Abstract
Endothelial cell dysfunction is associated with bioavailable nitric oxide deficiency and an excessive generation of reactive oxygen species. We modeled this condition by chronically inhibiting nitric oxide generation with subpressor doses of NG-monomethyl-l-arginine (L-NMMA) in C57B6 and Tie-2/green fluorescent protein mouse strains. L-NMMA-treated mice exhibited a slight reduction in vasorelaxation ability, as well as detectable abnormalities in soluble adhesion molecules (soluble intercellular adhesion molecule-1 and vascular cellular adhesion molecule-1, and matrix metalloproteinase 9), which represent surrogate indicators of endothelial dysfunction. Proteomic analysis of the isolated microvasculature using 2-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy revealed abnormal expression of a cluster of mitochondrial enzymes, which was confirmed using immunodetection. Aconitase-2 and enoyl-CoA-hydratase-1 expression levels were decreased in L-NMMA-treated animals; this phenotype was absent in nitric oxide synthase-1 and -3 knockout mice. Depletion of aconitase-2 and enoyl-CoA-hydratase-1 resulted in the inhibition of the Krebs cycle and enhanced pyruvate shunting toward the glycolytic pathway. To assess mitochondrial mass in vivo, co-localization of green fluorescent protein and MitoTracker fluorescence was detected by intravital microscopy. Quantitative analysis of fluorescence intensity showed that L-NMMA-treated animals exhibited lower fluorescence of MitoTracker in microvascular endothelia as a result of reduced mitochondrial mass. These findings provide conclusive and unbiased evidence that mitochondriopathy represents an early manifestation of endothelial dysfunction, shifting cell metabolism toward “metabolic hypoxia” through the selective depletion of both aconitase-2 and enoyl-CoA-hydratase-1. These findings may contribute to an early preclinical diagnosis of endothelial dysfunction.
An intense search for early markers of impending maladies is being conducted to gain insights into the mechanisms of progression and disease prevention. Specifically, in the era of “global risk assessment,” the genome-wide and proteomic screening of cardiovascular disease is intended to supplement the existing classical risk scores, such as Framingham and its modifications.1,2 In addition, traditional systems of risk assessment rely heavily on clinical presentations and provide3 little if any insights into the molecular profile of early disease processes.
Dozens of intuitive candidate markers of cardiovascular vulnerability, including serological, structural and functional alterations in the vasculature and myocardium, have been proposed based on the current concepts of evolution of atherogenesis.4 Some of these markers offer molecular insights into the pathogenesis of cardiovascular disease, but the preconceived character of these biomarkers limits their usefulness. For these reasons, an unbiased search for the markers of cardiovascular disease is gaining recognition as a potentially valuable tool to disclose hidden molecular pathology.
Analysis of gene expression in a model of atherosclerosis (apolipoprotein E knockout mice) has identified multiple gene families participating in the progression of aortic lesions.5 Similarly, the consequences of pericyte deficiency were examined in brain microvessels by microarray analysis in platelet derived growth factor-B-deficient mice.6 In a study of human coronary artery segments isolated from explanted hearts of cardiac transplant patients, King et al7 identified multiple effected gene pathways and further confirmed the microarray findings by immunohistochemical analysis of selected proteins. Downstream gene targets of nitric oxide (NO) have been detected by Bogdan et al8 however, no studies have been conducted to elucidate the molecular consequences of nitric oxide deficiency.
The need for such screening is most relevant to the pathogenesis of endothelial dysfunction, which, regardless of the initiating mechanisms, is associated with the deficiency of bioavailable NO and excessive generation of reactive oxygen species.9 Endothelial nitric oxide synthase (NOS)-deficient mouse model represents an ultimate lack of bioavailable NO, but it also lacks the concomitant generation of superoxide by the uncoupled enzyme. The strategy we elected was to chronically inhibit NO generation by subpressor doses of NG-monomethyl-l-arginine (L-NMMA),10 and to analyze the differential proteome of isolated microvasculature. By using difference gel electrophoresis (DIGE), we found a cluster of differentially expressed mitochondrial proteins, among others, that lead to the pre-clinical development of defective Krebs cycle and mitochondrial biogenesis.
