Abstract
TLR2 signaling by Mycobacterium tuberculosis 19-kDa lipoprotein (LpqH) inhibits IFN-γ-induced expression of CIITA by macrophages. Microarray analysis, quantitative RT-PCR, and Western blots showed that LpqH induced C/EBPβ and C/EBPδ in kinetic correlation with inhibition of CIITA expression. Of the C/EBPβ isoforms, liver inhibitory protein (LIP) was notably induced and liver-activating protein was increased by LpqH. Putative C/EBP binding sites were identified in CIITA promoters I and IV (pI and pIV). LpqH induced binding of C/EBPβ (LIP and liver-activating protein) to biotinylated oligodeoxynucleotide containing the pI or pIV binding sites, and chromatin immunoprecipitation showed that LpqH induced binding of C/EBPβ and C/EBPδ to endogenous CIITA pI and pIV. Constitutive expression of C/EBPβ LIP inhibited IFN-γ-induced CIITA expression in transfected cells. In summary, LpqH induced expression of C/EBPβ and C/EBPδ, and their binding to CIITA pI and pIV, in correlation with inhibition of IFN-γ-induced expression of CIITA in macrophages, suggesting a role for C/EBP as a novel regulator of CIITA expression.
Upon initial infection with Mycobacterium tuberculosis (Mtb),3 most individuals control infection, but remain asymptomatically infected with bacteria that survive inside macrophages. These bacteria are poised to reactivate disease under conditions of compromised immunity. It remains unclear how these bacteria are able to survive despite host immune responses, which include vigorous CD4+ T cell responses that are critical for control of infection.
Effective control of Mtb infection requires IFN-γ in both humans and mice (1–4), but some IFN-γ-induced genes are inhibited in macrophages infected with Mtb or exposed to Mtb components such as Mtb 19-kDa lipoprotein (LpqH) (5, 6). In particular, Mtb and LpqH inhibit IFN-γ-induced genes required for Ag processing and presentation to CD4+ T cells (6–9). We hypothesize that inhibition of macrophage MHC-II Ag presentation by Mtb may reduce presentation of Mtb Ags and the detection of infected macrophages by CD4+ T cells, allowing Mtb to evade immune surveillance. Because MHC-II transcription requires the expression of CIITA, and CIITA is inhibited by Mtb or LpqH, our studies focus on understanding the mechanisms by which LpqH inhibits CIITA expression.
CIITA transcription is regulated by three unique promoters. CIITA promoter I (pI) is constitutively active in dendritic cells, promoter III is active in B cells, and CIITA promoter IV (pIV) activity is induced in response to IFN-γ in various cell types, including macrophages and epithelial cells (10, 11). In addition, CIITA pI has recently been shown to be active in macrophages, where its activity is IFN-γ dependent (10). Although the mechanism of IFN-γ-induced CIITA pI activation remains relatively unexplored, the regulation of CIITA pIV is well characterized. IFN-γ induces activation of STAT1α and the subsequent induction of IFN regulatory factor (IRF) 1 expression, both of which are required for CIITA pIV transcriptional activation. Interestingly, STAT1α activation and IRF1 induction are not significantly inhibited by Mtb or LpqH (7, 8), indicating that proximal IFN-γ signaling mechanisms are intact and implicating regulation of distal transcriptional control mechanisms by Mtb and LpqH. Mtb-induced IL-6 inhibits IFN-γ-induced CIITA by a mechanism that involves de novo protein synthesis (12). LpqH, signaling through a TLR2/MAPK-dependent pathway, inhibits IFN-γ-induced chromatin remodeling of CIITA pIV (13). Together, these data suggest the hypothesis that LpqH initiates TLR2 signaling, inducing a transcription factor that selectively inhibits a subset of the genes that are induced by IFN-γ.
The C/EBP family of transcription factors includes C/EBPα, β, δ, γ, ε, and ζ, all of which contain a highly conserved C-terminal basic leucine zipper domain that allows homodimerization or heterodimerization of family members and subsequent binding to the promoters of target genes. Of the C/EBP family members, C/EBPβ and δ are induced during inflammation, suggesting roles in regulation of immune responses (14–16). In addition, C/EBPδ forms heterodimers with C/EBPβ, allowing these isoforms to interact in the regulation of target genes (16). C/EBPβ regulates a subset of IFN-γ-induced genes, including some that are STAT1 independent (17–19). There are three isoforms of C/EBPβ commonly referred to as liver-activating protein (LAP*), LAP, and liver inhibitory protein (LIP), which are translated from different AUG codons (alternative translation start sites) contained in a single mRNA sequence that lacks introns (20, 21). Thus, LAP*, LAP, and LIP share a C-terminal DNA binding domain, but vary in inclusion of sequence for the N-terminal trans activation domain. Depending on the promoter and mode of activation, C/EBPβ can act as a transcriptional activator or suppressor (22–25). The LIP isoform lacks the entire trans activation domain and therefore acts primarily as a dominant-negative regulator of transcription (20, 21). However, LAP represses transcription of some genes such as IL-12p35 and albumin (24, 26), and LIP activates transcription of IL-6 and α1-acid glycoprotein (22, 27). Importantly, LIP acts as a suppressor of transcription most favorably as a heterodimer with LAP and does so even at low stochiometric ratios (heterodimers will form and bind even with 5 times as much LAP present) (20). C/EBPδ is expressed as a single isoform that can activate (28, 29) or repress (30) gene transcription.
In this study, we demonstrate for the first time that C/EBPβ and C/EBPδ bind to CIITA promoters in correlation with inhibition of IFN-γ-induced CIITA transcription. TLR2 signaling by LpqH induced expression of LIP and C/EBPδ and enhanced expression of LAP at time points that correlated kinetically with the onset of inhibition of IFN-γ-induced CIITA expression. LpqH induced the binding of LAP, LIP, and C/EBPδ to both CIITA pI and CIITA pIV. In addition, constitutive expression of LIP by transfection suppressed IFN-γ-induced CIITA expression in the absence of LpqH, demonstrating a functional consequence of C/EBPβ expression for the control of CIITA expression. Studies with cells from C/EBPβ−/− mice demonstrated that LpqH-mediated inhibition of CIITA can occur in the absence of C/EBPβ, most likely due to redundant function of other C/EBP family members. In summary, we report a novel association of C/EBPβ and C/EBPδ with CIITA promoters that correlates with Mtb LpqH-mediated inhibition of CIITA expression, suggesting that C/EBPβ and C/EBPδ play novel roles in negative regulation of CIITA transcription.
Materials and Methods
Cells and medium
RAW264.7 cells (American Type Culture Collection) were maintained in standard medium composed of DMEM (BioWhittaker) with 10% heat-inactivated FBS, 50 μM 2-ME, 1 mM sodium pyruvate, 10 mM HEPES buffer, penicillin, and streptomycin. Primary macrophages were isolated from femur marrow from C57BL/6 mice (Jackson ImmunoResearch Laboratories); C/EBPβ−/− mice were generated, as described previously (31). Briefly, 129 ES cells with a targeted C/EBPβ deletion were delivered to CBA × C57BL/6 mice. Mice were bred over 20 generations, and wild-type (+/+) and knockout (−/−) mice were identified from C/EBPβ (+/−) female × C/EBPβ (+/−) male breeding. Bone marrow was cultured in standard medium supplemented with 20–25% LADMAC (32) cell-conditioned medium. After 5 days, nonadherent cells were removed. Adherent cells were harvested after 8–14 days in culture and replated for experimental use. Unless otherwise indicated, macrophages were activated with 2 ng/ml IFN-γ, and LpqH was used at 30 nM. All experiments were conducted at 37°C and 5% CO2.
Mtb culture and purification of LpqH
Mtb H37Ra (American Type Culture Collection) was grown to log phase in Middlebrook 7H9 medium (Difco). LpqH was prepared, as described (7), from Mtb H37Ra. Mtb H37Ra was resuspended in deionized water containing 7.5 mM EDTA, 0.7 μg/ml leupeptin, 0.7 μg/ml pepstatin A, 0.2 mM PMSF, 10 U/ml DNase, and 25 U/ml RNase. The suspension was passed through a French press two to three times, centrifuged to pellet cell wall components, resuspended in ice-cold 2% Triton X-114 (TX114) in 5 mM Tris-HCl, and centrifuged for 2 h at 100,000 × g. The supernatant was warmed to 37°C (above the cloud point), and detergent and aqueous phases were separated by centrifugation (2400 × g for 10–15 min). The TX114 layer was washed three to five times with cold 50 mM phosphate buffer, with the samples warmed to 37°C before each centrifugation. The TX114 layer was incubated overnight with cold acetone and then centrifuged at 2400 × g for 20–30 min. The pellet was dissolved in SDS-PAGE sample buffer (62.5 mM Tris (pH 6.8), 2% SDS, 10% glycerol, 700 μM 2-ME, and 0.01 μg/ml bromphenol blue), boiled for 5 min, and loaded onto a 12% SDS polyacrylamide gel. Electroelution was performed using a Model 491 Prep Cell (Bio-Rad) with fractions collected every 8 min for 20 h. Fractions were tested for LpqH and potential contaminants by SDS-PAGE with 12% polyacrylamide gels, followed by Western blot or silver staining (with a Bio-Rad Silver Stain Plus kit). For Western blot analysis, samples were transferred onto polyvinylidene difluoride membranes (Millipore). Membranes were incubated overnight at 4°C in 0.1% Tween 20 in PBS supplemented with 5% Carnation nonfat dry milk, and incubated for 1 h at room temperature with a polyclonal rabbit anti-Mycobacterium bovis antiserum that recognizes many mycobacterial constituents, including LpqH. Membranes were washed repeatedly, incubated for 1 h with HRP-labeled donkey anti-rabbit secondary Ab (Amersham), and developed with ECL detection kit (Amersham). Fractions determined to contain LpqH were pooled, extracted with TX114, precipitated in acetone, resuspended to 53 μM in DMSO, and stored at −80°C.
