Abstract
SEMA3F is a secreted semaphorin with potent antitumor activity, which is frequently downregulated in lung cancer. In cancer cell lines, SEMA3F overexpression decreases hypoxia-induced factor 1α protein and vascular endothelial growth factor mRNA, and inhibits multiple signaling components. Therefore, understanding how SEMA3F expression is inhibited in cancer cells is important. We previously defined the promoter organization of SEMA3F and found that chromatin remodeling by a histone deacetylase inhibitor was sufficient to activate SEMA3F expression. In lung cancer, we have also shown that ZEB-1, an E-box transcription repressor, is predominantly responsible for loss of E-Cadherin associated with a poor prognosis and resistance to epidermal growth factor receptor inhibitors. In the present study, we demonstrated that ZEB-1 also inhibits SEMA3F in lung cancer cells. Levels of ZEB-1, but not ZEB-2, Snail or Slug, significantly correlate with SEMA3F inhibition, and overexpression or inhibition of ZEB-1 correspondingly affected SEMA3F expression. Four conserved E-box sites were identified in the SEMA3F gene. Direct ZEB-1 binding was confirmed by chromatin immunoprecipitation assays for two of these, and ZEB-1 binding was reduced when cells were treated with a histone deacetylase inhibitor. These results demonstrate that ZEB-1 directly inhibits SEMA3F expression in lung cancer cells. SEMA3F loss was associated with changes in cell signaling: increased phospho-AKT in normoxia and increase of hypoxia-induced factor 1α protein in hypoxia. Moreover, exogenous addition of SEMA3F could modulate ZEB-1-induced angiogenesis in a chorioallantoic membrane assay. Together, these data provide further support for the importance of SEMA3F and ZEB-1 in lung cancer progression.
Introduction
SEMA3F was originally cloned from a recurrent 3p21.3 homozygous deletion in small cell lung carcinoma (SCLC), suggesting that it might be a tumor suppressor gene [1–3]. Class-3 semaphorins [4], including SEMA3F, are secreted proteins originally identified as mediators of growth cone repulsion [5], but their wide expression patterns suggested additional functions outside the nervous system [6]. Their involvement in cancer and angiogenesis was further described (see recent reviews [7–10]). Exogenous expression of SEMA3F in tumor cell lines reduced tumor formation in nude mice in several xenograft models [11–15]. The resulting tumors displayed a reduced density of blood vessels, implying that SEMA3F inhibits angiogenesis during tumor development. In addition, the SEMA3F-expressing tumor induced less metastases [11]. One possible explanation for the antiangiogenic activity of SEMA3F has been a competition between SEMA3F and vascular endothelial growth factor 165 (VEGF165) for binding to their common neuropilin receptor, as was shown for Sema3A [16].
Using a lung orthotopic model, we reported that SEMA3F blocked H157 lung cancer tumorigenesis [17]. This was associated with a SEMA3F-induced loss of activated αVβ3 integrin and impaired cell adhesion to extracellular matrix components [14,17]. Several signaling pathways were affected by SEMA3F, including decreased phosphoextracellular signal-regulated kinase 1/2, phospho-AKT, phospho-signal transducer and activator of transcription 3, and down-regulation of integrin-linked kinase activity [14]. In addition, SEMA3F negatively affected the level of hypoxia-induced factor 1α (HIF-1α) protein and, as a consequence, VEGF mRNA expression [14]. Therefore, we proposed a second explanation for the antiangiogenic effect of SEMA3F, i.e., VEGF165 down-regulation owing to HIF-1α loss. This effect is in accordance with our observations that SEMA3F is downregulated in a majority of human lung cancers and that loss of SEMA3F protein staining is significantly correlated with an advanced stage of disease and with VEGF165 overexpression [18].
Although SEMA3F is frequently downregulated in tumors, inactivating mutations have not been observed [15]. Therefore, it is important to understand how SEMA3F is regulated. Presently, little is known about SEMA3F regulation except that SEMA3F is a direct p53 target [12], and we reported that DNA methylation and chromatin remodeling by histone deacetylase inhibitors (HDACis) play a role in SEMA3F expression [19].
Previously, we defined the genomic organization of the SEMA3F promoter [19]. We identified several putative E-box sites (consensus palindromic sequence CANNTG) present in the SEMA3F promoter, as well as in introns 1 and 3. These sites bind basic helix-loop-helix proteins and other transcription factors with zinc fingers including ZEB-1, ZEB-2, Snail, and Slug, among others.We previously demonstrated that blocking ZEB-1 (also known as TCF8 and δEF1) with small interfering RNA (siRNA) in H661 lung cancer cells led to the up-regulation of E-Cadherin [20]. In addition, we reported that ZEB-1 expression and E-Cadherin loss are associated with resistance to epidermal growth factor receptor (EGFR) inhibitors and a poor prognosis in lung cancer [21,22]. ZEB-1 promotes tumor cell dedifferentiation with repression of regulators of epithelial polarity and is involved in metastasis [23–25]. ZEB-1 like ZEB-2 (also known as ZFXH1B and SMAD interacting protein 1 known as SIP1) have emerged as key factors that regulate induction of the epithelial-mesenchymal transition (EMT) playing a critical role in tumor progression, invasion, and metastasis [26–28]. Epithelial-mesenchymal transition is associated with E-Cadherin repression by direct ZEB-1 binding to E-Cadherin regulatory sequences. This regulation is facilitated by ZEB-1 interaction with the transcriptional corepressor CtBP, which can recruit HDACs leading to chromatin condensation [29–33]. Therefore, ZEB-1 could be a potential candidate for SEMA3F repression in lung cancer.
In the present study, we demonstrated that ZEB-1 levels are significantly correlated with down-regulation of SEMA3F in lung cancer cells. This negative correlation was verified by up-regulation or inhibition of ZEB-1. In addition, ZEB-1 binds to regulatory E-box sites in the SEMA3F gene, and this binding is impaired by HDACi. Furthermore, SEMA3F loss is associated with changes in cell signaling. When SEMA3F is added back after depletion by ZEB-1, it decreases ZEB-1-induced angiogenesis. In addition to providing a better understanding of SEMA3F regulation, these results support the importance of SEMA3F and ZEB-1 in lung cancer progression.
Materials and Methods
Cell Lines and Culture
A panel of 21 non-small cell lung carcinoma (NSCLC) cell lines, including NCI-H661 and NCI-H358 cell lines, were grown in RPMI-1640 supplemented by 10% fetal calf serum (Invitrogen, Cergy Pontoise, France). All the cell lines were previously described [21]. MCF7 breast cancer cells were grown in the same conditions. For HDAC inhibition, H661 cells were treated with 5 µM suberoylanilide hydroxamic acid (SAHA) for 16 hours before harvesting.
