Abstract
Hodgkin lymphoma (HL) is derived from preapoptotic germinal center B cells, although a general loss of B cell phenotype is noted. Using quantitative reverse transcription-polymerase chain reaction and miRNA microarray, we determined the microRNA (miRNA) profile of HL and compared this with the profile of a panel of B-cell non-Hodgkin lymphomas. The two methods showed a strong correlation for the detection of miRNA expression levels. The HL-specific miRNA included miR-17-92 cluster members, miR-16, miR-21, miR-24, and miR-155. Using a large panel of cell lines, we found differential expression between HL and other B-cell lymphoma-derived cell lines for 27 miRNA. A significant down-regulation in HL compared to non-Hodgkin lymphoma was observed only for miR-150. Next, we performed target gene validation of predicted target genes for miR-155, which is highly expressed in HL and is differentially expressed between HL and Burkitt lymphoma. Using luciferase reporter assays, we validated 11 predicted miR-155 target genes in three different HL cell lines. We demonstrated that AGTR1, FGF7, ZNF537, ZIC3, and IKBKE are true miR-155 target genes in HL.
Introduction
Hodgkin lymphoma (HL) is one of the most frequently occurring lymphomas, with an annual incidence rate of three to four new cases per 100,000 persons in the Western world. Two different subtypes of HL, i.e., classic (cHL) and nodular lymphocyte predominant (NLPHL), can be distinguished based on histology, infiltrating cells and tumor cell characteristics [1]. Classic HL is characterized by the appearance of Hodgkin and Reed-Sternberg (HRS) cells. These large multinuclear cells are derived from preapoptotic germinal center (GC) B cells and are characterized by a loss of B-cell phenotype [2]. The tumor cells of NLPHL, called lymphocytic and histiocytic cells, display a relatively normal B-cell phenotype and are also derived from GC B cells [3]. Using gene expression analysis, an overlap has been identified between cHL and primary mediastinal B-cell lymphoma (PMBL) [4]. Gene expression profiling of B-cell lymphoma cell lines classified cHL cell lines as a distinct entity, irrespective of cellular origin, indicating that a specific gene expression program is activated in cHL [2].
MicroRNA (miRNA) are short (approximately 22 nt) noncoding RNA molecules that inhibit gene expression by binding to complementary sequence at the 3′ untranslated region (UTR) of (target) genes, including oncogenes and tumor suppressor genes [5]. Nucleotides 2 to 7 from the 5′ end of the miRNA, the seed region, are considered crucial for miRNA-target gene interaction [6]. MiRNA expression has been shown to be tissue-specific as well as temporally regulated [7]. Furthermore, expression of specific miRNA can influence B-cell [8] and hematopoietic lineage differentiation [9] and also regulate the GC reaction [10]. Previously, we have shown that BIC and its mature miRNA product miR-155 are highly expressed in HL, PMBL, and diffuse large B-cell lymphoma cell lines (DLBCL) [11,12]. In contrast, Burkitt lymphoma (BL) is characterized by relatively low miR-155 expression levels, indicating a differential regulation of miR-155 expression between these B-cell lymphomas [13]. Germinal center B cells upregulate BIC/miR-155 [10] leading to down-regulation of target mRNA such as the activation-induced cytidine deaminase [14].
In this study, we established an HL-specific miRNA expression profile using stem-loop-specific quantitative reverse transcription-polymerase chain reaction (qRT-PCR) [15] and a miRNA microarray using the Agilent platform [16]. The specificity of miRNA that were abundantly or differentially expressed in HL was evaluated in cell lines derived from various B-cell non-Hodgkin lymphomas. In addition, we validated 11 predicted target genes of miR-155 in HL cell lines.
Materials and Methods
Cell Lines Included for Profiling of miRNA
The cHL cell lines (L428, KM-H2, and L1236), the NLPHL cell line (DEV), the EBV-transformed GC B-cell-derived lymphoblastoid cell lines (CB-LCL 5.B8 and CB-LCL 6.28 [17]), the PMBL cell lines (Karpas 1106P and MEDB-1), and the EBV-negative BL cell lines (Ramos, CA46, and DG-75) were cultured at 37°C in an atmosphere containing 5% CO2 in RPMI-1640 medium (Cambrex Biosciences, Walkersville, MD) supplemented with penicillin (100 U/ml), streptomycin (0.1 mg/ml), and ultraglutamine (2 mM; Cambrex Biosciences) and 5% (L428), 20% (DEV), or 10% (rest) fetal calf serum (Cambrex Biosciences). All cell lines included for subsequent qRT-PCR validation to determine which miRNA are HL-specific are listed in Table W1.
RNA Isolation
Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The quantity was measured on a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE), and the integrity of the RNA was checked on a 1% agarose gel. Good-quality RNA samples were used for subsequent analysis.
Quantitative RT-PCR
MiRNA expression was measured using a prerelease version of the TaqMan miRNA quantitative PCR assay including a total of 183 human miRNA from Applied Biosystems (Foster City, CA) as previously described [15].
Briefly, cDNA was made from 5 ng of total RNA in 15-µl reactions using (Multiscribe) MuLV RT and specific primers for each miRNA. The RT reaction was performed at 16°C for 30 minutes followed by 42°C for 30 minutes, 85°C for 5 minutes, and a final hold at 4°C. The PCR mix consists of 1.33 µl of cDNA from the RT product, 10 µl of Eurogentec Master Mix (Eurogentec, Liege, Belgium), and 2 µl of TaqMan MicroRNA Assay Mix containing primers and probe for the miRNA of interest, in a total volume of 20 µl. Cycle parameters for the PCR are 95°C for 10 minutes (AmpliTaq Gold enzyme activation), followed by 40 cycles of a denaturing step at 95°C for 15 seconds and an annealing/extension step at 60°C for 60 seconds. All reactions were run in triplicate. Mean cycle threshold (Ct) values for all miRNA were quantified with the sequence detection system software (SDS, version 2.1; Applied Biosystems). The miRNA expression was normalized to U6 mRNA expression resulting in a ΔCt from which the 2−ΔCt value was derived and depicted.
Cluster Analysis
Unsupervised clustering analysis of Ct data was performed using Genesis [18]. After median centering of the Ct data over all samples, complete similarity metrics were calculated for these analyses. Unsupervised clustering was performed using the Pearson correlation coefficient. MiRNA that were discriminative (t-test, P < .05) between cHL cell lines and EBV-transformed CB-derived lymphoblastoid cell lines, between cHL and PMBL cell lines, and between cHL and BL cell lines were depicted using a heat map.
