the actin monomer from different models, the superposition and comparisons were focused on the regions of actin nearest the interface. In particular, only residues that were within 12 Å of another residue in the opposing actin monomer were used for comparison; the longitudinal dimer structure reported here was used as the basis for this evaluation. There were 35 such residues from the top region of the lower monomer and 28 residues from the bottom region of the upper monomer. The overlapped coordinates are shown in Fig. 2 ▶(b).
The variations between different instances of the longitudinal interface were evaluated in a pairwise fashion by considering Cα atoms from the upper monomer and calculating the r.m.s. distance between corresponding atoms derived from two different models (Table 4 ▶). The interfaces derived from the six crystal structures are highly similar to each other overall. For example, the r.m.s. difference is only 0.5 Å between the interfaces visualized in the cross-linked actin dimer reported here and the structure of the cross-linked dimer reported previously in a different crystal form (Kudryashov et al., 2005 ▶). Close similarities are also seen involving structures in which longitudinally related actin monomers are not cross-linked together. For example, the interface between two monomers in the crystal structure of actin bound to sphinxolide B (PDB code 2aso) is only 0.7 Å different from that reported here. Some of the crystal structure interfaces differ by greater amounts. The greatest difference between two crystal structure interfaces in which the actin monomers are related by pure crystal lattice translations is 3.7 Å (Table 4 ▶). Greater differences are seen in comparisons involving the one instance (PDB code 2hmp) in which the longitudinally related monomers are not related by a lattice translation and so are permitted to have some twist between them. The greatest difference between this interface and another one derived from a crystal structure is 7.3 Å (Table 4 ▶). These values illustrate the range of similarities and differences in the multiple atomic resolution views of the longitudinal actin interface as it has been observed across numerous crystal forms.
Table 4. Deviations between different instances of the longitudinal actin interface as visualized in multiple crystal structures and in models of the F-actin filament.
The values reported are r.m.s. distances (in Å) between corresponding Cα atoms. Models A–F are labeled as in Table 3 ▶. Model G is the Holmes filament model (Holmes et al., 1990 ▶). Model H is a filament model based on an actin dimer crystal (Kudryashov et al., 2005 ▶).
| A | B | C | D | E | F | G | H | |
|---|---|---|---|---|---|---|---|---|
| A | 0.0 | 0.5 | 2.7 | 0.7 | 3.5 | 7.1 | 11.0 | 4.9 |
| B | 0.5 | 0.0 | 2.9 | 0.6 | 3.7 | 7.3 | 11.0 | 5.0 |
| C | 2.7 | 2.9 | 0.0 | 3.0 | 2.6 | 6.9 | 9.8 | 6.7 |
| D | 0.7 | 0.6 | 3.0 | 0.0 | 3.5 | 6.9 | 10.9 | 4.9 |
| E | 3.5 | 3.7 | 2.6 | 3.5 | 0.0 | 4.7 | 8.5 | 7.1 |
| F | 7.1 | 7.3 | 6.9 | 6.9 | 4.7 | 0.0 | 7.1 | 8.3 |
| G | 11.0 | 11.0 | 9.8 | 10.9 | 8.5 | 7.1 | 0.0 | 12.4 |
| H | 4.9 | 5.0 | 6.7 | 4.9 | 7.1 | 8.3 | 12.4 | 0.0 |
On the whole, the crystal structure views of the longitudinal interface are consistent with the arrangements of the actin monomers in models of the F-actin filament obtained from fiber diffraction and electron-microscopy image reconstructions. Taking the Holmes model as a reference, the differences between the longitudinal interface in that model and the various crystal structures described above range from 7.1 to 11 Å (Table 4 ▶). This range is comparable to the fiber-diffraction and electron-microscopy image resolutions on which such models have been based. The differences observed between crystal structures and helical filament models can be attributed to a combination of two factors: lower precision in the helical models and potential artifacts in the crystal structures, especially owing to the absence of twist in the latter. Interestingly, the interface in the Holmes model is closest to the one crystal interface (Klenchin et al., 2006 ▶) in which a twist is permitted between the two actin monomers. Another model differing somewhat from the original Holmes model was proposed recently based on an attempt to preserve the longitudinal interface observed in a crystal (Kudryashov et al., 2005 ▶). This filament model differs from the crystal structure interfaces by amounts ranging from 4.9 to 8.3 Å.
