Abstract
The Drosophila central nervous system is an excellent model system in which to resolve the genetic and molecular control of neuronal differentiation. Here we show that the wing selector vestigial is expressed in discrete sets of neurons. We track the axonal trajectories of VESTIGIAL-expressing cells in the ventral nerve cord and show that these cells descend from neuroblasts 1–2, 5–1, and 5–6. In addition, along the midline, VESTIGIAL is expressed in ventral unpaired median motorneurons and cells that may descend from the median neuroblast. These studies form the requisite descriptive foundation for functional studies addressing the role of vestigial during interneuron differentiation.
Keywords: Drosophila, vestigial; central nervous system; CNS; neuroblast; midline; ventral nerve cord; VNC; embryo
Introduction
The capacity of nervous systems to control simple and complex behaviors depends on the ability of thousands of neurons to adopt individual identities and to interconnect with each other in a reproducible manner. The precision with which researchers can dissect the genetic and molecular mechanisms that control the intricate wiring of the nervous system relies on the ability to map gene expression to individual cells and to assess the morphology and physiology of these cells in wild type and mutant backgrounds. The powerful genetic and embryological tools available in Drosophila make it an excellent model system in which to resolve the genetic and molecular control of neuronal differentiation.
Drosophila central nervous system (CNS) development initiates shortly after gastrulation when a set of neural precursor cells called neuroblasts segregate from the ventrolateral ectoderm into the interior of the embryo (reviewed in Skeath and Thor, 2003). Thirty neuroblasts form per hemisegment, each one uniquely identifiable based on its position and gene expression. Each neuroblast divides via a stereotyped cell lineage to produce a largely invariant clone/family of neurons. Together the neuroblasts produce the vast majority of interneurons, motorneurons, and glia found in the CNS. By the end of embryogenesis, neuroblasts, together with some midline precursors cells, have generated approximately 400 neurons per hemisegment. Most of these cells are interneurons (~90%), while glia and motorneurons each comprise about 5% of the cell population. By the initiation of the first larval stage these cells have become interconnected with each other in a precise and stereotyped pattern to produce a functional nervous system.
Recent work suggests that the stereotypy of neuronal connections results from the reproducible expression of distinct combinations of transcription factors in different neurons and the subsequent ability of these factors to effect neuronal differentiation by regulating downstream genes (Shirasaki and Pfaff, 2002; Thor and Thomas, 2002). However, our ability to characterize the regulatory networks through which these transcription factors guide neuronal differentiation is limited because we do not know the characteristic axonal trajectories of most neurons in the CNS. Thus, in most cells, we are unable to link transcription factor expression in a neuron with the morphology and physiology of that neuron. In addition, the characterization of transcriptional control of neuronal differentiation has focused mainly on motor neurons. Therefore, we know little about the transcriptional control of the differentiation of interneurons, even though these cells outnumber motor neurons ten to one in the Drosophila embryonic CNS.
We have begun to address the genetic control of neuronal differentiation by characterizing the gene expression profiles and axonal morphologies of subsets of neurons. Here we describe the subset of cells expressing the wing selector vestigial (vg). This gene has a well characterized role in wing development (Kim et al., 1996; Halder et al., 1998a; Simmonds et al., 1998), where it functions as a transactivator (Vaudin et al., 1999) with scalloped, which functions as a transcription factor to control the expression of genes throughout the wing genetic regulatory network (Halder et al., 1998b; Guss et al., 2001; Lunde et al., 2003). Although vg is expressed in the CNS (Williams et al., 1991), its role in this tissue is unclear. To address this question, we have characterized the expression of VG in the ventral nerve cord (VNC). Through coexpression studies and by mapping the axonal trajectories of VG-expressing neurons, we show that VG expression identifies discrete sets of interneurons descending from neuroblasts 1–2, 5-1 and 5–6. In addition, VG is expressed in all three midline ventral unpaired median motorneurons (mVUMs) and cells that are possibly a subset of progeny of the median neuroblast (MNB). These studies form the requisite descriptive foundation to investigate the function of vg in the differentiation of specific neurons.
