Abstract
Myosin binding protein C (MyBP-C) is a thick filament protein involved in the regulation of muscle contraction. Mutations in the gene for MyBP-C are the second most frequent cause of hypertrophic cardiomyopathy. MyBP-C binds to myosin with two binding sites, one at its C-terminus and another at its N-terminus. The N-terminal binding site, consisting of immunoglobulin domains C1 and C2 connected by a flexible linker, interacts with the S2 segment of myosin in a phosphorylation-regulated manner. It is assumed that the function of MyBP-C is to act as a tether that fixes the S1 heads in a resting position and that phosphorylation releases the S1 heads into an active state. Here, we report the structure and binding properties of domain C1. Using a combination of site-directed mutagenesis and NMR interaction experiments, we identified the binding site of domain C1 in the immediate vicinity of the S1–S2 hinge, very close to the light chains. In addition, we identified a zinc binding site on domain C1 in close proximity to the S2 binding site. Its zinc binding affinity (Kd of approximately 10–20 μM) might not be sufficient for a physiological effect. However, the familial hypertrophic cardiomyopathy-related mutation of one of the zinc ligands, glutamine 210 to histidine, will significantly increase the binding affinity, suggesting that this mutation may affect S2 binding. The close proximity of the C1 binding site to the hinge, the light chains and the S1 heads also provides an explanation for recent observations that (a) shorter fragments of MyBP-C unable to act as a tether still have an effect on the actomyosin ATPase and (b) as to why the myosin head positions in phosphorylated wild-type mice and MyBP-C knockout mice are so different: Domain C1 bound to the S1–S2 hinge is able to manipulate S1 head positions, thus influencing force generation without tether. The potentially extensive extra interactions of C1 are expected to keep it in place, while phosphorylation dislodges the C1–C2 linker and domain C2. As a result, the myosin heads would always be attached to a tether that has phosphorylation-dependent length regulation.
Abbreviations: MyBP-C, myosin binding protein C; FHC, familial hypertrophic cardiomyopathy; WTWT, wild type; IgI, immunoglobulin I; C1C2, C1–linker–C2; NOE, nuclear Overhauser enhancement; MD, molecular dynamics
Keywords: domain C1, NMR spectroscopy, model building, mutagenesis, protein structure
Introduction
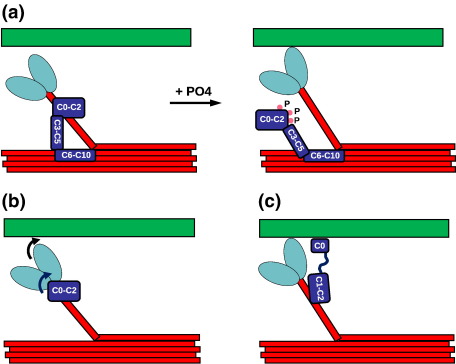
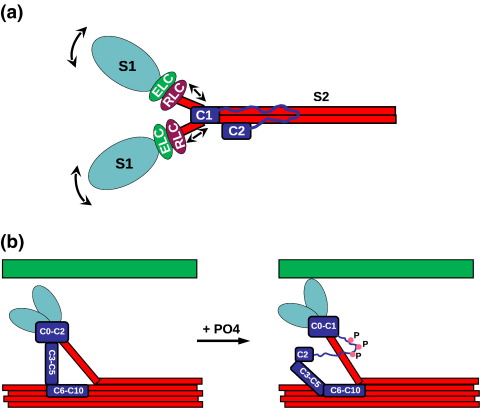
The regulation of muscle contraction depends on the precise interaction in time and space of a large number of proteins. In vertebrate cardiac muscle, the prime means of regulation is achieved by the well-established calcium-dependent steric blocking and unblocking of the myosin binding site on actin by the troponin–tropomyosin complex. Unlike skeletal muscle, where force is regulated by recruitment of activated fibres, modulation of the force levels in the electrically coupled cardiomyocytes occurs by posttranslational modification of regulatory proteins, especially phospholamban and troponin I.1 More recently, several proteins that provide an additional level of control in a more indirect and subtle manner have emerged. Among these is the cardiac isoform of myosin binding protein C (MyBP-C).2–4 MyBP-C belongs to the family of sarcomeric multidomain proteins that also includes titin, myomesin and obscurin, all of which are believed to play an important role in the assembly of different components of the sarcomere—modulation of contractile properties and mechanical strain sensing.5 For MyBP-C, more and more data suggesting its more prominent role in the regulation of muscle contraction are accumulating. Binding of the N-terminal portion of MyBP-C to myosin close to the S1 head6 is generally accepted to hold back the myosin heads and thus reduce their ability for interaction with actin as observed by X-ray fibre diffraction and electron microscopy7 (see Fig. 1a for illustration). Tethering of the S1 heads via MyBP-C to the rest of the thick filament reduces force development,8 which returns to normal once the tether is released upon phosphorylation of MyBP-C9,10 (see Fig. 1a). Malfunction of MyBP-C caused by missense or truncation mutations is a well-known cause of cardiomyopathies.11 After myosin itself, mutations in MyBP-C are the second most frequent cause of familial hypertrophic cardiomyopathy (FHC), suggesting that MyBP-C does indeed play a vital role in cardiac function.3 A vital yet subtle role for MyBP-C in the regulation of cardiac muscle was confirmed by the analysis of knockout mice.12 It was found that the overall structure of the sarcomere was unaffected, suggesting that MyBP-C is not required for sarcomere assembly, yet the knockout mice displayed symptoms of cardiac hypertrophy. Detailed studies of the structure of the thick filament revealed small rearrangements of the myosin heads but no significant change in diameter.13,14 The latter finding is surprising because large changes have been observed as a result of phosphorylation,15 which is thought to dislodge the MyBP-C N-terminus from its binding site on S2.9 If MyBP-C functions mainly as a tether, then the arrangements of myosin heads should be comparable in phosphorylated wild-type (WT) and knockout mice. The fact that they are very different suggests MyBP-C functionality beyond that of a mere tether. A more direct influence of the N-terminus of MyBP-C on muscle contraction is indeed supported by recent observations. Several studies showed that N-terminal fragments too short to perform the tethering function nevertheless had a significant effect on myocardial contraction16,17 (possible interpretations illustrated in Fig. 1b and c).
Fig. 1.
Cartoon depicting current understanding of MyBP-C function. (a) Illustration of the expected effect of phosphorylation of the N-terminal myosin binding site that dislodges MyBP-C, releasing the S2 coiled coil and subsequently promoting cross-bridge formation and muscle contraction. (b) A possible interpretation of the observation that MyBP-C constructs too short to act as a tether can influence muscle contraction. In this interpretation, the N-terminus directly affects the orientation of the myosin head group, indicated by the arrows. (c) Alternative interpretation where the effect of an N-terminal fragment of MyBP-C is caused by an interaction with the thin filament. The coiled-coil part of myosin is shown in red; S1, in light blue; MyBP-C, in dark blue; and F-actin, in green. For simplicity, several MyBP-C domains are grouped in a single box. Phosphorylated residues are indicated by pink dots.
The N-terminal myosin binding site of cardiac MyBP-C consists of two immunoglobulin I (IgI) domains,18 C1 and C2, connected by an ∼ 100-residue linker of unknown structure containing the phosphorylation sites. We recently determined the structure and mapped the binding site of domain C2 on myosin S2 using a shortened fragment of S2, S2Δ, giving a first glimpse of the mode of binding of these two proteins and how mutations related to FHC can interfere.19 Here, we extend our investigation of this interaction by determining the structure of domain C1 and locating its binding site on myosin S2. The results of our study on the binding of C1 and C2 to myosin S2 are combined in a model for the interaction that agrees well with current understanding of this system and its perturbation by mutations in MyBP-C or myosin S2. The location of domains C1 and C2 is also highly suggestive as an explanation of how the N-terminus of MyBP-C alone can affect myosin activity.16,17 Furthermore, our work shows that most FHC-related point mutations do not abolish the MyBP-C-myosin interaction but weaken it, reflecting the general observation that FHC-related mutations in MyBP-C lead to late-onset FHC with generally good prognosis.