Quantitative real-time RT-PCR
Macrophages (3–4 × 106) were incubated with or without LpqH (30 nM) and with IFN-γ (2 ng/ml) in the continued absence or presence of LpqH. RNA was isolated using RNeasy columns (Qiagen), as described by the manufacturer’s protocol. Total RNA yield was determined by spectrophotometer, and 1–2 μg of total RNA was used in a reverse-transcriptase reaction (SuperScript First Strand Synthesis System; Invitrogen Life Technologies) to convert RNA to cDNA. One-tenth of the resulting cDNA template was used for real-time PCR analysis with SYBR Green and the Bio-Rad iCycler fluorescence detection system. A standard curve for each gene was generated by serial dilution of amplified product standard of known starting concentration. The following primers were used: CIITA sense, 5′-ACG CTT TCT GGC TGG ATT AGT-3′; CIITA antisense, 5′-TCA ACG CCA GTC TGA CGA AGG-3′; C/EBPβ sense, 5′-AGC TTA GCG ACG AGT ACA AGA-3′; C/EBPβ antisense, 5′-GGC AGC TGC TTG AAC AAG T-3′; CIITA types I, III, and IV antisense, 5′-GGT CGG CAT CAC TGT TAA GGA-3′; CIITA type I sense, 5′-AAG AGC TGC TCT CAC GGG AAT-3′; CIITA type III sense, 5′-TCT TAC CTG CCG GAG TT-3′; CIITA type IV sense, 5′-GAG ACT GCA TGC AGG CAG CA-3′; GAPDH sense, 5′-AAC GAC CCC TTC ATT GAC-3′; GAPDH antisense, 5′-TCC ACG ACA TAC TCA GCA C-3′. Quantity was determined based on a standard curve of known concentration for each gene and normalized to GAPDH.
Preparation of nuclear extracts and Western blots
Macrophages (3–4 × 106) were plated in 60-mm petri dishes, incubated with or without LpqH for 18–24 h, and then incubated for various periods with or without IFN-γ in the continued presence or absence of LpqH. Cells were washed with ice-cold PBS, pelleted, and resuspended in buffer A (10 mM KCl, 0.1 mM EDTA, 1 mM EGTA, 10 mM HEPES (pH 7.9)) with a protease inhibitor mixture (P8340; Sigma-Aldrich) on ice for 10 min. Non-idet P-40 was added to a concentration of 0.2%, and the cells were passed through a 26-G needle three times. Nuclei were pelleted at 2310 × g for 5 min at 4°C and then resuspended in buffer B (20 mM HEPES (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 1 mM NaVO4 supplemented with a protease inhibitor mixture (Sigma-Aldrich P8340)). Repeat pipeting over 1 h was used to extract proteins from nuclei. Protein concentration was determined using the Bio-Rad protein detection kit. Lysates were boiled for 5 min in reducing sample buffer (No. 39001; Pierce). Equal quantities of total protein were electrophoresed on 10 or 12% SDS polyacrylamide gels and transferred onto polyvinylidene difluoride membranes. Membranes were washed in PBS/T (PBS with 0.1% Tween 20), incubated for 1 h at room temperature in 5% nonfat milk in PBS/T, incubated overnight at 4°C with primary Ab in 5% nonfat milk in PBS/T, washed, incubated for 1 h at room temperature with HRP-labeled donkey anti-rabbit (Amersham) or donkey anti-goat (Santa Cruz Biotechnology) secondary Ab, and developed with ECL detection kit (Pierce). Anti-C/EBPβ, anti-C/EBPδ, anti-C/EBPε, and anti-actin Abs were purchased from Santa Cruz Biotechnology.
CIITA pIV-oligodeoxynucleotide (ODN) pull-down assay
Macrophages were incubated with or without LpqH (30 nM) for 18–24 h and then incubated with IFN-γ (2 ng/ml) for 4–6 h. Nuclear extract was obtained as above, and 60–80 μg of protein was used for each precipitation. Thirty micrograms of protein per sample was reserved for Western blot to ensure equal input at the start of the immunoprecipitation. A biotinylated ODN containing 100 nt of the murine CIITA pIV sequence (−212 to −112; GenBank accession AF000008) that contains putative C/EBP binding sites was annealed to the complementary strand for 1 h at room temperature. This procedure was also performed for a negative control CIITA pIV ODN without putative C/EBP binding sites (CIITA pIV −17 to +83) and an ODN containing 100 nt of the murine CIITA pI sequence that contains one putative C/EBP binding site (−92 to +8; Gen-Bank accession AF000006). Nuclear extract samples were diluted to 400 μl with coimmunoprecipitation buffer (0.1% Triton X-100, 10 mM HEPES (pH 7.3), 2 mM EDTA, 1 mM EGTA, 10% glycerol, 1 mM NaF, 1 mM NaVO4 supplemented with a protease inhibitor mixture (Sigma-Aldrich; No. P8340)) and precleared with 60 μl of salmon sperm-agarose beads (Upstate Biotechnology) for 30 min at 4°C. Beads were removed by centrifugation, and the supernatant was incubated overnight at 4°C with 30 nM CIITA pIV-ODN in the presence of streptavidin-agarose beads (Upstate Biotechnology) washed three times with PBS before use. ODN-protein complexes were pelleted, washed three times, diluted in SDS-PAGE sample buffer, and boiled for 5 min. Boiled samples were centrifuged to remove beads, and the entire supernatant was loaded onto a 12% polyacrylamide gel. Western blots were performed, as described above.
Chromatin immunoprecipitation (ChIP)
Macrophages (7–8 × 106) were plated in 100-mm petri dishes and incubated with or without LpqH (30 nM) for 18–24 h. IFN-γ (2 ng/ml) was added for 4–6 h in the continued presence or absence of LpqH. ChIP was performed with reagents and protocol (item 17–295) from Upstate Biotechnology. Cells were fixed with 1% paraformaldehyde for 10 min at 37°C, washed with ice-cold PBS, detached by scraping, and centrifuged at 624 × g for 5 min. The pellet was resuspended for 10 min at 4°C in 450 μl of buffer A with a protease inhibitor mixture (P8340; Sigma-Aldrich). A total of 9 μl of 10% Nonidet P-40 was added, the suspension was passed through a 26-G needle, and nuclei were pelleted by centrifugation at 2310 × g for 5 min. Nuclei were resuspended in SDS lysis buffer (Upstate Biotechnology) and pulse sonicated for 80 s on ice to an average fragment size of 600 bp. Approximately one-third of the resulting suspension was used for each immunoprecipitation following the manufacturer’s protocol. A total of 40 μl of the suspension was set aside to determine the input quantity of DNA. After overnight immunoprecipitation at 4°C with antiC/EBPβ or anti-C/EBPδ (Santa Cruz Biotechnology), the precipitates were washed repeatedly, and cross-links were reversed overnight at 65°C. After proteinase K treatment, DNA was purified using DNeasy columns (Qiagen). The resulting DNA was quantified by real-time PCR, as described above, using the following primers: CIITA pIV sense, 5′-CTG CCT TGG AAT TCA GTT CTA-3′; CIITA pIV antisense, 5′-GAG TAT CTG TGG CGC TTT TC-3′; CIITA pI sense, 5′-CCC TAA CCC ATT TCC GTT CAT-3′; CIITA pI antisense, 5′-CTG CCT GGA GTC GCC TCT-3′; GAPDH sense, 5′-AGA CAA AAT GGT GAA GGT CGG-3′; GAPDH antisense, 5′-AGG TCA ATG AAG GGG TCG TT-3′. Quantity was determined based on a standard curve of known concentration for each gene and normalized to GAPDH of input DNA for each sample.