Transient Transfection Assays
Two micrograms of pCS2MT and pCS2MT-ZEB-1 plasmids (kindly provided by Dr. J. Richter) were transfected using 3 µl of Fugene (Roche Applied Sciences, Meylan, France) into 0.5 x 105 H661 cells plated 24 hours before transfection. Cells were harvested at 24, 48, 72, or 96 hours after transfection. Each transfection was tested in two independent experiments.
Stable Transfections
Establishment of H358 cell line with stably integrated ZEB-1, under the control of a tetracycline/doxycycline (Dox)-inducible promoter, was obtained following the Flip-In T-REX core kit instructions (Invitrogen). ZEB-1 was cloned with a 6 Myc tag at the 3′ end, into the EcoRV restriction site of pcDNA5/FRT/TO plasmid (Invitrogen). H358 cells transfected with empty vector (H358 FlpIn EV) or inducible ZEB-1 (H358 FlpIn ZEB-1) were selected with 100 µg/ml hygromycin and 5 µg/ml blasticidin (Invitrogen). ZEB-1 was induced for 3 days with Dox at 200 ng/ml.
Luciferase Reporter Assays
The SEMA3F promoter fragment [-5836 to -4013] relative to the start codon at position +1 was amplified by polymerase chain reaction (PCR) and cloned upstream of the Firefly luciferase gene in the pGL3basic vector (Promega, Charbonnières-les-Bains, France). For that purpose, XhoI and HindIII restriction sites were included in the primers described in Table W1E. Cloning was performed as previously described [19]. This construct (2 µg) was transfected into MCF7 and H661 cells with Fugene (Roche Applied Sciences). In each experiment, the pRL-TK plasmid (100 ng), encoding Renilla luciferase (Promega), was cotransfected to measure transfection efficiency and for normalization purposes. Luminescence was measured 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). The pGL3basic (promoterless) plasmid was used in each experiment to determine luciferase basal level. Reporter activity was normalized by calculating the ratio of Firefly/Renilla values. Each construct was tested in two independent transfections, each time in duplicate.
Site-Directed Mutagenesis
The bipartite element of the E-Box site in the SEMA3F promoter fragment was mutated using the Quick Change Site-Directed Mutagenesis Kit (Stratagene, Amsterdam, the Netherlands) following the protocol provided by the manufacturer except for the PCR conditions. Because the GC content is very high (82% GC) in this fragment, PCR was performed with the Platinum Pfx DNA polymerase (Invitrogen) with the recommended enhancer buffer. Primers are described in Table W1D.
Small Interfering RNA Transfection
To inhibit ZEB-1 expression, RNA interference inhibition was performed as described [20,34]. Double-stranded RNA oligonucleotides were prepared by using the Silencer kit from Ambion (Austin, TX). Primer sequences [20] are given in Table W1C. H661 cells (1 x 105) were transfected with 5, 50, or 100 nM of double-stranded RNA by Oligofectamine (Invitrogen). Controls were mock-transfected cells. H661 cells were transfected with 50 nM siRNA against SEMA3For nontargeting siRNA fromDharmacon (Brebieres, France; Table W1C; Reference Genome Smart pool M-0176644-02 and D-001210-02, respectively). Cells were harvested 48 hours after transfection. Each concentration of siRNA was tested in three independent experiments.
RNA Expression
Total RNA was extracted using the SV Total RNA isolation kit (Promega). Reverse transcription-PCR (RT-PCR) was performed with the Superscript II reverse transcriptase (Invitrogen) using the procedure supplied by the manufacturer. We assessed levels of SEMA3F, ZEB-1, ZEB-2, or E-Cadherin mRNA relative to GAPDHin cell lines by quantitative real-time PCR using the GeneAmp 7000 quantitative PCR system (Applied Biosystems, Courtaboeuf, France) with SYBR-Green chemistry [17]. The RT-PCR primers are described in Table W1A. The results are displayed in the relative expression (x100) compared to GAPDH expression.
Immunoblot Analysis
Immunoblots were done as reported previously [14,17]. The anti-Myc and anti-α tubulin were from Sigma-Aldrich (Saint Quentin Fallavier, France) and used at 1:2000 dilution. Anti-ZEB-1 (1:1000) was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-AKT and anti-phospho-Ser473-AKT were from Cell Signaling Technology (Danvers, MA) and used at 1:1000 dilution. Anti-HIF-1α (1:250) was from BD Transduction Laboratories (Erembodegem, Belgium). Horseradish peroxidase-conjugated antimouse and antirabbit secondary antibodies (1:5000) were fromPerkin-Elmer (Courtaboeuf, France). Detection was done with enhanced chemiluminescence (Perkin-Elmer).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assay was performed with the protocol described by Upstate (Millipore, Saint Quentin en Yvelines, France) with modifications for the last washes. Briefly, 5 x 106 cells per assay were cross-linked with 1% formaldehyde at 37°C for 5 minutes. Cells were resuspended in the SDS lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.1)] for 15 minutes on ice. The lysate was sonicated 10 times, 15 seconds on ice. After centrifuging at 13,000 rpm at 4°C for 10 minutes, the supernatant was diluted 10 times in the dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl (pH 8.1)] and was incubated for 1 hour on a rotating platform at 4°C with Protein-A Sepharose. One microgram of ZEB-1 antibodies (H102; Santa Cruz Biotechnology) or of IgG from a nonimmunized rabbit (i5006; Sigma) was incubated with the precleared chromatin on a rotating platform, overnight at 4°C. Immune complexes were collected with Protein-A Sepharose (Amersham, GE Healthcare, Orsay, France). They were washed three times with the LiCl Buffer [0.25 M LiCl, 1% Triton X-100, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris-HCl (pH 8.1)] and twice with the TE buffer [1 mM EDTA, 10 mM Tris-HCl (pH 8)] before phenol-chloroform extraction and ethanol precipitation. Quantitative real-time PCR was performed with SYBR-Green chemistry with primers described in Table W1B. For each PCR primer set, Ct values were obtained for purified DNA from the input, and ZEB-1 and IgG immunoprecipitated chromatin. Then, we determined ΔCtZEB-1 = CtZEB-1 - Ctinput and ΔCtIgG = CtIgG - Ctinput. The fold enrichment is obtained by the ratio 2-ΔCtZEB-1/2-ΔCtIgG. The percentage of input is shown in Figure W1 (B and C).