In order to illustrate the degree of similarity between the different instances of the longitudinal interface, the coordinates of the various models were analyzed by principal component analysis (PCA). This mathematical technique provides an optimal view of which interfaces are most similar to each other and which are most distinct. The eight models of the longitudinal interface discussed above are plotted in Fig. 2 ▶(c). The six crystal structure interfaces cluster mainly in one region of the diagram, with the exception of the interface in which the actin monomers are twisted relative to each other (PDB code 2hmp). This structure is somewhat distinct from the other crystal structure interfaces and is slightly closer to the original Holmes model of the filament, as noted above.
3.5. Conserved atomic interactions in the longitudinal interface
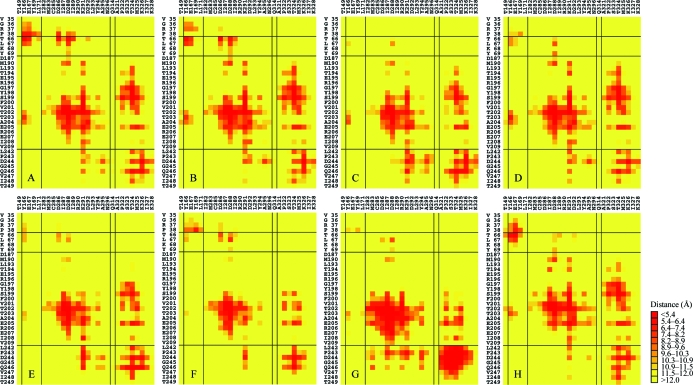
The comparisons provided above were based on the positions of the protein backbone in the longitudinal interface. A closer examination of the multiple structures suggests that the level of conservation in various interfaces is even greater when detailed interactions involving the amino-acid side chains are considered. Although the backbone differences reach 7 Å between some pairs of crystal structure interfaces, certain side-chain interactions are preserved across the crystal forms. This finding was analyzed by calculating the closest approach between amino-acid residues in the top region of the lower monomer and residues in the bottom region of the upper monomer. These results are displayed in the form of a contact map in Fig. 3 ▶. All eight instances of the longitudinal interface are shown.
Figure 3.
Contact maps highlighting the regions of interaction in the various instances of the longitudinal actin interface visualized in crystal structures and filament models. Interfaces from eight different models are shown, labeled A–H as in Tables 3 ▶ and 4 ▶. Regions of close contact are shown in red. Residues belonging to the top region of the lower monomer are indicated along the left side of each map, while residues belonging to the bottom region of the top monomer are indicated along the top. The six models from crystal structures (A–F) show especially strong similarities. The color map shown divides the range of contact distances between 4 and 12 Å into 11 bins, with a uniform spacing in the squared distance.
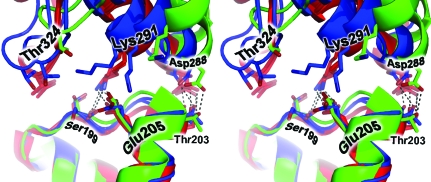
The extent of the similarity between the six crystal structure interfaces is striking. Numerous close interactions are conserved across all or most of the structures (Table 5 ▶). Residue pairs that came within 4.5 Å of each other in at least four of the six structures were tabulated. The list of highly conserved contacts includes six interactions, all between residues belonging to the 199–205 segment of the lower monomer and residues belonging to the 287–291 segment in the upper monomer (Table 5 ▶). Among this list are three interactions that are conserved across all six of the structures, despite the differences between the backbone coordinates described above. Among the closest and most conserved interactions is a hydrogen bond between Thr203 and Asp288. Fig. 4 ▶ illustrates some of the conserved interactions observed across the multiple crystal structures. Of the six contacts that are conserved in at least four crystal structures, three are present in the Holmes filament model (although there are deviations in the side-chain geometries) while three are not (Table 5 ▶).
Table 5. Contacts in the longitudinal interface that are strongly conserved across crystal structures.