Microarray Hybridization
Microarray analysis was performed by Agilent using the Agilent Oligo Microarray Kit (G4470A; Agilent, Santa Clara, CA) and the protocol provided by the company [16]. Briefly, 100 ng of total RNA, also used for qRT-PCR, was treated with calf intestine alkaline phosphatase for 30 minutes at 37°C before labeling. Samples (7 µl) were diluted with 5 µl of DMSO (D8418; Sigma, St. Louis, MO), denatured for 10 minutes at 100°C and labeled in a total volume of 20 µl at 16°C for 2 hours using pCp0-Cy3 in T4 RNA ligation buffer supplied in the company kit (5190-0408; Agilent). After purification with Bio-Rad MicroBio-Spin 6 columns (732-6221; Bio-Rad, Hercules, CA), labeled miRNA were kept on ice.
Samples were hybridized at 55°C for 20 hours in an Agilent SureHyb chamber (G2534A; Agilent) rotated at 20 rpm. The arrays were washed with Gene Expression Wash Buffer (Agilent) at 37°C under required ozone conditions before scanning with an Agilent microarray scanner (G2565BA; Agilent).
Microarray data analysis was performed using Agilent Feature Extraction Software (www.agilent.com/chem/fe) with the protocol available for miRNA expression analysis at www.agilent.com/chem/feprotocols.
MiR-155 Target Prediction
A list of potential miR-155 targets was created by combining predicted targets from mirBase, TargetBoost, TargetScanS, miRanda, and PicTar (reviewed by Watanabe et al. [19]). Potential targets were chosen based on gene function, number of predicted target sites, and target prediction by multiple algorithms and were subsequently validated using a luciferase reporter assay, as described below in detail.
MiR-155 Target Cloning
Putative miR-155 target sequences were amplified using primers, with NotI and SstI restriction sites (Table W2). Polymerase chain reaction products were ligated into a TOPO vector (Invitrogen, Breda, the Netherlands) and transformed into TOP10F′ competent cells (Invitrogen). Using NotI and SstI endonucleases (Invitrogen), the insert was cut out, purified, and ligated into the 3′ UTR of Renilla luciferase (RL) in a modified psiCHECK2 vector (Promega, Madison, WI). This vector also contains a firefly luciferase (FL) gene used to normalize for transfection efficiency. The constructs were sequenced to check for the proper insert.
Transfection of Hodgkin Cell Lines L428, L1236, and DEV
Using an Amaxa nucleofector device (AAD-1001; Amaxa, Gaithersburg, MD), cell lines were transfected with 2 µg of the modified psiCHECK2 plasmid containing a target 3′ UTR with or without 5.7 µM anti-miR-155 LNA-modified oligonucleotides (Custom LNA Oligonucleotides; Exiqon, Vedbaek, Denmark) in 100 µl of nucleofector solution (Amaxa) in triplicate. An additional control using an inhibitor against miR-220 (not expressed in HL cell lines) was performed for all putative targets. L1236 and DEV were transfected using kit V (protocols T01 and G16); L428 was transfected with kit L (protocol X01). Transfection efficiencies of living cells were tested using a green fluorescent protein plasmid and were 39% for L1236, 51% for DEV, and 88% for L428.
Generation of Seed Sequence Mutants
Mutated seed sequences were generated using psiCHECK2 plasmids as a template. The 7-nt miR-155 seed regions in the AGTR1 and FGF7 constructs were deleted using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as reported previously [20]. Briefly, a forward mutagenic deletion primer and a complementary reverse mutagenic deletion primer were synthesized for AGTR1 (forward: 5′-CTTCACTACCAAATGGCTACTTTTCAGAAT-3′; reverse: 5′-ATTCTGAAAAGTAGCCATTTGGTAGTGAAG-3′). Mutants were generated in a PCR experiment as described by the manufacturer. The amplified DNA was treated with DpnI restriction enzyme to eliminate the parental template, and the remaining DNA was used for transformation. The deletion of the miR-155 seed binding sequence was confirmed by sequencing. The seed sequence mutant for murine FGF7 (CCTCTGTGATTACAGGAATTA) was kindly provided by Dr. B. Mari (Faculte de Medecine Pasteur, Nice, France).
Luciferase Assay
Cell lysates were made 48 hours after transfection using the lysis buffer provided by the Promega Dual-Luciferase Reporter Assay System (E1910; Promega). In a 96-well plate, 40 µl of cell lysates was added to 100 µl of LARII (Promega) in duplicate. Firefly luciferase was measured in duplicate using a Luminoscan Ascent plate reader (Thermo Scientific, Waltham, MA). After the addition of 100 µl of Stop & Glo buffer, RL activity was measured on the Luminoscan Ascent plate reader. MiR-155 targeting was validated with an miR-155-specific inhibitor (Exiqon) and compared to transfection without an inhibitor (mock). An inhibitor against miR-220, which is not expressed in HL cell lines, was cotransfected as a control for nonspecific effects (not shown).
In this assay, RL expression is reduced on binding of miR-155 to the target sequences cloned into the 3′-UTR of the RL gene. Protein expression of the RL gene was normalized to the signal obtained from FL (endogenous transfection control) resulting in a relative luciferase expression. The RL/FL ratio of cells cotransfected with anti-miR-155 or anti-miR-220 (used as a negative control) was compared to cells transfected only with the construct of interest (set at 100%).
Statistical Analysis on Large Panel of Cell Lines
Differential expression between cell lines was established using a nonparametric Kruskal-Wallis test in Prism 5.0 (GraphPad Software Inc., San Diego, CA). P values <.05 were considered statistically significant. Dunn's multiple comparisons posttest was performed on significantly differentially expressed miRNA to determine which group of cell lines deviated significantly from HL. Asterisks indicate increasing levels of significance.
