Abstract
Intestinal cells grown in microgravity produce a three-dimensional tissue assembly or “organoid” similar to the human intestinal mucosa, making it an ideal model for enteric infections such as cryptosporidiosis.
Methods
HCT8 cells were grown in a reduced-gravity, low-shear, rotating wall vessel (RWV) system and infected with C. parvum oocysts. Routine and electron microscopy (EM), immunolabelling with fluorescein-labeled Vicia villosa lectin and phycoerythrin-labeled monoclonal antibody to a 15kD surface membrane protein and quantitative polymerase chain reaction (qPCR) using probes for 18s rRNA of C. parvum and HCT8 cells were performed.
Results
The RWV allowed development of columnar epithelium-like structures. Higher magnification revealed well-developed brush-borders at the apical side of the tissue. Incubation with C. parvum resulted to patchy disruption of the epithelium and localized infection with the organism at the surface of several epithelial cells. EM revealed irregular stunting of microvilli, foci of indistinct tight junctions, and areas of loose paracellular spaces. Quantitative PCR showed 1.85 logs (70-fold) progression of infection from 6 to 48 hours of incubation.
Conclusion
The HCT8 organoid displayed morphologic changes indicative of successful and quantifiable infection with C. parvum. The HCT8 organoid culture system may have application in interventional in vitro studies for cryptosporidiosis.
Keywords: Cryptosporidium, organoid, microgravity, in vitro model, cryptosporidiosis, 3D tissue culture
INTRODUCTION
In vitro and in vivo models of cryptosporidial infection have been limited by the lack of close approximation to human intestinal environment and variability in the physiologic responses to the pathogen. Molecular pathogenesis in cryptosporidiosis had been extensively studied in cell monolayers (1–3) as well as in a limited number of animal models such as piglet, calves and neonatal or immunodeficient mice (4–8). However, there is always the question of whether these current models of infection are physiologically relevant to humans. Furthermore, results of interventional studies in vitro may be drastically different and available animal models are cumbersome and again, may not be representative of what actually occurs in humans in vivo. Although human volunteer studies are ideal, the ethical issues involved and the low infectious dose for Cryptosporidium spp., which may be harmful to the unsuspecting immunocompromised hosts, further limit experimental human studies.
An alternative to current models of infection is the use of the low-shear modeled microgravity analogue culture system which produces a three-dimensional tissue assembly or “organoid”. It has been observed by the United States National Aeronautics and Space Agency scientists that cell lines grown during space flight tend to develop native tissue-like structures (9). The low-shear microgravity environment can be simulated in the laboratory by using bioreactors or rotating wall vessels (RWV) which keep cells in suspension during culture. This system subjects the cells to time-averaged gravitational field (vector-averaged gravity) to simulate low gravity conditions, where spatial co-location among and assembly of individual cells into large aggregates can occur (10). Small and large intestinal cells lines that have been cultured in this condition had shown cellular polarity, apical brush borders, intercellular junctions, and basal lamina (11;12). More recently, HCT-8 cell line grown in bioreactors was shown to form organoids with microvilli and desmosomes characteristic of normal tissue (13). Furthermore, cytokeratin, E-cadherin, simplekin, and villin staining patterns were noted to be more similar to human intestinal epithelium in the tissue assemblages grown in bioreactors than in cell monolayers grown in culture plates.
Few bacterial pathogens have been studied in cells grown in bioreactors. Salmonella enterica serovar Typhimurium in Int-407 and HT-29 cells, Pseudomonas aeruginosa in A549 lung epithelial cells, and enteropathogenic and enterohaemorrhagic Escherichia coli in HCT-8 cells are the only published studies as of this writing (13–16). In this paper, we report the first application of the low-shear microgravity culture system for a Category B parasite, Cryptosporidium parvum. We describe the development of the HCT-8 organoid model of cryptosporidiosis, effects of infection to the epithelial cells, and quantification of infection.