Materials and Methods
Reagents and Antibodies
The following antibodies were used: anti-Aconitase-2 (ABGEN AP1936, San Diego CA), anti-acetyl-CoA-acetyltransferase-1(ACAT-1; Cayman 100028, Ann Arbor MI), anti-enoyl-coenzyme A hydratase (ECHS-1; ProteinTech Group, Inc. 11305-1-AP, Chicago, IL), anti-ATP synthase (Affinity BioReagents MA1-930, Golden, CO), anti-Glutathione Peroxidase 3 (Abcam ab-27325, Cambridge, MA), anti-Annexin V (abcam ab14196, Cambridge, MA), anti-nitrosotyrosine (Millipore 05-233, Billerica, MA), anti-eNOS (BD 610297, San Jose, CA), anti-CD31 (sc-1506, Santa Cruz, CA), and anti-α-smooth muscle actin (Dako Cytomation 1A4 M0851, Glostrup Denmark). Monoclonal antibodies to β-actin were obtained from Sigma (St. Louis, MO). L-NMMA, NG-monomethyl-d-arginine monoacetate (D-NNMA), and NG, NG-dimethyl-l-arginine dihydrochloride (ADMA) were obtained from Alexis Biochemicals (San Diego, CA). MitoSox Red Mitochondrial Superoxide indicator (M22425) and MitoTracker (M36008) were bought from Invitrogen (Eugene, OR). All not mentioned chemicals have been purchased from Sigma.
Animal Studies
The animal study protocol was in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee. Experiments were performed in 20 12-week-old male C57Bl/6J mice, 8 NOS1 knock-out (B6;129S4-Nos1tm1Plh/J), 8 NOS3 knock-out (B6.129P2-Nos3tm1Unc/J), and 8 Tie-2/green fluorescent protein (GFP) mice obtained from Jackson Laboratory (Bar Harbor, ME). Animals were housed under a 12 hours light-dark cycle with free access to food and water. The study protocol was in conformity with the ethical guidelines of New York Medical College Care and Use committee and NIH guidelines for animal use. Animals received 0.3 mg/ml L-NMMA in drinking water, which was changed daily. Blood pressure was monitored regularly, at 2-week intervals, to detect early signs of developing hypertension. Twenty-four hour urine sampling was performed at 1 and 2 months for each group. At the time of sacrifice, mice were anesthetized with a combination of ketamine plus xylazine (2 μg/g and 23 μg/g body weight, respectively), the blood drawn via heart puncture and animals and the abdominal aorta was cannulated with ligatures placed above and below the renal arteries. The kidneys were perfused with 5 ml of low Ca2+ PSS solution (0.1 mmol/L CaCl2, 125.0 mmol/L NaCl, 5.0 mmol/L KCl, 1.0 mmol/L MgCl2, 10.0 mmol/L glucose, and 20.0 mmol/L HEPES), and 6% bovine serum albumin. Subsequently, the vasculature was stained by perfusing 2 ml of PSS, 6% bovine serum albumin, 1% Evan’s Blue. The kidneys were excised and microvasculature isolated ex tempore.
In some experiments, the descending thoracic aorta from C57B1/6J mice, treated or not treated with L-NMMA, was segmented into cylindrical segments that were mounted on a wire-myograph containing Krebs buffer gassed with 95% O2, 5% CO2, for recording of isometric tension11; the vessels were preconstricted with phenylephrine to 70% of maximal response and used for assessment of acetylcholine (0.001 to 100 μmol/L)-induced vasorelaxation in the absence and presence of Tempol (1 mmol/L), a superoxide dismutase mimetic.