Results
Structure of domain C1
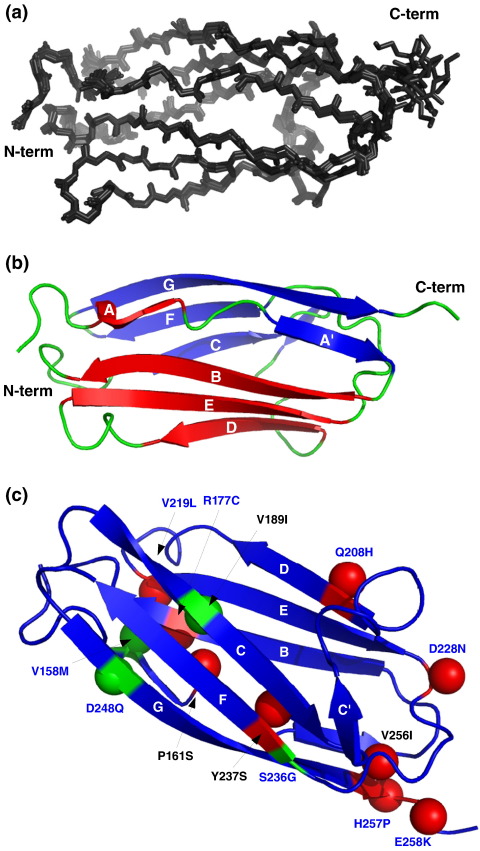
The overall quality of the NMR structure is apparent by the excellent agreement of the 29 structures shown superimposed in Fig. 2a, giving an overall backbone RMSD of 0.6 Å. The only region with modest disorder is around the last C-terminal residues, which are already part of the linker that connects C1 to C2. The domain folds as expected into an all-β-sheet structure of the IgI fold,18 apparent in Fig. 2b. Here, the structure is shown as a cartoon in the same orientation as in Fig. 2a. Four strands, ABED, form the first sheet and the remaining five, C′CFGA′, form the second sheet, which are assembled in a β-sandwich structure. The CD loop in domain C1 is slightly extended compared with other IgI domains in MyBP-C (see Fig. S1). The extra residues fold into a very compact structure, very close to a single turn of an α-helix. The turn is supported by an α-helix type of hydrogen bond between the amide group of Val203 and the backbone carbonyl oxygen of Leu199.
Fig. 2.
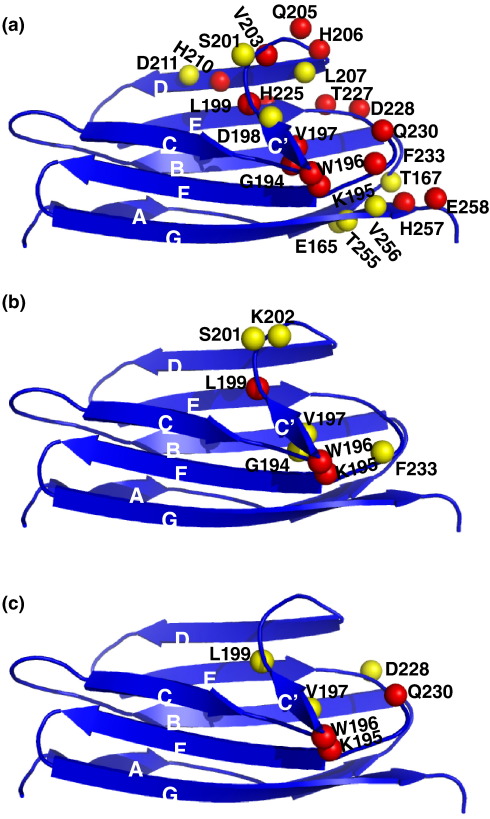
Structure of domain C1 of human cardiac MyBP-C. (a) Family of the best 29 structures resulting from the final round of structure calculation. (b) Cartoon view of the best structure of C1 from the family of structures in (a) in the same orientation as in (a). (c) FHC-linked point mutations found in C1 mapped on the structure of C1. Residues found mutated with a clear disease indication are shown in red, and those shown to be polymorphisms are shown in green. β-Carbons are shown as a Van der Waals sphere for all to make the side-chain orientation more evident. Labels for solvent-exposed residues are shown in blue, and those for non-exposed residues are shown in black. The β-strands are labelled in white. Strands A and A′ are hidden.
Zn binding of C1
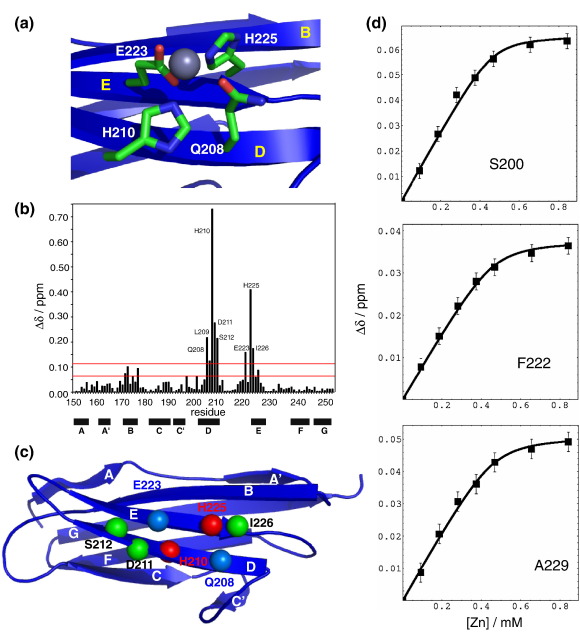
Close inspection of the structure of C1 revealed the presence of a zinc binding site formed by residues Gln208, His210, Glu223 and His225. These four residues are arranged in a neat square right in the centre of the A′BED β-sheet (Fig. 3a), two on β-strand D and two on β-strand E, in a manner similar to other zinc binding sites on the surface of β-sheets.20 Titrations of 15N-labelled protein with ZnCl2 were performed and monitored with the use of fast-heteronuclear multiple-quantum coherence spectra to experimentally ascertain the ability of domain C1 to bind zinc.21 Large chemical shift perturbations were observed (Fig. 3b) for two main regions, residues 208–213 and residues 223–230, covering very well the predicted zinc binding site. The largest chemical shift perturbations were seen for the two histidines in the predicted binding site, suggesting strong zinc coordination by these two residues (see Fig. 3c). A zinc atom was positioned in the binding site followed by energy minimisation in AMBER22 using standard zinc parameters23 to test the relevance for further ligands. The resulting distances suggest that the zinc is coordinated by four ligands—Gln208, His210, Glu223 and His225. Interestingly, the residues with the largest chemical shift perturbations are titrating in the slow-exchange regime, suggesting a slow off rate and thus reasonably tight binding (spectra for representative residues are shown in Fig. S2). Titration curves plotted for some residues close to the binding site in fast exchange are used for analysis of binding affinity, as shown in Fig. 3d. Fitting these curves leads to a dissociation constant in the range of 10–20 μM, in good agreement between these residues.
Fig. 3.
