SUMMARY
We have designed ‘split tetra-Cys motifs’ that bind the biarsenical fluorescein dye FlAsH across strands of a model β-rich protein. Our strategy was to divide the linear FlAsH-binding tetra-Cys sequence such that dye could be fully liganded only when the strands were arranged in space correctly by native protein conformational proximities. We introduced pairs of alternating cysteines on adjacent β-strands of cellular retinoic-acid binding protein (CRABP) to create FlAsH-binding sites in the native structure. Selective labeling occurred both in vitro and in vivo relative to sites with fewer than four Cys or with inappropriate geometry. Interestingly, two of the split tetra-Cys motif-carrying proteins bound FlAsH whether native or urea-unfolded, while one was capable of binding FlAsH only when native. This latter design exemplifies the potential of split motifs as structure sensors.
INTRODUCTION
Tsien and co-workers introduced specific sequence-encoded fluorophore-binding tetra-Cys motifs, which ligate biarsenical dyes via simultaneous formation of four covalent bonds (Griffin et al., 1998). Biarsenical-based fluorophores such as FlAsH have subsequently been widely used to specifically label proteins in vivo by introduction of the appropriate linear tetra-Cys binding sequence, Cys-Cys-Xaa-Yaa-Cys-Cys as a C- or N-terminal tag. We exploited a FlAsH-binding tetra-Cys motif internal to a protein sequence to probe in vivo stability and aggregation using as a model protein the intracellular lipid-binding protein, cellular retinoic acid-binding protein or CRABP (Ignatova and Gierasch, 2004; Ignatova et al., 2007). Our design incorporated a linear tetra-Cys motif in an Ω-loop and yielded a very useful fluorescence read-out of the folded/unfolded population because geometric constraints imposed on the tetra-Cys thiol ligands by the native structure lowered the FlAsH quantum yield substantially (Ignatova and Gierasch, 2004; Ignatova et al., 2007). In our subsequent work, we have attempted to design tetra-Cys motifs that would be even more sensitive to molecular architecture. Our strategy was to divide the binding motif into components that would only accommodate a fully liganded FlAsH dye when arranged in space correctly by native protein structure. We refer to the resulting discontinuous biarsenical dye-binding sequences as ‘split tetra-Cys motifs’.
A similar strategy was recently explored by Schepartz and coworkers (Luedtke et al., 2007). They added two cysteine pairs at the termini of avian pancreatic polypeptide and of Zip4, the first a helical hairpin and the latter a β-hairpin, and showed that only the native, folded state bound dye favorably. They also showed using dicysteine-tagged leucine zipper-forming sequences that intermolecular reconstitution of dye binding sites could be used to report on dimerization.
In the present study, we have extended the concept of structurally defined split-tetra-Cys motifs by designing FlAsH-binding sites within the folded interior of the β-clam protein, CRABP I. In this case, proper ligand arrangement requires alternating cysteine residues, Cys-Xaa-Cys. We incorporated these sequences into adjacent β-strands of CRABP I such that the four cysteines would come close in space and present in essence a recombined tetra-Cys motif in the native protein. In principle, this arrangement allows formation of the FlAsH-binding split tetra-Cys motif only in the folded protein, although populated states with native-like distances may be able to bind the fluorophore as well. We were able to demonstrate labeling of three different split tetra-Cys motif-carrying CRABP I proteins, both in vitro and in vivo. Selectivity for the well-designed motifs was indicated by the lack of labeling of sites with fewer than four cysteines or with non-optimal geometry. In vitro, achieving selective labeling of the split-tetraCys motif-containing proteins was highly dependent on solution conditions. Presence of competing thiol-based molecules such as β-mercaptoethanol, reduced glutathione (GSH) or ethanedithiol (EDT) led to greater selectivity of labeling in vitro. Furthermore, we observed that a combination of GSH, which is abundant in vivo, and a mixture of proteins (bovine serum albumin (BSA) and lysozyme) that mimics the cellular environment also resulted in specific labeling of the split tetra-Cys motif-containing proteins. Consistent with this observation, in vivo labeling was highly specific, and only proteins carrying geometrically optimized split tetra-Cys motifs were labeled.
Both the quantum yield of bound FlAsH and its binding affinity were found to be dependent on the location of the split tetra-Cys motif in the protein. We believe that the observed differences in quantum yield are a consequence of structural constraints on the geometry of the binding site from the protein architecture, which lead to non-optimal ligand arrangements. Any distortions from ideal geometry or structural deformations in the protein also lead to reduced stability of the dye-protein complex. Contrary to our expectations, only one of the sequence-distant split tetra-Cys motifs lost FlAsH-binding ability under denaturing solution conditions. While the reasons for the retention of FlAsH binding in the other cases are not clear, we speculate that the components of the split motif spend time in proximity even in the denatured state, leading to significant dye binding. These results provide insight into design of split tetra-Cys motifs that can be accommodated in a native structure and factors that enable them to report on the folding status of a β-sheet protein.
RESULTS
Design considerations in a cross-strand split tetra-Cys motif
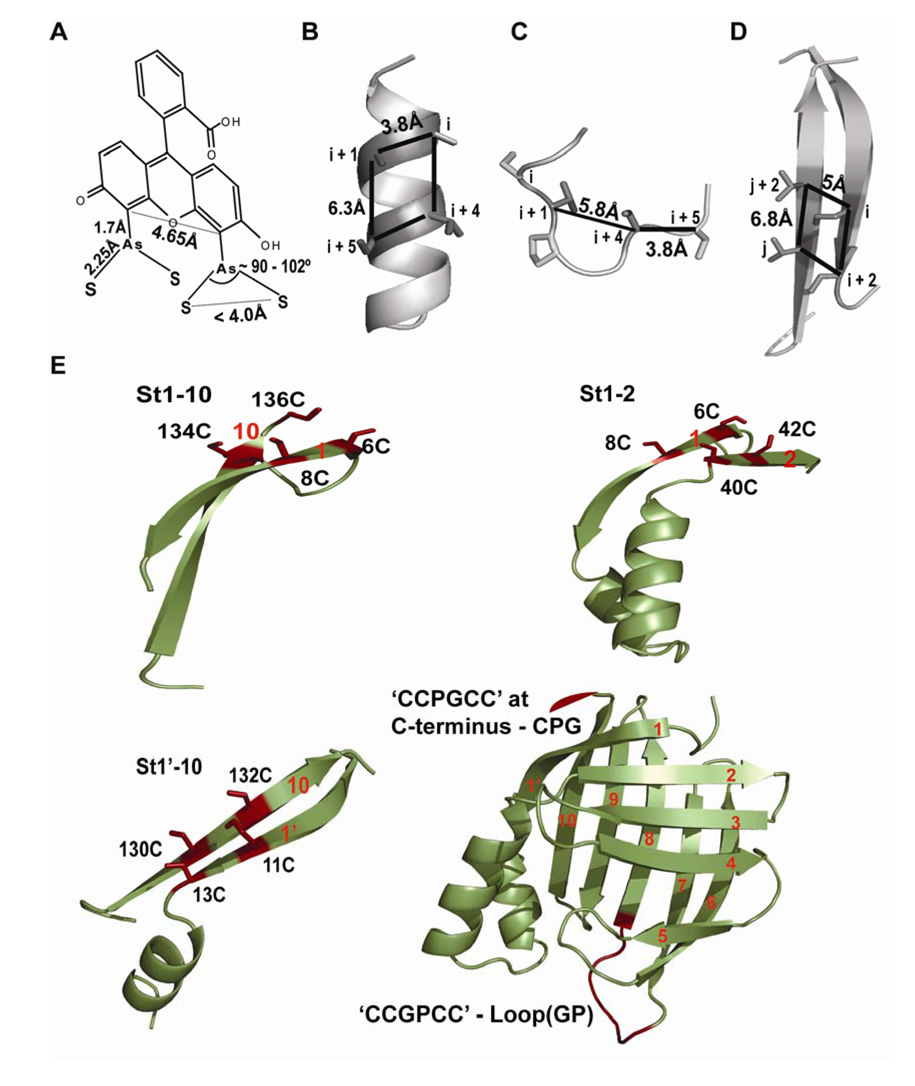
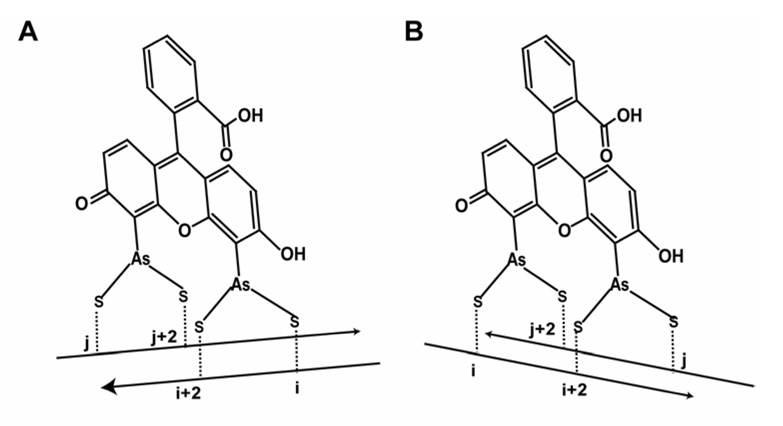
Rational design of a FlAsH dye-binding motif ideally requires knowledge of the structure of the biarsenical-bound tetra-Cys present in a model peptide or in a protein. However, due to the lack of any such structural data we decided to use available biochemical information, the NMR structure of As(III) binding to a di-cysteine in a helical peptide (Cline et al., 2003), fluorescence enhancement studies of FlAsH binding to di-cysteine loop-forming peptides (Adams et al., 2002; Stroffekova et al., 2001), and the crystal structures of As(III)-thiol-containing small molecules (Cruse and James, 1972; DiMaio and Rheingold, 1990; Shaikh et al., 2006a, b) and fluorescein (Korndorfer et al., 2003) to assist our design of split tetra-Cys motifs. The measured distances between the α-carbons of the cysteines forming FlAsH-binding motifs are indicated in Figures 1A – 1C. It can be seen that these distances are very similar to those between the α-carbons of a pair of residues at alternating positions on a β-strand along with a pair at the corresponding cross-strand positions (Figure 1D). It should be noted that a range of distances between the α-carbons could still enable the thiols to bind, given the flexibility in cysteine side chain orientations. Our design of placing the four cysteine residues in alternating positions on adjacent β-strands to form the tetra-Cys binding sequence was further supported by a recent report on As(III) binding to β-hairpin structures wherein the authors report that As(III) can bridge cysteines present on two anti-parallel β-strands (Ramadan et al., 2007).