Residues in column 1 are in the top region of the lower monomer, while those in column 2 are in the bottom region of the upper monomer.
| Residue 1 | Residue 2 | Interaction | Conservation† |
|---|---|---|---|
| Ser199 | Lys291 | Hydrogen bond | 4 |
| Val201 | Ile287 | van der Waals | 4‡ |
| Thr202 | Ile287 | van der Waals | 6‡ |
| Thr202 | Asp288 | van der Waals | 6 |
| Thr203 | Asp288 | Hydrogen bond | 6‡ |
| Glu205 | Lys291 | Salt bridge | 4 |
The value given is the number of crystal structure models (out of a maximum of six) in which the two residues listed are within 4.5 Å of each other. Only contacts with a conservation of four or higher are listed. The interaction types are based on the crystal structures in which they have been observed.
Residues that meet the 4.5 Å contact criterion in the Holmes model of the filament.
Figure 4.
A stereoview of some of the conserved atomic interactions in the longitudinal actin interface. Interfaces from the six independent crystal structures are shown. Shown in red are three structures (models A, B and D) that group together in the lower left quadrant of the principal component analysis plot (Fig. 2 ▶ c). Shown in blue are two structures (C and E) that fall in the lower right quadrant of the principal component analysis plot. Shown in green is the single crystal structure (F) that appears in the upper right quadrant of the principal component analysis plot. Highlighted interactions include the frequently observed hydrogen bond between Thr203 and Asp288.
4. Discussion and conclusions
Crystal structures often provide valuable information about the natural associations and interactions between macromolecules. However, macromolecules also contact each other in the crystalline state in fortuitous ways that may not relate to their natural biological forms. Interpreting interactions between molecules in the crystalline state is therefore a challenging problem. Considerable attention has been given to the problem of determining the biological relevance of protein interactions visualized in individual crystal structures (Ponstingl et al., 2000 ▶; Bahadur et al., 2004 ▶; Guharoy & Chakrabarti, 2005 ▶; Liu et al., 2006 ▶). Major advantages are gained when multiple crystal forms of a particular protein are available. When very similar interactions are observed across different crystal forms, it provides independent lines of evidence supporting the biological relevance of the interaction.
In this study, we report a series of new structures of cross-linked actin dimers. Actin dimers cross-linked in the lateral direction show no tendency to crystallize in a way that resembles the filamentous state of actin. The present findings on this point are consistent with previous observations (Dawson et al., 2003 ▶). It is noteworthy that in the two laterally cross-linked dimer structures reported here the actin monomers are arranged in a way that nearly matches the arrangement reported in the first structure of a laterally cross-linked trimer (Dawson et al., 2003 ▶). It is possible that such an arrangement also exists in solution, despite its incompatibility with the F-actin structure. This would not be without precedent; antiparallel actin dimers having arrangements that are incompatible with F-actin have been observed before in solution (Millonig et al., 1988 ▶; Hesterkamp et al., 1993 ▶; Bubb et al., 2002 ▶; Lassing et al., 2007 ▶; Steinmetz et al., 1997 ▶) and in crystals (Bubb et al., 2002 ▶; Lassing et al., 2007 ▶).
In contrast, a new crystal form of the longitudinally cross-linked actin dimer reveals monomers arranged in a way reminiscent of the way actin molecules are believed to be arranged along one protofilament of the two-stranded F-actin filament. The arrangement obtained here is highly similar to that observed previously in a different crystal form of the longitudinally cross-linked dimer and to several others that have now been reported for actin crystallized under various conditions. A systematic comparison of numerous such crystals shows that the longitudinal interaction between actin monomers is strongly conserved across crystal structures. Interestingly, a number of the specific atomic interactions between side chains in the interface are strongly conserved despite differences in the range of a few angstroms in the way the protein backbones are shifted in the various crystal forms. This observation further reinforces the relevance of the interactions visualized.
So far, all but one of the crystal structures showing the longitudinal interface have revealed actin monomers related to each other by a lattice translation, thereby precluding the native twist between monomers in a protofilament. Although this presents some difficulties for achieving a complete interpretation of the F-actin filament, it does not diminish the significance of the repeated observation of a highly conserved set of atomic interactions. The data are consistent with the presence of a specific intermolecular contact between longitudinally related actin monomers that is strong enough to persist under a wide range of conditions and to tolerate a range of subunit orientations. This requires a certain degree of flexibility within the actin structure. The ability to maintain intermolecular interactions under varying twists is consistent with structural data that emphasize the flexibility and dynamic structure of the actin molecule and its filament (Galkin et al., 2002 ▶; Otterbein et al., 2001 ▶; Page et al., 1998 ▶).