Results and Discussion
Distribution of vestigial-expressing cells in the ventral nerve cord
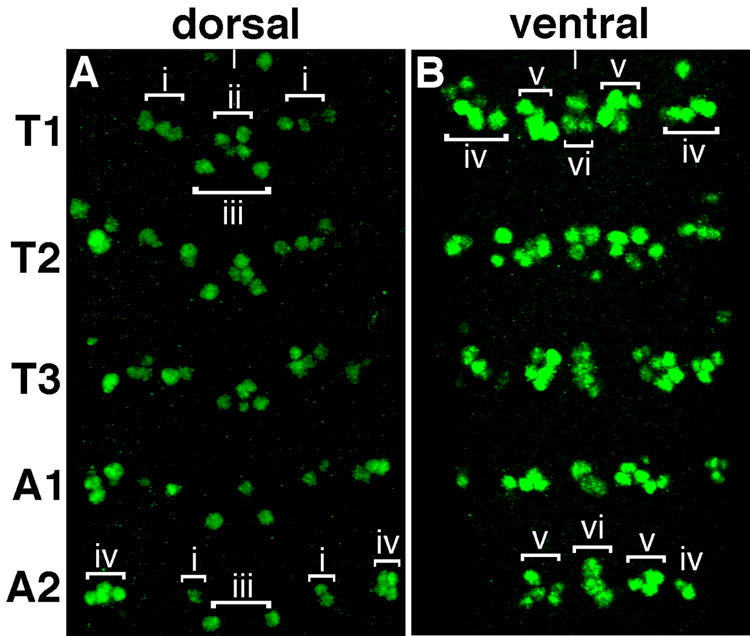
As a prerequisite to investigating the function of vg in the central nervous system, we have characterized VG expression in the VNC. VG expression is first detectable in cells in the ventral nerve cord in stage 12 embryos, when each thoracic and abdominal segment contains 10–12 VG+ cells (data not shown). By stage 16, each thoracic segment contains approximately 41–43 VG+ cells (Figure 1 and Figure 5). These cells are generally distributed into two planes, dorsal and ventral. Dorsally there are 12 VG+ cells that can be divided into two bilaterally symmetric clusters per hemisegment and one cluster located at the midline. Cluster i contains 3 cells and is located just lateral to the midline, while cluster iii contains 1 cell and located more medially. In addition, cluster ii contains 4 cells located at the midline. The remaining VG+ cells are found ventrally and are composed of a midline group of 3 cells (cluster vi) and two bilaterally symmetric clusters found laterally from the midline, with 6–7 cells in cluster iv and cells in cluster v.
Figure 1.
Vestigial expression in the VNC. Projection of 0.5 µM optical sections of a ventral nerve cord dissected from a stage 16 embryo showing VG expression (green) in three thoracic segments and first and second abdominal segments. A, Cells expressing VG in the dorsal and B, ventral planes of the VNC. At this stage, there are 41–43 VG+ cells in T1 and 27–29 VG+ cells in A2. The distribution of VG+ cells in clusters i-vi labeled in representative segments (T1 and A2) is also shown. In this specimen, the majority of cells in clusters iv in A2 are visible in the dorsal plane of the VNC. In all images, anterior is up and the midline is marked with a white line.