Chorioallantoic Membrane Assay
Fertilized Hubbard JA57 chick eggs were incubated under condition of constant humidity (70%) at 37.8°C. On the fourth day of incubation, a square window was opened in the shell after removal of 2.5 ml of albumin to detach the chorioallantoic membrane (CAM) from the shell. The window was closed with tape, and the eggs were returned to the incubator. At day 9, a pellet of 5 x 105 cells resuspended in an equivalent volume of Matrigel (BD Biosciences, Le Pont de Claix, France) was implanted on top of growing CAMs. For ZEB-1 induction, cells were grown with 0.2 µg/ml Dox for 10 days before implantation. To maintain ZEB-1 induction, 0.5 µg/ml Dox was added in Matrigel the day of implantation. Where indicated, recombinant mouse SEMA3F-Fc (RD Systems, Lille, France) was delivered at 0.7 pmol per plug. Blood vessels on the CAM were examined 6 days after cell implantation and photographed in ovo under a Nikon SMZ 1500 Microscope with a Nikon NS Fi1 camera (Nikon, Champigny sur Marne, France). Each egg was assigned a score number varying from 0 to 3, in which 0 is the no angiogenic response and 3 is the strongest response (Figure 5D). Blinded scoring was performed simultaneously by two observers. The mean was calculated by GraphPad Prism (version 5.01; GraphPad Software Incorporation, La Jolla, CA), and a nonparametric Kruskal-Wallis statistical test was performed.
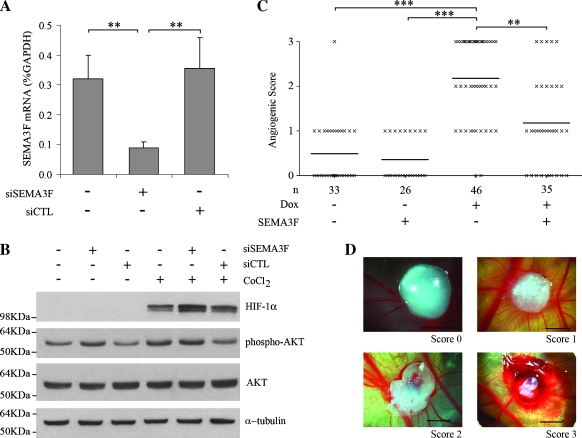
Figure 5.
Physiological effects of SEMA3F loss. (A, B) H661 cells were transfected with siRNA against SEMA3F (siSEMA3F) or nontargeting siRNA (siCTL). Control was mock-transfected cells. (A) SEMA3F mRNA was monitored by quantitative real-time RT-PCR 48 hours after transfection. Values for three independent experiments, done in duplicate, are expressed in percentage of GAPDH expression. Bars, SD. (B) H661 cells were transfected by siRNA in absence or presence of 100 µM CoCl2 during 2 hours, and effects on signaling were monitored by Western blot analysis for HIF-1α, phospho-Ser473-AKT, and total AKT. α-Tubulin was used as a loading control. (C) Chorioallantoic membrane assays were performed to test angiogenesis. H358 FlpIn ZEB-1 cells were treated or not with Dox and then loaded on chick embryos in Matrigel with either presence or absence of recombinant SEMA3F-Fc. Six days after implantation, angiogenesis was evaluated independently by two observers with scores ranging from 0 to 3 as shown in D (scale, 3 mm). Using a nonparametric Kruskal-Wallis test, scores were significantly different with ZEB-1-induced cells versus noninduced cells (***P < .001) and ZEB-1-induced cells versus ZEB-1-induced cells plus SEMA3F (**P µ .01). Bars, mean. n, number of eggs.
Results
ZEB-1 Expression Is Negatively Correlated with SEMA3F in Lung Cancer Cell Lines
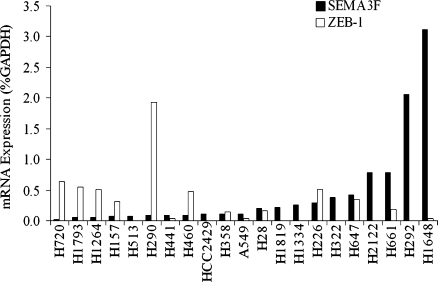
Using quantitative real-time RT-PCR, we screened 21 NSCLC cell lines for expression of SEMA3F, ZEB-1, and three additional E-box transcriptional repressors, ZEB-2, Snail, and Slug. ZEB-1 expression had a significant negative correlation with SEMA3F (Spearman correlation coefficient, r = -0.57, P = .0089; Figure 1). In contrast, no statistically significant correlations were observed between the expression of SEMA3F and ZEB-2, Snail, or Slug, nor were there any apparent trends (not shown).
Figure 1.
ZEB-1 expression is negatively correlated with SEMA3F in lung cancer cell lines. ZEB-1 (white bars) and SEMA3F (black bars) mRNA were measured by quantitative real-time RT-PCR in 21 lung cancer cell lines. Values are expressed as percentage of GAPDH expression. Because the variables are not normally distributed, we used the Spearman correlation coefficient (a nonparametric analog to the Pearson correlation coefficient) for statistical analysis.
ZEB-1 Induces Transcriptional Down-regulation of SEMA3F in the H661 NSCLC Cell Line
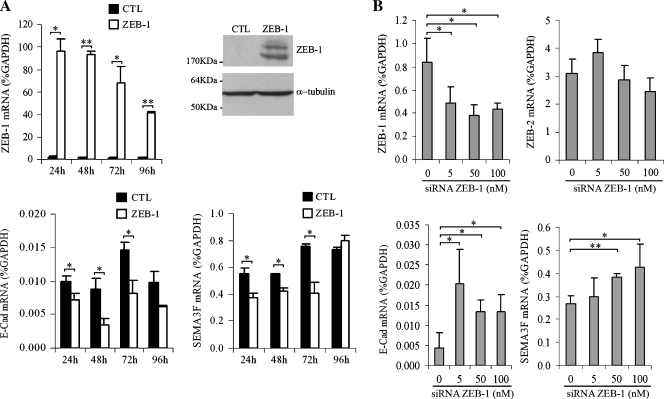
To determine whether ZEB-1 could regulate SEMA3F expression, H661 cells were transiently transfected with a Myc-tagged ZEB-1 expression construct. We first verified ZEB-1 expression by quantitative real-time RT-PCR and observed a 100-fold enrichment in expression (Figure 2A, top left). Western blot analysis using anti-ZEB-1 antibodies detected ZEB-1 protein in transfected cells (Figure 2A, top right). As a positive control, we examined E-Cadherin expression for two reasons: 1) ZEB-1 has been shown to bind E-box sites in the E-Cadherin promoter and to inhibit its expression; and 2) we previously demonstrated that blocking ZEB-1 with small interfering RNA (siRNA) in H661 cells led to up-regulation of E-Cadherin [20]. Indeed, 48 hours after ZEB-1 transfection, E-Cadherin mRNA was reduced to 40% (Figure 2A, bottom left) along with a 50% decrease for SEMA3F mRNA level at 72 hours (Figure 2A, bottom right). We noticed that E-Cadherin expression seemed to be still inhibited after 96 hours (however, the expression was not statistically different from the control), but SEMA3F was not.We believe that increasing ZEB-1 could modify ZEB-1 coregulator expression or other transcriptional factors affecting differently E-Cadherin and SEMA3F expression over time. To verify the relationship between endogenous ZEB-1 and SEMA3F, we used siRNA to inhibit ZEB-1 in H661 cells (Figure 2B). Compared with mock-transfected cells, the level of ZEB-1 mRNA was lowered by 50%, whereas E-Cadherin and SEMA3F expression increased accordingly. E-Cadherin and SEMA3F expressions seemed to be stimulated with ZEB-1 siRNA at very different concentrations (5 and 50 nM, respectively). Because we did not check siRNA concentrations between 5 and 50 nM, we cannot say that 10 times more siRNA were necessary to activate SEMA3F compared to E-Cadherin. In contrast, the expression of a nontarget gene, such as ZEB-2, was unaffected.