Figure 1. Distance-constraints for a tetra-Cys motif.
(A) The distances obtained from the crystal structures of fluorescein (PDB:1N0S) (Korndorfer et al., 2003) and As–S containing compounds (Cruse and James, 1972; DiMaio and Rheingold, 1990; Shaikh et al., 2006a, b).
(B) Positions of the cysteine residues (i, i+1, i+4 and i+5) of a tetra-Cys motif on an α-helix. The representative helix is the second helix in CRABP I (PDB:1CBI). The distances were measured between the α-carbon atoms of residues at positions i, i+1, i+4 and i+5.
(C) A ‘Pro-Gly’ containing loop of yeast iso-CytC (PDB: 1YCC) used to obtain distance between α-carbon atoms of the residues (i, i+1, i+4 and i+5) around the turn forming ‘Pro-Gly’ sequence.
(D) Positions of the cysteine residues on spatially adjacent β-strands used to form a cross-strand split tetra-Cys motif. Strands 1’ and 10 of CRABP I are shown. The distances between the α-carbons are similar to those measured for the tetra-Cys-forming residues on an α-helix.
(E) The structure of CRABP I (PDB:1CBI), bottom right, is shown with the numbering of the 10 anti-parallel β-strands, and the continuous tetra-Cys motifs are indicated in red. The positions of the split tetra-Cys motifs are illustrated in the surrounding fragments with the cysteine residues that form the binding site in red.
We used CRABP I, a 136-residue protein containing ten anti-parallel β-strands in a closed β-barrel, as a model protein to test our design. Three different pairs of β-strands—strands 1 and 2 (St1-2), strands 1’ and 10 (St1’-10), and strands 1 and 10 (St1-10)—were selected to host the split tetra-Cys motifs (Figure 1E). The split tetra-Cys motifs across these strand pairs encompass increasing lengths of intervening sequence (36 residues for St1-2, 117 residues for St 1’-10, and 126 residues for St1-10). In addition, we also carried out comparative FlAsH binding studies on CRABP I mutants containing continuous tetra-Cys motifs with a –CCPGCC– sequence at the C-terminus (CPG) or in the Ω-loop between strands 6 and 7, as previously reported (loop(GP)) (Ignatova and Gierasch, 2004). All the variants were checked by circular dichroism (CD) and found not to have suffered any structural perturbation as a consequence of incorporating the tetra-Cys motif (Figure S1).
In vitro FlAsH-labeling
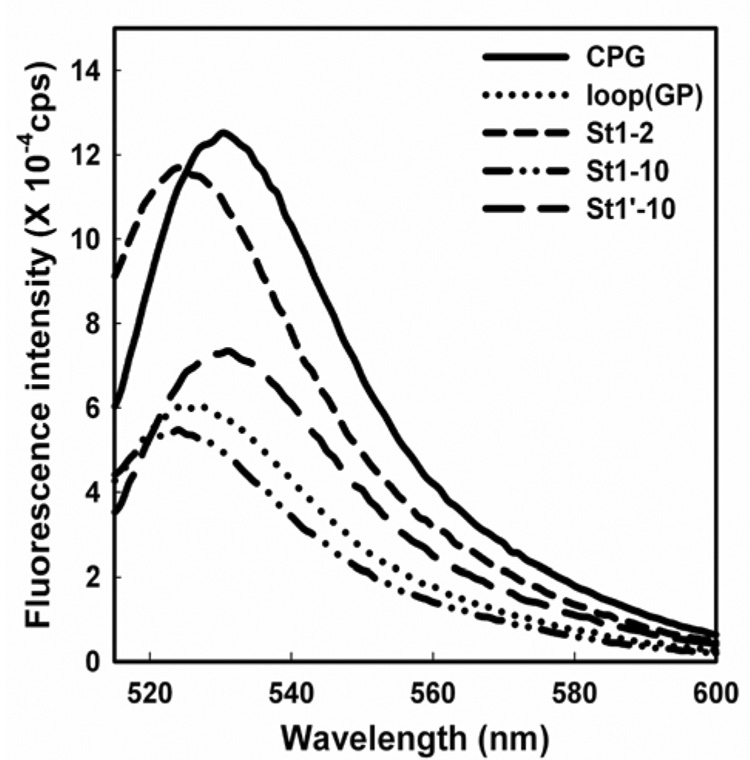
Initially, FlAsH-binding studies were carried out with purified split tetra-Cys-containing proteins under non-physiological, but controlled in vitro conditions in order to test and characterize dye binding to the split motifs. All the tetra-Cys variants could be labeled with FlAsH as purified native protein samples in solution, as indicated by an enhancement of FlAsH fluorescence upon binding (Figure 2). Importantly, far-UV CD spectra of the labeled proteins showed no significant perturbation of the protein structure upon labeling (Figure S1). The enhancements in FlAsH fluorescence upon binding to the different tetra-Cys motifs followed the trend, CPG > St1-2 > St1’-10 > St1-10 ~ loop(GP). This order reflects the quantum yields of the FlAsH-bound proteins, which were 0.35 ± 0.1 for CPG; 0.17 ± 0.1 for loop(GP); 0.3 ± 0.07 for St1-2; 0.16± 0.01 for St1-10 and 0.24± 0.09 for St1’-10. We have observed that the quantum yields of FlAsH bound to tetra-Cys motifs within this model protein are in general lower than those reported in earlier studies (0.44 – 0.71) (Adams et al., 2002; Luedtke et al., 2007; Wang et al., 2007). The other noticeable feature of the bound-FlAsH spectra was a 5 to 6 nm blue-shift of the emission maximum in the case of the St1-2 and St1-10 proteins, consistent with a hydrophobic environment around the bound dye. The cysteine residues on strand 1 for both of these split motifs flank a tryptophan residue (Trp7), which may form an apolar region close to the bound dye.
Figure 2. FlAsH fluorescence upon binding to the tetra-Cys containing proteins.
The fluorescence emission spectra of the various FlAsH-labeled tetra-Cys motif-containing proteins (0.2 µM solutions).