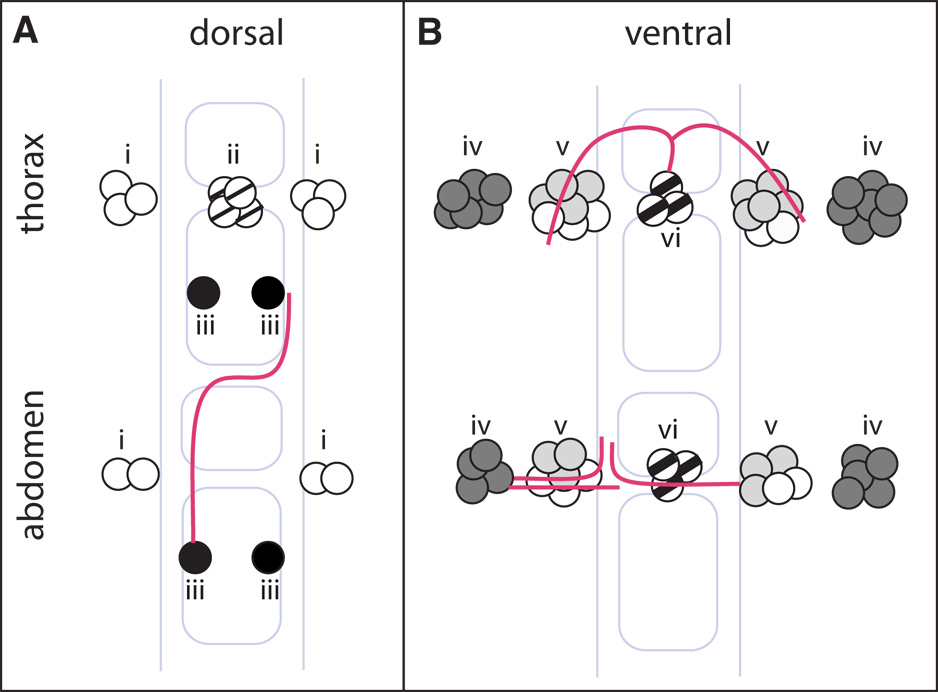
Figure 5.
Summary of distribution of VG+ cells and axonal morphologies in the dorsal and ventral regions of thoracic and abdominal segments in the VNC. The gray outline shows the commissures and longitudinal connectives, and red lines illustrate representative axonal trajectories described by this study. Neuroblast lineages of VG+ cells determined in this study are denoted by: 1–2, black; 5–1, light gray; 5–6, dark gray; likely MNB progeny, faint cross hatch; mVUM, bold cross hatch.
In contrast, each abdominal segment contains fewer cells in both the dorsal and ventral planes of the CNS (Figure 1 and Figure 5). The dorsal midline cluster ii is absent in abdominal segments. In addition, the medially and laterally located groups, in both the dorsal (cluster i) and ventral planes (clusters iv and v) contain fewer VG+ cells than the corresponding clusters in the thoracic segments.
A subset of VG+ interneurons descend from neuroblasts 1–2, 5-1 and 5–6
We next determined the classes of non-midline cells that express VG in the VNC. To determine whether these cells are motorneurons, we assessed VG coexpression with EVE and HB9, which collectively mark most motorneurons which do not arise from the midline (Broihier and Skeath, 2002; Broihier et al., 2004). Neither marker was coexpressed in VG+ cells (data not shown). In addition, the VG+ cells do not coexpress REPO, a marker for glia (Xiong et al., 1994). Together these results indicate that, with respect to non-midline cells, VG labels only interneurons.
Interneurons are characterized by neuroblast lineage, axonal morphology and physiology. To identify the lineage and axonal architecture of VG+ cells, we used a modified FLP/FRT system (Pignoni and Zipursky, 1997) This approach employs the GAL4/UAS system to generate clones of cells expressing a membrane-tethered form of GFP. By assessing coexpression of VG and GFP, the clonal relationships and axonal trajectories of VG+ cells in clones of GFP+ cells may be identified. Using this approach we characterized the neuroblast lineages of VG+ cells in clusters iii-v. This data is described here and summarized in Table 1.
Table 1.