Figure 2.
ZEB-1 downregulates SEMA3F transcription in H661 cells. (A) H661 cells were transiently transfected with control (CTL; black bars) or ZEB-1 expression vector (ZEB-1; white bars) and were harvested over time to measure ZEB-1, E-Cadherin, and SEMA3F expressions by quantitative real-time RT-PCR. Values are expressed in percentage of GAPDH expression. ZEB-1 protein was checked by Western blot analysis at 48 hours after transfection (top right). α-Tubulin was used as a loading control. The low E-Cadherin mRNA level in H661 cells is in accordance with previous results where H661 cells were selected among NSCLC for their undetectable E-Cadherin [20]. Reasons of the absence of E-Cadherin are the loss of Wnt7a/β-catenin pathway and ZEB-1 expression. (B) H661 cells were transfected with different amounts of ZEB-1 siRNA. ZEB-1, ZEB-2, E-Cadherin, and SEMA3F mRNA were monitored 48 hours after transfection. Values for three independent experiments (except for ZEB-2), done in duplicate, are expressed in percentage of GAPDH expression. Bars, SD. Statistical analysis was performed with Student's t-test: *P < .05, P < .01.
ZEB-1 Directly Binds E-box Sites in SEMA3F in H661 Cells and HDACi Inhibits This Binding
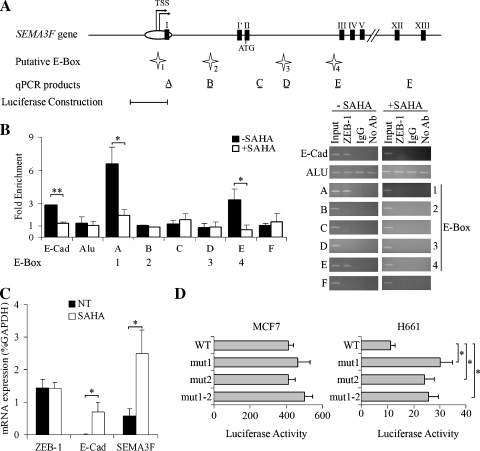
Four evolutionarily conserved E-box sites are present in the 5′ sequence of SEMA3F (Figure 3A). Site 1 is located within the CpG island promoter region, site 2 is in the first intron, and sites 3 and 4 are located within the third intron. Sites 1, 2, and 4 are bipartite elements, which have been reported to bind ZEB-1 in other target genes [35]. Direct in vivo analysis of ZEB-1 binding to these sites was carried out using ChIP assays, and changes in ZEB-1 binding were ascertained by quantitative real-time PCR. Chromatin immunoprecipitation assays were performed with IgG from a nonimmunized rabbit as a negative control. As a positive control, ZEB-1 binding was verified on the E-Cadherin promoter, and ZEB-1 overexpression was associated with a three-fold enrichment as expected (Figure 3B, black bars). The presence of PCR products was confirmed by electrophoresis (Figure 3B). In contrast, no enrichment was evident with Alu sequences used as a negative control. For SEMA3F, two of the four predicted E-box sites (i.e., sites 1 and 4) bound ZEB-1 (Figure 3B, black bars). Of note, site 1 is located in the CpG island approximately 300 bp from the experimentally determined transcriptional start sites [19]. To assess nonspecific binding, ChIP assays were performed for two E-box-negative regions located in intron 3 (product C) and intron 13 (product F; Figure 3A). As expected, no enrichment was noticed (Figure 3B, black bars).
Figure 3.
ZEB-1 binds directly to SEMA3F promoter and HDAC inhibition represses this binding in H661 cells. (A) Genomic organization of the SEMA3F promoter. Transcription start sites (TSS) are indicated by arrows. White oval indicates the CpG island, and black boxes indicate exons. Four putative E-box sites, numbered 1 to 4 (indicated by white stars) were predicted by computer analysis using Mulan and multiTF at NCBI DCODE.org (www.dcode.org) [51]. Horizontal black bars show the position of PCR products after ChIP experiments, with their name (A to F) indicated above. The SEMA3F promoter fragment [-5836 to -4013] tested in the luciferase reporter construction is indicated below. (B) Chromatin immunoprecipitation assays were done on H661 cells grown 16 hours in the absence (black bars) or in the presence (white bars) of 5 µM SAHA. E-Cadherin and Alu PCR, respectively, were used as positive and negative ChIP controls. The results of quantitative real-time PCR are expressed by fold enrichment, for three independent experiments with PCR reactions being performed in duplicate. Bars, SD. Semiquantitative analysis of PCR products obtained after ChIP assays was performed by electrophoresis on agarose gel after 30 cycles for Alu PCR and 33 cycles for all the other PCR. (C) ZEB-1, E-Cadherin, and SEMA3F mRNA levels were measured by quantitative real-time RT-PCR on H661 cells grown 16 hours in the absence (black bars) or in the presence (white bars) of 5 µM SAHA. This experiment was done three times, and each RT-PCR in duplicate. Values are expressed in percentage of GAPDH expression. Bars, SD. (D) The luciferase reporter constructions, with the [-5836 to -4013] SEMA3F promoter fragment, mutated or not (WT) for each or both elements of the E-box site 1 present in this fragment, were transfected into MCF7 and H661 cells. Firefly luciferase activity was measured and normalized to the Renilla luciferase activity of the cotransfected plasmid pRL-TK for two independent experiments done in duplicate. Bars, SD. Statistical analysis was performed with Student's t-test: *P < .05, P < .01.