The conserved interface discussed here involves only a subset of the interactions likely to exist in the native filament. The visualized interactions are restricted to subdomain 4 from the lower monomer and subdomain 3 from the upper monomer. Additional interactions are likely to exist in the native filament between subdomain 2 from the lower monomer and subdomain 1 from the upper monomer. Those interactions are not seen in the structures reported here, as subdomain 2 is largely disordered. As discussed earlier, this may relate to the absence of native twist in the crystal structures. The strongest interactions between actin monomers are likely to be those discussed here between subdomains 3 and 4. The interactions between subdomains 1 and 2 are believed to be dynamic and thus weaker (Orlova & Egelman, 1992 ▶, 1993 ▶) and may be sacrificed in order to allow the formation of an untwisted protofilament, as observed here, or to allow the formation of filaments with variable twist angles, as reported in electron-microscopy studies (Egelman et al., 1982 ▶; Stokes & DeRosier, 1987 ▶; Kim, Bobkova et al., 1998 ▶; Galkin et al., 2001 ▶).
The interactions visualized in the longitudinal interface place significant restraints on atomic models for the F-actin filament. On the basis of the first crystal structure of a longitudinally cross-linked actin dimer, a plausible model of the filament could be constructed in which the actin monomers were related by the native twist while still being nearly in contact about the interface visualized in the crystal (Kudryashov et al., 2005 ▶). In that first attempt to construct a filament, it was not yet clear which of the interactions were likely to be most critical. The results presented here are informative in that regard. From the multiple crystal views of the longitudinal interface, a set of atomic interactions that are particularly likely to be preserved in the native filament are now evident. It will be possible to test the veracity of the detailed interactions discussed here if higher resolution fiber-diffraction data or crystals of higher order actin oligomers can be obtained in the future. During preparation of this manuscript, a refinement of the helical F-actin structure was reported using fiber-diffraction data to a resolution of 7 Å (Oda et al., 2007 ▶). The likely interfacial regions highlighted in the present study, including residues 199–205 and 287–291, also appear to be in proximity in this most recent fiber-diffraction refinement.
Supplementary Material
PDB reference: cross-linked actin dimers, 2q1n, r2q1nsf
PDB reference: 2q31, r2q31sf
PDB reference: 2q36, r2q36sf
Acknowledgments
We thank Duilio Cascio for crystallographic assistance and the staff at the Advanced Light Source beamline 8.2.2 for technical support, Michael Levitt for comments on principal component analysis and Gerard Marriott for the gift of kabiramide C. This work was supported by National Institutes of Health Grant GM077190-28 and National Science Foundation Grant MCB0316269. MRS is supported by the Howard Hughes Medical Institute.
References
- Aebi, U., Millonig, R., Salvo, H. & Engel, A. (1986). Ann. NY Acad. Sci.483, 100–119. [DOI] [PubMed]
- Allingham, J. S., Zampella, A., D’Auria, M. V. & Rayment, I. (2005). Proc. Natl Acad. Sci. USA, 102, 14527–14532. [DOI] [PMC free article] [PubMed]
- Bahadur, R. P., Chakrabarti, P., Rodier, F. & Janin, J. (2004). J. Mol. Biol.336, 943–955. [DOI] [PubMed]
- Bubb, M. R., Govindasamy, L., Yarmola, E. G., Vorobiev, S. M., Almo, S. C., Somasundaram, T., Chapman, M. S., Agbandje-McKenna, M. & McKenna, R. (2002). J. Biol. Chem.277, 20999–21006. [DOI] [PubMed]
- Chik, J. K., Lindberg, U. & Schutt, C. E. (1996). J. Mol. Biol.263, 607–623. [DOI] [PubMed]
- Colovos, C. & Yeates, T. O. (1993). Protein Sci.2, 1511–1519. [DOI] [PMC free article] [PubMed]
- Dawson, J. F., Sablin, E. P., Spudich, J. A. & Fletterick, R. J. (2003). J. Biol. Chem.278, 1229–1238. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMol Molecular Graphics System. http://www.pymol.org.