Summary of clone data in this report. Abbreviations: ac, anterior commissure; pc, posterior commissure.
| VG+ cluster | Proposed Lineage | Number of VG+ cells in cluster | Number of clones marking this cluster | Number of VG+ cells/ total GFP+ cells in clone | Axon extended by VG+ cell(s)? | Axonal morphology |
|---|---|---|---|---|---|---|
| iii | 1–2 | 1 | 4 | 4 clones: 1 VG+/1 GFP+ | yes | Anterior in ipsilateral connective, crosses ac, continues anterior in contralateral connective |
| iv | 5–6 | 5 or 6 (thoracic) 4 or 5 (abdominal) | 7 | 1 clone: 4 or 5 VG+/ 9 GFP+ | yes | Turns anterior in ipsilateral connective at level of pc |
| 1 clone: 5 VG+/ 8 GFP+ | yes | Reaches midline at level of pc | ||||
| 1 clone: 3 VG+/ 3 GFP+ | yes | Reaches midline at level of ac | ||||
| 3 clones: 3-5 VG+/ 8 GFP+ | no | Reaches midline at level of pc | ||||
| 1 clone: 1-2 VG+/ 5 GFP+ | no | Reaches midline at level of ac | ||||
| v | Subset are 5-1 | 7 (thoracic) 5 or 6 (abdominal) | 2 | 2 clones: 3 VG+ / 7 or 8 GFP+ | unclear | crosses the midline in the pc, extends anteriorally in the contralateral connective |
| vi | mVUM | 3 | 4 | 4 clones: 1 VG+/2 GFP+ | yes | Axon extends to the anterior, branches, then to both sides of the midline |
The VG+ cell in cluster iii extends an axon anteriorally along the ipsilateral connective, which then crosses the midline within the anterior commissure and projects further anteriorally along one of the contralateral longitudinal connectives (Figure 2A–C). We observe that this VG-expressing cell is clonally related to a lateral group of approximately 10 VG- cells, which mainly comprise local interneurons and a single motoneuron (data not shown). The axonal morphology of this clone shows that the VG+ cell in cluster iii is a descendant of NB 1–2 (Bossing et al., 1996; Schmid et al., 1999).
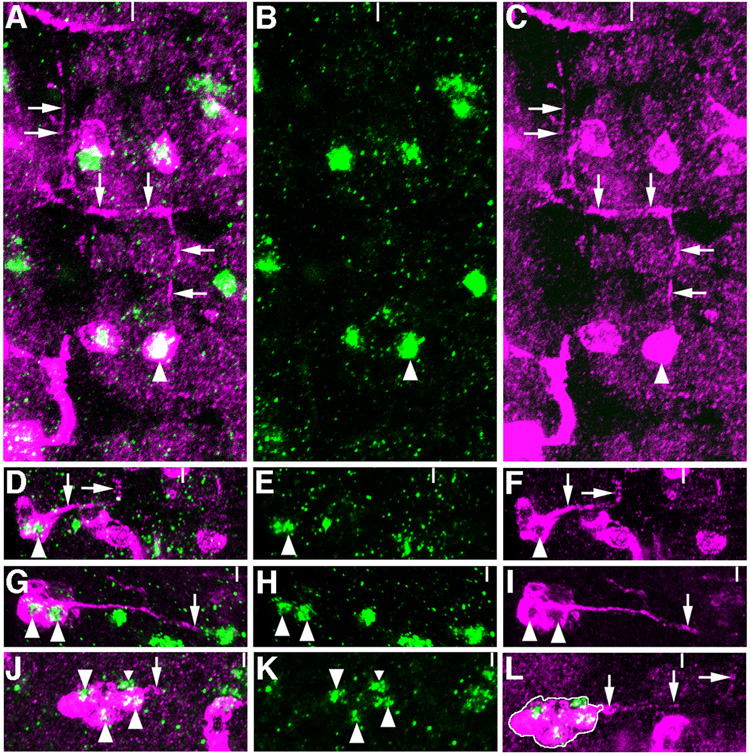
Figure 2.