METHODS
Cryptosporidium parvum excystation
C. parvum oocysts are in PBS when purchased commercially (Waterborne Inc., New Orleans, USA). The suspended oocysts are then subjected to 20% bleach (final volume) for 10 minutes, then spun down at 3000 rpm for 5 minutes before taking out but 100 μl of supernatant. Hank’s Balanced Salt Solution (Cambrex, MD) was then added to neutralize the bleach. Mixture was mixed thoroughly by vortexing then spun down at 3000 rpm for another 5 minutes. All but 100 μl of supernatant was discarded. These steps were done 4–5 times until the HBSS doesn’t change color. The pellet is then resuspended in HCT8 media prior to infection. Excystation rate is estimated by counting the number of oocysts before and after the procedure using a hemocytometer. The difference in the counts is the number of oocysts that have excysted.
Organoid culture system
Human colonic adenocarcinoma (HCT-8) cells (American Type Culture Collection, Manassas, MD) were cultured in 75 cm2 flasks using RPMI media supplemented with 10% horse serum and 100 u of penicillin-streptomycin. The cells were grown in a CO2 incubator at 37°C and were passed by enzymatic dissociation using Accutase (Chemicon, Billerica MA). The low shear microgravity environment was attained by using the rotary cell culture (RCCS) or rotating wall vessel (RWV) system or bioreactor (Synthecon, Houston Tx). The bioreactor was housed in a humidified incubator with 5% CO2 and with temperature maintained at 37°C. Dehydrated porcine small intestinal submucosa (SIS) grafts (Cook Bioteck, West Lafayette IN), 2 × 3 mm in size, were placed in 50 ml RCCS disposable vessel filled with culture media (RPMI media supplemented with 10% horse serum and Pen/Strep). After 2–4 hours of pre-incubation, the RCCS vessel was seeded with at 105 HCT-8 cells/ml. The initial rotation of the vessel was set at 10 r.p.m. and the speed was adjusted as the cell aggregates grew. The media was changed after 3 days and every 4 days thereafter. Bubbles in the vessels were aspirated as needed. After 8 days of incubation, one set of the organoids (n=4) was transferred to another rotating vessel and inoculated with excysted Cryptosporidium parvum oocysts (Waterborne, LO) at 105 oocysts/ml. One set (n=4) was transferred to another vessel and served as control. All organoids were initially incubated for 6 hours to allow excysted C. parvum to infect the HCT8 cells in the infected vessels. The media was then changed again. After 24 hours of incubation, the organoids were harvested for examination.
To quantify infection rates, another set of organoids were inoculated with excysted C. parvum 8 days after incubation. For comparison, HCT-8 monolayers (80–90% confluent) in culture plates, were also infected with C. parvum. Media was replaced after 6 hours and the HCT-8 cells were reincubated and harvested at 2, 6, 12, 24, and 48 hours post-infection. In another set of experiments aimed at quantifying infection rates at extended period of time, organoids (4 per timepoint) were harvested at 0, 24, 48, 72 and 96 hours post-infection. A corresponding set of uninfected organoids (4 per timepoint) were harvested as control. In this set-up, 10 ml RCCS vessels were used and seeded with HCT8 or C. parvum oocysts at the same concentrations as above.
Light microscopy
Upon harvest, all organoids were transferred to tissue cassettes and fixed in 4% paraformaldehyde. After 2 hours, they were immersed in 70% ethanol until embedded in paraffin blocks for thin sections. Organoid sections were stained with hematoxylin-eosin and Giemsa and examined under low, high, and oil immersion power magnification. Some slides were left unstained for immunolabelling.
Immunolabelling
Unstained sections were deparaffinized by 2 successive washings in xylene for 5 minutes each. The sections were then immersed in decreasing concentrations (100%, 95% and 75%) of ethanol for 2–5 minutes each and rinsed in deionized water. This was followed by incubation in 0.1% Triton x-100 for 20 minutes and blocking with 1% bovine serum albumin for 30 minutes. A monoclonal antibody against the 15kD membrane protein of intracellular C. parvum (Biodesign International, Saco ME) was labeled with phycoerythrin according to the manufacturer’s instructions (Zenon rabbit IgG labeling kit; Invitrogen, Carisbad CA) and placed on the slides containing the organoid sections. After 60 minutes, the slide was washed with PBS and incubated in fluorescein-labeled Vicia villosa lectin (Vector Laboratories, Burlingame CA) specific to C. parvum sporozoites. After fixation in 4% paraformaldehyde, the stained organoid slides were examined under epifluorescent microscope.