Results
Hodgkin Lymphoma-Specific Pattern of miRNA Expression
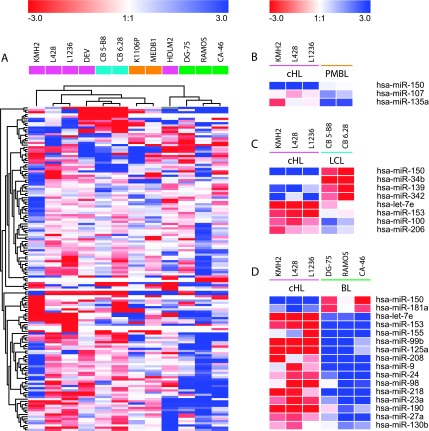
Quantitative RT-PCR for 183 miRNA was used to define an HL-specific pattern of miRNA expression. The 26 most abundantly expressed miRNA within the HL cell lines (L428, L1236, HDLM2, and KM-H2) included miR-92, miR-21, miR16, miR142-3p, mir-17-5p, and miR-155 (Table 1). Remarkably, within the group of 26 highly expressed miRNA, an overlap in genome position and seed sequence (seed families) was identified. Unsupervised clustering indicated that cHL cell lines clustered separately from CB-LCL, PMBL, and BL cell lines (Figures 1 and W1). The NLPHL cell line DEV clustered more closely to CB-LCL than to HL (Figure 1A). Burkitt lymphoma cell lines were located in a separate cluster, together with the T-cell HL cell line HDLM2. Of a total of 23 miRNA that were differentially expressed, 3 were found between cHL and PMBL cell lines, 8 between cHL and CB-LCL, and 17 between cHL and BL (Figure 1). In all three comparisons, most of the differentially expressed miRNA were upregulated in HL, including let-7e compared with LCL and BL (P = .0004 and P = .01), miR-155 compared with BL (P = .007), and miR-107 compared with PMBL (P = .05). Only a few miRNA were downregulated in HL, among which was miR-150 that was significantly downregulated in HL compared with LCL (P = .05), PMBL (P = .01), and BL (P = .02; Figure 1).
Table 1.
Abundantly Expressed miRNA in Classic Hodgkin Cell Lines.
| No. | miRNA | Chromosome | Seed Family* |
| 1 | miR-15b | 3q26.1 | AGCAGCA |
| 2 | miR-16 | 3q26.1/13q14.2 | AGCAGCA |
| 3 | miR-191 | 3p21.31 | |
| 4 | miR-565 | 3p21.31 | |
| 5 | miR-103 | 5q35.1/20p13 | |
| 6 | miR-25 | 7q22 | AUUGCAC |
| 7 | miR-93 | 7q22 | AAAGUGC |
| 8 | miR-106b | 7q22 | AAAGUGC |
| 9 | miR-29a | 7q32.3 | AGCACCA |
| 10 | miR-29b | 7q32.3/1q32.2 | AGCACCA |
| 11 | miR-29c | 1q32.2 | AGCACCA |
| 12 | miR-30b | 8q24.2 | |
| 13 | Let-7a | 9q22.32/11q24.1/22q13.31 | GAGGUAG |
| 14 | Let-7f | 9q22.32 | GAGGUAG |
| 15 | Let-7g | 3p21.1 | GAGGUAG |
| 16 | miR-17-5p | 13q31 | AAAGUGC |
| 17 | miR-19a | 13q31 | GUGCAAA |
| 18 | miR-19b | 13q31 | GUGCAAA |
| 19 | miR-20a | 13q31 | AAAGUGC |
| 20 | miR-92a | 13q31/Xq26.2 | AUUGCAC |
| 21 | miR142-3p | 17q23.2 | |
| 22 | miR142-5p | 17q23.2 | |
| 23 | miR-21 | 17q23.2 | |
| 24 | miR-155 | 21q21 | |
| 25 | miR-106a | Xq26.2 | AAAGUGC |
| 26 | miR-20b | Xq26.2 | AAAGUGC |
An miRNA family is composed of miRNA with the same seed region (positions 2 to 8 of the mature miRNA, also called seed+m8) (http://www.targetscan.org). MiRNA are grouped by location and seed family as indicated by altering shades of gray.
Figure 1.
Characterization of cell lines based on differential miRNA expression patterns. The Euclidian distance between cell lines and miRNA based on miRNA expression as found by an unsupervised hierarchical clustering (A). Differentially expressed miRNA identified by a t-test between cHL and PMBL (B), between cHL and CB-LCL (C), and between cHL and BL (D).
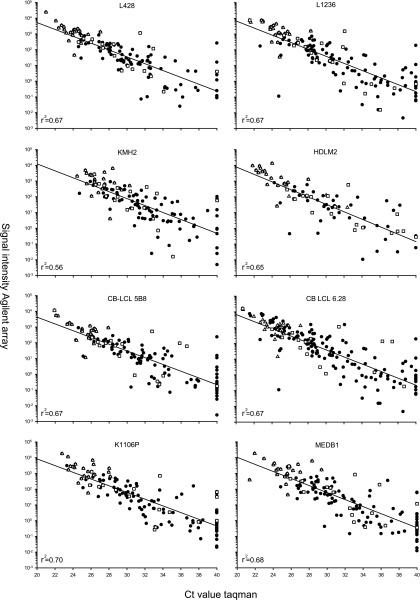
To underline these qRT-PCR data, miRNA microarrays were performed for cHL, CB-LCL, and PMBL cell lines. Ct values obtained with qRT-PCR correlated well with signal intensities on the array (0.56 ≤ r2 ≤ 0.70), indicating that the qRT-PCR data are consistent with the Agilent miRNA microarray (Figure 2). The 26 most abundant miRNA in cHL cell lines, as detected with qRT-PCR, were also among the most abundant as detected by miRNA microarray. Moreover, the miRNA differentially expressed between cHL and CB as well as cHL and PMBL did not deviate from the regression line, indicating that differential expression was based on accurate quantification.
Figure 2.
Correlation between miRNA microarray and qRT-PCR expression levels. The miRNA microarray contained 170/183 miRNA tested by qRT-PCR. Low Ct values (x-axis), obtained by qRT-PCR, correlate with high signal intensity (y-axis) observed with microarray. The high correlation coefficient (r2) indicates a positive relation between low Ct values and high signal intensity. Both platforms show the same miRNA abundantly expressed in HL (open triangles). MiRNA differentially expressed between cHL and CB and cHL and PMBL (open squares) do not deviate from this correlation. For some miRNA, no expression was observed by qRT-PCR (Ct value of 40), whereas low/average expression was observed on the microarray.