Comparative binding affinities of different split tetra-Cys motifs
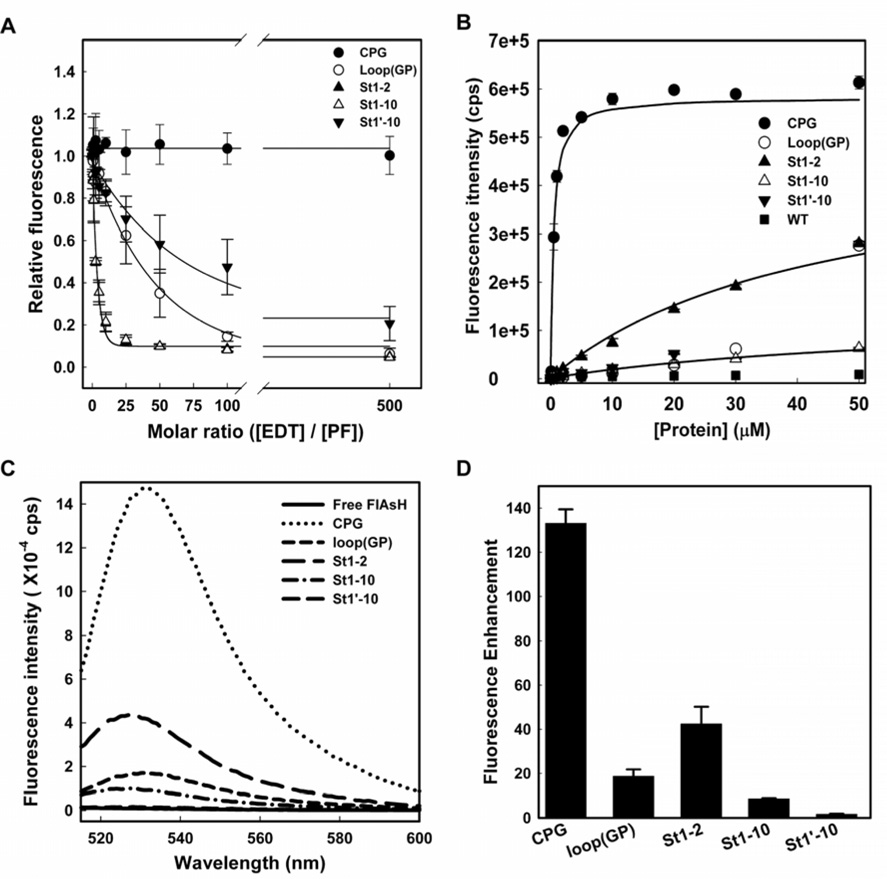
FlAsH binding affinity for the tetra-Cys motifs was determined using two methods: displacement of the bound dye by increasing concentrations of EDT followed by fluorescence titration (Adams et al., 2002) and direct measurement of the apparent dissociation constant (KDapp) by titrating FlAsH with increasing protein concentration in the presence of EDT. In the former, the fluorescence signal retained upon treating the FlAsH-labeled proteins with varying concentrations of EDT provided a direct measure of the stability of the FlAsH-bound species. We define the ‘EDT-50’ as the molar excess of EDT over the protein concentration at which the FlAsH fluorescence was reduced to half of its initial value in the absence of EDT. FlAsH bound to the CPG protein was most resistant to EDT displacement, while by contrast, FlAsH bound to all the other constructs was more readily displaced by EDT, with the EDT-50s following the trend St1’-10 > loop(GP) > St1-2 ~ St1-10 (Figure 3A). FlAsH was 50% displaced from the St1’-10 and loop(GP) motifs at about seventy-fold and fifty-fold molar excess of EDT respectively, while the motifs St1-2 and St1-10 were displaced by EDT at only about five-fold excess under similar conditions of labeling. The wide range of EDT-50 values for the different split motifs suggests that the stability of the bound dye is depends on the location of the binding motif within the protein. In addition, we noted that dye displacement occurs in two kinetic steps, which we interpret to indicate a slow (rate-limiting) initial release of one of the di-cysteine motifs, which then accelerates dislodging of the dye molecule from the second pair of cysteines in a second faster step. The details of the kinetic analyses are described in Figure S2.
Figure 3. Affinity of FlAsH for different tetra-Cys motifs.
(A) EDT titration of FlAsH-labeled proteins. Varying concentrations of EDT were added to 0.2 µM solutions of FlAsH-labeled proteins, and the decrease in fluorescence at 530 nm was used as a measure of displacement of bound FlAsH. (B) Equilibrium binding of FlAsH to the tetra-Cys motif-containing proteins. The apparent dissociation constant was determined from fitting the fluorescence enhancement at 530 nm upon addition of increasing protein concentration to a 50 nM of FlAsH solution. Panels (C) and (D) show the fluorescence emission spectra and the relative fluorescence increase with respect to the free dye fluorescence for 0.5 µM of the different tetra-Cys motif-containing proteins labeled with 1.0 µM of FlAsH, respectively. The error bars indicate the standard deviation from four independent experiments.
A direct measurement of KDapp in the presence of EDT (Figure 3B) shows that the CPG tetra-Cys motif has the highest affinity for FlAsH (KDapp = 0.4 µM) followed by the split motifs St1-2 (KDapp = 43 µM) and St1-10 (KDapp = 64 µM). The St1’-10 protein exhibited the lowest affinity for FlAsH (KDapp = 2.6 mM). In the case of the loop(GP) protein, the data were not fit well to a single binding equilibrium due to the formation of hyperfluorescent aggregates at higher protein concentration.
Taken together, the differing quantum yields and affinities led to a wide range of fluorescence enhancements for the labeled proteins at a fixed protein to dye concentration (a two-fold excess of FlAsH) (Figure 3C and D). Placing the CCGPCC motif at the C-terminus resulted in by far the greatest fluorescence enhancement (~130-fold), followed by one of the split tetra-Cys variants, St1-2. Under these conditions, FlAsH-labeled St1’-10 displayed the lowest increase in fluorescence. As a cautionary note, interpretation of FlAsH fluorescence of designed tetra-Cys motifs within the folded regions of a protein, whether split or continuous, requires that the potential variations in both quantum yield and affinity be acknowledged. Interestingly, the St 1’-10 split tetra-Cys motif shows weak direct binding affinity, but bound dye is quite resistant to EDT. This apparently paradoxical observation underlines the complexity of the binding reaction and the chemical nature of the thiol exchange process.
Is FlAsH binding to split tetra-Cys motifs selective?
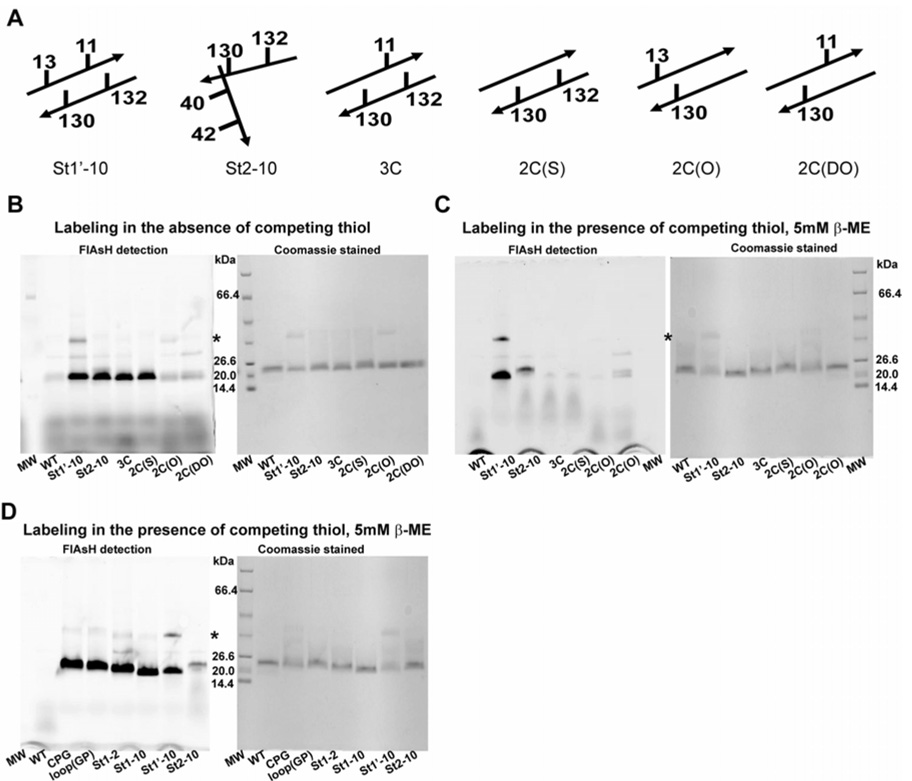
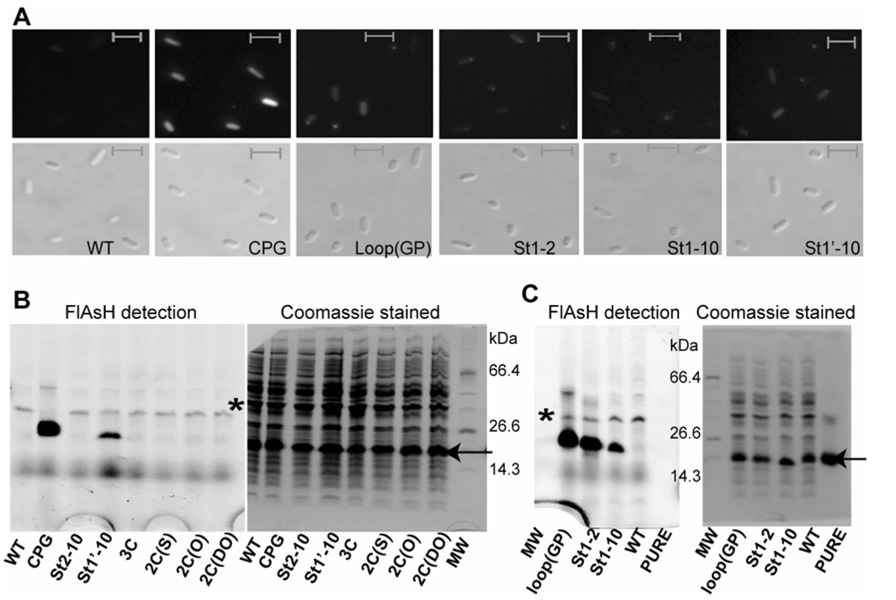
There is a real risk that the entropic cost of bringing the two components of a split tetra-Cys motif together to reconstitute a well-formed FlAsH binding site will lead to substantial loss of selectivity over adventitious sites, for example with fewer than four thiols. Non-specific biarsenical dye labeling in cells from binding to endogenous thiols and from the hydrophobic nature of the fluorophore has been reported (Adams et al., 2002; Stroffekova et al., 2001). FlAsH-binding to cysteines within protein sequences is based on arsenite chemistry, and arsenites have significant affinity for both mono- and di-thiol compounds (Rey et al., 2004; Spuches et al., 2005). We designed control variants of the St1’-10 split tetra-Cys motif containing protein carrying zero to three cysteines (Table S3, Figure 4A). We also included in our comparative studies as a negative control a tetra-Cys variant with a pair of cysteines on strands 2 and 10 such that the binding motif cannot form in the native protein. Under what we term ‘non-stringent’ conditions in vitro (i.e., in the absence of competing thiols), all of the purified control proteins bound FlAsH to varying extents (Figure 4B). In all cases, the observed fluorescence was similar or lower than the fluorescence associated with the proteins carrying the full tetra-Cys motifs, split or not. Also, increasing the labeling time led to increased labeling of all the proteins including the controls.