We previously reported that treatment of H460 NSCLC cells by Trichostatin A (TSA), an HDACi, induced SEMA3F expression [19]. As it is known that ZEB-1 recruits CtBP which can interact with HDACs [29–33] and because HDACis influence protein complexes on regulatory DNA sequences [36], we determined whether SAHA (vorinostat) treatment would cause ZEB-1 to dissociate from SEMA3F E-box sites. To examine ZEB-1 binding after treatment with an HDACi, we exposed H661 cells to 5 µM SAHA during 16 hours and first verified that SEMA3F and E-Cadherin mRNA were increased (≈4.3-fold and ≈7-fold, respectively) as anticipated (Figure 3C). In these conditions, the level of ZEB-1 mRNA was unaffected. By a ChIP assay, the binding of ZEB-1 to the E-Cadherin promoter was decreased (Figure 3B, white bar). Similarly, ZEB-1 dissociated completely from SEMA3F site 4, and its binding to site 1 was greatly decreased. These results support the direct binding of ZEB-1 in the transcriptional repression of SEMA3F and demonstrate that ZEB-1 dissociates from SEMA3F gene sequences after treatment with an HDACi.
Because site 1 is a bipartite element, located in the regulatory sequence in the CpG island, it seems to be an important regulatory sequence. To test this hypothesis, we performed a luciferase reporter assay by cloning the [-5836 to -4013] SEMA3F promoter fragment that contains site 1 and we introduced mutations of each part of the bipartite element separately (mut1, mut2) or together (mut1–2). These constructs were transfected into MCF7 breast cancer cells, chosen only as a positive control in this gene reporter assay because we previously demonstrated that the SEMA3F promoter fragment [-6310 to -4013] and [-5836 to -4013] are functional in these cells [19] (and data not shown). In addition, MCF7 cells express a high level of SEMA3F but almost no ZEB-1. The expressions of SEMA3F and ZEB-1 in H661 and MCF7 cells displayed in the relative expression (x100) compared to GAPDH expression were the following: SEMA3F = 0.7%, ZEB-1 = 0.2% in H661 cells and SEMA3F = 4.7%, ZEB-1 = 0.005% in MCF7 cells. Each mutation separately or in combination did not modify luciferase activity in MCF7 cells when compared to the wild type sequence (Figure 3D). In contrast, these mutations increased luciferase activity (≈2.5-fold) in H661 cells (Figure 3D). This reporter gene assay shows that site 1 is necessary to repress SEMA3F in H661 cells.
Tet-Induced ZEB-1 Binds SEMA3F E-box Sites in H358 Cells
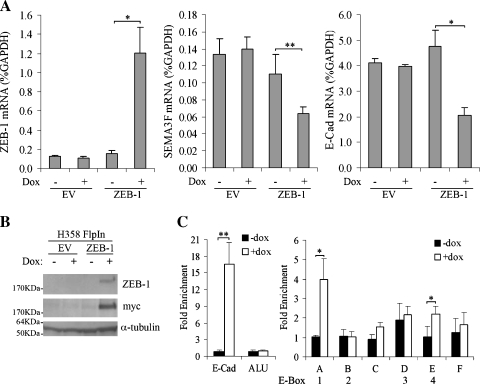
To demonstrate that ZEB-1 inhibition of SEMA3F was not limited to H661 cells, we engineered the H358 NSCLC cell line with the tet-inducible FlpIn system (Invitrogen) and introduced a 6 Myc-tagged ZEB-1 into the homologous recombination site. After 3 days of Dox induction (200 ng/ml), ZEB-1 mRNA was induced approximately eight-fold compared to noninduced or control cells containing an empty vector (Figure 4A). ZEB-1 protein was detected by Western blot with either ZEB-1 or Myc antibodies (Figure 4B). As expected, ZEB-1 induction was associated with a reduction in E-Cadherin mRNA (Figure 4A). Similarly, SEMA3F mRNA levels were reduced to approximately 50% of controls (Figure 4A). SEMA3F protein could not be detected because the commercial SEMA3F antibody (Chemicon, Hants, England) was raised against a peptide that corresponds to a variably expressed alternative exon [1,3], which we verified was not expressed in H358 cells (data not shown). By immunostaining, we also verified that induced ZEB-1 was localized in the nuclei (Figure W2A). Surprisingly, ZEB-1 induction was heterogeneous among cells within the same island (Figure W2B), and we noted that the cells expressing the highest levels of ZEB-1 protein were precisely those that had lost E-Cadherin staining (Figure W2B). These controls confirm that induced ZEB-1 is in the right cellular compartment and is functional.
Figure 4.
ZEB-1 binds directly to SEMA3F in ZEB-1-induced H358 cells. (A) H358 FlpIn EV or FlpIn ZEB-1 were treated (+) or not (-) with Dox for 3 days, and ZEB-1, SEMA3F, and E-Cadherin mRNA levels were determined by quantitative real-time RT-PCR. Values, for three independent experiments with each RT-PCR in duplicate, are expressed in percentage of GAPDH expression. Bars, SD. (B) ZEB-1 overexpression was checked by Western blot analysis in H358 FlpIn EV or FlpIn ZEB-1 induced by Dox in the same conditions as (A) with two different antibodies against Myc (as ZEB-1 is Myc-tagged) and ZEB-1. α-Tubulin was used as a loading control. (C) Chromatin immunoprecipitation assays were done with the ZEB-1 antibody on H358 FlpIn ZEB-1 cells grown 3 days in the absence (black bars) or in the presence (white bars) of Dox. E-Cadherin and Alu PCR primers, respectively, were used as positive and negative ChIP controls. Values for three independent experiments are expressed as those in Figure 3. Bars, SD. Statistical analysis was performed with Student's t-test: *P < .05, P < .01.
Chromatin immunoprecipitation assays were performed for ZEB-1 binding in vivo (Figure 4C). In H358 cells, which express abundant E-Cadherin, ZEB-1 binding to the E-Cadherin promoter was not detected as assessed by quantitative real-time PCR. However, on ZEB-1 induction, enrichment of E-Cadherin promoter sequences in the immunoprecipitate was strongly increased (19-fold). There was no enrichment of Alu sequences (used as a negative control) with ZEB-1 induction. Similarly, in the absence of Dox, there was no binding of endogenous ZEB-1 to the SEMA3F E-box sites, whereas on ZEB-1 induction, binding to sites 1 and 4 was significantly increased (4- and 2.2-fold enrichment, respectively), although not to the same degree that we observed with E-Cadherin.