Hodgkin Lymphoma-Specific miRNA Signature on Extended Cell Line Panel
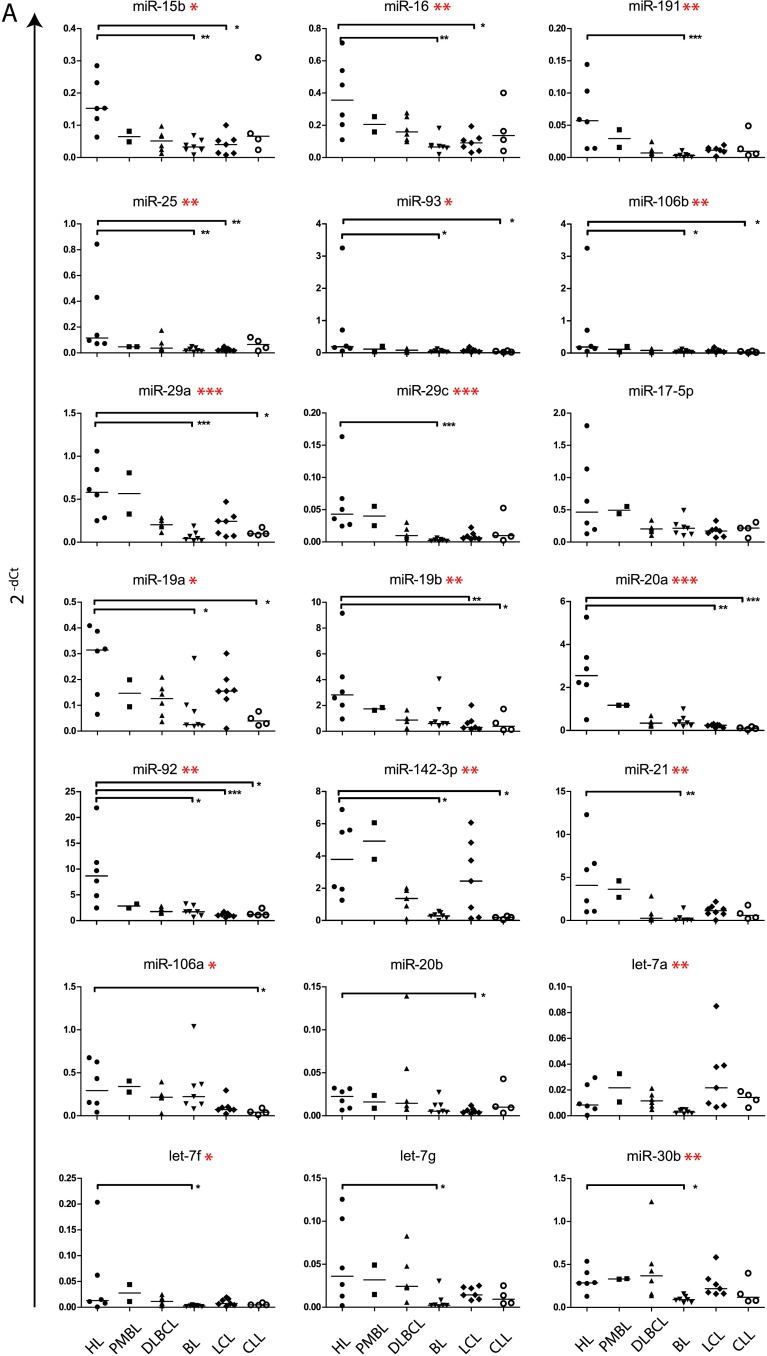
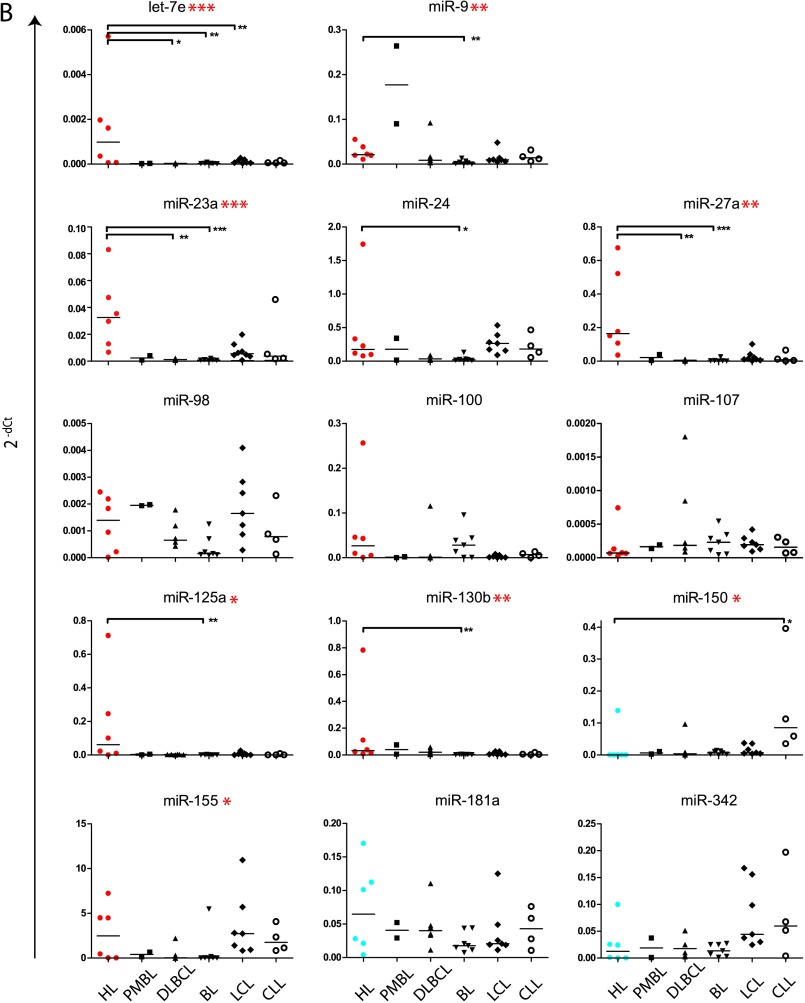
Of the 23 differentially expressed miRNA (Figure 1), we selected let-7e, miR-9, miR-23a, miR-24, miR-27a, miR-98, miR-100, miR-107, miR-125a, miR-130b, miR-150, miR-153, miR-155, miR-181a, miR-206, and miR-342 for further analysis. These 16 miRNA and 22 of the 26 highly expressed miRNA in HL cell lines (Table 1) were compared on an extended cell line panel, including EBV-transformed LCLs, BL cell lines, PMBL cell lines, DLBCL, and lymphocytic leukemia cell lines (CLL). Of the selected 16 differentially expressed miRNA, 8 were significantly different between HL and other cell lines, whereas of the 22 highly expressed miRNA, 19 miRNA significantly differed in expression levels (Figure 3). There was no differential expression of miR-155, miR-181a, miR-98 (HL vs BL), miR-107, miR-150 (HL vs PMBL), miR-342, miR-150, and miR-100 (HL vs LCL) in this larger cell line panel. For miR-155, high levels were found in the EBV-positive CLL and LCL cell lines, indicating a positive relation between EBV positivity and miR-155 expression. Hodgkin lymphoma cell lines significantly overexpressed a large number of well-known miRNA, including miR-21, miR-15b, and miR-16. Furthermore, there was a strong up-regulation of miR-17-92 cluster members, compared to BL (miR-19a, miR-19b, and miR-20a), DLBCL (miR-19b), LCL (miR-92), and CLL (miR-19a, miR-20a, and miR-92; Figure 3). Also, analogous cluster members located at chromosome Xq26 (miR-106a) and chromosome 7q22 (miR-25, miR-93, and miR-106b) were highly expressed in HL (Table 1). MiR-17-5p showed a trend for higher expression in HL (Figure 3A), whereas miR-18a was not differentially expressed and miR-17-3p showed significantly lower expression in HL compared to DLBCL cell lines. The most significant differences were generally found between HL and BL. No significant differences were found between HL and PMBL.