Figure 4. Tests for alternative FlAsH-binding sites in vitro.
(A) The schematic indicates the positions of the cysteine residues in the control constructs with sub-optimal FlAsH binding sequences. The various proteins illustrated in (A) were labeled in vitro in buffer (B) or in buffer containing 5 mM β-mercaptoethanol as a competing molecule (C). Panel (D) shows analogous labeling of the purified tetra-Cys motif-containing proteins in 5 mM β-mercaptoethanol. The left and the right panels in (B), (C) and (D) show detection of the same SDS-PAGE gel by FlAsH fluorescence (using a phosphorimager) and by Coomassie staining, respectively. Labeling under the stringent conditions in panel (C) demonstrates the selectivity of FlAsH-labeling for only the tetra-Cys proteins. The asterisk indicates the position of a CRABP I dimer. Note the strong fluorescence signal detected for the St1’-10 protein despite a lower protein amount based on Coomassie blue staining, indicating that it is well labeled by FlAsH.
On the other hand, in buffers containing 5 mM β-mercaptoethanol or in 5 mM glutathione redox buffer with a molar ratio of reduced to oxidized glutathione of 200 (similar to what would be present in E. coli cells (Koprowski and Kubalski, 1999; Messens and Collet, 2006)), the control proteins were significantly less well labeled with FlAsH than the tetra-Cys proteins (Figures 4C and D). Similar results were obtained when the FlAsH-labeling reaction was carried out in a glutathione redox buffer containing a mixture of BSA and lysozyme to simulate the presence of background proteins lacking a tetra-Cys sequence and to increase the complexity of the reaction mixture (Figure S4). The strong fluorescence associated with BSA is due to non-specific labeling or attachment of FlAsH, an observation that has been reported earlier (Cao et al., 2006). FlAsH displays a preferential interaction with expressed tetra-Cys proteins even when the amount of tetra-Cys protein is considerably lower than the amount of BSA, as indicated by a reduction in BSA-associated FlAsH fluorescence as the amount of expressed tetra-Cys protein increases (lanes 2 – 5 of Figure S4B). Our data clearly show that FlAsH labeling to favorably situated split tetra-Cys motifs is indeed selective over control proteins and competing background when the labeling reaction is carried out under stringent conditions.
Mode of thiol-binding to the arsenics in a split tetra-Cys motif
In the split tetra-Cys motifs that span spatially adjacent strands, there are two possible modes of FlAsH binding; the arsenic atoms can each bind across the interstrand space or along the strand (Figure 5). The fact that both the di-cysteine control protein with cysteines on the same strand, 2C(S), and the one with cysteines across two adjacent strands, 2C(O), bind significantly to the arsenic atoms of FlAsH (in the absence of competing thiol reductant) argues that either mode of arsenic-thiol liganding can occur. The 2C(O) protein with cysteines present across the strands exhibited about two-fold greater fluorescence upon FlAsH binding than did the protein carrying the di-Cys motif on the same strand (2C(S)) (Figure S5A). When the FlAsH-bound di-Cys motif proteins were analyzed by SDS-PAGE, the fluorescence of the 2C(O) protein diminished markedly relative to that of the 2C(S) protein (Figure S5B), presumably because of SDS-denaturation of the protein and resulting strand separation.
Figure 5. Two potential modes of arsenic binding to the cysteines of a split tetra-Cys motif.
(A) Arsenic atom linked to the cysteine residues on the same strand, and (B) arsenic atom bound to the cysteine residues from two adjacent strands.
In vivo FlAsH-labeling
Based on our finding that FlAsH binding is selective under stringent labeling conditions in vitro, we anticipated selective labeling of the tetra-Cys proteins expressed in vivo. We followed the protocol used previously to label continuous tetra-Cys containing proteins expressed in E. coli cells (Ignatova and Gierasch, 2004) with a few modifications. Note that no cellular toxicity was observed at the micromolar concentrations of FlAsH used for the in vivo labeling (data not shown). Significant FlAsH labeling of the split tetra-Cys proteins was indicated by uniform fluorescence throughout cells (Figure 6A) and confirmed by analysis of the cell lysates for FlAsH fluorescence by phosphorimaging, which showed that the major fraction of the observed in-cell fluorescence was indeed from FlAsH associated with the CRABP I tetra-Cys motif-containing protein (Figure 6B and C). A strong fluorescence signal was observed for both the continuous and the split tetra-Cys motif-containing proteins, and the fluorescence associated with the cells expressing the control proteins (with fewer than four cysteines or split tetra-Cys motifs on non-adjacent strands) was low, similar to that observed in cells expressing the wild type protein (devoid of a tetra-Cys motif). A difference in the efficiency of cell labeling was observed, with the best labeling (> 90% of cells) in the case of the CPG protein. Approximately seventy percent of the cells expressing St1-2, St1’-10 and loop (GP) were labeled, and only about 50% of cells expressing the St1-10 split motif protein were labeled.
Figure 6. In-cell FlAsH labeling of tetra-Cys proteins.
(A) Epi-fluorescence microscopy images of FlAsH-labeled E. coli cells expressing the various tetra-Cys motif-containing proteins (as described in Experimental Procedures). The upper and lower panels are the fluorescence and the differential interference contrast images, respectively. The scale bar is 2 µm. Lysates from the FlAsH-labeled cells expressing the control proteins for the split tetra-Cys motif (B) and the split tetra-Cys motif-containing proteins (C) were analyzed on a 10% tricine SDS-PAGE. The left panels in (B) and (C) represent the FlAsH fluorescence as a phosphorimage of the Coomassie-stained gel shown in the right panel. The Coomassie-blue staining indicates similar expression levels for all the proteins. The lane labels correspond to the lysate of cells expressing the specified protein. The arrow and the asterisk indicate the position of CRABP I and SlyD (an E. coli protein that has recently been identified to be a high-affinity biarsenical binding protein by Wang et al. (2007)), respectively.
Using a split tetra-Cys motif to discriminate native and non-native protein conformation
We anticipated that the ability of a split tetra-Cys motif to bind FlAsH could be used to report on the presence of native-like structure, reasoning that this structure was necessary to bring the liganding residues into the arrangement required for binding. We expected therefore that the ability of all of the designed split tetra-Cys-containing variants of CRABP I to bind the FlAsH dye would be dependent on the presence of natively folded species under a given set of conditions. We were surprised to find in a comparison of FlAsH binding to the three native and urea-unfolded split tetra-Cys-containing proteins (carried out in the presence of mM EDT, see Experimental Procedures) (Figure S6A) that only the St1’-10 split tetra-Cys-containing protein displayed this sensitivity of FlAsH fluorescence to native structure. For comparison purposes, we also show the previously described behavior of the FlAsH-labeled loop(GP) variant, which has proven very useful for in-cell urea melts (Ignatova and Gierasch, 2004; Ignatova et al., 2007), where the FlAsH fluorescence intensity is higher in the unfolded state. A decrease in FlAsH fluorescence for the St1’-10 split tetra-Cys containing protein was also observed when it was expressed in E. coli cells and then the resulting cell lysate was urea-treated (Figure S6B). The St1’-10 protein thus exemplifies the anticipated potential of split tetra-Cys motifs as folding sensors (see below).