Zinc binding of C1. (a) Detailed view of the zinc binding site in the structure of C1. Only side chains of Gln208, His210, Glu223 and His225 are shown. A zinc atom has been modelled in the binding site. After energy minimisation, the zinc-ligand distances are 2.27 Å for His210(Ne2), 2.26 Å for His225(Ne2), 1.60 Å for Glu223(Oe1), 1.81 Å for Glu223 (Oe2) and 2.22 Å for Gln208(Oe1). (b) Plot of chemical shift perturbation against the protein sequence. The first red line represents the 〈Δδ〉tot level, and the second red line is 〈Δδ〉tot + 1∗σ. Residues with chemical shift perturbations above 〈Δδ〉tot + 1∗σ are explicitly labelled. (b) Titration curves for residues in fast exchange for estimating binding affinity and stoichiometry. (c) Mapping of chemical shift perturbations on the three-dimensional structure of C1. Residues with chemical shift perturbations above 〈Δδ〉tot + 1∗σ are shown as spheres. The residues expected to coordinate the zinc are shown in red (histidines) and blue (glutamate/glutamine), and those with significant perturbations not expected to be directly involved are shown in green. (d) Titration curves for residues in fast exchange for estimating binding affinity and stoichiometry.
Binding of C1 to myosin S2Δ
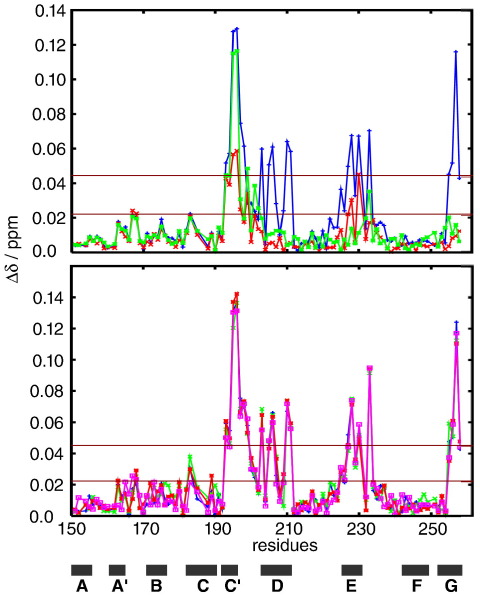
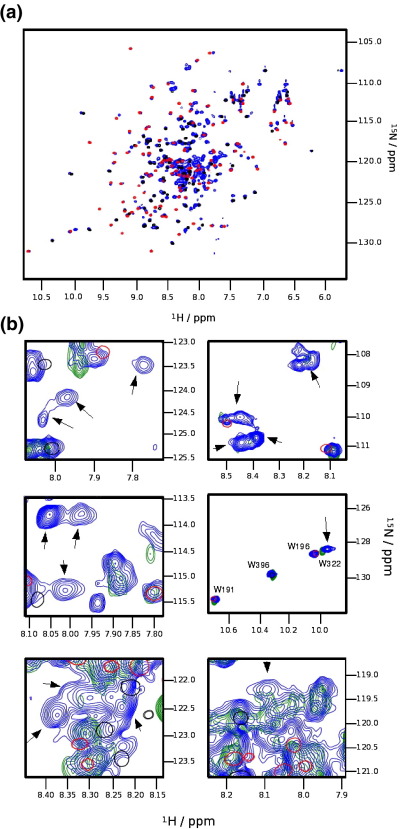
To probe the binding of C1 to S2Δ, we followed essentially the same approach previously described for domain cC2.19 A transverse relaxation-optimized spectroscopy two-dimensional 15N–1H spectrum of C1 was recorded in the absence and in the presence of a 4-fold excess of S2Δ (examples are shown for several representative residues in Fig. S3). To identify the binding site of S2Δ on C1, we plotted the chemical shift differences for all assigned residues against the sequence of C1 (Fig. 5, top, blue curve). In this plot, three regions of chemical shift perturbations clearly stand out: the region around strands C, C′, the CD loop and parts of strand D; the EF loop and part of β-strand E; and the last three residues of β-strand G. Most prominent as individual residues are Lys195 and Trp196. Mapping of the significant chemical shift perturbations on the surface of the structure of C1 (Fig. 6a) shows that the three regions, although disconnected in the sequence, come together in the three-dimensional structure. These residues form a continuous surface covering most of the C-terminal half, from the CD loop onward.
Fig. 5.
Chemical shift perturbations in titrations of 15N-labelled C1 with unlabelled S2Δ. Shown are combined 15N and 1HN chemical shift perturbations against the sequence of C1. Top: C1 WT + S2Δ WT (blue), C1 WT + S2Δ E846K (red) and C1 D228N + S2Δ WT (green). Bottom: C1 WT + S2Δ WT (blue), C1 WT + S2Δ E924K (green), C1 WT + S2Δ E936K (red) and C1 WT + S2Δ E894G (pink). The 〈Δδ〉tot and 〈Δδ〉tot + 1∗σ levels (0.021 and 0.042 ppm, respectively) for C1 WT + S2Δ WT are shown as dark red horizontal lines. The position of β-strands is indicated by black bars.
Fig. 6.
The S2Δ binding site on C1 mapped by chemical shift perturbations on the three-dimensional structure. (a) Binding of C1 to S2Δ. (b) Binding of C1 D228N to S2Δ. (c) Binding of C1 to S2Δ E846K. All residues with chemical shift perturbations above 〈Δδ〉tot are labelled and have their N atoms displayed as spheres. Residues with Δδ values between 〈Δδ〉tot and 〈Δδ〉tot + 1∗σ are shown in yellow, and those above 〈Δδ〉tot + 1∗σ are shown in red. The β-strands are labelled in white.
Effects of FHC-related mutations in C1 on binding to myosin
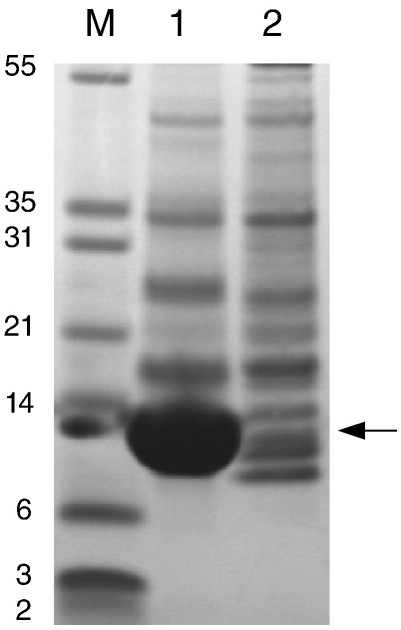
Several point mutations linked to FHC have been identified in domain C1 of cardiac MyBP-C. They are shown mapped on the structure of the domain in Fig. 2d. For completeness, known polymorphisms are also indicated. Among these is residue 236, which in the construct used in this work is actually a glycine.24 It is worth noting that most polymorphisms in human cardiac C1 are found in other isoforms or species, confirming their compatibility with the structure and function of C1 (Fig. S1). In contrast, the majority of residues where mutations correlate with disease are highly conserved, at least in the cardiac isoform (e.g., Arg177, Tyr237 or Val256). Most of the FHC-related mutations affecting exposed amino acids cluster toward the C-terminus of the domain (on the right side of Fig. 2c). A smaller cluster of FHC-related mutants is formed on the ABED sheet, close to the β-bulge in strand A and far away from the S2Δ binding site. The correlation of most FHC-linked mutations with the location of the binding site of S2Δ suggests that at least the solvent-exposed amino acids might contribute to FHC by interfering with protein interactions. To investigate this hypothesis, we selected two mutants from this region, Asp228 to asparagine, representing exposed charged residues in the binding site for S2Δ, and Tyr237 to serine, representing mutations of non-exposed amino acids close to the binding site. Tyr237 is strongly conserved, and part of the hydrophobic core and its replacement by a serine would remove substantial hydrophobic contacts, leading to destabilisation of the fold. In contrast, there is no indication that the replacement of Asp228 with asparagine will affect the structure. The different expectations for the effects of both mutants on the structure of C1 were confirmed by expression trials of both mutants (Fig. 4). C1 Asp228Asn expressed extremely well as a soluble protein (Fig. 4, lane 1). In contrast, Tyr237Ser did not express as a soluble protein at all (Fig. 4, lane 2). Efforts to refold this mutant from inclusion bodies failed repeatedly. As a result, only Asp228Asn could be studied with respect to binding to S2Δ.