Figure 3.
Differential expression of selected miRNA in a large panel of cell lines. Abundant miRNA (A) and differentially expressed miRNA (B) selected from qRT-PCR profiling were evaluated on a panel of lymphoma cell lines. MiR-155, shown in (B), was both highly and differentially expressed in the initial profile. In (B), HL cell lines with a significantly higher miRNA expression in the initial screen on differential expression (Figure 1) are marked in red, whereas cell lines that contained a significantly lower expression of the given miRNA are marked in blue. Data for miR-206 and miR-153 were not shown because their expression was too low (Ct values >35) to be quantified reliably. Significant differences in a comparison between all groups were obtained by a Kruskal-Wallis test and are marked with asterisks next to the miRNA name. Asterisks within the plots indicate significant differences between Hodgkin cell lines and other categories specifically as found by Dunn's multiple comparison test. For both tests: *P < .05, **P < .01, and ***P < .001.
MiR-155 Target Gene Validation
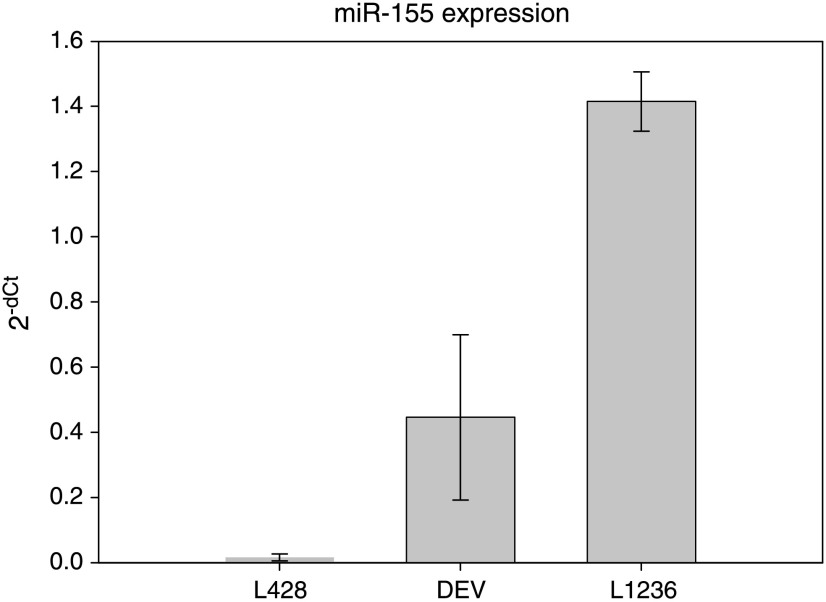
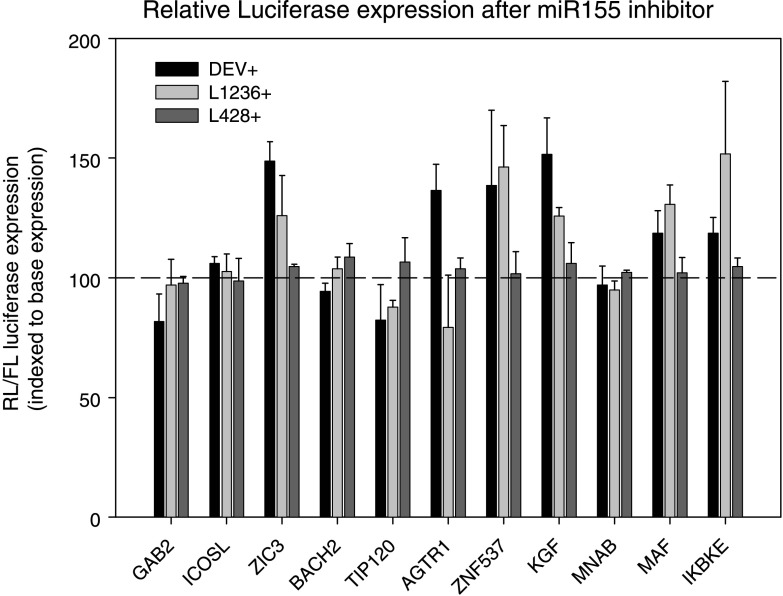
On the basis of the abundant and differential expression of miR-155 in HL and the reported interest for miR-155, both by our group and other groups, we chose to further validate putative target genes of miR-155. Putative target genes identified by one or more of five different target prediction algorithms (PicTar, TargetBoost, TargetScanS, MiRanda, and miRbase) were screened for the location and number of putative binding sites as well as their biologic relevance. Eleven putative targets were selected, cloned into the 3′ UTR of an RL reporter gene of a modified psiCHECK2 vector containing and transfected into HL cell lines L428, L1236, and DEV for further target validation (Figure W2). Selection of these cell lines was based on differences in miR-155 levels as previously demonstrated by Northern blot analysis [12]. Using qRT-PCR, we showed that miR-155 levels are high in DEV and L1236 and relatively low in L428 (Figure 4). After cotransfection with anti-miR-155 oligonucleotides, significantly increased relative luciferase expression levels were obtained for AGTR1, ZNF537 (TSHZ3), FGF7 (KGF), ZIC3, MAF, and IKBKE, implying that these genes are true miR-155 targets (Figure 5). The RL/FL ratios did not coincide with the level of repression observed after the addition of anti-miR-155 oligonucleotides, indicating that the ratio itself is not a good measure for the extent of miRNA-based repression (Figure W3). Cotransfection with oligonucleotides against miR-220, a miRNA that is not expressed in the HL cell lines, did not alter luciferase expression (data not shown)-moreover, transfection with AGTR1 and FGF7, containing mutated seed sequences, confirmed the effects obtained by the anti-miR-155 oligonucleotides (data not shown).