Axon morphologies of clones including VG+ cells. A–C, One-cell GFP clone (magenta) in T3 includes the VG+ cell (green) of cluster iii (arrowhead). This cell projects an axon to the anterior in the ipsilateral connective in a stage 15 embryo. This axon turns at the anterior commissure, crosses the midline, and again projects anteriorally in the contralateral connective (arrows). D–F, GFP+ clone in A5 of a stage 17 embryo contains VG+ cells of cluster iv. This clone contains 9 cells, 4 of which are VG+. Two axons are extended by cells in this clone. An axon extended by a VG+ cell (arrowhead) reaches the outer edge of the longitudinal connective and turns to the anterior (arrows) just anterior to the level of the posterior connective. G–I, Second example of a clone including VG+ cells of cluster iv. This 8-cell GFP+ clone in T3 contains 5 VG+ cells. A single thick axon extended by two VG+ cells (arrowheads) reaches the midline at the level of the posterior commissure (arrows). J–L, Eight-cell GFP+ clone in A4 includes 3 VG+ cells of cluster v. An axon extended by this clone of cells crosses the midline and projects anteriorally in the contralateral connective (arrowheads). L is a photomontage to show the relationship of the clone shown in J with its axonal trajectory. One VG+ cell in cluster v is not included in this clone (J, small arrowhead).
In the ventral plane of the VNC, cells in clusters iv are descendants of NB 5–6. We observe clones with three classes of axonal morphology. In one clone, two VG+ cells extend axons that are either tightly apposed or one of these axons ends early, so that a single axon reaches the outer edge of the longitudinal connective at the level of the posterior connective and then turns to the anterior (Figure 2D–F). More frequently, we observe axons extended by VG- cells in the clone, which extend across the midline at the posterior connective (Figure 2G–I). We also observed two cases in which the axon crosses the midline at the level of the anterior commissure (data not shown). These three types of axonal architectures and the position of cluster iv within the hemisegment are consistent with observations for descendants of NB 5–6 (Schmidt et al., 1997; Schmid et al., 1999). In addition, VG+ cells in cluster iv express wg-lacZ, further substantiating this conclusion (Doe, 1992).
With respect to the cells in cluster v, the VG+ cells do not extend axons, nor are all of the VG+ cells in this cluster clonally related. We have determined the neuroblast lineage for at least a subset of VG+ cells in this cluster, however. First, three to four of the VG+ cells in cluster v express wg-lacZ (data not shown), suggesting that these cells descend from one of NBs 5-1 through 5–6 (Doe, 1992). Second, cells clonally related to VG+ cells in cluster v extend an axon which crosses the midline just posterior to the anterior edge of the posterior commissure and then projects to the anterior in the contralateral connective (Figure 2J–L). In addition, both of these clones are associated with epidermal subclones. Based on these observations, we propose a subset of VG+ cells in cluster v are descendants of NB 5-1.
VG expression in midline cells marks mVUMs
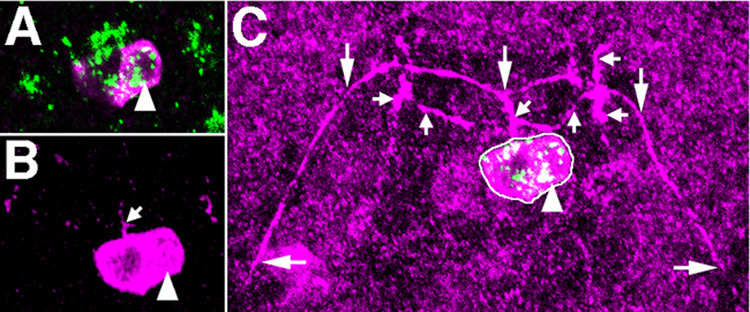
VG+ expressing cells in cluster vi are mVUMs, based on coexpression data and axon morphology. These cells coexpress UAS tau myc EGFP driven by sim-Gal4, which marks all midline cells (Figure 4A–C; Xiao et al., 1996). These 3 VG+ cells also express UAS tau myc EGFP driven by MzVUM-Gal4, which drives expression in mVUMs (Landgraf et al., 2003; Kearney et al., 2004; Figure 4D–F). Using the FLP/FRT system described above, we recovered four 2-cell clones marking cluster vi, each of which includes one VG+ cells and one VG- cell (Figure 3). We propose that the VG+ cell is a mVUM and extends the axons marked by large arrows in Figure 3C, and the VG- cell is its sibling iVUM, which extends the axons marked by small arrows. The architectures of axons extended by these two cells are very similar to those reported for mVUM/iVUM pairs (Bossing and Technau, 1994). We also recovered a number of 1-cell clones that include only one of the VG+ cells in cluster vi, and these cells do not extend axons (data not shown). Our results suggest that only one of the three cells marked by MzVUM-Gal4 expression extends an axon.