Serum samples were examined for creatinine concentration (Cayman, Ann Arbor MI) and cytokines (multiplex biomarker analysis using Linco Inc, St. Louis, MO). For the determination of plasma l-lactate, the Roche Diagnostic assay was used (Roche Hitachi Modular P800 analyzer, Roche Diagnostics, Indianapolis, IN). The method uses an enzymatic reaction (microbial lactate oxidase) to convert lactate to pyruvate. The hydrogen peroxide produced in this reaction is then used in a horseradish peroxidase enzymatic color reaction the intensity of which is proportional to the l -lactate concentration. Urine albumin and creatinine concentrations were measured using enzyme-linked immunosorbent assay and creatinine kit (Cayman, Ann Arbor, MI) to obtain the protein:creatinine ratio.
The multiplex mouse cardiovascular diseases biomarkers panels 1 and the 13 Cytokines/Chemokines panel were used (MCVD1-67AK 4 plex, MCyt/Chem 13Plex, Linco Inc, St. Louis, MO) for the simultaneous quantification of the following analytes: soluble E-Selectin (sE-Selectin), soluble intercellular adhesion molecule-1 (sICAM-1), soluble vascular cellular adhesion molecule-1 (sVCAM), matrix metalloproteinase-9 (MMP-9), and 13 soluble cytokines (macrophage inflammatory protein [MIP-1α], GMCFS, monocyte chemotactic protein [MCP1], KC, RANTES, interferon [IFN]γ, interleukin [IL]1β, IL1α, granulocyte colony-stimulating factor [GCSF], inositol phosphate [IP]10, IL-10, tumor necrosis factor [TNF]-α). All examined analytes had been tested individually and in combination to ensure that there were no cross-reactions. Briefly, the multibiomarkers and cytokine standards were resuspended in the assay buffer and then differentially serially diluted. Twenty-five μl of standard, quality controls, or sample were added to each well of a 96-well plate with 25 μl of the bead solution. Each plate was sealed, covered with aluminum foil and incubated overnight (16 to 18 hours) with agitation on a shaker platform at 4°C. The plates were washed twice with 200 μl/well of wash buffer, removing buffer by vacuum filtration between each wash. This was followed by addition of 25 μl of a detection antibody cocktail into each well and incubation at room temperature for 1.5 hours. Streptavidin-phycoerythrin solution (25 μl) was added to each well and incubated at room temperature for 30 minutes. The multianalyte composition was then analyzed using a LuminexIS100 analyzer (Luminex Inc, Austin TX). The data were evaluated as median fluorescence intensity using appropriate curve-fitting software (Luminex 100IS software version 2.3). A five-parameter logistic method with weighting was used. All measurements were performed in duplicate.
Isolation of Renal Microvasculature
Renal microvasculature was isolated according to the previously published technique.12 Briefly, kidneys were removed, decapsulated, and gently passed through a sieve (75 μm) into an ice-cold container with ice-cold low Ca2+ PSS solution. The retained vessels were selected, freed of any tubulointerstitial elements, and isolated from the rest of the retentate under a stereomicroscope.