Figure 4.
Quantitative RT-PCR on miR-155 expression in HL cell lines. Triplicate measurements were performed for three (two for L1236) separate RNA isolations. Constant miR-155 levels were observed for cells that were harvested at different time points.
Figure 5.
Relative luciferase expression after miR-155-specific inhibition was indexed relative to the ratio obtained without inhibitor. Relative luciferase ratios are given for the cell lines transfected with psiCHECK2 constructs containing 3′ UTR sequences of potential miR-155 target genes. Error bars indicate the SD for three replicate experiments. AGTR1 (DEV), ZNF537 (DEV, L1236), FGF7 (DEV, L1236), ZIC3 (DEV), MAF (L1236), and IKBKE (L1236) show increased expression after miR-155-specific inhibition.
Discussion
MicroRNA profiling has been used to classify human cancers according to their developmental lineage and differentiation state [21]. In lymphoid malignancies, miRNA profiles have been used to successfully characterize BL and CLL [22,23]. In addition, Navarro et al. [24] have used miRNA profiling on total HL tissue specimens to distinguish between different HL subtypes, and Nie et al. [25] have shown that Blimp-1 is a target of miR-9 and let-7a in HL cell lines. Up until now, no studies have reported on a complete miRNA expression profile for HRS cells of HL.
In this study, we compared the miRNA expression profile of HL to that of other B-cell-derived lymphoma cell lines using qRT-PCR and miRNA microarrays to screen 183 and 470 different human microRNA, respectively. The results obtained from these independent platforms were significantly correlated, and verification on a large panel of B-cell-derived cell lines revealed that 19 of 22 selected highly expressed miRNA were consistent and that 8 of 16 selected differentially expressed miRNA were differentially expressed between HL and a large panel of other B-cell lymphoma-derived cell lines (Figure 3). The lack of differential expression for 8 of the miRNA is most likely caused by the heterogeneity in miRNA profile of the HL cell lines used in the larger cell line panel (including the non-cHL cell line, DEV, the T-cell-derived HL cell line, HDLM2, and the EBV-positive HL cell line, L591). The differences in miRNA profiles can be seen for two of these cell lines in Figure 1A. Although half of the selected miRNA within the larger panel were not significantly differentially expressed, consistent results were obtained for the three cHL cell lines used to identify differentially expressed miRNA.
Compared to other B-cell lymphoma cell lines, overexpression of the miR-17-92 cluster members miR-17-5p, miR-19a, miR-19b, miR-20a, and miR-92, is prominent in HL. Furthermore, a high expression of analogous cluster members miR-106a, miR-25, and miR-93, derived from chromosome 7 and X, implies an important role of these clusters of miRNA with partially overlapping seed sequences in the pathophysiology of HL [26]. Although most miR-17-92 cluster members (but not miR-17-3p and miR-18a) are highly expressed in HL, we show that differences in expression levels between miRNA derived from this single cluster exist. This implies not only that expression depends on the primary transcript levels but also that some cluster members are more or less stable or specifically regulated, as has previously been shown for pre-miR-18a [27]. Moreover, highly expressed miRNA were not specifically located within aberrant chromosomal regions, present in the tested HL cell lines [28,29].
MiR-15 and miR-16 are both abundantly and differentially expressed in HL. Reduced levels of both these miRNA, which are known to target BCL2, have been shown to protect cells against apoptosis in CLL [30]. High Bcl-2 protein levels in HRS cells have been reported as a marker for unfavorable prognosis in HL cases [31]. Because not only Bcl-2 but also miR-15 and miR-16 expression levels are high in HL, miR-15. and miR-16-mediated repression of BCL2 does not seem to occur in HL. Whereas elevated levels of miR-15 and miR-16 decrease Bcl-2 expression, elevated levels of miR-21 have been associated with increased Bcl-2 protein levels in breast cancer cell lines [32]. If BCL2 is indeed a target of both miR-21 and miR-15/-16 in HL, high miR-21 expression might, presumably indirectly, counteract repression of BCL2 translation, induced by miR-15 and miR-16.
Although most miRNA had significantly higher expression levels in HL compared to other B-cell lymphoma cell lines, the level of miR-150 expression was significantly lower. MiR-150 is expressed in mature B and T cells and it has been shown to target the transcription factor c-Myb [8,33]. In our cell line panel, the only HL with abundant miR-150 levels was HDLM2, a T-cell-derived HL cell line. We found the largest difference in miR-150 expression levels between HL and CLL cell lines. In CLL, we previously reported that miR-150 expression levels were strong and homogeneous for neoplastic cells outside proliferation centers and absent or very low for the CLL cells in the proliferation centers, and this staining pattern was inversely correlated to miR-155 expression levels [34].
In HL cell lines and cases, the primary miR-155 gene BIC and miR-155 are abundantly expressed [12], whereas in BL, BIC and miR-155 expression are almost absent [13]. The lack of significant differences for miR-155 between HL and BL cell lines is probably caused by inclusion of the three EBV-positive BL cell lines Jiyoye, Namalwa, and Raji, which express high levels of miR-155. We have previously reported on the putative relation between latency type III EBV-positive BL cell lines and miR-155 expression [13]. In line with this and consistent with observations by others [35], the EBV-transformed LCL cell lines used in our study were also miR-155-positive.