Electron microscopy
Uninfected and infected organoids for electron microscopy were transferred to tissue cassettes and fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3–7.4). Further processing was performed at the Advanced Microscopy Section of the University of Virginia. Post fixation was done with 1% osmium tetroxide for 1 hour. The samples were dehydrated with acetone and infiltrated with epoxy resin. When infiltrated with 100% epoxy resin was complete the samples were baked at 60°C for 48 hours to harden. Half a micron and 60 nanometer sectioned were cut on a Leica Ultracut E ultramicrotome. The thin sections were examined with the microscopist with a JEOL 1230 Transmission Electron Microscope.
Real-time PCR
DNA extraction was done on infected and uninfected HCT8 cells and pure C. parvum oocysts following the manufacturer’s procedure (Qiagen DNeasy® Tissue Kit, Valencia CA). Organoids were harvested and placed in an Eppendorf tube containing 100 μl of Accutase. Twenty μl of proteinase K, followed by 200 μl of Buffer AL was then added. The mixture was mixed and placed in a 70°C water bath for 10 minutes. Two hundred μl of 100% ethanol was added and the mixture was filtered at 8000 rpm for 1 minute. The filter was then washed twice with buffer, first at 8000 rpm for 1 minute and the second at 14000 rpm for 3 minutes. Recovery of the DNA was done with 200 μl of elution buffer spun at 8000 rpm for 1 minute. Quantification of infection was performed by real-time PCR (BioRad iCycler, Hercules CA) using primers for the 18s rRNA gene of C. parvum (Cp18SR931 [5′-CTGCGAATGGCTCATTATAACA-3′], Accession X64341) and 18s rRNA gene of HCT8 cells (housekeeping gene) [5′-GGTTCGAAGACGATCAGA-3′], Accession NR_003286) as described previously (17). The reaction mixture was composed of 12.5 μl of BioRad iQ™ SYBR® Green Supermix, 5.5 μl of Fisher nuclease-free water, and 1 μl each of the forward and reverse primers for every sample. Two reaction mixtures were prepared, one for C. parvum detection, the other for HCT8 cells detection. Twenty μl of the reaction mixture was placed at the bottom of the wells of a 96-well PCR plate. Five μl of DNA sample was then added each well. Amplification consisted of 15 min at 95°C followed by 40 cycles of 15 sec at 95°C, 30 sec at 60°C, and 30 sec at 70°C. Amplification and detection were performed using the iCycler real-time detection system (Bio-Rad). Fluorescence was measured during the annealing step of each cycle. Cycle numbers of each run was compared to a standard curve of known amounts of Cryptosporidium DNA and converted into parasite count per HCT8 cell.
RESULTS
The HCT8 organoid
Incubation of HCT-8 cells in RWV allowed development of stratified cuboidal to columnar epithelium-like structures around the bio-scaffold (Figure 1). In some areas, the epithelium folded and formed villus-like structures with loose cells underneath the organized layer of cell aggregates. The outermost layer of cells showed polarization, with the nuclei tending to be basally oriented. At higher magnification, these cells revealed well-developed brush-borders at the apical side of the tissue. Electron microscopy showed well-developed microvilli, tight junctions and other intercellular junctional apparatus (Figure 3).
Figure 1.
Low power microscopy of HCT-8 cells organoid showing villus-like epithelial folds around the bioscaffold (A). At high power magnification, the HCT-8 cells formed layers of cells with the outermost layer showing polarization of cells and development of apical brush borders. Underneath the HCT-8 cell layers were loosely organized undifferentiated HCT-8 cells (B). Infected organoid showing localized infection and detachment of both uninfected and infected cells in the affected area (C). Oil power magnification showed disruption of apical brush borders (arrows) and looseness of intercellular attachments (arrowheads) (D). There were several detached cells, including possibly apoptotic cells, within the vicinity.