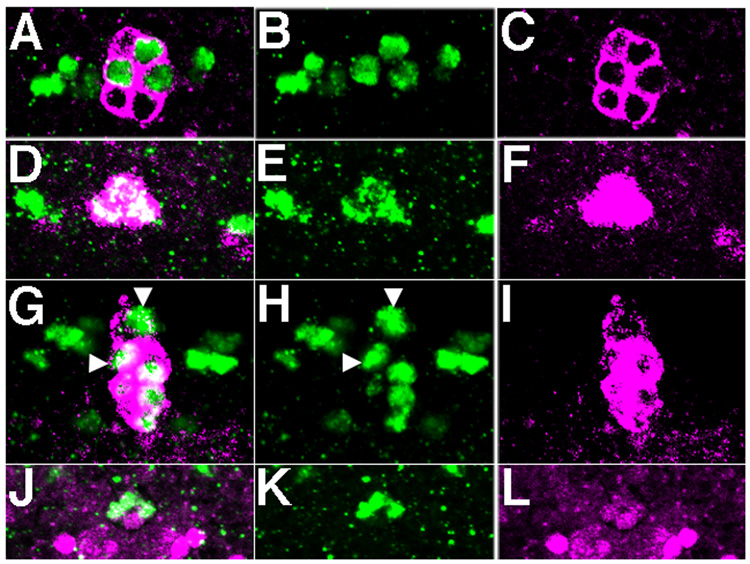
Figure 4.
Midline lineages of VG+ cells in clusters ii and vi. A–C and G–I, Coexpression of VG (green) and UAS tau myc EGFP (magenta) driven by sim-GAL4 in cluster vi (A–C) and cluster ii (G–I). The arrowheads in G, H mark cluster vi cells that are visible in this focal plane. The remaining four VG+, GFP+ cells compose cluster ii. D–F, Coexpression of VG (green) and UAS GFP driven by MzVUM-Gal4 by cells in cluster vi. J–L, Coexpression of VG (green) and castor-lacZ (magenta) in cluster ii cells.
Figure 3.
VG+ cells in cluster vi are mVUMs. A, Two-cell GFP+ clone in A8 of a stage 17 embryo includes one VG+ cell of cluster vi (arrowhead). B, The axon projected by this clone extends to the anterior (small arrow), and branches twice. C, Photomontage of clone cell bodies shown in B and axonal morphology of this clone. The first branching event produces two projections with complex arborizations at the lateral ends (small arrows). The second branching event results in long axons that extend on either side of the midline, each forming a broad ‘U’ (large arrows). We propose that the large axon marked by the large arrows is extended by the VG+ cell (arrowhead, B), which is MzVUM, and the shorter axons are extended by its sibling iVUM, which does not express VG.
A subset of MNB progeny may express VG
We did not recover GFP clones that marked cluster ii cells. However, we have determined the probable lineage of these cells via coexpression of molecular markers. The appearance of these cells at stage 13 (data not shown) and the coexpression of castor-lacZ and UAS tau myc EGFP driven by sim-Gal4 (Figure 4G–L) indicate that these cells are likely a subset of MNB progeny (Wheeler et al., 2006). We have eliminated other possible midline lineages for cluster ii cells, because these VG+ cells do not coexpress WRAPPER (a marker for midline glia; Noordermeer et al., 1998), pale-Gal4 (a marker for the H-cell; Friggi-Grelin et al., 2003), or period-Gal4 (a marker for H-sib cell and iVUMs; Plautz et al., 1997). However, it is possible that these cells are of unknown origin, because they do not express ENGRAILED or show nuclear localization of PROSPERO, both of which are markers for MNB progeny (Wheeler et al., 2006).