Values obtained in ChIP experiments for ZEB-1 binding on E-Cadherin and SEMA3F were different in H661 and H358 cells. More ZEB-1 bound on the E-Cadherin promoter than on the SEMA3F promoter in H358 cells (Figure 4C), whereas the opposite was found in H661 cells (Figure 3B). We cannot exclude that when modifying the endogenous level of a transcription factor, organization of regulatory complexes could be changed. In addition, H358 and H661 cells could present differences in their repertoire of transcriptional regulators. Epigenetic modifications in E-Cadherin and SEMA3F regulatory sequences in H358 and H661 cells could explain different accessibility to ZEB-1.
SEMA3F Loss Is Associated with Cell Signaling Changes in H661 Cells
We recently reported that SEMA3F overexpression reduced HIF-1α protein in hypoxia in three cell lines (H157 and H460 lung cancer cells and COS cells) [14]. Because ZEB-1 represses SEMA3F in H661 cells, we asked whether SEMA3F inhibition by siRNA would lead to the opposite effect. We first verified SEMA3F inhibition by siRNA (Figure 5A). Then, HIF-1α protein levels were monitored after treating the cells with the hypoxia mimetic, CoCl2. As shown in Figure 5B, HIF-1α levels were higher when SEMA3F was inhibited compared to control or to mock-transfected cells (Figure 5B). Furthermore, phospho-AKT was increased in normoxia when SEMA3F was inhibited (Figure 5B). These results demonstrate that SEMA3F repression has functional consequences on cell signaling.
ZEB-1-Induced Angiogenesis Is Reduced by Exogenous SEMA3F
In addition to direct effects on tumor cells, SEMA3F has been reported to be a potential antiangiogenic protein [11–14]. Because ZEB-1 inhibits SEMA3F, we wondered whether ZEB-1 induction would increase angiogenesis. This was tested using a chick CAM assay. Uninduced or Dox-induced H358 FlpIn ZEB-1 cells in Matrigel plug were implanted into the CAMs. Six days after implantation, the presence and orientation of newly formed blood vessels were evaluated (Figure 5, C and D). With uninduced cells, little angiogenesis was observed, whereas a significant angiogenic response was noticed when ZEB-1 was induced. However, this angiogenic response was reduced when recombinant SEMA3F was added in the plug. Therefore, we conclude that ZEB-1 induction is associated with increased angiogenesis that can be at least partially overcome by SEMA3F.
Discussion
SEMA3F impairs tumor formation in several xenograft models and has been shown to be an antiangiogenic and antimetastatic molecule [11–15]. Because of its tumor suppressor activity and its loss of expression in human lung cancers, it is important to understand how SEMA3F expression is regulated. Screening 21 NSCLC cell lines by quantitative real-time RT-PCR revealed a significant inverse correlation between SEMA3F and ZEB-1 expression suggesting that this E-box transcriptional repressor negatively regulates SEMA3F. To verify this model, we chose H661 lung cancer cells in which we previously demonstrated E-Cadherin down-regulation by ZEB-1 [20]. In the present study, we found that transiently transfected ZEB-1 decreased SEMA3F expression. On the contrary, when ZEB-1 was inhibited by siRNA, the expression of both SEMA3F and E-Cadherin was enhanced. With the presence of four conserved putative E-box sites in SEMA3F, we performed ChIP assays and found that two of these (site 1 in the CpG island and site 4 in intron 3) bound ZEB-1 (summary in Figure W1A). In addition, treatment with SAHA (vorinostat), an HDACi, increased SEMA3F expression and decreased ZEB-1 binding to these two sites, without affecting ZEB-1 expression. To verify that ZEB-1 binds SEMA3F in another lung cancer cell line, we performed ChIP assays in ZEB-1-inducible H358 FlpIn cells. As in H661 cells, ZEB-1 bound to sites 1 and 4 (Figure W1A) and reduced SEMA3F mRNA. Because site 1 is bipartite, composed of a CACCTG and CAGGTG sequence separated by 104 nucleotides, we mutated each site separately or together and observed increased expression using a luciferase reporter assay in H661 cells. Together, these results demonstrate that ZEB-1 is capable of repressing SEMA3F in lung cancer cells by directly binding to conserved E-box sites in the 5′region of the gene.
Snail, another E-box binding factor, which regulates the induction of EMT, is known to induce ZEB-1 expression by directly binding to ZEB-1 promoter [37]. Snail is induced by VEGF [38], and this induction involves suppression of the glycogen synthase kinase-3 after binding of VEGF to neuropilin in breast cancer cells [39]. Interestingly, in a microarray analysis, SEMA3F was downregulated in Snail-induced colon cancer cells [40]. In agreement with the down-regulation of SEMA3F by ZEB-1, we observed that Snail overexpression simultaneously induced ZEB-1 and repressed SEMA3F in H358 cells (data not shown). These data suggest that Snail indirectly regulates SEMA3F through ZEB-1 in these cells, although our studies do not rule out the possibility that Snail could also regulate the SEMA3F promoter perhaps at different stages of lung cancer development or progression.
We previously reported that SEMA3F, when overexpressed, reduced phospho-AKT in normoxia and HIF-1α protein in hypoxia in several cell lines. In this study, we demonstrated that SEMA3F loss in another cell line (H661 cells) has the opposite effect. We also report, in a CAM assay, that SEMA3F is able to modulate the angiogenic activity induced by ZEB-1. To our knowledge, angiogenic activity of ZEB-1 has not been previously reported. Because Snail and E47-expressing MDCK induced angiogenesis by the host stromal tissue in a transplantation assay [41] and angiogenic-related genes such as Jagged1 (JAG1) were upregulated in these cells [42], there is a precedent for ZEB-1 to have a similar effect. In addition, the consequences of SEMA3F loss and previous data published by us and others after SEMA3F overexpression suggest that loss of SEMA3F confers survival to tumor cells and is involved in angiogenesis.