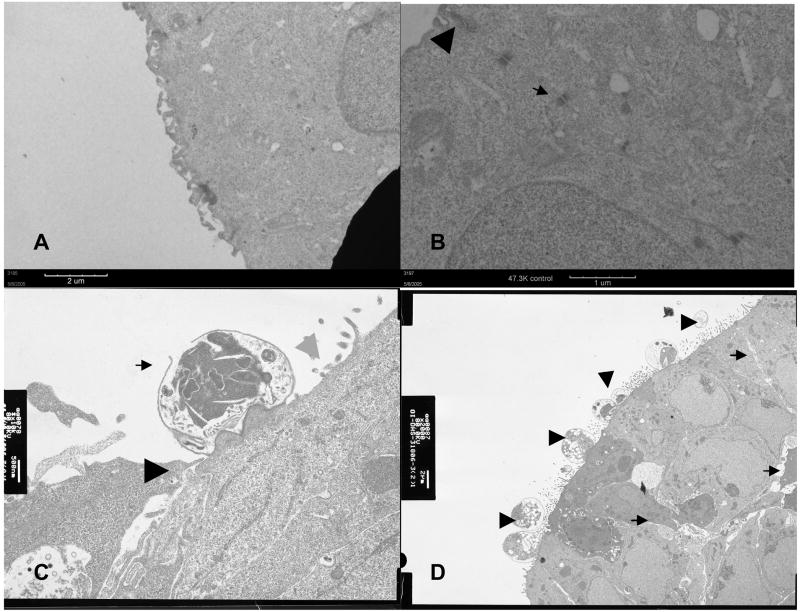
Figure 3.
Electron micrographs of uninfected organoids showed well-developed microvilli (Panel A) and distinct junctional apparatuses, including tight junction (arrowhead) and desmosomes (arrow) (Panel B). Panel C shows infected organoids with a meront (arrow), probably in the process of releasing merozoites, indistinct tight junction (black arrowhead) and denudation of the apical surface (gray arrowhead). Panel D shows an area of multiple infection with C. parvum at different stages of development (arrowheads) and loosening of the paracellular spaces (arrows). Of note, the tight junctions and desmosomes became inapparent in the infected organoids.
C. parvum-infected HCT8 organoids
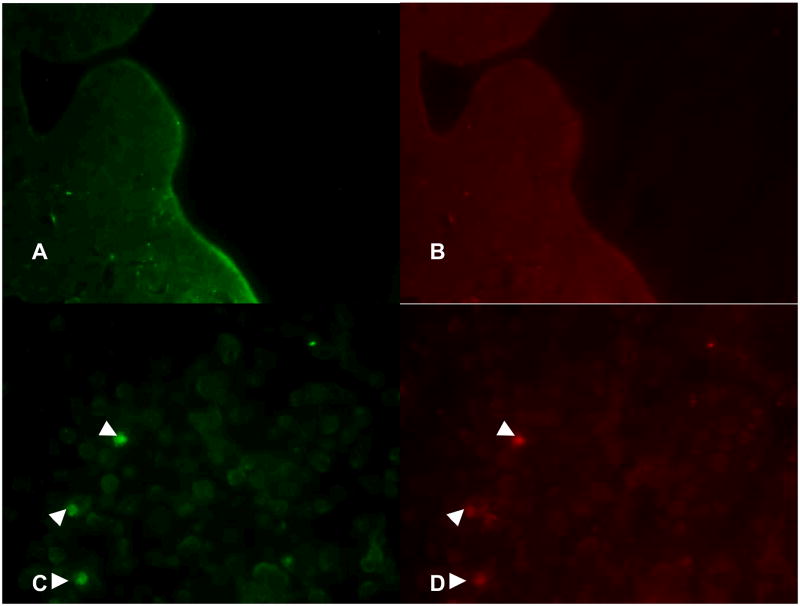
Incubation with C. parvum resulted to patchy disruption of the epithelium interrupted by areas of intact epithelium (Figure 1). There was localized infection with trophozoites at the surface of several epithelial cells. Some infected and uninfected cells were seen detached from the affected epithelium. Immunolabelling confirmed intracellular localization of C. parvum in infected cells (Figure 2). Light microscopy and immunolabelling also demonstrated the organism at different stages of its life cycle. Parasitophorous vacuoles were evident in electron microscopy (Figure 3). Transmission electron micrograph also revealed irregular stunting of microvilli seen even in some uninfected adjacent cells. In contrast to uninfected controls, there were foci of indistinct tight junctions, and areas of loose paracellular spaces in infected organoids. Junctional apparatus such as desmosome and zona adherens became inapparent in most organoid sections.