Plasmids and transfection
To generate the pCMV-LIP plasmid, total RNA was isolated from rat liver cells and cDNA was generated using reverse transcription. The cDNA sequence encoding the LAP isoform (from second AUG to the stop codon) of rat C/EBPβ (GenBank NM024125) was amplified by PCR and subcloned into pcDNA3. Target cDNA was amplified by PCR with the following primers: sense, 5′-CCG CCG AAG CTT GCC GCC TTT AGA CCC ATG-3′ and antisense, 5′-CAA ACC AAT CTA GAC GGG CTA GCA GTG ACC-3′. Restriction enzyme sites for HindIII and XbaI were introduced to the ends of the PCR-amplified fragment and used to subclone the PCR-amplified fragment into pcDNA3 (Invitrogen Life Technologies). The NcoI fragment in the coding sequence of C/EBPβ was deleted to generate pCMV-LIP, which directs expression of only the LIP isoform of C/EBPβ (from third AUG to the stop codon). The subcloned sequence of LIP was verified by DNA sequencing. RAW264.7 cells were transfected with 2 μg of pCMV-LIP or vector control (pcDNA3) using the Superfect Transfection system (Qiagen). Stably transfected cells were selected over 3–4 wk with 500 μg/ml geneticin and were maintained as a polyclonal population. For experimental use, cells were treated with 2 ng/ml IFN-γ for 8 or 24 h. Cells were lysed, RNA was extracted, and CIITA mRNA expression was determined, as described above.
Results
LpqH inhibits IFN-γ-induced CIITA expression and induces expression of C/EBPβ
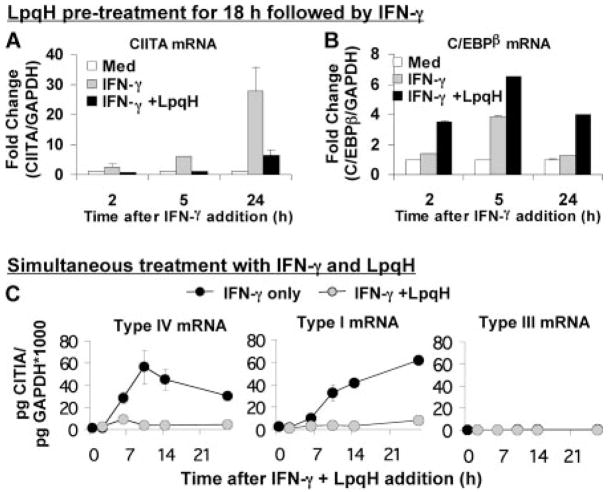
Previous microarray gene expression analyses showed inhibition of a subset of IFN-γ-induced genes, including CIITA, in response to prolonged exposure to LpqH (6). However, these studies also revealed a set of genes for which expression was significantly enhanced by LpqH. Of these, C/EBPβ was of particular interest due to its role in IFN-γ-induced gene regulation. To confirm and extend the microarray analyses, we performed quantitative real-time RT-PCR studies of C/EBPβ expression by bone marrow-derived macrophages that were incubated with LpqH for 18 h and with IFN-γ for various subsequent periods in the continued presence of LpqH. In agreement with previous studies, LpqH inhibited IFN-γ-induced expression of CIITA (Fig. 1A). In addition, LpqH induced the expression of C/EBPβ (Fig. 1B), consistent with the microarray data. Thus, LpqH increases C/EBPβ expression and inhibits IFN-γ-induced expression of CIITA. These data suggested the hypothesis that C/EBPβ negatively regulates IFN-γ-induced expression of CIITA.
FIGURE 1.
LpqH inhibits IFN-γ-induced CIITA, but increases expression of C/EBPβ. Quantitative RT-PCR analysis of CIITA (A) and C/EBPβ (B). Macrophages were incubated with or without 30 nM LpqH for 18 h and then with 2 ng/ml IFN-γ for an additional 2, 5, or 24 h (Med, indicates no IFN-γ or LpqH). Quantitative real-time RT-PCR was performed using a standard curve for each gene and normalization to GAPDH. Data are expressed as fold change compared with untreated cells (Med). C, Macrophages were incubated with 2 ng/ml IFN-γ and 30 nM LpqH (added simultaneously) or with IFN-γ only. Data are expressed as amount of CIITA mRNA (normalized to GAPDH) per μg of total mRNA. Data are expressed as means and SDs of triplicate samples and are representative of at least three independent experiments. Where error bars cannot be seen they are smaller than the symbol width. CIITA type III expression was so low that symbols for both treatment conditions overlap along the x-axis.
CIITA expression is regulated by three promoters (pI, pIII, and pIV) that drive expression of unique mRNA transcripts with different exon 1 sequences (11). Using quantitative real-time RT-PCR with primers specific for CIITA type I, type III, or type IV, we analyzed CIITA expression in macrophages in response to IFN-γ only or simultaneous treatment with both IFN-γ and LpqH (Fig. 1C). Expression of each of the three isoforms of CIITA was induced in macrophages by IFN-γ, but to varying degrees. CIITA type IV was induced rapidly (30-fold by 6 h), and its expression then declined over time. Expression of CIITA type I was induced by IFN-γ with somewhat slower kinetics (Fig. 1C). CIITA type III expression was minimal in macrophages; although it was induced by IFN-γ, its expression always remained 100-fold or more less than either CIITA type I or IV. We conclude that CIITA types I and IV account for most, if not all, of IFN-γ-induced CIITA expression in macrophages. To determine the extent to which LpqH inhibits the three CIITA isoforms, we treated cells simultaneously with IFN-γ and LpqH. LpqH inhibited IFN-γ-induced expression of all three CIITA isoforms, with CIITA types I and IV showing the most dramatic inhibition (Fig. 1C).
LpqH-induced expression of C/EBPβ is kinetically correlated with inhibition of CIITA expression
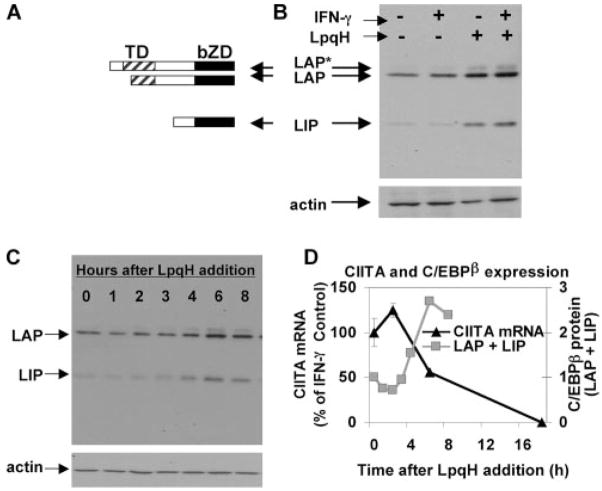
There are three isoforms of C/EBPβ, LIP, LAP, and LAP*, each of which is translated from a unique AUG start codon within a common mRNA transcript (Fig. 2A). Therefore, it is impossible to distinguish one isoform from the others by RT-PCR. To study induction of specific C/EBPβ isoforms, macrophages were incubated with or without LpqH and stimulated with IFN-γ. Western blot analysis of nuclear extracts was used to assess C/EBPβ protein expression (Fig. 2B). There was little or no expression of LIP in the absence of LpqH (with or without IFN-γ), but LIP was induced by LpqH. LAP was expressed constitutively, but its expression was moderately increased by LpqH. LAP* was expressed minimally and only in response to LpqH. To assess the kinetics of C/EBPβ induction by LpqH, nuclear extracts were prepared from macrophages after incubation with LpqH for various periods and probed for C/EBPβ expression by Western blot. The expression of LIP (and, to a lesser extent, LAP) increased by 4 h of exposure to LpqH and continued to increase until 24 h (Fig. 2C and data not shown). To assess the kinetics of CIITA inhibition, macrophages were treated simultaneously with IFN-γ and LpqH, and CIITA expression was analyzed by quantitative real-time RT-PCR. CIITA expression was inhibited by 6 h and continued to decrease over time (Fig. 2D). Thus, induction of C/EBPβ (both LAP and LIP) by LpqH was kinetically correlated with inhibition of IFN-γ-induced CIITA expression, suggesting a negative regulatory role for C/EBPβ in the inhibition of IFN-γ-induced CIITA expression by LpqH.
FIGURE 2.
LpqH induces expression of C/EBPβ isoforms LAP and LIP. A, Diagram depicting the protein sequence of each of the three C/EBPβ isoforms (LAP*, LAP, and LIP). TD, Trans activation domain. bZD, Basic leucine zipper domain. B, Induction of C/EBPβ by LpqH. Macrophages were incubated with or without 30 nM LpqH for 18 h, followed by treatment with 2 ng/ml IFN-γ for 5 h in the continued presence or absence of LpqH. Nuclear extracts were prepared and analyzed by Western blot with Abs to C/EBPβ or actin (to ensure equal loading). C, Kinetics of C/EBPβ induction by LpqH. Macrophages were incubated with 30 nM LpqH for 0–8 h. Nuclear extracts were prepared, and equal amounts of protein were probed with anti-C/EBPβ or anti-actin Abs. D, Macrophages were incubated for various times with 2 ng/ml IFN-γ with or without 30 nM LpqH (added simultaneously). Quantitative real-time RT-PCR analysis was performed and CIITA expression was normalized to GAPDH. Data are represented as percentage of CIITA expression by cells treated with IFN-γ alone at the same time point, and are expressed as means and SDs of triplicate samples (error bars cannot be seen because they are smaller than the symbols). D, Also includes densitometric data from the blot in C (the sum of densitometric values for LAP and LIP normalized to the 0-h time point).