As discussed earlier, ZEB-1 regulates the induction of EMT, which plays a critical role in tumor progression and invasion leading to metastasis [26]. Loss of E-Cadherin is one hallmark of EMT. E-Cadherin is a cell surface transmembrane protein that plays a major role in epithelial cell adhesion and connects the extracellular environment to the contractile cytoskeleton. Interestingly, E-Cadherin inhibition has been shown to result in a gradual EMT in A431 cells, although changes in E-box transcription factors were not reported [43]. Impaired E-Cadherin expression or function can alter the pattern of cell growth, differentiation, and invasiveness influencing survival in patients with cancer [22,44,45]. In lung cancer, we reported that expression of ZEB-1 and the loss of E-Cadherin correlate with resistance to gefitinib (an EGFR inhibitor), and with a poor prognosis [21,22]. We previously noted that MCF7 breast cancer cells, when treated with conditioned medium as a source of SEMA3F, reduced their contacts with delocalized E-Cadherin and β-catenin [46]. Therefore, we examined E-Cadherin expression and localization in islands of SEMA3F-induced H358-FlpIn cells.We observed both E-Cadherin and SEMA3F labeling at the membrane without E-Cadherin loss of intensity (Figure W3). The different results obtained with MCF7 and H358 cells might be explained by the different intrinsic adhesive properties of the two cell lines, the use of SEMA3F-conditioned medium (exocrine effects) versus induction (autocrine effects) or by the differences in the signaling response to SEMA3F [14]. Interestingly, very recently we transfected ZEB-1 into MCF7 cells (these cells express high levels of endogenous SEMA3Fand almost no ZEB-1). ZEB-1 was induced to a level similar to that of H661 lung cancer cells. However, we did not notice any difference in SEMA3F and E-Cadherin expression (data not shown) suggesting that in MCF7 cells, SEMA3F and E-Cadherin are not direct ZEB-1 targets or that a ZEB-1 coregulator/partner is absent in these cells. Therefore, there is some evidence that SEMA3F regulation and signaling involve different mechanisms in MCF7 breast cancer cells than in lung cancer cells.
In H661 cells, we found that HDAC inhibition was associated with ZEB-1 dissociation from its binding sites in E-Cadherin and SEMA3F, allowing the expression of these two genes that play a critical role in the inhibition of tumor development or progression. Similar effects with SAHA on transcription factor binding were reported for Myc and the p21 promoter in multiple myeloma cells [47]. Also, butyrate and trichostatin A led to decreased Sp1 and increased Sp3 binding to the major vault protein promoter in Hep3B cells [48], and trichostatin A nearly abolished TF-κB binding to the human tissue factor promoter in endothelial cells and monocytes [49]. Also, induction of TRAIL by HDACi was associated with the down-regulation of promoter-associated enzymes and acetylation of resident and de novo recruitment of Sp1 and Sp3 [50].
To our knowledge, SEMA3F represents the second tumor suppressor gene in lung cancer affected by ZEB-1 in addition to E-Cadherin. SEMA3F has been shown by us and others to have potent antitumor effects, and loss of E-Cadherin is associated with a poor prognosis and resistance to EGFR inhibitors. Together, these data indicate that ZEB-1 plays a critical role in the biology of lung cancer and represents an important therapeutic target.
Supplementary Material
Acknowledgments
The authors thank Anne Cantereau for helpful and efficient technical assistance for confocal microscopy studies performed in the confocal microscopy core at the University of Poitiers and Joanna Wdzieczak-Bakala, Neila Hajem, and Jian-Miao Liu from Institut de Chimie des Substances Naturelles, CNRS, Gif-sur-Yvette, France, for their help with the CAM assay.
Abbreviations
- CAM
chorioallantoic membrane
- ChIP
chromatin immunoprecipitation
- Dox
doxycycline
- E-Cad
E-Cadherin
- EGFR
epidermal growth factor receptor
- EMT
epithelial-mesenchymal transition
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- HIF
hypoxia-induced factor
- NSCLC
non-small cell lung carcinoma
- SCLC
small cell lung carcinoma
- VEGF
vascular endothelial growth factor
Footnotes
Financial support: This work was supported by La Ligue Contre le Cancer (J.C., J.R., and V.P.), l'Association pour la Recherche sur le Cancer (J.C., J.R., and V.P.), and National Institutes of Health CA58187 (Colorado Lung Cancer Specialized Program of Research Excellence to H.D., R.G., and V.P.).
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Roche J, Boldog F, Robinson M, Robinson L, Varella-Garcia M, Swanton M, Waggoner B, Fishel R, Franklin W, Gemmill R, et al. Distinct 3p21.3 deletions in lung cancer and identification of a new human semaphorin. Oncogene. 1996;12:1289–1297. [PubMed] [Google Scholar]
- 2.Sekido Y, Bader S, Latif F, Chen JY, Duh FM, Wei MH, Albanesi JP, Lee CC, Lerman MI, Minna JD. Human semaphorins A(V) and IV reside in the 3p21.3 small cell lung cancer deletion region and demonstrate distinct expression patterns. Proc Natl Acad Sci USA. 1996;93:4120–4125. doi: 10.1073/pnas.93.9.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang RH, Hensel CH, Garcia DK, Carlson HC, Kok K, Daly MC, Kerbacher K, van den Berg A, Veldhuis P, Buys CH, et al. Isolation of the human semaphorin III/F gene (SEMA3F) at chromosome 3p21, a region deleted in lung cancer. Genomics. 1996;32:39–48. doi: 10.1006/geno.1996.0074. [DOI] [PubMed] [Google Scholar]
- 4.Semaphorin Nomenclature Committee, author. Unified nomenclature for the semaphorins/collapsins. Cell. 1999;97:551–552. doi: 10.1016/s0092-8674(00)80766-7. [DOI] [PubMed] [Google Scholar]
- 5.Luo Y, Raible D, Raper JA. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- 6.Yazdani U, Terman JR. The semaphorins. Genome Biol. 2006;7:211–00. doi: 10.1186/gb-2006-7-3-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielenberg DR, Klagsbrun M. Targeting endothelial and tumor cells with semaphorins. Cancer Metastasis Rev. 2007;26:421–431. doi: 10.1007/s10555-007-9097-4. [DOI] [PubMed] [Google Scholar]
- 8.Neufeld G, Kessler O. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat Rev Cancer. 2008;8:632–645. doi: 10.1038/nrc2404. [DOI] [PubMed] [Google Scholar]