Fig. 4.
Expression trials of C1 mutants D228N and Y236S. Pilot expression was performed in 5-mL cultures in 20-mL tubes using BL21∗ cells. Cells were grown at 37 °C until reaching induction levels of cell density, after which the temperature was dropped to 15 °C and protein expression was induced overnight. Cells were harvested by centrifugation and opened by sonication. Samples for gel electrophoresis were taken of the soluble fraction after centrifugation of the cell extracts. M, molecular mass marker (Mark12, Invitrogen; molecular masses are given in kilodaltons); 1, C1 D228N; 2, C1 Y237S. Expected position of C1 is marked by an arrow.
The Asp228Asn mutant of C1 is still capable of binding to S2Δ (Fig. 5, top, green curve), with almost the full chemical shift perturbations around Trp196 as in WT. In contrast, in the other two regions of largest chemical shift perturbations in WT, residues around His210 on strand D and those in the EF loop around Gln230, there is very little response to binding. Also, the three amino acids at the C-terminus show hardly any response (Fig. 6b). Asp228 is located right in the middle of the EF loop, such that its mutation to asparagine, leading to the loss of negative charge, might lead to an almost complete lack of binding ability in this part of the interface. The Asp228Asn mutant of C1 is still able to bind, albeit with only part of its interface and consequently substantially reduced affinity.
Effects of FHC-related mutations in S2Δ on binding to MyBP-C domain C1
A large number of point mutations contributing to the development of FHC have been identified in myosin (for databases, see Materials and Methods; a summary on the structure of S2Δ is given in Fig. S4). Previously, use of point mutations has been successful in mapping the binding site of MyBP-C domain C2 in the vicinity of Arg870.19 We therefore used the same approach in the hope that an FHC-related point mutation might be linked to the disease by interfering with the binding of domain C1. The initial focus was on the cluster of eight mutations of exposed amino acids between residues 921 and 935. The majority are either charge inversions or charge changes, suggesting the cluster to be a potential protein–protein interaction site. Furthermore, the S2Δ mutant Glu924Lys was shown to abolish binding to an MyBP-C fragment containing domain C1.6 However, binding experiments of isolated C1 with two representative mutants, Glu924Lys and Glu935Lys, led to chemical shift perturbations indistinguishable from those of WT S2Δ (Fig. 5, bottom). As this region was the only clearly discernible cluster of mutations in addition to the region around Arg870, we opted to systematically mutate exposed residues mutated in FHC. The first mutations tested were Glu846Lys and Glu894Gly, the former being an FHC-related mutation in S2Δ and the latter being a representative of a mini-cluster of mutants immediately following the C2 binding site. Figure 5, top and bottom, shows that the chemical shift perturbations of C1 in a mixture with S2Δ Glu894Gly are virtually identical with those observed with the WT of S2Δ. In marked contrast, mutant Glu846Lys led to a significant reduction in the chemical shift perturbations. This reduction is non-uniform, with perturbations around residues 190–210 comprising the CD loop and the D-strand dropping significantly, even to almost noise level for some residues in the D-strand (Fig. 6c). In contrast, perturbations around the EF loop were much closer to those observed for the WT. C1 thus binds to S2Δ around residue Glu846, but binding of C1 to the S2Δ E846K mutant is not completely abolished but severely compromised.
Model of C1 bound to S2Δ
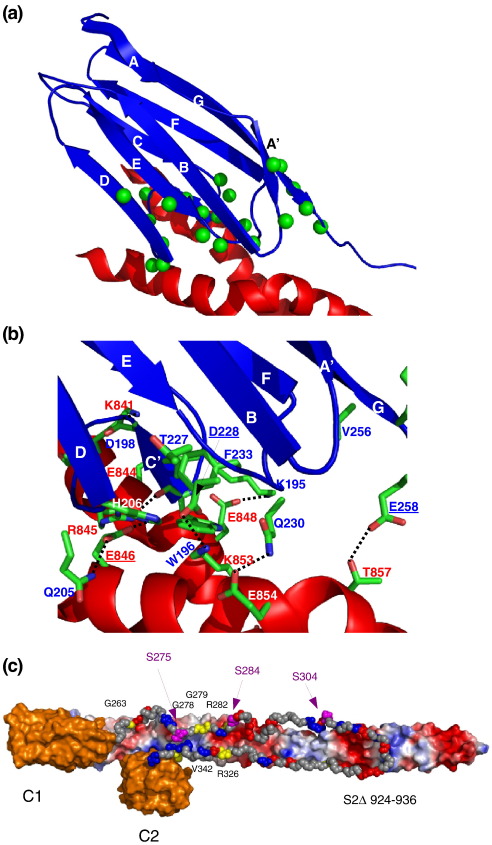
Expanding the previous model of C2 bound to S2Δ,19 we produced a model of the complex of C1–linker–C2 (C1C2) with S2Δ. Use of the entire minimal binding site of MyBP-C for S2 should aid in identifying a realistic binding mode. Based on the FHC-related mutation S2Δ Glu846Lys, which led to a significant reduction in binding, domain C1 was modelled to be in contact with this amino acid on S2Δ, very close to the start of the S2 region at Pro838. The interface in the model is formed by C1 aligning with its β-strands almost in line with the main axis of the coiled coil. The slope created by the crossing helices is well covered in shape by the extended loops at the C-terminus of C1, which generate an interface of complementary shape (Fig. 7a). This arrangement covers most of the amino acids with significant chemical shift perturbations in the C1–S2Δ interaction experiments very well. At the heart of the interface sits the residue with the largest chemical shift perturbations, Trp196. Together with Ala232, it forms a small hydrophobic patch that packs to Phe856 and Met852 on S2Δ. Furthermore, the backbone oxygen of Trp196 makes a hydrogen bond with the side chain of Arg845. The rest of the interface is composed of polar interactions, even though the charge distribution is much more mixed compared with the C2–S2Δ interface.19 Right next to the Trp196 is Lys195, which forms an ionic bridge and/or a hydrogen bond with Glu848. The previous residue, Gly194, forms a hydrogen bond with its backbone oxygen to the side chain of Lys847. On the other side of the central tryptophan, Asp198 interacts with Lys841 via an ionic bridge and/or a hydrogen bond. Lys202 in the following single turn of α-helix forms another ionic bridge and/or a hydrogen bond with Glu844. At the end of the CD loop, Gln205 makes a hydrogen bond with Glu846 and potentially with Lys847. The final residue on this stretch of protein involved in the interface is His206, which forms a hydrogen bond with Glu846. In the EF loop, Gln230 forms hydrogen bonds with Glu854. Importantly, Asp228 forms an ionic bridge and/or a hydrogen bond with Lys853, which will be significantly weakened in the FHC-causing Asp228Asn mutant. Finally, the very last residue of C1, Glu258, is positioned to make a hydrogen bond with Thr857.
Fig. 7.
Model of the complex of C1 and S2Δ. (a) Overview of the position of C1 (blue) on S2Δ (red). Amino acids in C1 with chemical shift perturbations larger than 〈Δδ〉tot are marked by green spheres on their N positions. (b) Detailed view of the interactions of C1 and S2Δ in the model. C1 is shown in blue, and S2Δ is shown in red. Important side chains in the interaction are coloured by atom type (carbon, green; oxygen, red; nitrogen, blue), and labels are coloured by protein. (c) Depiction of the overall assembly of C1 and C2 and the linker on S2Δ. Domains C1 and C2 of MyBP-C are shown in orange, and the linker between them is shown in gray. The three phosphorylation sites in the linker are shown in purple, residues mutated in FHC are shown in yellow with labels and charged residues are coloured according to their charge. S2Δ is shown with solvent-accessible surface coloured by a simple electrostatic potential with the N-terminus on the left (hidden by C1) and the C-terminus on the right. The position of the C-terminal cluster of FHC-related point mutations in S2Δ is indicated by the residue numbers (924–936).