2-D DIGE of Microvascular Preparations
The freshly collected microdissected vessels were washed (10 mmol/L Tris-HCl, 5 mmol/L magnesium acetate, pH 8.0) thrice to remove the traces of blood and homogenized in a lysis buffer (for 10 mg of tissue we added 200 μl 2-D cell lysis buffer of the following composition: 30 mmol/L Tris-HCl, pH 8.8, containing 7 mol/L urea, 2 mol/L thiourea, and 4% 3-[(3-cholamidopropyl) dimethylammonio] propanesulfonate [CHAPS]). The tissue was subjected to sonication at 4°C, followed by agitation on a shaker for 30 minutes at room temperature. Samples were centrifuged for 30 minutes at 16,000 rpm. The supernatants were collected and the protein concentration measured using Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). To label proteins (analytical gel), 30 μg of each protein samples were incubated with 0.7 μl of diluted CyDye (1:5 diluted in dimethylformamide from 1 nmol/μstock, GE Health care, Piscataway, NJ) at 4°C for 30 minutes. The labeling was stopped by adding 0.7 μl of 10 mmol/L l-Lysine and incubating at 4°C for 15 minutes. The labeled samples were mixed together, and equal volume of 2 × 2-D sample buffer (8 mol/L urea, 4% CHAPS, 20 mg/ml dithiothreitol [DTT], 2% pharmalytes and trace amount of bromophenol blue) and 100 μl of de-streak solution (GE Health care) were added. The total sample volume was adjusted to 260 μl by adding rehydration buffer (7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 20 mg/ml DTT, 1% pharmalytes, and trace amount of bromophenol blue). The samples were incubated at room temperature for 10 minutes on a shaker and centrifuged for 10 minutes at 16,000 × g. The supernatants were loaded onto 13 cm immobilized pH gradient strip holder (GE Health care). Thirteen cm immobilized pH gradient strips (pH 3 to 10) were put on the loaded samples and 1 ml of mineral oil was added. Isoelectric focusing were run following the protocol provided by the manufacturer (GE Health care). On completion of the Isoelectric focusing, the strips were equilibrated in buffer 1 (50 mmol/L Tris-HCl, pH 8.8, containing 6 mol/L urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue, and 10 mg/ml DTT) for 15 minutes and buffer 2 (50 mmol/L Tris-HCl, pH 8.8, containing 6 mol/L urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue, and 45 mg/ml DTT) for 10 minutes with gentle agitation. The immobilized pH gradient strips were then rinsed in the SDS-gel running buffer, transferred to SDS-gel (10.5% SDS-gel prepared using low florescent glass plates) and sealed with 0.5% (w/v) agarose solution (in SDS-gel running buffer). The electrophoresis was performed at room temperature until the dye fronts run out of the gels. Image scans were performed immediately following the SDS-polyacrylamide gel electrophoresis using Typhoon TRIO (Amersham BioSciences, Piscataway, NJ). The scanned images were then analyzed by Image Quant software (version 5.0, Amersham BioSciences), and subjected to in-gel analysis and cross-gel analysis using DeCyder software version 6.0 (Amersham BioSciences). The ratio change of the protein differential expression was obtained from in-gel DeCyder software analysis. The analytical gels were run in duplicate; a total of 120 μg of proteins were loaded.
Spot Picking, Digestion, and Matrix-Assisted Laser Desorption/ionization Time-of-Flight Mass Spectroscopy
Preparatory gel with total 600 μg of proteins along with labeled proteins was run to pick spots. Protein spots were selected based on cut-off of 1.5 fold change on the 2D- gels were picked by Ettan Spot Picker (Amersham BioSciences) and subjected to in-gel trypsin digestion, peptide extraction, and desalting before matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MS; ABI 4700, Applied Biosystems, Foster City, CA). The GPS Explorer workstation was used to search the database to match both MS and MS/MS data to proteins in database.
Immunoblotting
Isolated microvessels were washed with washing buffer (10 mmol/L Tris-HCl, 5 mmol/L magnesium acetate, pH 8.0) three times to remove the traces of blood and homogenized in a lysis buffer (150 mmol/L NaCl, 50 mmol/L Tris, 1% Triton X-100 supplemented with 5 mmol/L of proteolysis inhibitor mix [Roche, Mannheim, Germany] and 1 mmol/L pepstatin (Sigma) on ice. The tissue was subjected to sonication at 4°C, followed by agitation on a shaker for 30 minutes at 4°C. Samples were centrifuged for 10 minutes at 13,000 rpm. After centrifugation, soluble protein content was measured using a Bio-Rad assay kit. Thirty μg of protein from each sample were diluted 1:1 in SDS-polyacrylamide gel electrophoresis loading buffer and electrophoresed on 12% SDS-polyacrylamide gels (Invitrogen). Proteins were transferred to ImmobilonP membranes (Millipore) and blocked for 2 hours at room temperature with blocking buffer (4% nonfat milk powder) in TBST (20 mmol/L Tris base; 500 mmol/L NaCl; 0.05% Tween 20, pH 7.4). Protein bands were immunolocalized on transblots with above-mentioned primary antibodies followed by horseradish peroxidase-conjugated secondary anti-rabbit or anti-mouse antibody and developed by SuperSignal West Pico Chemiluminescent Substrate (Pierce). Quantification of protein bands was performed in triplicate for each sample by determination of the relative optical density (ImageJ).