- 9.Sun Q, Nawabi-Ghasimi F, Basile JR. Semaphorins in vascular development and head and neck squamous cell carcinoma-induced angiogenesis. Oral Oncol. 2008;44:523–531. doi: 10.1016/j.oraloncology.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Potiron VA, Roche J, Drabkin HA. Semaphorins and their receptors in lung cancer. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.05.032. In press, online July 14, 2008. doi:10.1016/j.canlet.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielenberg DR, Hida Y, Shimizu A, Kaipainen A, Kreuter M, Kim CC, Klagsbrun M. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J Clin Invest. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Futamura M, Kamino H, Miyamoto Y, Kitamura N, Nakamura Y, Ohnishi S, Masuda Y, Arakawa H. Possible role of semaphorin 3F, a candidate tumor suppressor gene at 3p21.3, in p53-regulated tumor angiogenesis suppression. Cancer Res. 2007;67:1451–1460. doi: 10.1158/0008-5472.CAN-06-2485. [DOI] [PubMed] [Google Scholar]
- 13.Kessler O, Shraga-Heled N, Lange T, Gutmann-Raviv N, Sabo E, Baruch L, Machluf M, Neufeld G. Semaphorin-3F is an inhibitor of tumor angiogenesis. Cancer Res. 2004;64:1008–1015. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 14.Potiron VA, Sharma G, Nasarre P, Clarhaut JA, Augustin HG, Gemmill RM, Roche J, Drabkin HA. Semaphorin SEMA3F affects multiple signaling pathways in lung cancer cells. Cancer Res. 2007;67:8708–8715. doi: 10.1158/0008-5472.CAN-06-3612. [DOI] [PubMed] [Google Scholar]
- 15.Xiang R, Davalos AR, Hensel CH, Zhou XJ, Tse C, Naylor SL. Semaphorin 3F gene from human 3p21.3 suppresses tumor formation in nude mice. Cancer Res. 2002;62:2637–2643. [PubMed] [Google Scholar]
- 16.Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M. Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. J Cell Biol. 1999;146:233–242. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusy S, Nasarre P, Chan D, Potiron V, Meyronet D, Gemmill RM, Constantin B, Drabkin HA, Roche J. Selective suppression of in vivo tumorigenicity by semaphorin SEMA3F in lung cancer cells. Neoplasia. 2005;7:457–465. doi: 10.1593/neo.04721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brambilla E, Constantin B, Drabkin H, Roche J. Semaphorin SEMA3F localization in malignant human lung and cell lines: a suggested role in cell adhesion and cell migration. Am J Pathol. 2000;156:939–950. doi: 10.1016/S0002-9440(10)64962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusy S, Potiron V, Zeng C, Franklin W, Brambilla E, Minna J, Drabkin HA, Roche J. Promoter characterization of semaphorin SEMA3F, a tumor suppressor gene. Biochim Biophys Acta. 2005;1730:66–76. doi: 10.1016/j.bbaexp.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, Zeng C, Baron A, Bemis L, Erickson P, et al. WNT7a induces E-Cadherin in lung cancer cells. Proc Natl Acad Sci USA. 2003;100:10429–10434. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66:944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 22.Bremnes RM, Veve R, Gabrielson E, Hirsch FR, Baron A, Bemis L, Gemmill RM, Drabkin HA, Franklin WA. High-throughput tissue microarray analysis used to evaluate biology and prognostic significance of the E-Cadherin pathway in non-small-cell lung cancer. J Clin Oncol. 2002;20:2417–2428. doi: 10.1200/JCO.2002.08.159. [DOI] [PubMed] [Google Scholar]
- 23.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, Jung A, Kirchner T, Brabletz T. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 26.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 27.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 28.Thiery JP, Sleeman JP. Complex networks orchestrate epithelialmesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 29.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furusawa T, Moribe H, Kondoh H, Higashi Y. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor deltaEF1. Mol Cell Biol. 1999;19:8581–8590. doi: 10.1128/mcb.19.12.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian T, Chinnadurai G. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 2003;540:255–258. doi: 10.1016/s0014-5793(03)00275-8. [DOI] [PubMed] [Google Scholar]
- 34.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 35.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC, Huylebroeck D. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 37.Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 38.Yang AD, Camp ER, Fan F, Shen L, Gray MJ, Liu W, Somcio R, Bauer TW, Wu Y, Hicklin DJ, et al. Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 2006;66:46–51. doi: 10.1158/0008-5472.CAN-05-3086. [DOI] [PubMed] [Google Scholar]
- 39.Wanami LS, Chen HY, Peiro S, Garcia de Herreros A, Bachelder RE. Vascular endothelial growth factor-A stimulates Snail expression in breast tumor cells: Implications for tumor progression. Exp Cell Res. 2008;314:2448–2453. doi: 10.1016/j.yexcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Craene B, Gilbert B, Stove C, Bruyneel E, van Roy F, Berx G. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–6244. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- 41.Peinado H, Marin F, Cubillo E, Stark HJ, Fusenig N, Nieto MA, Cano A. Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo. J Cell Sci. 2004;117:2827–2839. doi: 10.1242/jcs.01145. [DOI] [PubMed] [Google Scholar]
- 42.Moreno-Bueno G, Cubillo E, Sarrio D, Peinado H, Rodriguez-Pinilla SM, Villa S, Bolos V, Jorda M, Fabra A, Portillo F, et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res. 2006;66:9543–9556. doi: 10.1158/0008-5472.CAN-06-0479. [DOI] [PubMed] [Google Scholar]
- 43.Andersen H, Mejlvang J, Mahmood S, Gromova I, Gromov P, Lukanidin E, Kriajevska M, Mellon JK, Tulchinsky E. Immediate and delayed effects of E-cadherin inhibition on gene regulation and cell motility in human epidermoid carcinoma cells. Mol Cell Biol. 2005;25:9138–9150. doi: 10.1128/MCB.25.20.9138-9150.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 45.Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 46.Nasarre P, Kusy S, Constantin B, Castellani V, Drabkin HA, Bagnard D, Roche J. Semaphorin SEMA3F has a repulsing activity on breast cancer cells and inhibits E-cadherin-mediated cell adhesion. Neoplasia. 2005;7:180–189. doi: 10.1593/neo.04481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steiner E, Holzmann K, Pirker C, Elbling L, Micksche M, Berger W. SP-transcription factors are involved in basal MVP promoter activity and its stimulation by HDAC inhibitors. Biochem Biophys Res Commun. 2004;317:235–243. doi: 10.1016/j.bbrc.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Mahmud SA, Bitterman PB, Huo Y, Slungaard A. Histone deacetylase inhibitors suppress TF-kappaB-dependent agonist-driven tissue factor expression in endothelial cells and monocytes. J Biol Chem. 2007;282:28408–28418. doi: 10.1074/jbc.M703586200. [DOI] [PubMed] [Google Scholar]
- 50.Nebbioso A, Clarke N, Voltz E, Germain E, Ambrosino C, Bontempo P, Alvarez R, Schiavone EM, Ferrara F, Bresciani F, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 51.Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.