Figure 2.
Fluorescent imaging of infected and uninfected HCT-8 cells. Panels A and B show uninfected cells stained with fluorescein-labeled Vicia villosa lectin specific to intracellular C. parvum sporozoites and phycoerythrin-labeled monoclonal antibody against the 15 kD membrane protein of C. parvum, respectively. Panels C and D showed intracellular C. parvum-positive cells (arrowheads) in the infected organoids.
Quantification of infection
The excystation rates of the of C. parvum used in these experiments were approximately 30–35%. The inocula of oocysts were incubated in the RCCS vessels for 6 hours to allow excysted oocysts to attach and infect the HCT-8 cells. Unattached sporozoites are removed by changing the media. Using pure oocysts as standard and HCT-8 cell housekeeping gene, real-time PCR showed 1.85 logs (70-fold) progression of infection from 6 to 48 hours of incubation for the organoids, while only a 1.04 logs (10.7-fold) progression for the monolayers. However, the infection rates were higher in the monolayers than the organoids. At 6, 12, 24, and 48 hours post-infection, the infection rates were 0.040, 0.266, 0.498, and 2.848 in the organoids compared with 0.464, 0.351, 1.072, and 4.977 in the monolayers (Figure 4A-C). Peak of infection appeared to be at 48 hours during extended period of incubation (initial parasite count of 22 at 0 hour to 247 at 48 hours). Interestingly, infection was decreased to baseline by 72 and to 0 by 96 hours possibly because of apoptosis of infected HCT-8 cells and detachment of even viable cells during infection. PCR of supernatant at 96 hours revealed 933 detectable C. parvum suggesting continued viability of the organism but release to the surrounding media due to the friability and cell death occurring in the infected organoids. Uninfected organoids had HCT-8 cell counts of 1.9 × 105 at 0, 8 × 105 at 24, 2.3 × 105 at 48 and 2 × 105 at 96 hours. No cryptosporidial DNA was detected in uninfected organoids.
Figure 4.
Comparison of infection rates between HCT-8 organoids and monolayers using quantitative PCR with C. parvum and HCT-8 cell probes. The organoids showed a steady increase in infection from 6 to 48 hours. The infection rates were higher in monolayers (Panel B) than in organoids (Panel A) but the rate of increase was more pronounced in the organoids than in the monolayers. On extended incubation, after the peak of infection at 48 hours, a steady decrease in parasite burden is noted starting at 76 hours post-infection (Panel C). Decrease in HCT-8 cells occurred at 96 hours suggesting cell detachment from the bioscaffold in infected organoids compared with the more stable amount of cells in the intact, uninfected organoids.
DISCUSSION
The development of HCT-8 organoid using RCCS was successfully replicated and applied for the first time in protozoal infection in this study. HCT-8 cells formed layers of cells with the outermost layer similar to the intestinal epithelium in humans and clearly showed well-developed microvilli and intercellular junctions. It has been previously reported that villin, a cytoskeletal protein of the intestinal epithelial microvilli, was expressed predominantly in the apical surface of the organoids consistent with normal human small intestine (13). Furthermore, tight junction proteins, E-cadherin, ZO-1, and simplekin were also shown to follow the patterns seen in normal human tissue more in the organoids than in monolayers. These findings suggest that the HCT-8 organoid may be an ideal model for cryptosporidiosis, a parasitic infection that causes both malabsorption and increased intestinal permeability (1;18;19). Indeed, the C. parvum-infected organoids showed blunting of microvilli and disruption of tight junctions consistent with what has been described in biopsy specimens of infected patients (20–22).