LpqH induces C/EBPβ binding to CIITA pIV and pI
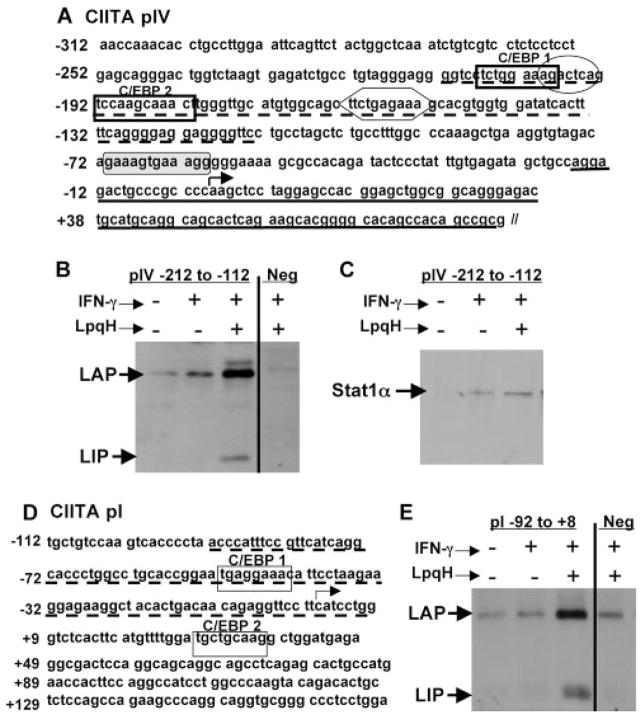
C/EBPβ has not previously been associated with control of CIITA expression or the function of CIITA promoter elements. Using TRANSFEC software (www.cbil.upenn.edu/tess), we analyzed CIITA pIV to determine putative binding site(s) for C/EBPβ. The program searched the entire promoter sequence for any partial or total matches to published transcription factor-binding sequences. This analysis identified two putative C/EBPβ binding sites located within CIITA pIV at −207 to −199 (TCTGGAAAG) and −192 to −182 (AAGCAAAC) (Fig. 3A). The former is a good match to the widely accepted C/EBP consensus sequence T(T/G)NNGNAA(T/G), and the latter matches a lesser known C/EBP-binding sequence TNNGCAAAC found in human alcohol dehydrogenase 3 and murine xanthine dehydrogenase promoters (33–35). Thus, we report the presence of putative C/EBPβ-binding sequences in CIITA pIV.
FIGURE 3.
LpqH induces C/EBPβ-LAP and LIP binding to CIITA pIV and CIITA pI. A, CIITA pIV nucleotide sequence from −312 to +83 with two putative C/EBPβ binding sites (bold rectangles) and putative binding sites for STAT1 (hexagon), AP1 (oval), and IRF-1 (shaded rectangle with rounded corners). ODN sequences used for pull-down experiments are underlined. B and C, CIITA pIV ODN pull-down experiment. Macrophages were treated with or without 30 nM LpqH for 18 h and then with 2 ng/ml IFN-γ for an additional 5 h. Nuclear extracts were prepared, and 70 μg of protein from each sample was incubated overnight with a synthetic biotinylated ODN containing both putative C/EBP binding sites (pIV −212 to −112, dotted underline in A) or a negative control ODN without a putative C/EBP binding site (pIV −17 to +83, Neg; solid underline in A). ODNs were precipitated using streptavidin-conjugated agarose beads. Protein was eluted with reducing sample buffer and analyzed by Western blot with anti-C/EBPβ (B) or anti-STAT1α Abs (C). D, Schematic of CIITApI. Putative C/EBP binding sites are enclosed in rectangles. The CIITA pI ODN sequence used for the pull-down experiment in E is indicated with the broken underline. E, CIITA pI pull-down. The experiment was performed as described for B using biotinylated ODNs containing pI sequence (−92 to +8) with a putative C/EBP binding site and a negative control ODN without a putative C/EBP binding site (pIV sequence −17 to +83, Neg). Results are representative of at least three independent experiments (two independent experiments for E).
To test binding of C/EBPβ to CIITA pIV, we performed pull-down assays using biotinylated ODNs containing portions of the murine CIITA pIV sequence inclusive of the putative C/EBPβ binding sites (−212 to −112, broken underline in Fig. 3A). Macrophages were incubated with or without LpqH for 18 h and then exposed to IFN-γ for 5 h in the continued presence or absence of LpqH. Nuclear extracts were prepared and incubated with biotinylated ODNs, which were then precipitated with streptavidin-conjugated Sepharose beads. Western blot analysis was used to detect C/EBPβ protein that was associated with the ODNs. The addition of LpqH induced C/EBPβ-LIP binding and increased C/EBPβ-LAP binding (Fig. 3B). IFN-γ alone produced a small signal for binding of LAP to CIITA pIV, but this signal was far less than that produced by LpqH plus IFN-γ, and subsequent ChIP studies did not show binding of C/EBPβ to pIV in cells after stimulation of macrophages with IFN-γ alone (below). The pull-down assay detected little or no binding of LIP to CIITA pIV with IFN-γ alone (Fig. 3B). An ODN containing the −17 to +83 sequence of CIITA pIV, which contains no putative C/EBPβ binding sites, was used as a negative control, and we observed no binding of C/EBPβ to this sequence (Fig. 3B, Neg). As an additional control, samples pulled down with the pIV ODN containing the C/EBPβ binding sites (−212 to −112) were additionally probed with anti-STAT1α Ab, because a known STAT1 binding site exists within this sequence. As previously reported, STAT1α binding was induced by IFN-γ, and this binding was not inhibited by LpqH (Fig. 3C). These data confirm the discovery of novel C/EBPβ binding sites in CIITA pIV.
Like CIITA pIV, activation of CIITA pI by IFN-γ is inhibited by treatment of macrophages with LpqH (7, 10). Therefore, we analyzed the CIITA pI sequence and identified two putative C/EBPβ-binding sequences at −52 to −40 and +29 to +37 (Fig. 3D). Both sequences perfectly match the C/EBP consensus sequence T(T/G)NNGNAA(T/G). ODN pull-down experiments were performed, and results similar to those seen for CIITA pIV were observed with an ODN sequence from CIITA pI (−92 to +8) that included the first putative C/EBP binding site. LpqH induced the binding of C/EBPβ-LIP and C/EBPβ-LAP to this CIITA pI promoter sequence, but not a negative control ODN (Fig. 3E). We conclude that LpqH induced C/EBPβ-LIP and C/EBPβ-LAP that were capable of binding to CIITA pI and pIV. LAP and LIP may function together to bind and potentially regulate CIITA promoters, because LAP-LIP heterodimers have been reported as important in regulation of other genes (see Discussion).
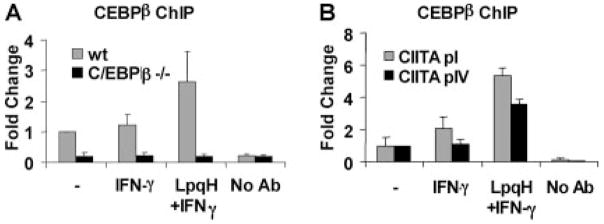
ChIP assays were used to directly test whether C/EBPβ binds to CIITA pIV in living cells. Macrophages were incubated with or without LpqH for 18–24 h and then stimulated with IFN-γ for 5 h. Proteins were cross-linked to DNA using paraformaldehyde. Chromatin was sheared and incubated with anti-C/EBPβ Ab. Protein A-conjugated agarose beads were used to precipitate C/EBPβ-associated chromatin/DNA fragments. The amount of CIITA pIV that was associated with C/EBPβ was determined by quantitative real-time PCR with primers specific for CIITA pIV. C/EBPβ binding to CIITA pIV was induced by LpqH and IFN-γ, but not by IFN-γ alone (Fig. 4A). This assay did not discriminate LAP from LIP (the anti-C/EBPβ Ab binds to both). To eliminate the possibility of cross-reactivity or nonspecific binding of our Ab, we performed the same experiment with C/EBPβ−/− macrophages. No C/EBPβ binding to CIITA pIV was detected in these cells (Fig. 4A). These results establish the binding of C/EBPβ to CIITA pIV in living cells and demonstrate that binding of C/EBPβ to CIITA pIV is induced by LpqH.
FIGURE 4.