Discussion
Structure of domain C1
The structure of C1 conforms to the well-established pattern for the IgI fold,18 including the once controversial C′-strand.25 A single-turn α-helix is introduced through a small extension of the CD loop, a feature unique to domain C1 and seen in the NMR structure of the slow skeletal isoform of domain C1 as well [Protein Data Bank (PDB) code 2dav; Fig. S5]. The structure overall is rather extended, much longer than, for example, the adjacent domain C219 and very similar to the recently published crystal structure.26 Comparison with DaliLite gives an RMSD of 1.8 Å and a z score of 13.2. The only structural differences are found in the termini and some loops (e.g., the BC loop). In the crystal structure, the first five residues extend away from the protein, stabilised by crystal contacts while they pack tightly to the rest of the protein in the NMR structure. The most similar structures found through a DALI search27 in the PDB are those of the slow skeletal muscle isoform of human MyBP-C, the well known telokin and the first and second IgI domains of human cardiac titin. Cardiac and slow skeletal isoforms of domain C1 share a sequence identity of 53% (for more details, see Table S1) such that a highly similar structure is not surprising. Main sequence differences exist in the BC loop, the EF loop and close to the C-terminus (see Fig. S1). These variations are reflected in the structure where the BC loop is packing very closely to the N-terminus in the cardiac isoform, whereas it is shifted toward the DE hairpin in the slow skeletal isoform. Overall, however, the structures are extremely similar such that the substantial sequence divergence between the isoforms has no impact on the detailed structure. This is not surprising, as most of the conserved amino acids are the key hydrophobic residues important for determining the fold.18 Most of the divergent amino acids are found on the surface of the protein, suggesting subtle functional differences (e.g., with regard to binding partners).
Zinc binding of C1
The zinc binding site exhibits very similar structure and topology in employing two amino acids each on adjacent β-strands (Fig. 3a) to a variety of otherwise completely unrelated structures.20 The arrangements of ligands are identical in the NMR and X-ray structures. Interestingly, the composition of amino acids in the binding site, His–His–Glu–Gln, is typical of zinc binding in metalloenzyme active sites. Taken together with the appreciable binding affinity in the lower micromolar range, this makes C1 a bona fide zinc binding domain. Zinc binding in enzyme active sites occurs with affinities from picomolar to micromolar,28 while it is even higher when it fulfils a structural role.29 In the context of cardiac muscle, however, the presence of a zinc binding site with micromolar affinity is surprising. Measurements of free zinc concentration in rat cardiac muscle show values of less than 1 nM, even under extreme cell stress,30 such that the affinity of C1 is too low to bind zinc in vivo. One could make some allowance for the underestimation of binding affinities by NMR spectroscopy and for different zinc concentrations in human cardiac muscle compared with rat, but a variation by several orders of magnitude seems unlikely. Given the usual specificity of zinc binding sites, it is also unlikely that this binding site could bind other metals. Instead, we expect the zinc binding site to become physiologically relevant by the mutation of Gln208 to histidine, which is linked to FHC by an as yet unknown mechanism.31 Gln208 is one of the four zinc-coordinating ligands and shows significant chemical shift perturbations in the zinc titration. However, glutamines are known to contribute much less as zinc ligands, such that a replacement by histidine is expected to strongly increase the affinity, which would allow for zinc binding in cardiac muscle. Because of the very low concentration of free zinc, even a modest sequestration of zinc might have pathological consequences. The effect could also be more direct because the zinc binding site partly overlaps with the S2Δ binding site. As a result, having zinc bound in part of the S2Δ binding site might influence the interaction.
Interaction of C1C2 and S2Δ
The results of the interaction experiments between C1 and S2Δ, including the respective mutations, are well summarised in the model of the complex. The amino acids of C1 showing the largest chemical shift perturbations are well covered by the interface (Fig. 7a) and involved in hydrogen bonds/ionic interactions (Fig. 7b). This is only a model based on chemical shift perturbations combined with mutagenesis. Yet, many details agree well with the NMR observations and can explain why a number of residues mutated in FHC may cause the disease (Fig. 7b). Residue Glu846 is involved in a hydrogen bond and/or an ionic bridge and when replaced by a lysine will weaken the interaction considerably, in particular around the C-terminus of the CD loop (residues 202–208), while the core interactions around W196 and the EF loop are maintained (Fig. 7b). Also, the last residue in the C1 construct, Glu258, is an FHC-related residue when mutated to lysine. In our model, Glu258 forms a hydrogen bond with Thr857, which will be abolished by a lysine. Even though this interaction was not investigated experimentally because Glu258 is not very well defined in the structure and not part of domain C1 but of the C1–C2 linker, we can clearly reveal its importance in our model. It can be assumed that the nearby mutation His257Pro would have similar consequences. We do not see any direct interactions of His257 with S2Δ, but a proline in its place would distort the backbone at this position, resulting in repositioning Glu258 in a manner not conducive to the interaction with Thr857. This mutation could also have long-range effects, because it is located in a place where C1 and C2 are almost in touching distance, exemplified by the interaction of Arg858 of S2Δ with Asp431 in C2, right next to the interaction of Thr857 with Glu258 in C1.
The effects of the FHC-related mutations in C1 that were experimentally investigated here, Tyr237Ser and Asp228Asn, can also be explained in this context. While the mutation of Tyr237 to serine led to the unfolding of the domain, the other point mutation, Asp228Asn, did not interfere with folding at all, as expected. Instead, the Asp228Asn mutant severely weakened the interaction of C1 with S2Δ, as would be expected based on our complex model. Asp228 forms an ionic bridge and/or a hydrogen bond with Lys853, which will be much weakened by replacing the negatively charged aspartate with an uncharged asparagine. The latter would still be able to form a hydrogen bond, but this interaction would be much weaker. This is confirmed by the chemical shift perturbations measured in the Asp228Asn mutant, where almost all chemical shift perturbations in the EF loop are absent, while binding is still maintained by the interactions made by the CD loop, as well as other parts of the binding site.
Intriguingly, two further solvent-exposed amino acids that are FHC related when mutated, Arg177Cys and Val219Leu, are distant from the S2Δ binding site. Yet, being on the surface, it is unlikely that they would interfere with the domain stability like Tyr237Ser. This suggests that C1 could be involved in other interactions, as these residues are close together on a part of the surface pointing away from S2Δ, which is thus accessible for other ligands. In our model, this part of the protein points toward the S1–S2 junction close to the regulatory light chains. Hence, C1 could directly or indirectly be involved in an interaction with the myosin head group. This may provide us with a possible explanation why untethered N-terminal MyBP-C fragments have significant effects in motility assays,16,17 as illustrated in Fig. 9a.
Fig. 9.
Summary of the results. (a) Potential effects of C1 binding close to the hinge and the light chains. Interactions of C1 could either bring the heads closer or drive them farther apart as indicated by the arrows. These effects could occur symmetrically or asymmetrically. (b) As a consequence of C1 binding to the S1–S2 hinge domain, C1 and the region further N-terminal might remain bound even when MyBP-C gets phosphorylated. C2 binds weaker than C1 and could be dislodged with the C1–C2 linker, thus only lengthening the leash without ever completely unleashing the S1 heads. The coiled-coil part of myosin is shown in red; S1, in light blue; MyBP-C, in dark blue; and F-actin, in green. For simplicity, several MyBP-C domains are summarised in a single box. Phosphorylated residues are indicated by pink dots.