Previously, AGTR1 [20,36], IKBKE [36,37], BACH1 [36,38], Fos [36], Pu.1 [39], ZIC3 [38], and TP53INP1 [40] were shown to be miR-155 target genes in various cell lines. We demonstrate effective targeting in HL by miR-155 for AGTR1, IKBKE, ZNF537, ZIC3, and FGF7. Remarkably, the extent of target inhibition in the HL cell lines seemed to correlate with the miR-155 levels. We show that miR-155-mediated repression of these miR-155 target genes in HL cell lines depends on the cellular context and possibly the amount of available miRNA (Figures 4 and 5). This has been suggested previously for miR-150 by Xiao et al. [33] who have shown that small changes in miR-150 expression profoundly affects the expression of its target gene c-Myb in hematopoietic cell lineages specifically. On the basis of conditional and partial ablation of c-Myb, effective repression of c-Myb by miR-150 is determined within a narrow window of expression of both miR-150 and c-Myb [33]. The lack of luciferase suppression, as observed in the miR-155-low cell line L428 (which has the best transfection efficiency), might be due to low levels of miR-155 or to repression by miRNA other than miR-155. Differences in repression between DEV and L1236, both with high levels of miR-155, indicate that it is important to study miRNA function in cell lines with relevant endogenous miRNA expression levels and not in cell lines of other cell types.
IKBKE (IKKε)-mediated phosphorylation of cRel leads to dissociation of the IκBα-cRel complex resulting in nuclear accumulation of cRel and subsequent NF-κB activation [41]. In HL, cREL is often amplified [42] resulting in the activation of NF-κB. The high levels of miR-155 in HL cell lines might represent a counter action to reduce activated NF-κB levels [37]. This is supported by our observation on cREL amplification without increased mRNA levels in HL cell lines [28]. The angiotensin type II receptor 1 (AGTR1/AT1) is expressed on B cells and monocytes [43]. Recently, a single nucleotide polymorphism in the 3′ UTR of AGTR1 (A1166C) has been shown to affect miR-155-mediated repression [44]. ZNF537 (TSHZ3) is a highly conserved human variant of the teashirt (tsh) gene. TSHZ3 might act as a transcriptional repressor, which in Drosophila can be activated by FGF8 [45]. The potential miR-155 target gene FGF7 (KGF) is a canonical fibroblast growth factor family member known to activate Akt [46]. In HL, Akt is activated, possibly through CD40, CD30, and RANK [47]. However, there is no data on the expression and function of AGTR1, TSH23, ZIC3, and FGF7 in HRS cells. Expression of MAF has only been detected on the mRNA level in HL cell lines L1236 and KM-H2, whereas no protein expression was found by immunohistochemistry in HL patients [48].
We conclude that HL is characterized by high expression levels of various miRNA. The miRNA expression profile of HL cell lines can be used to distinguish HL from other B-cell-derived cell lines because HL cell lines cluster separately based on miRNA expression. We also show that IKBKE, ZNF537, ZIC3, FGF7, and AGTR1 are functional targets of miR-155 in HL.
Supplementary Material
Acknowledgments
The authors thank B. Mari (Faculté de Médecine Pasteur, Nice, France) for providing the FGF7 constructs. The authors thank M.J. Dyer (University of Leicester, UK) for the primary mediastinal cell line Karpas 1106; R. Küppers and D. Siemer (University of Duisburg-Essen, Germany) for the CB-LCL cell lines; and A. Epstein (UCLA, Los Angeles, CA) for providing the DLBCL cell lines SU-DHL-4 and SU-DHL-6.
Abbreviations
- BL
Burkitt lymphoma
- CB
centroblast
- CB-LCL
germinal center B-cell-derived lymphoblastoid cell line
- CLL
chronic lymphocytic leukemia
- DLBCL
diffuse large B-cell lymphoma
- EBV
Eppstein-Barr virus
- FL
firefly luciferase
- cHL
classic Hodgkin lymphoma
- GC
germinal center
- HL
Hodgkin lymphoma
- HRS
Hodgkin and Reed-Sternberg
- miRNA
microRNA
- NLPHL
nodular lymphocyte predominant Hodgkin lymphoma
- PMBL
primary mediastinal B-cell lymphoma
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- RL
Renilla luciferase
- UTR
untranslated region
Footnotes
This project is supported by the Dutch Cancer Foundation (2006-2634) and the VanderEs Foundation.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Harris NL. Hodgkin's disease: classification and differential diagnosis. Mod Pathol. 1999;12:159–175. [PubMed] [Google Scholar]
- 2.Schwering I, Brauninger A, Klein U, Jungnickel B, Tinguely M, Diehl V, Hansmann ML, la-Favera R, Rajewsky K, Kuppers R. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed Sternberg cells of Hodgkin lymphoma. Blood. 2003;101:1505–1512. doi: 10.1182/blood-2002-03-0839. [DOI] [PubMed] [Google Scholar]
- 3.Brauninger A, Wacker HH, Rajewsky K, Kuppers R, Hansmann ML. Typing the histogenetic origin of the tumor cells of lymphocyte rich classical Hodgkin's lymphoma in relation to tumor cells of classical and lymphocytepredominance Hodgkin's lymphoma. Cancer Res. 2003;63:1644–1651. [PubMed] [Google Scholar]
- 4.Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De LL, Kurtin P, Dal CP, Ladd C, Feuerhake F, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- 5.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 8.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluiver J, Kroesen BJ, Poppema S, van den Berg A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–1936. doi: 10.1038/sj.leu.2404387. [DOI] [PubMed] [Google Scholar]
- 10.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 11.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 13.Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, van den Berg A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 14.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechtel D, Kurth J, Unkel C, Kuppers R. Transformation of BCRdeficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood. 2005;106:4345–4350. doi: 10.1182/blood-2005-06-2342. [DOI] [PubMed] [Google Scholar]
- 18.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe Y, Tomita M, Kanai A. Computational methods for microRNA target prediction. Methods Enzymol. 2007;427:65–86. doi: 10.1016/S0076-6879(07)27004-1. [DOI] [PubMed] [Google Scholar]