LpqH induces binding of C/EBPβ to CIITA pI and pIV in intact cells. Macrophages were incubated with or without 30 nM LpqH and then with IFN-γ, as in Fig. 3. Cells were fixed to cross-link proteins to DNA, and sonication was used to shear chromatin into 300- to 1000-bp fragments. The resulting lysate was incubated overnight with anti-C/EBPβ Ab. Protein A-agarose beads were used to precipitate Ab-C/EBPβ-DNA complexes. DNA was purified for quantitative real-time PCR to determine the coimmunoprecipitation of C/EBPβ and CIITA promoter fragments. A, Binding of C/EBPβ to CIITA pIV in wild-type (wt) or C/EBPβ−/− macrophages. B, Binding of C/EBPβ to CIITA pI or pIV in wild-type macrophages. Graphs depict fold change in PCR signal in samples relative to the control without LpqH or IFN-γ (labeled “-”). All samples were first normalized to input DNA. No Ab, Indicates results with Ab omitted during ChIP with a sample from cells incubated with LpqH and IFN-γ. Data are expressed as means and SDs of triplicate samples and are representative of at least three independent experiments (two independent experiments with C/EBPβ−/− macrophages).
Because activation of CIITA pI by IFN-γ is also inhibited by treatment of macrophages with LpqH (Fig. 1C) (7, 10), we tested whether LpqH induces C/EBPβ binding to CIITA pI. ChIP experiments were performed, as described above, with Ab to C/EBPβ and primers specific for CIITA pI. Similar to results with CIITA pIV, LpqH induced C/EBPβ binding to CIITA pI (Fig. 4B). These data suggest that C/EBPβ plays a role in suppression of both CIITA pI and pIV in response to LpqH.
Transfection of the LIP isoform of C/EBPβ inhibits IFN-γ-induced CIITA expression
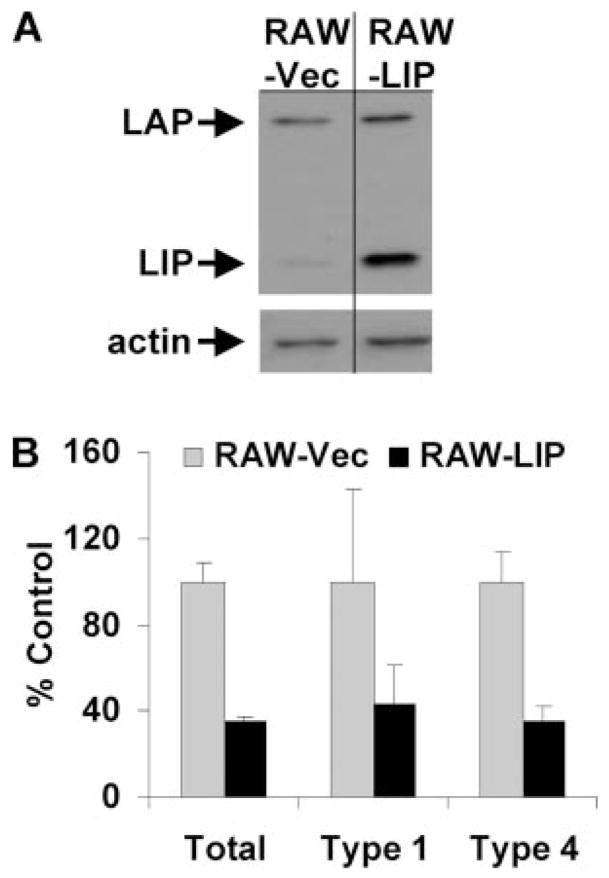
To test the ability of C/EBPβ to inhibit CIITA expression, RAW264.7 cells were transfected to achieve constitutive expression of LIP. RAW264.7 cells were chosen due to the feasibility of their use in transfection studies and the prior determination that these cells recapitulate core observations reported above for primary macrophages (inhibition of CIITA and induction of LIP expression in response to LpqH) (13) (data not shown). LIP was chosen as the C/EBPβ isoform for these studies. There is an established role for LIP as a negative regulator of gene expression due to its lack of a trans activation domain, although LAP may also inhibit gene expression. Moreover, LpqH inhibition of CIITA expression correlated with the kinetics of LIP induction by LpqH and the dependence of LIP expression on LpqH. LAP expression also increased in response to LpqH, but LAP was constitutively expressed in the absence of LpqH. Therefore, LIP was a candidate to test for potential function as a regulator of CIITA expression (perhaps in concert with LAP in the context of LAP-LIP heterodimers).
RAW264.7 cells were stably transfected with a LIP expression plasmid or a control vector and selected over 2 wk or more with geneticin to produce RAW-LIP or RAW-Vec cells, respectively. Nuclear extracts were prepared from each cell population, and Western blot analysis was used to determine expression of C/EBPβ in the transfected cells. LIP was constitutively expressed in RAW-LIP, but not in RAW-Vec cells (Fig. 5A). The cells were incubated with IFN-γ and analyzed for CIITA expression by quantitative real-time RT-PCR. After 8 h of incubation with IFN-γ, RAW-LIP cells expressed ~50–60% less CIITA than the control RAW-Vec cells (Fig. 5B). Types I and IV CIITA were equally susceptible to this inhibition. Although the achieved level of LIP expression did not completely inhibit IFN-γ-induced CIITA, these results indicate that LIP is an inhibitor of CIITA expression. The lack of complete inhibition as observed with LpqH may be due to other factors induced by TLR2 signaling that could participate in the inhibitory mechanism, because multicomponent mechanisms may not be fully reconstituted by single gene transfection strategies. In addition, TLR2 signaling may induce posttranslational modifications of C/EBPβ that enhance its ability to affect CIITA expression. It is possible that the LAP isoform of C/EBPβ also inhibits CIITA expression, but that possibility was not assessed in these studies. These data indicate that LIP can inhibit IFN-γ-induced expression of CIITA in macrophages. In the context of our other results, this finding suggests that LIP plays a role in the suppression of CIITA in response to LpqH, possibly in concert with other C/EBP proteins.
FIGURE 5.
C/EBPβ is sufficient to inhibit IFN-γ-induced CIITA. RAW264.7 cells were transfected with empty control vector or a C/EBPβ LIP expression plasmid driven by a constitutively active CMV promoter to produce RAW-Vec or RAW-LIP cells, respectively. A, Western blot analysis of nuclear extracts prepared from RAW-Vec or RAW-LIP cells using anti-C/EBPβ. B, Transfected cells were treated with 2 ng/ml IFN-γ for 8 h. Quantitative real-time RT-PCR was used to determine relative expression of IFN-γ-induced CIITA (total, type 1 and type 4) in RAW-LIP cells relative to RAW-Vec cells. All samples were first normalized to GAPDH. Data are expressed as means and SDs of triplicate samples and are representative of at least three independent experiments.
C/EBPβ−/− macrophages are sensitive to LpqH-mediated inhibition of CIITA, suggesting the existence of redundant factors
The preceding experiments established that C/EBPβ, particularly LIP, inhibits IFN-γ-induced CIITA expression in macrophages. To determine whether C/EBPβ is essential for LpqH-mediated inhibition of IFN-γ-induced CIITA, macrophages from C/EBPβ−/− and wild-type mice were incubated with LpqH for 18 h and then stimulated with IFN-γ for various times in the continued presence or absence of LpqH. C/EBPβ−/− and wild-type macrophages showed similar induction of CIITA by IFN-γ and similar inhibition of CIITA by LpqH (Fig. 6), indicating that C/EBPβ is not essential for LpqH-mediated inhibition of CIITA. Knockout cells, however, are problematic for dissection of systems that may involve redundancy, and knockout animals may compensate for loss of some essential factors that are required under physiological conditions by enhancing expression or function of factors with redundant or overlapping function. Because our other data strongly suggested a role for C/EBPβ in inhibition of CIITA in wild-type cells, we considered the hypothesis that other members of the C/EBP family of transcription factors may contribute to inhibition of CIITA expression.
FIGURE 6.
LpqH inhibits CIITA mRNA expression in C/EBPβ−/− macrophages. Macrophages from wild-type or C/EBPβ−/− mice were incubated with or without 30 nM LpqH for 18 h plus an additional 6 h with 2 ng/ml IFN-γ. Quantitative real-time RT-PCR was used to determine CIITA expression relative to the no IFN-γ control for each cell type. Samples were first normalized to GAPDH. Data are expressed as means and SDs of triplicate samples and are representative of two independent experiments.
LpqH induces expression of C/EBPδ and its binding to CIITA pI and pIV, suggesting functional redundancy with C/EBPβ
Several studies have suggested that other C/EBP family members, including C/EBPα and C/EBPδ, may have functional redundancies with C/EBPβ. Similarity of the DNA binding domain accounts for the ability of most family members to recognize identical DNA target sequences (36), and homology within the basic leucine zipper domain accounts for heterodimerization of family members. For example, C/EBPβ/δ heterodimers have been linked to the suppression of cartilage-derived retinoic acid-sensitive protein (CD-RAP) (30). Furthermore, C/EBPδ expression is induced by inflammatory mediators known to induce C/EBPβ, such as IL-1β and LPS. Unlike C/EBPβ, C/EBPδ is expressed as a single protein containing both a C-terminal DNA binding domain and N-terminal trans activation domain. C/EBPδ induces expression of certain genes, but it has also been implicated as a transcriptional repressor of some target genes, including CD-RAP, which is inhibited by either C/EBPβ or C/EBPδ (30). Overall, data from multiple studies establish the ability of both C/EBPδ and C/EBPβ to repress transcriptional activity of certain promoters.