Although, there was no attempt to quantify degree of epithelial cell injury, infection rate was measured with real-time PCR using pure oocyst and HCT-8 housekeeping gene for standardization. The use of housekeeping gene was critical as both organoids and monolayers have variable amounts of host cells. When compared with monolayers, the proportion of infected cells was lower in the organoids, but the rate of increase of infection was higher and more persistent in the organoids than in the monolayers. This finding may be consistent with what has been observed in 3-D assemblages of Int-407 cells and HT-29 cells infected with Salmonella Typhimurium where invasion was noted to be decreased (15;16). Similarly, P. aeruginosa was observed to attach and penetrate 3-D aggregates of A549 lung epithelial cells significantly less than monolayers and this was attributed to the enhanced formation of tight junction complexes (14). These observations may suggest that higher degree of cellular differentiation in the organoids offers greater resistance to invading pathogens.
The ability to identify epithelial injury—in terms of denuded microvilli and junctional apparatus abnormalities, and to quantify infection, opens the possibility of applying the model to preventive and therapeutic interventional studies. We hypothesize that the physiologic response to drugs, vaccines and micronutrients may be more akin to the human intestinal cell response in vivo because as discussed above, the rotatory culture system led to the development of features more similar to human tissue. Indeed, primary cell lines cultivated in the simulated microgravity condition maintain key metabolic functions and probably respond to pathogens in the same manner as in vivo (23;24). Moreover, the low shear, low turbulence environment of microgravity analogue culture is similar to that found in certain areas of the body like in utero and in the protected environment between the brush border microvilli of epithelial cells in human (25–28). The later environment is relevant to that encountered by microbial pathogens and commensals during their life cycles in the gastrointestinal, respiratory, and urogenital tracts. In this setting, the pathogen’s response during infection including gene expression, physiology, and pathogenesis seem to be affected by these mechanical forces (29–32). Whether these findings could be generalized to all bacteria or to other pathogens, e.g. protozoa such as Cryptosporidium, warrants further investigations.
In summary, we have successfully developed an organoid model of cryptosporidial infection. The HCT8 organoid displayed morphologic changes indicative of successful and quantifiable infection with C. parvum. Because of its closer approximation to the human intestinal morphologic and physiologic environment, the low shear microgravity cell culture system may be a significant tool in assessing human intestinal epithelial cell response to infection and evaluation of potential targets for protective and therapeutic interventions in cryptosporidiosis.
Acknowledgments
Funding for this study was provided by the Middle Atlantic Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research. This study has been presented at the 44th Annual Meeting of the Infectious Disease Society of America (Abstract No. 352); Toronto, Ontario, Canada; October 2006.
Funding for this study was provided by the Middle Atlantic Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research. We thank the Advanced Microscopy Facility, Research Histology Core, and Digestive Health Research Center Morphology/Imaging Core of University of Virginia for technical assistance.
Footnotes
All authors do not have any commercial or other association that might pose a conflict of interest.
Reference List