- Diamond, R. (1974). J. Mol. Biol.82, 371–391. [DOI] [PubMed]
- Egelman, E. H., Francis, N. & DeRosier, D. J. (1982). Nature (London), 298, 131–135. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Galkin, V. E., Orlova, A., Lukoyanova, N., Wriggers, W. & Egelman, E. H. (2001). J. Cell Biol.153, 75–86. [DOI] [PMC free article] [PubMed]
- Galkin, V. E., VanLoock, M. S., Orlova, A. & Egelman, E. H. (2002). Curr. Biol.12, 570–575. [DOI] [PubMed]
- Guharoy, M. & Chakrabarti, P. (2005). Proc. Natl Acad. Sci. USA, 102, 15447–15452. [DOI] [PMC free article] [PubMed]
- Hanein, D., Volkmann, N., Goldsmith, S., Michon, A. M., Lehman, W., Craig, R., DeRosier, D., Almo, S. & Matsudaira, P. (1998). Nature Struct. Biol.5, 787–792. [DOI] [PubMed]
- Hegyi, G., Mak, M., Kim, E., Elzinga, M., Muhlrad, A. & Reisler, E. (1998). Biochemistry, 37, 17784–17792. [DOI] [PubMed]
- Hesterkamp, T., Weeds, A. G. & Mannherz, H. G. (1993). Eur. J. Biochem.218, 507–513. [DOI] [PubMed]
- Hodgkinson, J. L., Marston, S. B., Craig, R., Vibert, P. & Lehman, W. (1997). Biophys. J.72, 2398–2404. [DOI] [PMC free article] [PubMed]
- Holmes, K. C., Angert, I., Kull, F. J., Jahn, W. & Schroder, R. R. (2003). Nature (London), 425, 423–427. [DOI] [PubMed]
- Holmes, K. C., Popp, D., Gebhard, W. & Kabsch, W. (1990). Nature (London), 347, 44–49. [DOI] [PubMed]
- Kabsch, W. (1976). Acta Cryst. A32, 922–923.
- Kabsch, W., Mannherz, H. G., Suck, D., Pai, E. F. & Holmes, K. C. (1990). Nature (London), 347, 37–44. [DOI] [PubMed]
- Kim, E., Bobkova, E., Hegyi, G., Muhlrad, A. & Reisler, E. (2002). Biochemistry, 41, 86–93. [DOI] [PubMed]
- Kim, E., Bobkova, E., Miller, C. J., Orlova, A., Hegyi, G., Egelman, E. H., Muhlrad, A. & Reisler, E. (1998). Biochemistry, 37, 17801–17809. [DOI] [PubMed]
- Kim, E., Phillips, M., Hegyi, G., Muhlrad, A. & Reisler, E. (1998). Biochemistry, 37, 17793–17800. [DOI] [PubMed]
- Klenchin, V. A., Allingham, J. S., King, R., Tanaka, J., Marriott, G. & Rayment, I. (2003). Nature Struct. Biol.10, 1058–1063. [DOI] [PubMed]
- Klenchin, V. A., Khaitlina, S. Y. & Rayment, I. (2006). J. Mol. Biol.362, 140–150. [DOI] [PubMed]
- Klenchin, V. A., King, R., Tanaka, J., Marriott, G. & Rayment, I. (2005). Chem. Biol.12, 287–291. [DOI] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1994). CCP4/ESF–EACBM Newsl. Protein Crystallogr.31, 9–14.