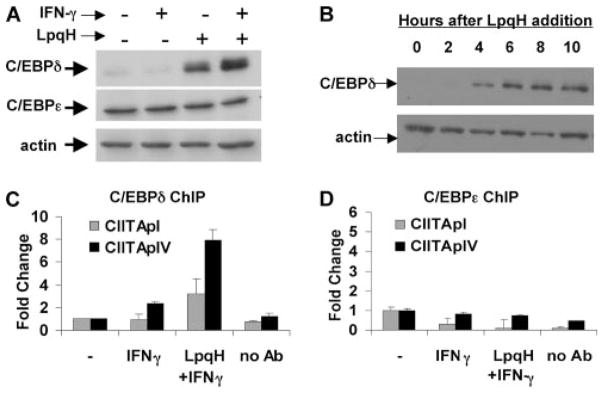
To address the hypothesis that other C/EBP family members may contribute to LpqH-mediated regulation of CIITA, we analyzed nuclear extracts of macrophages stimulated with IFN-γ in the presence or absence of LpqH by Western blot for expression of C/EBPα, γ, δ, and ε. Of these proteins, only C/EBPδ was significantly induced by LpqH (Fig. 7A and data not shown). C/EBPδ was induced beginning ~4 h after exposure to LpqH (Fig. 7B), similar to C/EBPβ. C/EBPε was also detected by Western blot, but its expression was not increased in response to LpqH (Fig. 7A). We were not able to detect C/EBPα or γ protein in response to any of our stimuli (data not shown). Of these C/EBP family members, C/EBPδ was the only one that was regulated in parallel with C/EBPβ, suggesting that C/EBPδ may also regulate CIITA in response to LpqH.
FIGURE 7.
LpqH-induced C/EBPδ binds to CIITA pI and pIV. A, LpqH induces expression of C/EBPδ, but not C/EBPε. Macrophages were incubated with 30 nM LpqH for 18 h and an additional 5 h with or without 2 ng/ml IFN-γ. Nuclear extracts were prepared and analyzed by Western blot for expression of C/EBPδ, C/EBPε, or actin (as a loading control). B, Macrophages were incubated with 30 nM LpqH for 0–10 h. Nuclear extracts were prepared, and equal amounts of protein were probed with antiC/EBPδ or anti-actin (as a loading control). C and D, ChIP assays demonstrate that LpqH induces binding of C/EBPδ (C), but not C/EBPε (D) to CIITA pI and pIV. Macrophages were incubated with or without 30 nM LpqH for 18 h and an additional 5 h with IFN-γ. Samples were processed for ChIP as in Fig. 4 using anti-C/EBPδ or anti-C/EBPε Ab. Quantitative real-time PCR was performed to determine coimmunoprecipitation of CIITA pI- or pIV-specific DNA fragments. Data are represented as fold change relative to untreated cells (without IFN-γ or LpqH, labeled “-”). No Ab, Indicates samples from LpqH-stimulated cells with Ab omitted in the immunoprecipitation step. Data are expressed as means and SDs of triplicate samples. Results are representative of at least three independent experiments for A and C, and two independent experiments for B and D.
To determine whether C/EBPδ binds to CIITA pI or pIV, we performed ChIP assays using anti-C/EBPδ Ab. LpqH induced C/EBPδ binding to both CIITA pI and pIV (Fig. 7C). ChIP analysis using anti-C/EBPε Ab revealed no binding of C/EBPε to the CIITA promoters under any of our experimental conditions (Fig. 7D), suggesting that regulation of CIITA transcription is controlled by C/EBPβ and C/EBPδ, but not C/EBPε. Together, these data indicate that C/EBPβ and C/EBPδ are unique among C/EBP family members in their induction by LpqH and their ability to bind CIITA pIV and CIITA pI. Thus, C/EBPβ and C/EBPδ may contribute to LpqH-mediated inhibition of IFN-γ-induced expression of CIITA by macrophages.
Discussion
Several studies have shown that Mtb or Mtb lipoproteins, including LpqH, signal through TLR2 and inhibit macrophage expression of a subset of IFN-γ-induced genes, including CIITA (5–9, 12, 13, 37–39). Proximal IFN-γ signaling, including STAT1 activation and function, is not inhibited by Mtb or LpqH, suggesting that the inhibition occurs by distal mechanisms that affect expression of this subset of genes. LpqH inhibits expression of CIITA pIV reporter constructs in stably, but not transiently transfected macrophages (7, 13), consistent with the observation that inhibition of IFN-γ-induced CIITA expression is dependent on native chromatin structure (7, 13) and involves altered chromatin remodeling (13). To further examine the mechanisms by which LpqH may affect chromatin remodeling and gene expression, we studied the regulation of transcription factors by LpqH, focusing particularly on C/EBP.
C/EBP family members have been implicated in both induction and suppression of multiple target genes. LIP is particularly recognized as an inhibitory isoform of C/EBPβ, because it lacks a trans activation domain. Due to its high affinity for C/EBP binding sites, LIP acts as a dominant-negative transcription factor, even when bound as a LAP-LIP heterodimer (15, 21). Therefore, even minimal induction of LIP may inhibit CIITA transcription. Although LAP and C/EBPδ activate transcription of some genes, they may also act as transcriptional repressors. For example, transcriptional repression of a constitutively active CD-RAP reporter plasmid was reported with tranfection with any of the C/EBPβ isoforms or C/EBPδ (30). Overall, these observations indicate that the functions of C/EBP family members must be evaluated in a gene-specific manner.
Our studies provide a novel implication that C/EBP family members may be involved in the regulation of CIITA transcription. First, there is a kinetic relationship between inhibition of CIITA and the induction of C/EBPβ (LIP induced from a very low baseline level; LAP enhanced above constitutive expression) and C/EBPδ in response to LpqH. Furthermore, LpqH induces binding of C/EBPβ (LIP and LAP) and C/EBPδ to CIITA pIV and pI both in vitro (assessed by pull-down assays) and in intact cells (assessed by ChIP). Although C/EBPβ−/− macrophages remain sensitive to LpqH-mediated inhibition of CIITA, C/EBPδ may compensate for the absence of C/EBPβ in these cells. Finally, constitutive expression of C/EBPβ LIP in transfected cells inhibited IFN-γ-induced CIITA, confirming the ability of C/EBP to regulate CIITA transcription. Although expression of C/EBPβ LIP in transfected RAW cells did not produce a complete inhibition, we consistently observed that LIP expression caused 50–60% inhibition of CIITA expression (LpqH signaling may also induce other transcriptional inhibitors or cause posttranslational modifications that activate inhibitory transcription factors, resulting in even greater inhibition). Thus, our data provide the novel observation that C/EBPβ LIP can inhibit CIITA expression and suggest that C/EBPδ and C/EBPβ LAP may also contribute to inhibition of CIITA expression.
Functional overlap of C/EBPβ and C/EBPδ is suggested by our studies and many others. Due to the conserved DNA binding domain, C/EBP family members bind similar target DNA sequences. Furthermore, C/EBPβ and C/EBPδ share properties that are not shared by other C/EBP family members and may share redundant functions. C/EBPβ and C/EBPδ function either as homo- or heterodimers to bind and regulate identical target sequences (16, 30). Several groups have shown functional redundancy between C/EBPβ and C/EBPδ in the activation of genes such as IL-6, IL-10, and MCP-1 (40–43). Additionally, C/EBPβ and C/EBPδ knockout animals have revealed redundant functions between these two proteins (23, 44, 45). C/EBPβ and C/EBPδ are often coregulated. Both C/EBPβ and C/EBPδ are induced by stimuli such as LPS or IL-1β (14–16), suggesting that they both serve roles in mediating inflammatory responses. Our studies demonstrate that LpqH induces expression of both C/EBPβ and C/EBPδ and their binding to CIITA pI and pIV, suggesting that C/EBPβ and C/EBPδ may both regulate CIITA expression. Our model for redundancy of C/EBPβ and C/EBPδ in control of CIITA expression is consistent with multiple reports of functional redundancy of C/EBPβ and C/EBPδ in other systems, lending further plausibility to the model.