At present, we have no explanation for the retention of dye-binding ability of the denatured state of the St1-2 and St1-10 split tetra-Cys-containing proteins other than to suggest that the unfolded state of CRABP I retains native-like proximities between some regions due to sequence-dependent biases that are not weakened by urea. For example, we previously showed that an isolated peptide fragment of CRABP 1 (residues 10 – 32) populates to a substantial extent the native-like Schellman motif, which would bring strands 1 and 2 into proximity in the unfolded state (Sukumar and Gierasch, 1997). By contrast, the likelihood that the two components of the St1-10 split tetra-Cys motif come into proximity in the urea-unfolded state is expected to be small. More intensive study of the conformational ensemble present in the urea-unfolded state of CRABP I may provide insight into the FlAsH-binding abilities of these different split tetra-Cys variants.
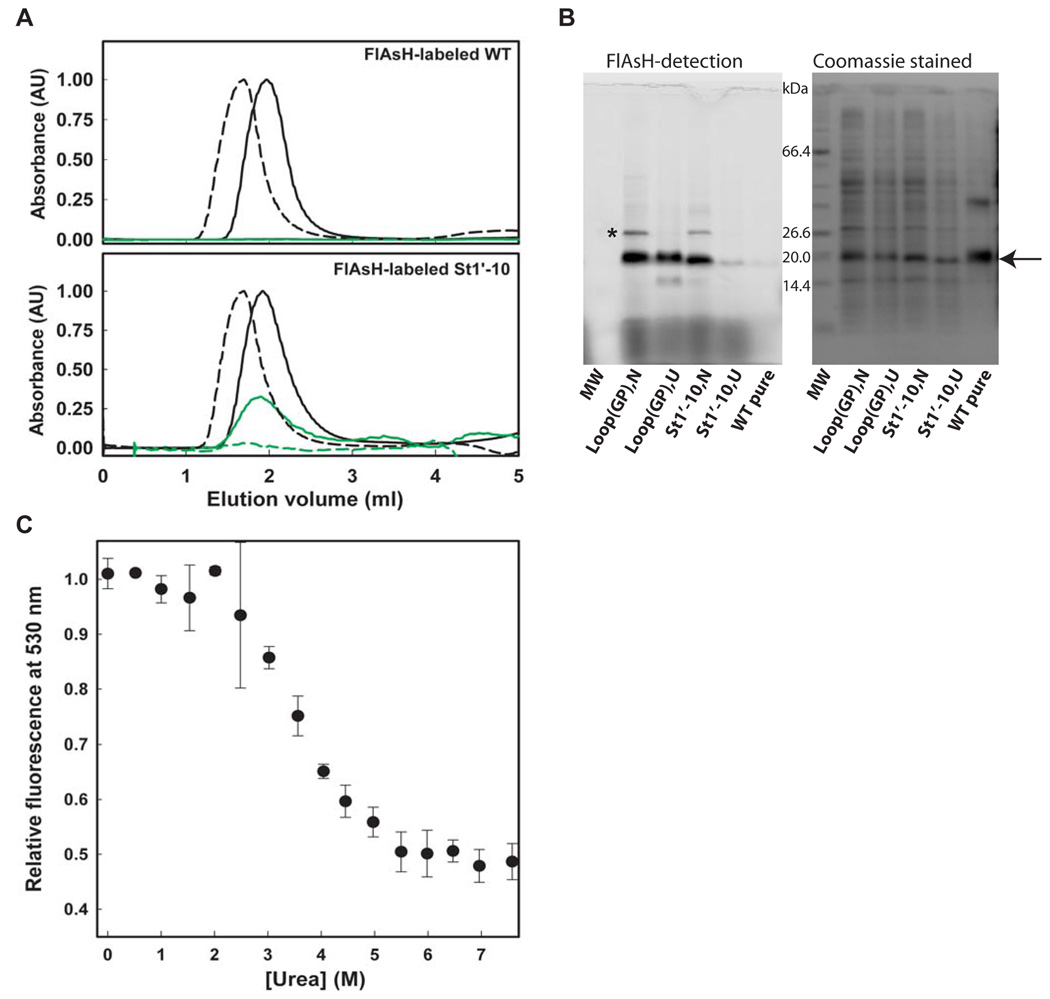
Through more detailed analysis, we confirmed that the urea dependence of the St1’-10 split tetra-Cys motif protein FlAsH fluorescence arises from loss of binding affinity as structure is disrupted at higher urea concentration. Note that this is distinct from our original design of the loop(GP) tetra-Cys variant where the quantum yield of bound FlAsH is urea-dependent, and not the binding itself. Analyzing the FlAsH-labeled pure St1’-10 protein on a desalting column (Figure 7A) or the FlAsH-labeled expressed St1’-10 protein in cell lysate by SDS-PAGE (Figure 7B) confirms a lower extent of labeling under denaturing conditions. Urea titrations of FlAsH-labeled St1’-10 protein either purified or in cell lysate revealed a reduction of FlAsH fluorescence with increasing urea concentration (Figure S6C and Figure 7C). It is difficult to interpret the apparent Cm from these urea titrations of FlAsH-labeled St1’-10 protein because they are carried out in the presence of a significant concentration of EDT, which will displace the intramolecular FlAsH Cys ligands as structure is disrupted. Thus, the apparent Cm is a measure of how readily EDT can displace dye from the tetra-Cys motif as a function of urea concentration and not strictly thermodynamic stability. For comparison purposes, we show a urea titration of the St1’-10 protein monitored by Trp fluorescence, which yields a higher Cm (5.4 M, Figure S6C). Nonetheless, under the same solution conditions, the apparent Cm’s from FlAsH binding will be useful comparative indicators of stability. Caution must be exercised in moving to different solution conditions, as they may yield different apparent Cm’s. For example, we show in Figure S6C that the apparent Cm from FlAsH fluorescence observed with purified protein in the presence of 0.5 mM EDT is lower than that observed in cell lysate, presumably because there are many proteins present in lysate that bind EDT to some extent and diminish its ability to displace the FlAsH dye.
Figure 7. Structure dependence of FlAsH binding of split tetra-Cys motif-containing CRABP I variants.
(A) Urea dependence of FlAsH binding probed by gel filtration of the FlAsH-labeled St1’-10 protein. The solid line and the dashed line represent the traces at 0 M and 7 M urea, respectively, followed by absorbance at 280 nm (black line, protein) and or 500 nm (green line, FlAsH). No binding of dye is seen to the wild-type control protein lacking a tetra-Cys mofif (top panel). By contrast, co-elution of the dye with the St1’-10 protein in 0 M urea but not in 7 M urea shows that dye binds to the native protein and not to the unfolded protein. (B) SDS-PAGE analysis of FlAsH binding to the loop(GP) and the St1’-10 tetra-Cys containing proteins in cell lysate in either their native (N) or urea-unfolded (U) states. The reduced fluorescence seen for the unfolded state of the St1’-10 split tetra-Cys motif-containing protein indicates that unfolded protein binds FlAsH poorly. The arrow indicates the position of CRABP I, and the asterisk the position of the non-specifically FlAsH-binding protein, SlyD. (C) Urea titration of St1’-10 split tetra-Cys containing protein in cell lysate measured by FlAsH fluorescence at 530 nm. The error bars for the cell lysate samples were estimated from three independent measurements.
DISCUSSION
Because of their facile cell permeability, their low fluorescence in the unbound form, and their specificity for a short genetically encoded sequence motif, biarsenical dyes such as FlAsH offer one of the most elegant in-cell labeling strategies. We previously exploited these traits along with a fortuitous structure-dependent quantum yield of the protein-bound FlAsH to monitor the urea-induced denaturation of CRABP I inside E. coli cells (Ignatova and Gierasch, 2004; Ignatova et al., 2007). The FlAsH binding tetra-Cys motif in our earlier study was embedded in the middle of the sequence of CRABP I and placed in a site (an Ω-loop) that we thought would tolerate mutation but also perhaps show structure-dependent fluorescence of bound FlAsH. The success of this work established the approach of designing FlAsH-binding sequence locations such that FlAsH fluorescence properties report on the structure and folding of the host protein. The responsiveness of the FlAsH fluorescence to the state of folding of the Ω-loop tetra-Cys protein arose from high sensitivity of the FlAsH quantum yield to the optimal arrangement of thiol ligands in the tetra-Cys motif. This sensitivity was key to the success of this past design but also confounds the design of such structure-sensitive binding sites in other proteins. Our hope to design a more direct strategy for structure-sensitive FlAsH binding and thus to simplify the transfer of this design concept to other systems was the genesis of the present study: We proposed to introduce pairs of cysteine residues in different locations of a protein, not continuous in sequence, such that the residues are in close proximity only in a particular conformational state, such as in the native form. Fluorescence enhancement observed upon FlAsH-binding to such a split tetra-Cys motif could then be used as an indicator of the proximity of the split tetra-Cys residues. While we were implementing this approach, a similar strategy was successfully developed by Schepartz and coworkers (Luedtke et al., 2007). Their choice was to use the split tetra-Cys components (which they term ‘bipartite tetracysteine’ motifs) as tags on the termini of proteins to be tested. In their case, the increased FlAsH binding and fluorescence reported that the termini had come together to form a FlAsH-binding site either through folding or through dimerization.