- 1.Adams RB, Guerrant RL, Zu S, Fang G, Roche JK. Cryptosporidium parvum infection of intestinal epithelium: morphologic and functional studies in an in vitro model. J Infect Dis. 1994 Jan;169(1):170–7. doi: 10.1093/infdis/169.1.170. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths JK, Moore R, Dooley S, Keusch GT, Tzipori S. Cryptosporidium parvum infection of Caco-2 cell monolayers induces an apical monolayer defect, selectively increases transmonolayer permeability, and causes epithelial cell death. Infect Immun. 1994 Oct;62(10):4506–14. doi: 10.1128/iai.62.10.4506-4514.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Upton SJ, Tilley M, Brillhart DB. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol Lett. 1994 May 15;118(3):233–6. doi: 10.1111/j.1574-6968.1994.tb06833.x. [DOI] [PubMed] [Google Scholar]
- 4.Laurent F, McCole D, Eckmann L, Kagnoff MF. Pathogenesis of Cryptosporidium parvum infection. Microbes Infect. 1999 Feb;1(2):141–8. doi: 10.1016/s1286-4579(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 5.Seydel KB, Zhang T, Champion GA, et al. Cryptosporidium parvum infection of human intestinal xenografts in SCID mice induces production of human tumor necrosis factor alpha and interleukin-8. Infect Immun. 1998 May;66(5):2379–82. doi: 10.1128/iai.66.5.2379-2382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argenzio RA, Lecce J, Powell DW. Prostanoids inhibit intestinal NaCl absorption in experimental porcine cryptosporidiosis. Gastroenterology. 1993 Feb;104(2):440–7. doi: 10.1016/0016-5085(93)90412-6. [DOI] [PubMed] [Google Scholar]
- 7.Argenzio RA, Armstrong M, Rhoads JM. Role of the enteric nervous system in piglet cryptosporidiosis. J Pharmacol Exp Ther. 1996 Dec;279(3):1109–15. [PubMed] [Google Scholar]
- 8.Abrahamsen MS, Lancto CA, Walcheck B, Layton W, Jutila MA. Localization of alpha/beta and gamma/delta T lymphocytes in Cryptosporidium parvum-infected tissues in naive and immune calves. Infect Immun. 1997 Jun;65(6):2428–33. doi: 10.1128/iai.65.6.2428-2433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unsworth BR, Lelkes PI. Growing tissues in microgravity. Nat Med. 1998 Aug;4(8):901–7. doi: 10.1038/nm0898-901. [DOI] [PubMed] [Google Scholar]
- 10.Hammond TG, Hammond JM. Optimized suspension culture: the rotating-wall vessel. Am J Physiol Renal Physiol. 2001 Jul;281(1):F12–F25. doi: 10.1152/ajprenal.2001.281.1.F12. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin TJ, Jessup JM, Wolf DA. Morphologic differentiation of colon carcinoma cell lines HT-29 and HT-29KM in rotating-wall vessels. In Vitro Cell Dev Biol. 1992 Jan;28A(1):47–60. doi: 10.1007/BF02631079. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin TJ, Schroeder WF, Wolf DA, Moyer MP. Rotating-wall vessel coculture of small intestine as a prelude to tissue modeling: aspects of simulated microgravity. Proc Soc Exp Biol Med. 1993 Feb;202(2):181–92. doi: 10.3181/00379727-202-43525. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho HM, Teel LD, Goping G, O’Brien AD. A three-dimensional tissue culture model for the study of attach and efface lesion formation by enteropathogenic and enterohaemorrhagic Escherichia coli. Cell Microbiol. 2005 Dec;7(12):1771–81. doi: 10.1111/j.1462-5822.2004.00594.x. [DOI] [PubMed] [Google Scholar]
- 14.Carterson AJ, Honer zu BK, Ott CM, et al. A549 lung epithelial cells grown as three-dimensional aggregates: alternative tissue culture model for Pseudomonas aeruginosa pathogenesis. Infect Immun. 2005 Feb;73(2):1129–40. doi: 10.1128/IAI.73.2.1129-1140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honer zu BK, Ramamurthy R, Ott CM, et al. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes Infect. 2006 Jun;8(7):1813–25. doi: 10.1016/j.micinf.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Nickerson CA, Goodwin TJ, Terlonge J, et al. Three-dimensional tissue assemblies: novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun. 2001 Nov;69(11):7106–20. doi: 10.1128/IAI.69.11.7106-7120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parr JB, Sevilleja JE, Amidou S, et al. Detection and quantification of Cryptosporidium in HCT-8 cells and human fecal specimens using real-time polymerase chain reaction. Am J Trop Med Hyg. 2007 May;76(5):938–42. [PMC free article] [PubMed] [Google Scholar]