Although LIP has commonly been implicated as a negative regulator of gene transcription, LIP and LAP may function together (and with C/EBPδ) in the regulation of CIITA and other genes. The greater dependence of LIP expression on LpqH and its established negative regulatory roles for other genes suggest that LIP may play an important role in negative regulation of CIITA expression, but even if LIP is central to this inhibition, it may perform this function in heterodimers. LIP forms heterodimers with LAP or C/EBPδ (20, 30). Even in the presence of excess levels of LAP, LAP-LIP heterodimers are favored relative to LAP-LAP or LIP-LIP homodimers (20). In ChIP studies, C/EBPβ binding to CIITA pI and CIITA pIV was induced by LpqH plus IFN-γ, but little or no binding was induced with IFN-γ alone (Fig. 4; a slight increase was observed with IFN-γ alone at CIITA pI only). This assay did not discriminate LAP from LIP (the anti-C/EBPβ Ab binds to both). ChIP studies also showed that C/EBPδ binding to CIITA pI and CIITA pIV was induced by LpqH plus IFN-γ, but not by IFN-γ alone (Fig. 7B), excepting a slight binding of C/EBPδ to CIITA pIV seen with IFN-γ alone. Together, these results suggest importance of LIP or C/EBPδ (possibly as a heterodimer with LAP), because induction of LIP and C/EBPδ expression and binding of C/EBPβ and C/EBPδ to CIITA pIV required LpqH, whereas LAP was expressed constitutively and moderately increased by LpqH. Alternatively, LpqH may induce a posttranslational modification of LAP that increases its binding to CIITA pIV. Therefore, C/EBP proteins may bind and regulate CIITA promoters in the form of LIP-LAP or LIP-C/EBPδ or C/EBPδ-LAP heterodimers, although homodimers of LIP, LAP, or C/EBPδ may contribute, as may heterodimers of LIP, LAP, or C/EBPδ with other transcription factors.
We observed a slight increase of LAP expression (Fig. 2) and its binding in vitro to the biotinylated CIITA pIV-ODN sequence (Fig. 3) in response to IFN-γ, but our ChIP assays consistently showed little or no binding of C/EBPβ to CIITA pI or pIV in living cells in response to IFN-γ alone (Fig. 4). Only in the presence of LpqH did ChIP demonstrate substantial C/EBPβ binding to the CIITA promoters, despite the presence of LAP in both untreated and IFN-γ-stimulated cells. There are several possible explanations for these results. First, MAPK-dependent phosphorylation of C/EBPβ has been shown to increase its binding to target promoters (46–50). Because our previous studies demonstrated that LpqH-mediated inhibition of CIITA is MAPK dependent (13), LpqH may increase both expression and posttranslational modification of C/EBPβ to enhance its activity. Second, C/EBPβ-LIP and C/EBPδ were expressed only after stimulation with LpqH. Therefore, binding may be dependent on a particular composition of C/EBP heterodimers, e.g., LAP-LIP or C/EBPβ-C/EBPδ heterodimers. Third, LpqH-induced binding of C/EBP to CIITA promoters may depend on an unidentified protein that is induced by TLR2 signaling and that acts as a cofactor to recruit C/EBP to CIITA promoters. Regardless of other factors located at the promoter, our data show a clear correlation between the presence of C/EBPβ and δ and inhibition of CIITA transcription.
C/EBPβ may exert an inhibitory effect on CIITA transcription by several different mechanisms. First, LIP may act in its classical role as a dominant-negative inhibitor due to its missing N terminus trans activation domain. However, binding of LAP or C/EBPδ may also inhibit IFN-γ-induced CIITA transcription (similar to the inhibitory effects of these proteins on expression of some other genes), perhaps by interfering with other proteins that must assemble at the promoter to drive gene transcription (possibly a competitive inhibitor function for C/EBP proteins). Alternatively, C/EBP proteins may recruit transcriptional repressor proteins to the CIITA promoters. For example, C/EBPβ has been shown to interact with the histone deacytelase 1, recruiting it to the PPARβ promoter and resulting in transcriptional repression (51). This model fits nicely with our previously published data that LpqH inhibits histone acetylation at the CIITA promoter (13). Future investigations will address these various mechanisms.
Our results suggest a novel regulatory role for C/EBPβ and possibly C/EBPδ in control of CIITA expression in macrophages. Signaling by the TLR2 agonist LpqH induced expression of C/EBPβ and its binding to CIITA pI and pIV. Transfection studies established that C/EBPβ LIP inhibits IFN-γ-induced expression of CIITA in macrophages. Furthermore, LpqH induced C/EBPδ and its binding to CIITA pI and pIV, and C/EBPδ may be functionally redundant with C/EBPβ in regulation of CIITA expression. These data suggest that C/EBPβ and C/EBPδ play novel roles in the negative regulation of CIITA transcription.
Footnotes
This work was supported by National Institutes of Health Grants AI035726 and AI034343 (to C.V.H.) and AI027243 and HL055967 (to W.H.B.).
Abbreviations used in this paper: Mtb, Mycobacterium tuberculosis; CD-RAP, cartilage-derived retinoic acid-sensitive protein; ChIP, chromatin immunoprecipitation; IRF, IFN regulatory factor; LAP, liver-activating protein; LIP, liver inhibitory protein; LpqH, Mtb 19-kDa lipoprotein; ODN, oligodeoxynucleotide; pI, CIITA promoter I; pIV, CIITA promoter IV; TX114, Triton X-114.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme MI. Disseminated tuberculosis in interferon γ gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamijo R, Gerecitano J, Shapiro D, Green SJ, Aguet M, Le J, Vilcek J. Generation of nitric oxide and clearance of interferon-γ after BCG infection are impaired in mice that lack the interferon-γ receptor. J Inflamm. 1995;46:23–31. [PubMed] [Google Scholar]
- 3.Jouanguy E, Lamhamedi-Cherradi S, Lammas D, Dorman SE, Fondaneche MC, Dupuis S, Doffinger R, Altare F, Girdlestone J, Emile JF, et al. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 4.Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, Levin M, Blanche S, Seboun E, Fischer A, Casanova JL. Interferon-γ-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 5.Ting LM, Kim AC, Cattamanchi A, Ernst JD. Mycobacterium tuberculosis inhibits IFN-γ transcriptional responses without inhibiting activation of STAT1. J Immunol. 1999;163:3898–3906. [PubMed] [Google Scholar]
- 6.Pai RK, Pennini ME, Tobian AA, Canaday DH, Boom WH, Harding CV. Prolonged Toll-like receptor signaling by Mycobacterium tuberculosis and its 19-kilodalton lipoprotein inhibits γ interferon-induced regulation of selected genes in macrophages. Infect Immun. 2004;72:6603–6614. doi: 10.1128/IAI.72.11.6603-6614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pai RK, Convery M, Hamilton TA, Boom WH, Harding CV. Inhibition of IFN-γ-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol. 2003;171:175–184. doi: 10.4049/jimmunol.171.1.175. [DOI] [PubMed] [Google Scholar]
- 8.Kincaid EZ, Ernst JD. Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-γ without inhibiting STAT1 function. J Immunol. 2003;171:2042–2049. doi: 10.4049/jimmunol.171.4.2042. [DOI] [PubMed] [Google Scholar]
- 9.Noss EH, Pai RK, Sellati TJ, Radolf JD, Belisle J, Golenbock DT, Boom WH, Harding CV. Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19 kD lipoprotein of Mycobacterium tuberculosis. J Immunol. 2001;167:910–918. doi: 10.4049/jimmunol.167.2.910. [DOI] [PubMed] [Google Scholar]
- 10.Pai RK, Askew D, Boom WH, Harding CV. Regulation of class II MHC expression in APCs: roles of types I, III, and IV class II transactivator. J Immunol. 2002;169:1326–1333. doi: 10.4049/jimmunol.169.3.1326. [DOI] [PubMed] [Google Scholar]
- 11.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagabhushanam V, Solache A, Ting LM, Escaron CJ, Zhang JY, Ernst JD. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-γ. J Immunol. 2003;171:4750–4757. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- 13.Pennini ME, Pai RK, Schultz DC, Boom WH, Harding CV. Mycobacterium tuberculosis 19-kDa lipoprotein inhibits IFN-γ-induced chromatin remodeling of MHC2TA by TLR2 and MAPK signaling. J Immunol. 2006;176:4323–4330. doi: 10.4049/jimmunol.176.7.4323. [DOI] [PubMed] [Google Scholar]
- 14.Alam T, An MR, Papaconstantinou J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J Biol Chem. 1992;267:5021–5024. [PubMed] [Google Scholar]