In our design strategy, we have instead incorporated the split tetra-Cys moieties internal to a protein structure. We tested one of the plausible split tetra-Cys arrangements by placing pairs of cysteines in alternating positions (Cys-Xaa-Cys) on two adjacent β-strands, which situates the four thiols to form a FlAsH-binding site on the protein in its native form. Three such split motifs were grafted into the β-clam protein CRABP I, used as a test case, to gain insight into the optimal design of such split motifs. The St1-2 motif, with cysteines present at the beginning of frayed ends of weakly hydrogen-bonded strands 1 and 2 (Figure S7A), showed the highest affinity for FlAsH. This was followed by the St1-10 motif, which has the cysteine pairs introduced at the end of the β-sheet formed by strands 1 and 10 (Figure S7B). The cysteine residues incorporated into strand 10 in this case are very near to the flexible C-terminus of the protein. The third motif, St1’-10, places the binding motif across a regular well-hydrogen bonded pair of β-strands 1’ and 10 (Figure S7C). Three properties of these sites were used for comparison purposes and to assess their utility for example as conformational probes: the binding affinity measured by apparent dissociation constant of the FlAsH-protein complex (KDapp) at a given EDT concentration, the ease of displacement of bound FlAsH by added EDT, and the quantum yield of bound FlAsH. As a reference, we used CRABP I with a C-terminal continuous tetra-Cys tag.
All of the engineered split tetra-Cys motifs and the internal Ω-loop continuous tetra-Cys motif have lower FlAsH affinity, greater ease of FlAsH displacement, and lower quantum yield of bound FlAsH than the C-terminally tagged tetra-Cys protein. We attribute these properties to the non-optimal geometry of the designed binding sites in combination with the energetic cost that must be invested to rearrange the discontinuous sites to a more favorable geometry. Higher affinity binding was observed for the two cross-strand sites that deviate from regular β-sheet structure (St1-2 and St1-10). Binding to the St1’-10 site, which is embedded in a very regular sheet region of the native structure, is weaker. Interestingly, displacement from the St1’-10 site requires higher EDT. We speculate that this parameter is sensitive to the inherent flexibility of the site, since a thiol exchange reaction must be initiated for displacement to occur. While definitive interpretation of the origin of the differing properties of these designed motifs must await three-dimensional structure determination for FlAsH-bound proteins, the ability of a site to adopt optimal geometry for binding to the FlAsH arsenics seems to influence both affinity and quantum yield. Thus, from a design perspective, to simply optimize binding a split tetra-Cys motif might best be placed in flexible regions, like the terminal tagging strategy used by Luedtke et al. (2007). However, there may be situations were incorporation of FlAsH-binding split tetra-Cys motifs into the well-structured regions of proteins presents advantages. Our successful design of split motifs based on the initial distance estimates of residues on adjacent β-strands suggests the general applicability of using split binding motifs on non-sequence contiguous, spatially adjacent β-strands as fluorescent tagging motifs in proteins.
The results of this study also shed light on the initial design goal: use of split tetra-Cys motifs as structure-sensitive probes, with the initial idea being that binding would occur much more avidly to the native than to the non-native state. We anticipated that the non-native state would release its FlAsH dye rapidly allowing re-equilibration, only a small fraction of the protein will be bound, and hence the dye in this case would serve as a reporter of the conformational distribution in the population without significantly altering the distribution. Indeed, FlAsH-binding to the St1’-10 protein was essentially absent in the urea-unfolded state (in the presence of moderate concentration of reductant and sub-stoichiometric quantitites of FlAsH dye). This protein therefore epitomized our original design and serves as a suitable subject for a FlAsH-based protein-folding assays, either in vitro or in vivo. For reasons that are not immediately apparent, the other two of the split motif-carrying CRABP I variants, St1-2 and St1-10, retained substantial FlAsH binding capacity in the ensemble of urea-unfolded conformations. We speculate that this observation is due to the sampling of structures in the unfolded state that have native-like proximities of the cysteine residues in the region around the split motifs.
We suggest that the simplest and quickest screen for structure-dependent FlAsH binding of a given design for a split tetra-Cys motif location is to measure FlAsH fluorescence in SDS polyacrylamide gels on samples labeled in native or denatured conditions. This approach is independent of structure-dependent quantum yield variations, as the proteins are denatured in the gel in any case. A concern that must be considered in any of these designs is the potential effect of FlAsH binding to a split tetra-Cys motif on the stability of the protein. The presence of the cross-linkages created by FlAsH binding to a split tetra-Cys motif will certainly alter the energetic difference between the folded and unfolded state. Our preliminary measurements of apparent Tm for the FlAsH-labeled proteins in the absence of added EDT show modest destabilization upon dye binding (Figure S8).
Background fluorescence has been a complicating issue in in-vivo FlAsH labeling studies (Stroffekova et al., 2001; Adams et al., 2002). Happily, we observed that FlAsH-labeling of the split tetra-Cys motifs retained high selectivity (i.e., good binding required all four thiols) under stringent in vitro conditions or in vivo. It is likely that FlAsH binding in complex mixtures of thiols, as presented by the cellular milieu and added exogenous reductant EDT, occurs by a “transfer mechanism”, similar to the previously reported mechanism of arsenite binding to GSH (Rey et al., 2004; Schmidt et al., 2007; Spuches et al., 2005) and the transfer of arsenite from glutathione to higher affinity dithiols (Delnomdedieu et al., 1993). Reduced glutathione along with other cellular proteins, and of course added EDT, likely serve as the early recipients of FlAsH, and upon expression of the recombinant tetra-Cys proteins, the dye is transferred from its low affinity substrates. The extent of transfer and hence the background will depend on the relative affinities. Therefore, high background signals may also be indicative of low affinity FlAsH-binding sites on the protein. As suggested by Stroffekeva et al. (2001), a higher expression level of the recombinant protein will also promote selective labeling, and our simple rate expression (Supplemental Material) also indicates that high ratios of protein concentrations to dye concentration are expected to maximize the labeling efficiency. We recommend that these issues be kept in mind in the use of any tetra-Cys motif containing protein, but particularly when utilizing split tetra-Cys motif-containing proteins where affinity will be reduced by entropic factors.
SIGNIFICANCE
We have shown that tetra-Cys motifs designed to bind biarsenical dyes like FlAsH can be incorporated into the folded interior of a protein as two separate moieties. FlAsH binding across strands can occur without perturbing the native structure of a β-sheet protein. Our detailed analysis of the sensitivity of both FlAsH fluorescence enhancement (i.e., quantum yield) and affinity to the nature of the designed binding site underlines the necessity to tune the split tetra-Cys motifs for a particular application. Our results indicate that regions of proteins capable of conformational adjustment to optimize ligand geometry are the most suitable for incorporation of FlAsH-binding split tetra-Cys motifs with high affinity and quantum yield. We have demonstrated that it is possible to design a split tetra-Cys motif that is only capable of FlAsH binding in the native protein. However, the surprising ability of other split tetra-Cys motifs to retain FlAsH binding in the denatured state, even when the component parts are well separated in sequence, argues that populated states with close approach of the tetra-Cys moieties may allow dye to bind significantly, in turn pulling the equilibrium towards the binding-capable states. The results we have obtained with split tetra-Cys motifs introduced across strands in a β-sheet within a protein suggest that intermolecular cross-strand interactions could also be favorable in vivo targets for split tetra-Cys motif incorporation by positioning a pair of cysteines on interacting β-strands of protein partners. This strategy complements the recent report from the Schepartz lab (Luedtke et al., 2007) where ‘bipartite’ tetra-Cys motifs at termini of helical coiled-coil binding partners enabled observation of specific dimerization in vivo.
Experimental Procedures
Materials
EDT was obtained from Aldrich. FlAsH-EDT2 was synthesized as described (Griffin et al., 2000).