Summary
Neuroblast identity is controlled in part by the differential expression of transcription factors. Similarly, the identity of the progeny of a single neuroblast is also controlled either through the de novo or continued expression of a combination of transcription factors in individual neurons. A specific combination of transcription factors appears to determine the specific axonal morphology and physiological characteristics of a single neuron. Our ability to dissect the genetic mechanisms that control neuron development has been limited by the descriptive understanding of neuronal morphology and transcription factor expression.
Here we show that VG is expressed in discrete subsets of interneurons and midline motorneurons. This map of VG-expressing cells provides the foundation for the dissection of the genetic mechanisms that control the differentiation of these cells. The next steps will be to identify more genes expressed in these cells, followed by the assessment of how these genes alone, and in combination, control the development of these neurons. In addition, the wing selector scalloped is expressed in the VNC (K.A.G., A. Kopp and K. Brondell, unpublished observations), presenting the intriguing possibility that vg and scalloped may function together during neuron differentiation, as they do in the wing.
Methods
Drosophila melanogaster stocks
Beta-galactosidase driven by wg-lacZ was detected in embryos of genotype +/CyO wg-lacZ (Bloomington Drosophila Stock Center). Other stocks used in these studies were sim-Gal4 (Bloomington Drosophila Stock Center), castor-lacZ (Chris Doe) and MzVUM-Gal4; UAS-GFP-lacZ (Scott Wheeler).
Antibodies
Rabbit anti-VG antibody was the gift of Sean Carroll (University of Wisconsin-Madison), and was diluted to 1:25 and preadsorbed before use. To enhance detection of anti-VG in the embryo, we employed biotinyl tyramide amplification (TSA™ Biotin System, Perkin Elmer). Additional antibodies used were goat anti-GFP (1:500), mouse anti-GFP (1:1000; Molecular Probes), mouse anti-beta-galactosidase (1:1000; Promega), and Alexa Fluor-conjugated secondary antibodies (1:300; Molecular Probes).
Immunofluorescence methods
Embryos were collected, washed with 0.4% NaCl, 0.1% Triton X-100, dechorionated with 100% bleach for 2.5 minutes, washed with deionized water, and fixed by one of two methods. Embryos were agitated for either 2 minutes in 2 ml 37% formaldehyde with 2 ml heptane, or for 25 minutes in 4 ml fix buffer (6 mM K2HPO4, 4 mM KH2PO4, 0.14 M NaCl, 9.25% formaldehyde), with 4 ml heptane. Both fixation methods were followed by removal of the aqueous phase, and washing the embryos several times with methanol. For immunofluorescence, embryos were then rehydrated into phosphate buffered Tris with 3% bovine serum albumin, which was used for all antibody incubations and washes. Following processing, embryos were suspended in 0.02M 1,4 diazabicyclo (2, 2,2) octane (DABCO; Sigma) in glycerol.
Generation of clones
We used a modified FLP/FRT system (Pignoni and Zipursky, 1997) to trace the lineage and projections of neurons that express VG. Males of genotype w; UAS tau myc EGFP/CyO; flp out GAL4/TM3 +/− were crossed to virgin females of genotype hsflp;;. Embryos were collected for four hours, then aged at room temperature for 5 hours, then subjected to a heat shock at 32°C for 7 minutes. Embryos were then allowed to incubate overnight (approximately 14 hours) at 18°C and fixed for immunofluorscence the following day. Stage 15–17 embryos (Campos-Ortega and Hartenstein, 1997) were identified, scored for the presence of clones via GFP expression, and then for coexpression of VG and GFP.