In this work, we have modelled the position of both IgI domains of MyBP-C's N-terminal binding site on myosin that begs the question of the location of the linker (MyBP-C motif). Recently, the linker was proposed to be an IgI domain as well,32 which appears very unlikely, given the low sequence similarity of the linker sequence to Ig sequences. Furthermore, these observations are in direct contrast to our own NMR data on the whole C1C2 fragment, which show the linker to be completely unstructured (Fig. 8a). All residues of the linker are in the random-coil region of the spectrum. Additionally, in a heteronuclear nuclear Overhauser enhancement (NOE) experiment, the highest value for residues in the linker was 0.2, while the residues of domains C1 and C2 had values around 0.8 (Fig. 8b). One would expect residues in a rigid, well-folded domain to have values between 0.7 and 0.9, while values close to zero are indicative of almost complete freedom of motion as expected of an unfolded polypeptide chain. As both the small-angle X-ray scattering and our NMR experiments were performed with bacterially expressed protein, methodological differences in protein folding can be excluded. We therefore propose that the linker is unfolded in solution but adopts a compact shape, similar to other unstructured peptides (e.g., the extensively characterised α-synuclein). This protein is intrinsically unstructured yet has been shown by a variety of NMR methods to adopt a compact overall shape.33,34 This would enable it to adopt a more defined conformation upon binding to myosin. We do not have experimental data as yet about the binding mode of the linker but at least endeavoured to ensure that the positions of C1 and C2 in the model allow a location of the linker in accord with published interaction data. In our extended model (Fig. 7c), the positions of C1, C2 and the linker on S2 are indicated. The positions of C1 and C2 are such that the linker runs along S2 to reach the area around residue 930 of S2Δ. Here, we find a cluster of FHC-related point mutations, one of which, E924K, makes binding of C1C2 to S2Δ undetectable by isothermal titration calorimetry.6 As neither C2 binding nor C1 binding to S2Δ is affected by this mutation, this part of S2Δ likely interacts with part of the linker. The linker can easily reach this part of S2 in our model, even though we cannot make comments about detailed interactions. It is conceivable, however, that the linker does not adopt a precise, stable and rigid conformation with very specific interactions. Instead, the contribution from the linker to binding might come from general charge complementarity. Retaining a certain degree of mobility might add to complex stability by reducing the entropic penalty otherwise unavoidable for folding upon binding. This mode of binding is not unusual and has been observed (e.g., in the interaction of microbial proteins with host receptors).35 The general charge matching would also suggest an explanation for the effect of phosphorylation upon binding: all three serines known to be phosphorylated36 are positioned in the vicinity of negatively charged patches on the surface of S2. Phosphorylation would subsequently lead to increased charge repulsion and consequent destabilisation of the complex in accordance with experimental observations.9 That the linker/MyBP-C motif might bind alternative ligands when undergoing phosphorylation-induced conformational changes is another possibility to explore.
Fig. 8.
NMR of 15N-labelled construct C1C2, corresponding to residues 151–451 of human cardiac MyBP-C. (a) Heteronuclear single-quantum coherence spectrum of C1C2 (blue) superimposed with the spectra of C1 (black) and C2 (red). (b) Sections of a heteronuclear 1H–15N NOE experiment (green) of C1C2 superimposed on its reference experiment (blue) together with the spectra of C1 (black) and C2 (red). Peaks of the linker showing very weak NOEs around zero are indicated by arrows.
Conclusions
Our combined NMR and mutagenesis data place the N-terminus of MyBP-C right on top of the S1–S2 hinge and thus in immediate vicinity of the S1 head and the light chains. In this location, MyBP-C is able to directly interact with the hinge, potentially influencing the positioning of the S1 heads directly or indirectly via interactions with the light chains (Fig. 9a). Such an interaction could also involve the as yet relatively uncharacterised domain C0, which is specific for the cardiac isoform. The ability to manipulate the S1–S2 junction would give MyBP-C the potential to directly adjust the position of the S1 heads and thus subtly influence muscle contraction. With further interactions possible for domains C1 and C0 in the S1/light chain region, one could expect that phosphorylation of the C1–C2 linker does not dislodge the MyBP-C N-terminus completely. While the linker and possibly domain C2 will dissociate, the more N-terminal part might remain attached (Fig. 9b). The dissociation of a substantial part of polypeptide chain will allow the release of the myosin head and facilitate actomyosin cross-bridge formation. At the same time, the remaining part of MyBP-C is well positioned to exert a subtle influence on the position of the S1 heads. Maintaining a loosely tethered state would also be useful through increasing the probability of rebinding once the C1–C2 linker is dephosphorylated and the S1 heads need to return to their resting positions. The myosin heads are thus never taken off the leash, but the length of the leash is adjusted by phosphorylation. As such, our data provide a good explanation for the observations that MyBP-C has functions beyond those of a simple tether and that its influence on the structure of the S1 heads is more complex than can be explained by a simple on/off model driven by phosphorylation.
Materials and Methods
Sample preparation
Domain C1 of human cardiac MyBP-C (residues 151–258 of UniProt entry Q14896/MYBPC3_HUMAN) was cloned as previously described24 into pET8c vector. Protein expressed in this vector had a non-removable histidine tag of sequence MHHHHHHSS attached to the original sequence. For the zinc binding experiments, a histidine-tag free construct of C1 was produced by cloning into pETM-11 (EMBL Protein Expression Laboratory) with the original construct as template using PCR and adding NcoI/BamHI restriction sites to the construct. The constructs for S2Δ were identical with those used previously in the crystal structure determination.37 All mutants of C1 and S2Δ used in this work were created using QuikChange (Stratagene), following the standard protocol. Proteins were overexpressed as described previously24 using the commercial bacterial host BL21∗ (Invitrogen). Proteins were purified using FastFlow6 histidine binding resin (GE Healthcare) in gravity flow mode as capture step followed by gel filtration on a preparative Sephadex 75 16/60 column (GE Healthcare). Samples were exchanged into NMR buffer using either dialysis with Slidalyzer cassettes (Pierce; volume = 3–12 mL; cutoff = 3 kDa) or NAP10 gel-filtration columns (GE Healthcare). Samples were concentrated for measurement using VivaSpin 20 concentrators (VivaScience) with a 5-kDa (C1) or 10-kDa (S2Δ) molecular mass cutoff.
Structure determination of C1
Protein concentrations were 0.8 mM. Structure calculation and all other NMR work reported here are based on the previously published NMR assignment,24 available from BMRB with accession code 6015. Structure calculation was based on high-quality 15N- and 13C-resolved three-dimensional NOE spectroscopy spectra recorded with a mixing time of 100 ms on a 600-MHz Bruker DMX spectrometer. The spectra were recorded on a protein sample of 0.8 mM in a buffer of 20 mM sodium phosphate, pH 7.0, 50 mM NaCl, 2 mM DTT, 1 mM ethylenediaminetetraacetic acid and 0.02% NaN3. Constraints used in the structure calculation are listed in Table 1. Dihedral constraints were produced with TALOS,38 and distance constraints were taken from manually picked peaks in the NOESY spectra. Structure calculation was performed using a protocol identical with that of domain C219 using ARIA 1.2.39 The coordinates of the structure are available from the PDB with accession code 2AVG.
Table 1.