Mutagenesis, Protein Expression and Purification
CRABP I with a stabilizing R131Q mutation (Zhang et al., 1992) and an N-terminal (His)10 extension was cloned in pET16b as previously described (Clark et al., 1998) and used as a template for all mutagenesis. All mutant forms of CRABP I (Table S3) were generated using the QuikChange procedure (Stratagene, La Jolla, CA) following the manufacturer’s protocol. All of the proteins were expressed in BL21(DE3) cells and purified from the soluble fraction of the cell extract using a Ni-NTA column (Qiagen, Valencia, CA) as described (Clark et al., 1998). Mutations were verified by both DNA sequencing and mass spectrometry of the purified proteins. The protein concentrations for the wild type and the reduced cysteine variants were determined from the absorbance at 280 nm using a molar extinction coefficient of 20,970 M−1cm−1. In the case of the St1’-10 protein, we typically observed an apparent overestimation of protein concentration from A280 measurements as indicated by a lower amount of monomeric protein visualized upon Coomassie blue staining of the SDS-PAGE. This may be due to a greater tendency of this mutant protein to form dimers (suggested by a persistent dimer band on SDS-PAGE).
In vitro Labeling of Purified Proteins with FlAsH
Purified proteins were reduced overnight in 1 mM TCEP at room temperature (RT). FlAsH labeling of the TCEP-reduced, purified tetra-Cys proteins was carried out using either a 2-fold or 10-fold excess of protein in a reaction volume of 0.5 ml for 4 h at RT. The labeled proteins were desalted on a PD10 desalting column (GE Health care) to remove any free dye and EDT. The concentration of labeled protein in the desalted fraction was determined from the absorbance measured at 508 nm assuming a molar extinction coefficient for bound FlAsH of 41,000 M−1cm−1 for all the proteins. Fluorescence emission spectra (515–600 nm) were collected on 0.2 µM solutions of the labeled protein samples on a Photon Technologies International, Inc. (Birmingham, NJ) QM1 spectrofluorimeter with excitation at 508 nm. The excitation and emission bandwidths were each set at 2 nm.
Quantum Yield (ϕ) Measurement
Samples with OD508 < 0.05 were used for the quantum yield measurements at 20 °C. Fluorescence measurements were carried out as described earlier. The quantum yield of the protein-FlAsH complex (PF) was determined using the relation: ϕpF = ϕFlu * (IPF / IFlu) * ( ODFlu / ODPF) * (n2PF / n2Flu), where Flu represents fluorescein, which was used as a standard with ϕFlu = 0.95, I represents the integrated fluorescence intensity obtained from the fluorescence emission spectrum setting the wavelength of excitation at 508 nm for the FlAsH-protein complex and 496 nm for fluorescein, OD is the optical density of the samples at 508 nm and 496 nm for PF and fluorescein, respectively, and n represents the refractive index of the samples. The excitation and emission bandwidths for the fluorescence measurements were set at 2 nm each.
In vitro Labeling of Proteins Using a Fixed FlAsH Concentration
FlAsH labeling of TCEP reduced protein at a 2-fold excess of dye (1.0 µM) was carried out in a 150 µl reaction volume using 0.5 µM of the reduced protein in 50 mM MOPS, pH 7.4 containing 150 mM sodium chloride and 1.0 mM TCEP at RT for 4 h. Fluorescence measurements were carried out as described earlier.
For labeling of proteins under cellular conditions, the buffer included 5 mM β-mercaptoethanol or a total glutathione concentration of 5 mM (with a 200:1 molar ratio of reduced to oxidized glutathione) and 5 µg each of BSA and lysozyme. The buffer was pre-incubated with FlAsH and EDT at concentrations of 10 µM and 50 µM respectively at RT for 45 min. Purified protein was then added to the reaction mix at a final concentration of 5 µM, and the labeling reaction was carried out at RT for 2 h. The samples were then analyzed on 10% tricine SDS-PAGE. The gel was scanned for FlAsH fluorescence using the blue laser (473 nm) of the phosphorimager (Fuji) followed by Coomassie staining. As a control, the reaction was performed in buffer without glutathione and the BSA, lysozyme protein mixture as described above.
EDT Titration of FlAsH-Labeled Proteins
Purified proteins labeled with FlAsH were de-salted as described above. EDT titration was performed with 0.2 µM of labeled protein and increasing concentrations of EDT, up to a 500-fold molar excess, and the samples were incubated at room temperature for 4 h. The decrease in FlAsH-fluorescence emission at 530 nm (λex 508 nm) was measured. EDT-50, the molar excess of EDT at which the fluorescence at 530 nm in the absence of EDT was reduced by half, was used as a measure of apparent affinity for FlAsH. The kinetics of EDT displacement of FlAsH were fit to a bi-exponential expression, and the fluorescence intensities at saturation, at time t=0 s obtained from the extrapolation of the fits, and the rate constants were compared for the various constructs.
Determination of Apparent Dissociation Constant
The apparent dissociation constant (KDapp) for FlAsH binding to the proteins was determined by monitoring the increase in the FlAsH fluorescence at 530 nm (λex = 508 nm) using a fixed dye concentration of 50 nM and increasing protein concentrations up to 50 µM. The titrations were performed in 50 mM MOPS, pH 7.4 containing 150 mM sodium chloride, 1 mM TCEP and 0.1 mM EDT. The samples were incubated at room temperature for 4 h prior to the measurements. The band widths were set to 5 nm each. The data were fit to a 1:1 binding model (equation below) to obtain the KDapp.
Where, Fobs is the observed fluorescence, Fmin and Fmax are the minimum and the maximum fluorescence value. The Fmax value was obtained by fitting the data to a single exponential expression.
FlAsH Labeling Expressed Proteins in E. coli Cells
FlAsH labeling of the recombinant proteins in vivo was carried out using a protocol similar to that previously described (Ignatova and Gierasch, 2004). In brief, freshly transformed BL21(DE3) E. coli cells were used for in-cell labeling. An overnight culture (50 µl) was used to inoculate a 5 ml culture, and cells were grown at 37 °C. At an OD600 ~ 0.5, 0.5 ml of the culture was withdrawn and FlAsH, EDT, and IPTG were added to a final concentration of 2.94 µM, 0.1 mM, and 0.5 mM, respectively. Ninety to 120 min after induction, cells after were pelleted at RT, 6000×g, for a minute, and re-suspended in 0.2 ml LB. The cell suspension was used for fluorescence measurement or fluorescence imaging. After fluorescence measurements, cells were lysed, and the lysate was electrophoresed on 10% tricine-PAGE and analyzed on a phosphorimager as described above. In-gel analysis of in vivo samples reports on extent of labeling, as the FlAsH signal arising from the denatured proteins in gel may be directly compared. In all in-cell labeling of expressed protein, the gel was also stained with Coomassie brilliant blue to check the expression level of different constructs.
Fluorescence Microscopy
For fluorescence microscopy studies, a 3 µl sample of cell suspension was imaged on a Nikon E600 microscope, with excitation at 488 nm and using a 510-nm emission barrier filter. A 100X oil objective was used. The images were processed using Spot advanced software. Observing fluorescence by microscopy in cells reports on the protein solubility and provides a rough estimate of the efficiency of labeling under cellular conditions.
In Vitro FlAsH Labeling of Urea-unfolded Proteins
FlAsH-labeled proteins (as prepared for the quantum yield measurements) were subjected to urea unfolding in buffer (50 mM MOPS pH 7.4,150 mM sodium chloride and 1 mM TCEP) containing 7 M urea at room temperature for one hour. EDT was added to the native and unfolded proteins to a final concentration of 1 mM and the reaction continued for 4 h. Fluorescence measurements were carried out as described earlier. Relative fluorescence intensities at 530 nm were calculated with respect to native state fluorescence for each of the constructs. Similar results were obtained when the experiment was done with labeling of the native and 7M urea-unfolded forms of the protein in the presence of 0.5 or 1.0 mM EDT at RT for 4 h. It is important to note that in the presence of high concentrations of EDT, the EDT-bound form of free FlAsH is practically non-fluorescent, and hence any significant fluorescence signal present in such a case arises only from FlAsH bound to the protein.
FlAsH-labeling of Urea-unfolded Proteins in Cell Lysate
Proteins were expressed in BL21(DE3) cells following a protocol similar to that used for protein purification. Typically, 3 ml of the cells were pelleted at various times after induction and re-suspended in 0.5 ml of 50 mM MOPS, pH 7.4 containing 150 mM sodium chloride. After sonication, the cell lysate was spun at 16,000×g for 15 min at 4 °C to separate the soluble and the insoluble fractions. Protein present in the soluble fraction was used for FlAsH labeling. The labeling reaction was performed in a reaction volume of 150 µl using a 5 –7 fold dilution of the soluble fraction of the cell lysate. The cell lysate was equilibrated in buffer in the absence and presence of 7 – 8 M urea for 2 h. FlAsH and EDT were added to the equilibrated samples at concentration of 0.2 µM and 1.0 µM, respectively. Labeling was carried out for one hr at RT. The samples were analyzed on a fluorimeter as well as by SDS-PAGE as previously described.