- 18.Goodgame RW, Kimball K, Ou CN, et al. Intestinal function and injury in acquired immunodeficiency syndrome-related cryptosporidiosis. Gastroenterology. 1995 Apr;108(4):1075–82. doi: 10.1016/0016-5085(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 19.Lima AA, Silva TM, Gifoni AM, et al. Mucosal injury and disruption of intestinal barrier function in HIV-infected individuals with and without diarrhea and cryptosporidiosis in northeast Brazil. Am J Gastroenterol. 1997 Oct;92(10):1861–6. [PubMed] [Google Scholar]
- 20.Genta RM, Chappell CL, White AC, Jr, Kimball KT, Goodgame RW. Duodenal morphology and intensity of infection in AIDS-related intestinal cryptosporidiosis. Gastroenterology. 1993 Dec;105(6):1769–75. doi: 10.1016/0016-5085(93)91075-s. [DOI] [PubMed] [Google Scholar]
- 21.Lumadue JA, Manabe YC, Moore RD, Belitsos PC, Sears CL, Clark DP. A clinicopathologic analysis of AIDS-related cryptosporidiosis. AIDS. 1998 Dec 24;12(18):2459–66. doi: 10.1097/00002030-199818000-00015. [DOI] [PubMed] [Google Scholar]
- 22.Orenstein JM. Cryptosporidiosis. In: Connor DH, Chandler FW, Manz HJ, Schwartz DA, Lack EE, editors. Pathology of Infectious Diseases. Stamford, CT: Appleton & Lange; 1997. pp. 1147–68. [Google Scholar]
- 23.Dabos KJ, Nelson LJ, Bradnock TJ, et al. The simulated microgravity environment maintains key metabolic functions and promotes aggregation of primary porcine hepatocytes. Biochim Biophys Acta. 2001 May 3;1526(2):119–30. doi: 10.1016/s0304-4165(01)00097-6. [DOI] [PubMed] [Google Scholar]
- 24.Margolis LB, Fitzgerald W, Glushakova S, et al. Lymphocyte trafficking and HIV infection of human lymphoid tissue in a rotating wall vessel bioreactor. AIDS Res Hum Retroviruses. 1997 Nov 1;13(16):1411–20. doi: 10.1089/aid.1997.13.1411. [DOI] [PubMed] [Google Scholar]
- 25.Beeson JG, Rogerson SJ, Cooke BM, et al. Adhesion of Plasmodium falciparum-infected erythrocytes to hyaluronic acid in placental malaria. Nat Med. 2000 Jan;6(1):86–90. doi: 10.1038/71582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo P, Weinstein AM, Weinbaum S. A hydrodynamic mechanosensory hypothesis for brush border microvilli. Am J Physiol Renal Physiol. 2000 Oct;279(4):F698–F712. doi: 10.1152/ajprenal.2000.279.4.F698. [DOI] [PubMed] [Google Scholar]
- 27.Stock UA, Vacanti JP. Cardiovascular physiology during fetal development and implications for tissue engineering. Tissue Eng. 2001 Feb;7(1):1–7. doi: 10.1089/107632701300003241. [DOI] [PubMed] [Google Scholar]
- 28.Cai Z, Xin J, Pollock DM, Pollock JS. Shear stress-mediated NO production in inner medullary collecting duct cells. Am J Physiol Renal Physiol. 2000 Aug;279(2):F270–F274. doi: 10.1152/ajprenal.2000.279.2.F270. [DOI] [PubMed] [Google Scholar]
- 29.Nickerson CA, Ott CM, Wilson JW, et al. Low-shear modeled microgravity: a global environmental regulatory signal affecting bacterial gene expression, physiology, and pathogenesis. J Microbiol Methods. 2003 Jul;54(1):1–11. doi: 10.1016/s0167-7012(03)00018-6. [DOI] [PubMed] [Google Scholar]
- 30.Hammond TG, Benes E, O’Reilly KC, et al. Mechanical culture conditions effect gene expression: gravity-induced changes on the space shuttle. Physiol Genomics. 2000 Sep 8;3(3):163–73. doi: 10.1152/physiolgenomics.2000.3.3.163. [DOI] [PubMed] [Google Scholar]
- 31.Johanson K, Allen PL, Lewis F, Cubano LA, Hyman LE, Hammond TG. Saccharomyces cerevisiae gene expression changes during rotating wall vessel suspension culture. J Appl Physiol. 2002 Dec;93(6):2171–80. doi: 10.1152/japplphysiol.01087.2001. [DOI] [PubMed] [Google Scholar]
- 32.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. Microbial responses to microgravity and other low-shear environments. Microbiol Mol Biol Rev. 2004 Jun;68(2):345–61. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]