Structure calculation statistics for the NMR structures of domain C1
| NMR constraints | |
| Total NOEs | 2920 |
| Initial unambiguous distance constraints | 320 |
| Final list of distance constraints from ARIA | 1796 |
| Hydrogen-bond restraints | 42 |
| Dihedral restraints | 132 |
| Number of violations | |
| NOE distance restraints > 0.5 Å | 0.0 |
| Hydrogen-bond restraints > 0.3 Å | 1.43 ± 1.05 |
| Dihedral restraints > 5 | 2.02 ± 0.75 |
| RMSD from idealized geometry | |
| Bonds (Å) | 0.0067 ± 0.0004 |
| Angles (°) | 0.71 ± 0.03 |
| Dihedrals (°) | 23.5 ± 0.6 |
| Improper (°) | 0.80 ± 0.05 |
| Energy | |
| Van der Waals (kcal/mol) | − 865.3 ± 119.0 |
| Electrostatic (kcal/mol) | − 4015.9 ± 86.3 |
| Ramachandran analysis (%) | |
| Residues in most favored regions | 78.6 |
| Residues in allowed regions | 17.3 |
| Residues in generously allowed regions | 3.8 |
| Residues in disallowed regions | 0.3 |
| Coordinate precision (all residues, Å) | |
| Backbone atoms | 0.60 ± 0.08 |
| Heavy atoms | 0.99 ± 0.09 |
Sequence analysis
Sequences similar to domain C1 of the cardiac isoform of human MyBP-C were identified using BLAST40 to search in the UniProt Knowledgebase database with standard search parameters. Hits representing MyBP-C were selected, and the corresponding regions were taken from the respective full-length database entries. Selected sequences were aligned using ClustalW41 and displayed using ClustalX.42 Colouring was done using the same thresholds as previously described.19
Mutants linked to FHC were extracted from publicly available databases: the DNA Mutation Database for Familial Hypertrophic Cardiomyopathy† and the more general Sarcomere Protein Gene Mutation Database‡.
C1–S2Δ interaction
All NMR experiments were measured in 40 mM phosphate, pH 7.0, 50 or 100 mM NaCl, 2 mM DTT and 0.02% NaN3. Sample concentration was 200 μM for 15N-labelled C1 and was 800 μM for unlabelled S2Δ, at a ratio of 1:4. 1H–15N transverse relaxation-optimized spectroscopy spectra43 were recorded for free C1 and the mix of C1 and S2Δ 1:4 on a Bruker DMX spectrometer at 800 MHz equipped with a cryoprobe. Acquisition and processing were done with TOPSPIN 1.3 and 2.0. The appropriate volumes of C1 and S2Δ stocks were pipetted in a VivaSpin 20 concentrator with a 5-kDa cutoff and concentrated down to a volume of 400 μL, which was then transferred into a 5-mm NMR tube (Shigemi) without plunger, to prepare samples of C1–S2Δ mixtures. Deuterium lock was achieved using 5% D2O in the sample. Prior to each experiment, the pH value was adjusted to < 0.1 deviation of the target of 7.0. Peaks were automatically picked in all spectra and assigned using the published assignment of C1. Chemical shift differences, Δδ, were measured for both 15N and 1H and combined using a scaling factor of 1.0 for 1H and that of 0.15 for 15N. NMR analysis was performed with the analysis software of the CCPN initiative.44 The average of the combined Δδ values for the interaction of C1 WT with S2Δ WT was calculated to give the standard deviation 1∗σ.
Zinc binding
NMR titrations were performed in 40 mM cacodylate, pH 6.4, 100 mM NaCl, 2 mM DTT and 0.02% NaN3. Protein concentration was 450 μM, and chemical shift changes were monitored on a Bruker DMX spectrometer at 600 MHz using fast-heteronuclear multiple-quantum coherence experiments.21 The zinc was used as ZnCl2 in a stock solution of 10 mM in the respective buffer and pH level of which the appropriate amount was added at each step of the titration to the sample. The sample had a volume of 400 μL in an NMR tube (Shigemi) minus the insert. Steps used in the titration were (C1/Zn concentration ratios): 1.0:0.0, 1.0:0.2, 1.0:0.4, 1.0:0.6, 1.0:0.8, 1.0:1.0, 1.0:1.2, 1.0:1.4 and 1.0:1.8. Spectra were processed with TOPSPIN 2.0 and analysed in CCPN analysis.44 After peak picking and assignment, a series of experiments was created. With the use of the “follow shift change” function, peaks of the same amino acid along the titration were automatically grouped. Tables of chemical shift change versus zinc concentration or concentration ratio were then constructed and exported for further analysis in Mathematica 5.0 (Wolfram Research). Chemical shift perturbation versus zinc concentration curves were fitted against a standard binding isotherm:
Chemical shift mapping was performed by calculating combined chemical shift differences, Δδ, for both 15N and 1H using a scaling factor of 1.0 for 1H and that of 0.15 for 15N based on the first experiment and the last experiment in the series. The average of the combined Δδ values over all residues was calculated to give the standard deviation 1∗σ.
Model building
The model of the C1–S2Δ interaction was modelled in the context of the whole fragment, C1C2 bound to S2Δ. The position of C2 on S2Δ was used as published previously.19 The model of the C1C2 complex with S2Δ was generated in three consecutive stages: (1) knowledge-based molecular docking using HADDOCK,45 (2) molecular dynamics (MD) simulation trajectory calculations using AMBER22 and (3) energy minimisation of the final model structures using AMBER and ranking of the complex models. This protocol scheme was used in an iterative manner to reach the best final complex model:
Stage 1: We produced the complex models of C1C2–S2Δ using the previously reported C2–S2Δ complex interface to lock C2 on S2Δ;19 all the binding site residues of C1 were used as follows: active and passive residues (shown as red and yellow spheres, respectively, in Fig. 6a); all residues of the linker between C1 and C2 were used as active and flexible; the residue region used as the binding site on S2Δ was from P383 to E861; and all S2Δ residues except the region between D945 and A962 were used as flexible.
Stage 2: We calculated short MD trajectories, 200–400 ps, for the best three to five complex models obtained from stage 1. This provides more structural rearrangements between C1C2 and S2Δ and better conformational space sampling of the complex. All the MD simulations were performed in explicit TIP3P water, the complex charge was neutralized by adding Na+ ions, the time step was 1.5 ps and the temperature was 300 K.
Stage 3: We minimised the complex structure models and ranked them according to the number of residues that are involved in the interface between C1 and S2Δ by making hydrogen bonds and/or salt bridges. The final best two or three complex models are then subjected to another full run through stage 1 to stage 3.
We repeated this cycle five times before no further improvement was achieved. The final best complex model was used for further structural analysis, as shown in Results.
NMR of C1C2
NMR experiments were recorded on a 0.4 mM 15N-labelled sample of C1C2 in 20 mM phosphate, pH 7.3, 50 mM NaCl, 2 mM DTT, 1 mM ethylenediaminetetraacetic acid, 0.02% NaN3 at 800 MHz and a temperature of 303 K. In-house-modified Bruker standard pulse programs were used to record the heteronuclear single-quantum coherence and the heteronuclear NOE experiments. The latter was recorded with a 3.5-s saturation period in a 5-s relaxation delay. Saturation and non-saturation spectra were collected in interleaved mode.
Accession code
The coordinates of the structure are available from the PDB with accession code 2AVG.
Acknowledgements
This work was supported by a project grant of the British Heart Foundation (PG99/121) to M.P. and M.G. M.P. was a recipient of a Royal Society University Research Fellowship. We thank Dr. F. Muskett for helping with the NMR experiments, Drs. T. Stevens and W. Boucher for helping with CCPN analysis and Prof. Merz for allowing use of his AMBER software.