Urea Titration of FlAsH-labeled St1’-10 In Vitro and in Cell Lysates
Purified St1’-10 protein was incubated overnight in the presence of the reductant TCEP, then labeled with FlAsH (at a protein to dye ratio of 10:1) in 50 mM MOPS, pH 7.4 containing 150 mM sodium chloride for 2 – 4 h. Labeled protein was added to buffer containing varying urea concentrations and 0.5 mM EDT and the samples incubated at 25 °C for an hour. Fluorescence measurements were carried out as described above. We do not advise using longer incubations of FlAsH-labeled pure protein in urea as we saw significant changes in dye fluorescence over time.
Urea titrations of protein present in the cell lysate were carried out by equilibrating the cell lysate containing St1’-10 (5-fold dilution of the cell lysate) in varying urea concentrations in 50 mM MOPS, pH 7.4 containing 150 mM sodium chloride for 8 h at 25°C followed by FlAsH labeling using 2 µM of the dye in the presence of 0.5 mM EDT for 10 min. The short labeling time was used to reduce any non-specific labeling and was confirmed to be sufficient based on the gel filtration data (Figure 7A). Fluorescence of the labeled cell lysate was measured as described above. In all the urea titration experiments, TCEP was excluded to reduce the complications from the effect of TCEP on protein stability (unpublished data).
Gel Filtration Chromatography for the FlAsH-labeled Protein
The extent of dye binding was estimated by gel filtration on a 5 ml HiTrap desalting column (GE Healthcare); the amount of dye co-eluting with the protein at the void volume of the column yielded an estimate of bound dye. Protein samples (18 µM St1’-10 or 47 µM WT) were equilibrated in native (0 M urea) or unfolding (7 M urea) buffer in a reaction volume of 0.5 ml for at least 2 h. FlAsH labeling was carried out on the equilibrated samples using 2 µM dye in the presence of 0.5 mM EDT for 10 min followed by immediate desalting on the pre-equilibrated HiTrap column on a AKTA purifier unit (GE Healthcare). Desalting was carried out at room temperature and the column run at 2 ml/min flow rate. The chromatogram traces were monitored for protein at 280 nm and 220 nm, and for FlAsH at 500 nm.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Aneta Szymanska, Pranorm Saejueng and D. Venkatacharam for helping with the FlAsH synthesis, and Krastyu Ugrinov and Patricia Clark for providing FlAsH for our early experiments. We thank Nick Renzette, Steve Sandler and Dale Callaham for help with fluorescence microscopy. We acknowledge the contributions from Virginie Sjoelund and Bing Gong in initiating the work. We appreciate critical reading of the manuscript by Joanna Swain, Eugenia M. Clerico, Zoya Ignatova, Harekrushna Sahoo, Jenny Maki, and Ivan Budyak. This research was supported by National Institute of Health grants (GM027616 and GM034962) and an NIH Director’s Pioneer Award (DP1 OD000945).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Cao H, Chen B, Squier TC, Mayer MU. CrAsH: a biarsenical multi-use affinity probe with low non-specific fluorescence. Chem. Commun. (Camb) 2006;28:2601–2603. doi: 10.1039/b602699k. [DOI] [PubMed] [Google Scholar]
- Clark PL, Weston BF, Gierasch LM. Probing the folding pathway of a β-clam protein with single-tryptophan constructs. Fold. Des. 1998;3:401–412. doi: 10.1016/s1359-0278(98)00053-4. [DOI] [PubMed] [Google Scholar]
- Cline DJ, Thorpe C, Schneider JP. Effects of As(III) binding on α-helical structure. J. Am. Chem. Soc. 2003;125:2923–2929. doi: 10.1021/ja0282644. [DOI] [PubMed] [Google Scholar]
- Cruse WBT, James MNG. The crystal structure of the arsenite complex of dithiothreitol. Acta. Cryst. 1972;B28:1325–1331. [Google Scholar]
- Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ. Transfer of arsenite from glutathione to dithiols: a model of interaction. Chem. Res. Toxicol. 1993;6:598–602. doi: 10.1021/tx00035a002. [DOI] [PubMed] [Google Scholar]
- DiMaio AJ, Rheingold AL. Arsenic-sulfur heterocycle formation via metal coordination. Synthesis and molecular structure of cyclo-(CH3AsS)n (n = 3, 4), [(CO)3Mo][η3-cyclo-(CH3As)6S3], and the triple-decker-sandwich complex [η5-(C5H5)2Mo2(η2,µ-As3)(η2,µ-AsS)] Inorg Chem. 1990;29:798–804. [Google Scholar]
- Griffin BA, Adams SR, Jones J, Tsien RY. Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 2000;327:565–578. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Ignatova Z, Gierasch LM. Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc. Natl. Acad. Sci. U S A. 2004;101:523–528. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatova Z, Krishnan B, Bombardier JP, Marcelino AM, Hong J, Gierasch LM. From the test tube to the cell: exploring the folding and aggregation of a β-clam protein. Biopolymers. 2007;88:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski P, Kubalski A. Glutathione (GSH) reduces the open probability of mechanosensitive channels in Escherichia coli protoplasts. Pflugers Arch. 1999;438:361–364. doi: 10.1007/s004240050921. [DOI] [PubMed] [Google Scholar]
- Korndorfer IP, Beste G, Skerra A. Crystallographic analysis of an "anticalin" with tailored specificity for fluorescein reveals high structural plasticity of the lipocalin loop region. Proteins. 2003;53:121–129. doi: 10.1002/prot.10497. [DOI] [PubMed] [Google Scholar]
- Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Surveying polypeptide and protein domain conformation and association with FlAsH and ReAsH. Nat Chem. Biol. 2007;3:779–784. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messens J, Collet JF. Pathways of disulfide bond formation in Escherichia coli. Int. J. Biochem. Cell Biol. 2006;38:1050–1062. doi: 10.1016/j.biocel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Ramadan D, Cline DJ, Bai S, Thorpe C, Schneider JP. Effects of As(III) binding on β-hairpin structure. J. Am. Chem. Soc. 2007;129:2981–2988. doi: 10.1021/ja067068k. [DOI] [PubMed] [Google Scholar]
- Rey NA, Howarth OW, Pereira-Maia EC. Equilibrium characterization of the As(III)-cysteine and the As(III)-glutathione systems in aqueous solution. J. Inorg. Biochem. 2004;98:1151–1159. doi: 10.1016/j.jinorgbio.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Schmidt AC, Koppelt J, Neustadt M, Otto M. Mass spectrometric evidence for different complexes of peptides and proteins with arsenic(III), arsenic(V), copper(II), and zinc(II) species. Rapid Commun. Mass Spectrom. 2007;21:153–163. doi: 10.1002/rcm.2823. [DOI] [PubMed] [Google Scholar]
- Shaikh TA, Parkin S, Atwood DA. Synthesis and characterization of a rare arsenic trithiolate with an organic disulfide linkage and 2-chloro-benzo-1,3,2-dithiastibole. J. Organomet. Chem. 2006a;691:4167–4171. [Google Scholar]
- Shaikh TA, Ronald C, Bakus II, Parkin S, Atwood DA. Structural characteristics of 2-halo-1,3,2-dithiarsenic compounds and tris-(pentafluorophenylthio)-arsen. J. Organomet. Chem. 2006b;691:1825–1833. [Google Scholar]
- Spuches AM, Kruszyna HG, Rich AM, Wilcox DE. Thermodynamics of the As(III)-thiol interaction: arsenite and monomethylarsenite complexes with glutathione, dihydrolipoic acid, and other thiol ligands. Inorg. Chem. 2005;44:2964–2972. doi: 10.1021/ic048694q. [DOI] [PubMed] [Google Scholar]
- Stroffekova K, Proenza C, Beam KG. The protein-labeling reagent FlAsH-EDT2 binds not only to CCXXCC motifs but also non-specifically to endogenous cysteine-rich proteins. Pflugers Arch. 2001;442:859–866. doi: 10.1007/s004240100619. [DOI] [PubMed] [Google Scholar]
- Sukumar M, Gierasch LM. Local interactions in a Schellman motif dictate interhelical arrangement in a protein fragment. Fold. Des. 1997;2:211–222. doi: 10.1016/S1359-0278(97)00030-8. [DOI] [PubMed] [Google Scholar]
- Wang T, Yan P, Squier TC, Mayer MU. Prospecting the proteome: identification of naturally occurring binding motifs for biarsenical probes. ChemBioChem. 2007;8:1937–1940. doi: 10.1002/cbic.200700209. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu ZP, Jones TA, Gierasch LM, Sambrook JF. Mutating the charged residues in the binding pocket of cellular retinoic acid-binding protein simultaneously reduces its binding affinity to retinoic acid and increases its thermostability. Proteins. 1992;13:87–99. doi: 10.1002/prot.340130202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.