- 15.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinoshita S, Akira S, Kishimoto T. A member of the C/EBP family, NF-IL6β, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc Natl Acad Sci USA. 1992;89:1473–1476. doi: 10.1073/pnas.89.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Meng Q, Roy SK, Raha A, Zhang J, Hashimoto K, Kalvakolanu DV. A novel transactivating factor that regulates interferon-γ-dependent gene expression. J Biol Chem. 2002;277:30253–30263. doi: 10.1074/jbc.M202679200. [DOI] [PubMed] [Google Scholar]
- 18.Ramana CV, Gil MP, Han Y, Ransohoff RM, Schreiber RD, Stark GR. Stat1-independent regulation of gene expression in response to IFN-γ. Proc Natl Acad Sci USA. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy SK, Wachira SJ, Weihua X, Hu J, Kalvakolanu DV. CCAAT/enhancer-binding protein-β regulates interferon-induced transcription through a novel element. J Biol Chem. 2000;275:12626–12632. doi: 10.1074/jbc.275.17.12626. [DOI] [PubMed] [Google Scholar]
- 20.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 22.Nishio Y, Isshiki H, Kishimoto T, Akira S. A nuclear factor for interleukin-6 expression (NF-IL6) and the glucocorticoid receptor synergistically activate transcription of the rat α1-acid glycoprotein gene via direct protein-protein interaction. Mol Cell Biol. 1993;13:1854–1862. doi: 10.1128/mcb.13.3.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBPβ-deficient mice. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorgoni B, Maritano D, Marthyn P, Righi M, Poli V. C/EBPβ gene inactivation causes both impaired and enhanced gene expression and inverse regulation of IL-12 p40 and p35 mRNAs in macrophages. J Immunol. 2002;168:4055–4062. doi: 10.4049/jimmunol.168.8.4055. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh AK, Bhattacharyya S, Mori Y, Varga J. Inhibition of collagen gene expression by interferon-γ: novel role of the CCAAT/enhancer binding protein β (C/EBPβ) J Cell Physiol. 2006;207:251–260. doi: 10.1002/jcp.20559. [DOI] [PubMed] [Google Scholar]
- 26.Geiger T, Andus T, Klapproth J, Hirano T, Kishimoto T, Heinrich PC. Induction of rat acute-phase proteins by interleukin 6 in vivo. Eur J Immunol. 1988;18:717–721. doi: 10.1002/eji.1830180510. [DOI] [PubMed] [Google Scholar]
- 27.Hu HM, Tian Q, Baer M, Spooner CJ, Williams SC, Johnson PF, Schwartz RC. The C/EBP bZIP domain can mediate lipopolysaccharide induction of the proinflammatory cytokines interleukin-6 and monocyte chemoattractant protein-1. J Biol Chem. 2000;275:16373–16381. doi: 10.1074/jbc.M910269199. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez MA, Åkerblad P, Sigvardsson M, Rosen ED. Critical role of Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCauslin CS, Heath V, Colangelo AM, Malik R, Lee S, Mallei A, Mocchetti I, Johnson PF. CAAT/enhancer-binding protein δ and cAMP-response element-binding protein mediate inducible expression of the nerve growth factor gene in the central nervous system. J Biol Chem. 2006;281:17681–17688. doi: 10.1074/jbc.M600207200. [DOI] [PubMed] [Google Scholar]
- 30.Okazaki K, Li J, Yu H, Fukui N, Sandell LJ. CCAAT/enhancer-binding proteins β and δ mediate the repression of gene transcription of cartilage-derived retinoic acid-sensitive protein induced by interleukin-1β. J Biol Chem. 2002;277:31526–31533. doi: 10.1074/jbc.M202815200. [DOI] [PubMed] [Google Scholar]
- 31.Croniger CM, Millward C, Yang J, Kawai Y, Arinze IJ, Liu S, Harada-Shiba M, Chakravarty K, Friedman JE, Poli V, Hanson RW. Mice with a deletion in the gene for CCAAT/enhancer-binding protein β have an attenuated response to cAMP and impaired carbohydrate metabolism. J Biol Chem. 2001;276:629–638. doi: 10.1074/jbc.M007576200. [DOI] [PubMed] [Google Scholar]
- 32.Sklar MD, Tereba A, Chen BD, Walker WS. Transformation of mouse bone marrow cells by transfection with a human oncogene related to c-myc is associated with the endogenous production of macrophage colony stimulating factor 1. J Cell Physiol. 1985;125:403–412. doi: 10.1002/jcp.1041250307. [DOI] [PubMed] [Google Scholar]
- 33.Chow CW, Clark MP, Rinaldo JE, Chalkley R. Multiple initiators and C/EBP binding sites are involved in transcription from the TATA-less rat XDH/XO basal promoter. Nucleic Acids Res. 1995;23:3132–3140. doi: 10.1093/nar/23.16.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart MJ, Shean ML, Paeper BW, Duester G. The role of CCAAT/enhancer-binding protein in the differential transcriptional regulation of a family of human liver alcohol dehydrogenase genes. J Biol Chem. 1991;266:11594–11603. [PubMed] [Google Scholar]
- 35.Van Ooij C, Snyder RC, Paeper BW, Duester G. Temporal expression of the human alcohol dehydrogenase gene family during liver development correlates with differential promoter activation by hepatocyte nuclear factor 1, CCAAT/enhancer-binding protein α, liver activator protein, and D-element-binding protein. Mol Cell Biol. 1992;12:3023–3031. doi: 10.1128/mcb.12.7.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 37.Ehrt S, Schnappinger D, Bekiranov S, Drenkow J, Shi S, Gingeras TR, Gaasterland T, Schoolnik G, Nathan C. Reprogramming of the macrophage transcriptome in response to interferon-γ and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J Exp Med. 2001;194:1123–1140. doi: 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gehring AJ, Rojas RE, Canaday DH, Lakey DL, Harding CV, Boom WH. The Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits γ interferon-regulated HLA-DR and FcγR1 on human macrophages through Toll-like receptor 2. Infect Immun. 2003;71:4487–4497. doi: 10.1128/IAI.71.8.4487-4497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sibley LD, Adams LB, Krahenbuhl JL. Inhibition of interferon-γ-mediated activation in mouse macrophages treated with lipoarabinomannan. Clin Exp Immunol. 1990;80:141–148. doi: 10.1111/j.1365-2249.1990.tb06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu HM, Baer M, Williams SC, Johnson PF, Schwartz RC. Redundancy of C/EBPα, -β, and -δ in supporting the lipopolysaccharide-induced transcription of IL-6 and monocyte chemoattractant protein-1. J Immunol. 1998;160:2334–2342. [PubMed] [Google Scholar]
- 41.Hungness ES, Pritts TA, Luo GJ, Hershko DD, Robb BW, Hasselgren PO. IL-1β activates C/EBP-β and δ in human enterocytes through a mitogen-activated protein kinase signaling pathway. Int J Biochem Cell Biol. 2002;34:382–395. doi: 10.1016/s1357-2725(01)00129-7. [DOI] [PubMed] [Google Scholar]
- 42.Liu YW, Tseng HP, Chen LC, Chen BK, Chang WC. Functional cooperation of simian virus 40 promoter factor 1 and CCAAT/enhancer-binding protein β and δ in lipopolysaccharide-induced gene activation of IL-10 in mouse macrophages. J Immunol. 2003;171:821–828. doi: 10.4049/jimmunol.171.2.821. [DOI] [PubMed] [Google Scholar]
- 43.Gutierrez S, Javed A, Tennant DK, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 46.Hu J, Roy SK, Shapiro PS, Rodig SR, Reddy SP, Platanias LC, Schreiber RD, Kalvakolanu DV. ERK1 and ERK2 activate CCAAAT/enhancer-binding protein-β-dependent gene transcription in response to interferon-γ. J Biol Chem. 2001;276:287–297. doi: 10.1074/jbc.M004885200. [DOI] [PubMed] [Google Scholar]
- 47.Meng Q, Raha A, Roy S, Hu J, Kalvakolanu DV. IFN-γ-stimulated transcriptional activation by IFN-γ-activated transcriptional element-binding factor 1 occurs via an inducible interaction with CAAAT/enhancer-binding protein-β. J Immunol. 2005;174:6203–6211. doi: 10.4049/jimmunol.174.10.6203. [DOI] [PubMed] [Google Scholar]
- 48.Piwien Pilipuk G, Galigniana MD, Schwartz J. Subnuclear localization of C/EBPβ is regulated by growth hormone and dependent on MAPK. J Biol Chem. 2003;278:35668–35677. doi: 10.1074/jbc.M305182200. [DOI] [PubMed] [Google Scholar]
- 49.Raymond L, Eck S, Mollmark J, Hays E, Tomek I, Kantor S, Elliott S, Vincenti M. Interleukin-1β induction of matrix metalloproteinase-1 transcription in chondrocytes requires ERK-dependent activation of CCAAT enhancer-binding protein-β. J Cell Physiol. 2006;207:683–688. doi: 10.1002/jcp.20608. [DOI] [PubMed] [Google Scholar]
- 50.Roy SK, Hu J, Meng Q, Xia Y, Shapiro PS, Reddy SP, Platanias LC, Lindner DJ, Johnson PF, Pritchard C, et al. MEKK1 plays a critical role in activating the transcription factor C/EBP-β-dependent gene expression in response to IFN-γ. Proc Natl Acad Sci USA. 2002;99:7945–7950. doi: 10.1073/pnas.122075799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di-Poi N, Desvergne B, Michalik L, Wahli W. Transcriptional repression of peroxisome proliferator-activated receptor β/δ in murine keratinocytes by CCAAT/enhancer-binding proteins. J Biol Chem. 2005;280:38700–38710. doi: 10.1074/jbc.M507782200. [DOI] [PubMed] [Google Scholar]