Edited by M. F. Summers
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2008.09.065
Appendix A. Supplementary data
References
- 1.Kobayashi T., Solaro R.J. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 2.Winegrad S. Myosin-binding protein C (MyBP-C) in cardiac muscle and contractility. Adv. Exp. Med. Biol. 2003;538:31–40. doi: 10.1007/978-1-4419-9029-7_3. Discussion 40-1. [DOI] [PubMed] [Google Scholar]
- 3.Oakley C.E., Hambly B.D., Curmi P.M.G., Brown L.J. Myosin binding protein C: structural abnormalities in familial hypertrophic cardiomyopathy. Cell Res. 2004;14:95–110. doi: 10.1038/sj.cr.7290208. [DOI] [PubMed] [Google Scholar]
- 4.Winegrad S. Myosin binding protein C, a potential regulator of cardiac contractility. Circulation. 2000;86:6–7. doi: 10.1161/01.res.86.1.6. [DOI] [PubMed] [Google Scholar]
- 5.Lange S., Ehler E., Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Gruen M., Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy abolish the interaction with the regulatory domain of myosin binding protein C. J. Mol. Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 7.Levine R., Weisberg A., Kulikovskaya I., McClellan G., Winegrad S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophys. J. 2001;81:1070–1082. doi: 10.1016/S0006-3495(01)75764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClellan G., Kulikovskaya I., Winegrad S. Changes in cardiac contractility related to calcium-mediated changes in phosphorylation of myosin-binding protein C. Biophys. J. 2001;81:1083–1092. doi: 10.1016/S0006-3495(01)75765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunst G., Kress K.R., Gruen M., Uttenweiler D., Gautel M., Fink R.H.A. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circulation. 2000;86:51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 10.Gruen M., Prinz S., Gautel M. cAPK-phosphorylation controls the interactions of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on–off fashion. FEBS Lett. 1999;453:254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 11.Alcalai R., Seidman J.G., Seidman C.E. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J. Cardiovasc. Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 12.Harris S.P., Bartley C.R., Hacker T.A., McDonald K.S., Douglas P.S., Greaser M.L. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 13.Zoghbi M.E., Woodhead J.L., Moss R.L., Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl Acad. Sci. USA. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kensler R.W., Harris S.P. The structure of isolated cardiac myosin thick filaments from cardiac myosin binding protein-C knockout mice. Biophys. J. 2008;94:1707–1718. doi: 10.1529/biophysj.107.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisberg A., Winegrad S. Alteration of myosin cross-bridges by phosphorylation of myosin-binding protein C in cardiac muscle. Proc. Natl Acad. Sci. USA. 1996;93:8999–9003. doi: 10.1073/pnas.93.17.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herron T.J., Rostkova E., Kunst G., Chaturvedi R., Gautel M., Kentish J.C. Activation of myocardial contraction by the N-terminal domains of myosin binding protein-C. Circ. Res. 2006;98:1290–1298. doi: 10.1161/01.RES.0000222059.54917.ef. [DOI] [PubMed] [Google Scholar]
- 17.Razumova M., Shaffer J.F., Tu A.Y., Flint G.V., Regnier M., Harris S.P. Effects of the N-terminal domains of myosin binding protein C in an in vitro motility assay. J. Biol. Chem. 2006;281:35486–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 18.Harpaz Y., Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 19.Ababou A., Gautel M., Pfuhl M. Dissecting the N-terminal myosin binding site of human cardiac myosin-binding protein C. Structure and myosin binding of domain C2. J. Biol. Chem. 2007;282:9204–9215. doi: 10.1074/jbc.M610899200. [DOI] [PubMed] [Google Scholar]
- 20.Saarinen S., Kato H., Uchiyama T., Miyoshi-Akiyama T., Papageorgiou A.C. Crystal structure of Streptococcus dysgalactiae-derived mitogen reveals a zinc-binding site and alterations in TcR binding. J. Mol. Biol. 2007;373:1089–1097. doi: 10.1016/j.jmb.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Schanda P., Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 22.Weiner J.K., Kollman P.A. AMBER: assisted model building with energy refinement. A general program for modeling molecules and their interaction. J. Comput. Chem. 1981;2:287–296. [Google Scholar]
- 23.Suarez D., Diaz N., Merz K.M., Jr Molecular dynamics simulations of the dinuclear zinc-beta-lactamase from Bacteroides fragilis complexed with imipenem. J. Comput. Chem. 2002;23:1587–1600. doi: 10.1002/jcc.10157. [DOI] [PubMed] [Google Scholar]
- 24.Ababou A., Zhou L., Gautel M., Pfuhl M. Sequence specific assignment of domain C1 of the N-terminal myosin binding site of myosin binding protein C. J. Biomol. NMR. 2004;29:431–432. doi: 10.1023/B:JNMR.0000032510.03606.63. [DOI] [PubMed] [Google Scholar]
- 25.Mayans O., Wuerges J., Canela S., Gautel M., Wilmanns M. Structural evidence for a possible role of reversible disulphide bridge formation in the elasticity of the muscle protein titin. Structure. 2001;9:331–340. doi: 10.1016/s0969-2126(01)00591-3. [DOI] [PubMed] [Google Scholar]
- 26.Fisher S.J., Helliwell J.R., Khurshid S., Govada L., Redwood C., Squire J.M., Chayen N.E. An investigation into the protonation states of the C1 domain of cardiac myosin-binding protein C. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:658–664. doi: 10.1107/S0907444908008792. [DOI] [PubMed] [Google Scholar]
- 27.Holm L., Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 28.Hernandez Valladares M., Felici A., Weber G., Adolph H.W., Zeppezauer M., Rossolini G.M. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-beta-lactamase activity and stability. Biochemistry. 1997;36:11534–11541. doi: 10.1021/bi971056h. [DOI] [PubMed] [Google Scholar]
- 29.Blindauer C.A. Metallothioneins with unusual residues: histidines as modulators of zinc affinity and reactivity. J. Inorg. Biochem. 2008;102:507–521. doi: 10.1016/j.jinorgbio.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 30.Ayaz M., Turan B. Selenium prevents diabetes-induced alterations in [Zn2+]i and metallothionein level of rat heart via restoration of cell redox cycle. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H1071–H1080. doi: 10.1152/ajpheart.00754.2005. [DOI] [PubMed] [Google Scholar]
- 31.Morita H., Seidman J., Seidman C.E. Genetic causes of human heart failure. J. Clin. Invest. 2005;115:518–526. doi: 10.1172/JCI200524351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeffries C.M., Whitten A.E., Harris S.P., Trewhella J. Small-angle X-ray scattering reveals the N-terminal domain organization of cardiac myosin binding protein C. J. Mol. Biol. 2008;377:1186–1199. doi: 10.1016/j.jmb.2008.01.080. [DOI] [PubMed] [Google Scholar]
- 33.Bernado P., Bertoncini C.W., Griesinger C., Zweckstetter M., Blackledge M. Defining long-range order and local disorder in native alpha-synuclein using residual dipolar couplings. J. Am. Chem. Soc. 2005;127:17968–17969. doi: 10.1021/ja055538p. [DOI] [PubMed] [Google Scholar]
- 34.Sung Y.H., Eliezer D. Residual structure, backbone dynamics, and interactions within the synuclein family. J. Mol. Biol. 2007;372:689–707. doi: 10.1016/j.jmb.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penkett C.J., Dobson C.M., Smith L.J., Bright J.R., Pickford A.R., Campbell I.D., Potts J.R. Identification of residues involved in the interaction of Staphylococcus aureus fibronectin-binding protein with the 4F15F1 module pair of human fibronectin using heteronuclear NMR spectroscopy. Biochemistry. 2000;39:2887–2893. doi: 10.1021/bi992267k. [DOI] [PubMed] [Google Scholar]
- 36.Gautel M., Zuffardi O., Freiburg A., Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14:1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blankenfeldt W., Thomä N., Wray J., Gautel M., Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc. Natl Acad. Sci. USA. 2006;103:17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 39.Linge J.P., O'Donoghue S., Nilges M. Assigning ambiguous NOEs with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 40.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J.D., Higgins D.G., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pervushin K., Riek R., Wider G., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez C., Boelens R., Bonvin A.M.J.J. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.