- 20.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Getz G, Miska EA, varez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 22.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 24.Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, Gel B, Abrisqueta P, Lopez-Guillermo A, Artells R, et al. MicroRNA expression profiling in classical Hodgkin lymphoma. Blood. 2008;111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 25.Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LH, Chadburn A, Tuschl T, Knowles DM, Tam W. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 27.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 28.Kluiver J, Kok K, Pfeil I, de Jong D, Blokzijl T, Harms G, van der Vlies P, Diepstra A, Atayar C, Poppema S, et al. Global correlation of genome and transcriptome changes in classical Hodgkin lymphoma. Hematol Oncol. 2007;25:21–29. doi: 10.1002/hon.804. [DOI] [PubMed] [Google Scholar]
- 29.Mader A, Bruderlein S, Wegener S, Melzner I, Popov S, Muller-Hermelink HK, Barth TF, Viardot A, Moller P. U-HO1, a new cell line derived from a primary refractory classical Hodgkin lymphoma. Cytogenet Genome Res. 2007;119:204–210. doi: 10.1159/000112062. [DOI] [PubMed] [Google Scholar]
- 30.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sup SJ, Alemany CA, Pohlman B, Elson P, Malhi S, Thakkar S, Steinle R, Hsi ED. Expression of bcl-2 in classical Hodgkin's lymphoma: an independent predictor of poor outcome. J Clin Oncol. 2005;23:3773–3779. doi: 10.1200/JCO.2005.04.358. [DOI] [PubMed] [Google Scholar]
- 32.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 33.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Tan L, Dijkstra M, van LK, Robertus JL, Harms G, Blokzijl T, Kooistra K, van TM, Rosati S, et al. miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J Pathol. 2008;215:13–20. doi: 10.1002/path.2333. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Lee EJ, Schmittgen TD. Increased expression of microRNA- 155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer. 2006;45:103–106. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- 36.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR- 125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 38.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vigorito E, Perks KL, breu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris J, Oliere S, Sharma S, Sun Q, Lin R, Hiscott J, Grandvaux N. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J Immunol. 2006;177:2527–2535. doi: 10.4049/jimmunol.177.4.2527. [DOI] [PubMed] [Google Scholar]
- 42.Barth TF, Martin-Subero JI, Joos S, Menz CK, Hasel C, Mechtersheimer G, Parwaresch RM, Lichter P, Siebert R, Mooller P. Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin lymphoma. Blood. 2003;101:3681–3686. doi: 10.1182/blood-2002-08-2577. [DOI] [PubMed] [Google Scholar]
- 43.Rasini E, Cosentino M, Marino F, Legnaro M, Ferrari M, Guasti L, Venco A, Lecchini S. Angiotensin II type 1 receptor expression on human leukocyte subsets: a flow cytometric and RT-PCR study. Regul Pept. 2006;134:69–74. doi: 10.1016/j.regpep.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Manfroid I, Caubit X, Marcelle C, Fasano L. Teashirt 3 expression in the chick embryo reveals a remarkable association with tendon development. Gene Expr Patterns. 2006;6:908–912. doi: 10.1016/j.modgep.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Ray P, Devaux Y, Stolz DB, Yarlagadda M, Watkins SC, Lu Y, Chen L, Yang XF, Ray A. Inducible expression of keratinocyte growth factor (KGF) in mice inhibits lung epithelial cell death induced by hyperoxia. Proc Natl Acad Sci USA. 2003;100:6098–6103. doi: 10.1073/pnas.1031851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Georgakis GV, Li Y, Rassidakis GZ, Medeiros LJ, Mills GB, Younes A. Inhibition of the phosphatidylinositol-3 kinase/Akt promotes G1 cell cycle arrest and apoptosis in Hodgkin lymphoma. Br J Haematol. 2006;132:503–511. doi: 10.1111/j.1365-2141.2005.05881.x. [DOI] [PubMed] [Google Scholar]
- 48.Atayar C, Poppema S, Blokzijl T, Harms G, Boot M, van den Berg A. Expression of the T-cell transcription factors, GATA-3 and T-bet, in the neoplastic cells of Hodgkin lymphomas. Am J Pathol. 2005;166:127–134. doi: 10.1016/S0002-9440(10)62238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Schaadt M, Diehl V, Stein H, Fonatsch C, Kirchner HH. Two neoplastic cell lines with unique features derived from Hodgkin's disease. Int J Cancer. 1980;26:723–731. doi: 10.1002/ijc.2910260605. [DOI] [PubMed] [Google Scholar]
- 2.Atayar C, Poppema S, Blokzijl T, Harms G, Boot M, van den Berg A. Expression of the T-cell transcription factors, GATA-3 and T-bet, in the neoplastic cells of Hodgkin lymphomas. Am J Pathol. 2005;166:127–134. doi: 10.1016/S0002-9440(10)62238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanzler H, Hansmann ML, Kapp U, Wolf J, Diehl V, Rajewsky K, Kuppers R. Molecular single cell analysis demonstrates the derivation of a peripheral blood-derived cell line (L1236) from the Hodgkin/Reed-Sternberg cells of a Hodgkin's lymphoma patient. Blood. 1996;87:3429–3436. [PubMed] [Google Scholar]
- 4.Diehl V, Pfreundschuh M, Fonatsch C, Stein H, Falk M, Burrichter H, Schaadt M. Phenotypic and genotypic analysis of Hodgkin's disease derived cell lines: histopathological and clinical implications. Cancer Surv. 1985;4:399–419. [PubMed] [Google Scholar]
- 5.Atayar C, Kok K, Kluiver J, Bosga A, van den BE, van d V, Blokzijl T, Harms G, Davelaar I, Sikkema-Raddatz B, et al. BCL6 alternative breakpoint region break and homozygous deletion of 17q24 in the nodular lymphocyte predominance type of Hodgkin's lymphoma-derived cell line DEV. Hum Pathol. 2006;37:675–683. doi: 10.1016/j.humpath.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Copie-Bergman C, Boulland ML, Dehoulle C, Moller P, Farcet JP, Dyer MJ, Haioun C, Romeo PH, Gaulard P, Leroy K. Interleukin 4-induced gene 1 is activated in primary mediastinal large B-cell lymphoma. Blood. 2003;101:2756–2761. doi: 10.1182/blood-2002-07-2215. [DOI] [PubMed] [Google Scholar]
- 7.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 8.Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, van den Berg A. Lack of BIC and microRNA miR- 155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 9.Bechtel D, Kurth J, Unkel C, Kuppers R. Transformation of BCRdeficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood. 2005;106:4345–4350. doi: 10.1182/blood-2005-06-2342. [DOI] [PubMed] [Google Scholar]
- 10.Bende RJ, Jochems GJ, Frame TH, Klein MR, van Eijk RV, van Lier RA, Zeijlemaker WP. Effects of IL-4, IL-5, and IL-6 on growth and immunoglobulin production of Epstein-Barr virus-infected human B cells. Cell Immunol. 1992;143:310–323. doi: 10.1016/0008-8749(92)90028-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.