- Knight, P. & Offer, G. (1978). Biochem. J.175, 1023–1032. [DOI] [PMC free article] [PubMed]
- Knight, P. & Offer, G. (1980). Biochemistry, 19, 4682–4687. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Kudryashov, D. S., Sawaya, M. R., Adisetiyo, H., Norcross, T., Hegyi, G., Reisler, E. & Yeates, T. O. (2005). Proc. Natl Acad. Sci. USA, 102, 13105–13110. [DOI] [PMC free article] [PubMed]
- Lal, A. A., Korn, E. D. & Brenner, S. L. (1984). J. Biol. Chem.259, 8794–8800. [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Lassing, I., Schmitzberger, F., Bjornstedt, M., Holmgren, A., Nordlund, P., Schutt, C. E. & Lindberg, U. (2007). J. Mol. Biol.370, 331–348. [DOI] [PubMed]
- Liu, S., Li, Q. & Lai, L. (2006). Proteins, 64, 68–78. [DOI] [PubMed]
- Lorenz, M., Popp, D. & Holmes, K. C. (1993). J. Mol. Biol.234, 826–836. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- McLaughlin, P. J., Gooch, J. T., Mannherz, H. G. & Weeds, A. G. (1993). Nature (London), 364, 685–692. [DOI] [PubMed]
- Milligan, R. A., Whittaker, M. & Safer, D. (1990). Nature (London), 348, 217–221. [DOI] [PubMed]
- Millonig, R., Salvo, H. & Aebi, U. (1988). J. Cell Biol.106, 785–796. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Oda, T., Stegmann, H., Schroder, R. R., Namba, K. & Maeda, Y. (2007). Adv. Exp. Med. Biol.592, 385–401. [DOI] [PubMed]
- Orlova, A. & Egelman, E. H. (1992). J. Mol. Biol.227, 1043–1053. [DOI] [PubMed]
- Orlova, A. & Egelman, E. H. (1993). J. Mol. Biol.232, 334–341. [DOI] [PubMed]
- Orlova, A., Galkin, V. E., VanLoock, M. S., Kim, E., Shvetsov, A., Reisler, E. & Egelman, E. H. (2001). J. Mol. Biol.312, 95–106. [DOI] [PubMed]
- Otomo, T., Tomchick, D. R., Otomo, C., Panchal, S. C., Machius, M. & Rosen, M. K. (2005). Nature (London), 433, 488–494. [DOI] [PubMed]
- Otterbein, L. R., Graceffa, P. & Dominguez, R. (2001). Science, 293, 708–711. [DOI] [PubMed]
- Page, R., Lindberg, U. & Schutt, C. E. (1998). J. Mol. Biol.280, 463–474. [DOI] [PubMed]
- Ponstingl, H., Henrick, K. & Thornton, J. M. (2000). Proteins, 41, 47–57. [DOI] [PubMed]
- Rizvi, S. A., Tereshko, V., Kossiakoff, A. A. & Kozmin, S. A. (2006). J. Am. Chem. Soc.128, 3882–3883. [DOI] [PubMed]
- Rould, M. A., Wan, Q., Joel, P. B., Lowey, S. & Trybus, K. M. (2006). J. Biol. Chem.281, 31909–31919. [DOI] [PubMed]
- Sablin, E. P., Dawson, J. F., VanLoock, M. S., Spudich, J. A., Egelman, E. H. & Fletterick, R. J. (2002). Proc. Natl Acad. Sci USA, 99, 10945–10947. [DOI] [PMC free article] [PubMed]
- Schutt, C. E., Myslik, J. C., Rozycki, M. D., Goonesekere, N. C. & Lindberg, U. (1993). Nature (London), 365, 810–816. [DOI] [PubMed]
- Steinmetz, M. O., Goldie, K. N. & Aebi, U. (1997). J. Cell Biol.138, 559–574. [DOI] [PMC free article] [PubMed]
- Stokes, D. L. & DeRosier, D. J. (1987). J. Cell Biol.104, 1005–1017. [DOI] [PMC free article] [PubMed]
- Strong, M., Sawaya, M. R., Wang, S., Phillips, M., Cascio, D. & Eisenberg, D. (2006). Proc. Natl Acad. Sci. USA, 103, 8060–8065. [DOI] [PMC free article] [PubMed]
- Tirion, M. M., ben-Avraham, D., Lorenz, M. & Holmes, K. C. (1995). Biophys. J.68, 5–12. [DOI] [PMC free article] [PubMed]
- Vorobiev, S., Strokopytov, B., Drubin, D. G., Frieden, C., Ono, S., Condeelis, J., Rubenstein, P. A. & Almo, S. C. (2003). Proc. Natl Acad. Sci. USA, 100, 5760–5765. [DOI] [PMC free article] [PubMed]
- Vriend, G. & Sander, C. (1991). Proteins, 11, 52–58. [DOI] [PubMed]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: cross-linked actin dimers, 2q1n, r2q1nsf
PDB reference: 2q31, r2q31sf
PDB reference: 2q36, r2q36sf