Image collection
Embryos were examined using an Olympus Fluoview 500 inverted confocal microscope with argon (488 nm) and krypton/argon (568 nm) laser excitation. Data was collected using sequential scans to avoid artifacts from overlapping emission spectra, using image slices of 0.2-0.5 µM. Z-series and projections were generated using Fluoview® software version 4.3 and images were merged and/ or adjusted for brightness, contrast, and levels, and cropped using Adobe Photoshop version 8.0.
Acknowledgements
We thank Beth Anne Wilson for technical assistance, Sean Carroll for anti-VG antibody, and the Bloomington Drosophila Stock Center and Chris Doe for fly stocks. Scott Wheeler also provided fly stocks and helpful conversation regarding midline lineages. Several antibodies used in the course of these studies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This paper was also improved by suggestions of the anonymous reviewers; thank you.
Grant Sponsors: Research & Development Committee, Dickinson College and NSF MRI/RUI 0320606 to K.A.G and NINDS RO1-NS-036570 to J.B.S.
References
- Bossing T, Technau GM. The fate of the CNS midline progenitors in Drosophila as revealed by a new method for single cell labelling. Development. 1994;120:1895–1906. doi: 10.1242/dev.120.7.1895. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Kuzin A, Zhu Y, Odenwald W, Skeath JB. Drosophila homeodomain protein Nkx6 coordinates motoneuron subtype identity and axonogenesis. Development. 2004;131:5233–5242. doi: 10.1242/dev.01394. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Skeath JB. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Germany: Springer; 1997. p. 405. [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Guss KA, Nelson CE, Hudson A, Kraus ME, Carroll SB. Control of a genetic regulatory network by a selector gene. Science. 2001;292:1164–1167. doi: 10.1126/science.1058312. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998a;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Carroll SB. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in response to signaling proteins. Genes and Development. 1998b;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JB, Wheeler SR, Estes P, Parente B, Crews ST. Gene expression profiling of the developing Drosophila CNS midline cells. Dev Biol. 2004;275:473–492. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Sanchez-Soriano N, Technau GM, Urban J, Prokop A. Charting the Drosophila neuropile: a strategy for the standardised characterisation of genetically amenable neurites. Dev Biol. 2003;260:207–225. doi: 10.1016/s0012-1606(03)00215-x. [DOI] [PubMed] [Google Scholar]
- Lunde K, Trimble JL, Guichard A, Guss KA, Nauber U, Bier E. Activation of the knirps locus links patterning of morphogenesis of the second wing vein in Drosophila. Development. 2003;130:235–248. doi: 10.1242/dev.00207. [DOI] [PubMed] [Google Scholar]
- Noordermeer JN, Kopczynski CC, Fetter RD, Bland KS, Chen WY, Goodman CS. Wrapper, a novel member of the Ig superfamily, is expressed by midline glia and is required for them to ensheath commissural axons in Drosophila. Neuron. 1998;21:991–1001. doi: 10.1016/s0896-6273(00)80618-2. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Liu X, Soanes KH, Krause HM, Irvine KD, Bell JB. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes and Development. 1998;12:3815–3820. doi: 10.1101/gad.12.24.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath JB, Thor S. Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB. Motor neuron specification in worms, flies and mice: conserved and 'lost' mechanisms. Curr Opin Genet Dev. 2002;12:558–564. doi: 10.1016/s0959-437x(02)00340-4. [DOI] [PubMed] [Google Scholar]
- Vaudin P, Delanoue R, Davidson I, Silber J, Zider A. TONDU (TDU) a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development. 1999;126:4807–4816. doi: 10.1242/dev.126.21.4807. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Bell J, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes and Development. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- Xiao H, Hrdlicka LA, Nambu JR. Alternate functions of the single-minded and rhomboid genes in development of the Drosophila ventral neuroectoderm. Mech Dev. 1996;58:65–74. doi: 10.1016/s0925-4773(96)00559-x. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]