Measurement of Mitochondrial Mass in Vivo
To visualize mitochondria in endothelial cells of the renal peritubular capillaries, a mitotracker solution was intrarenally perfused into both control (untreated) and L-NMMA-treated Tie2-GFP mice, according to the previously reported protocol adapted to the kidney.13 Tie2-GFP mice were anesthetized with an intraperitoneal bolus injection of ketamine and xylazine (2 μg/g and 23 μg/g body weight, respectively). Left kidney was exposed via a flank incision. The renal artery was cannulated with a 33-gauge needle (Hamilton, Reno, NV) connected to a 1 ml syringe. Mitotracker Red 580 FM (Molecular Probes Inc, Eugene, OR) was infused for 10 minutes into the left renal artery at a final concentration of 150 μmol/L M. Vehicle for Mitotracker Red 580 FM delivery was 20 mmol/L HEPES (pH 7.4), 150 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L CaCl2, 1 mmol/L MgSO4, 4% dextran, and 1% fetal bovine serum.
Quantitative Real-Time PCR for Mitochondrial DNA
Mitochondrial DNA (mtDNA) content was quantified by real-time reverse transcriptase polymerase chain reaction (RT-PCR) on isolated vessels according to the modified protocols of Duncan et al.14 Briefly, mtDNA was extracted from the kidney microvasculature and real-time RT-PCR was performed using the Stratagen MX 3000 as previously reported.15 Three different mtDNA sequences were assayed. Two different housekeeping genes (UPC and YWHAZ) were used for normalization. As this assay is performed using genomic DNA and mtDNA, any possible changes in gene transcription as a result of treatments are not an issue. mtDNA per nuclear genome was calculated as the ratio of cytochrome b (mitochondrial) DNA to uncoupling protein 1 (nuclear) DNA. Efficiencies of RT-PCR reactions were determined using a dilution series of a standard vascular sample. Quantification was performed using the ΔΔCT method.
Intravital Microscopy
Experiments were performed in animals surgically prepared as described above. During microscopy, animals were placed on a thermal plate (Tokai Hit, Japan) on the stage of an intravital microscope and the left kidney immobilized in a kidney cup. Intravital fluorescence microscopy of the peritubular capillaries was performed using a Nikon Y-FL epifluorescence intravital microscope (Nikon Inc., Melville, NY) equipped with an intensified CoolSNAP HQ tube camera (Photometrics, Tucson, AZ), a filter wheel and a beam splitter (Photometrics Tucson, AZ). Using magnification of ×600, peritubular microvessels were observed in the superficial cortex. Endothelial cell GFP and Mitotracker Red 580 FM fluorescence were recorded and fluorescence intensity of mitochondria was quantified within endothelial cells using MetaVue software (Universal Imaging, Downingtown, PA).
d-Glucose-to-Galactose Switch Assay in Cultured Endothelial Cells
Previously characterized murine endothelial cells, EOMA16 were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 4 mmol/L l-glutamine, 4500 mg/L glucose or 4500 mg/L galactose, 1 mmol/L sodium pyruvate, and 1500 mg/L sodium bicarbonate at 37°C in a 95% air-5% CO2 humidified atmosphere. Where indicated, Dulbecco’s Modified Eagle’s Medium content was switched from d-glucose to the equimolar galactose with L-NMMA (100 μmol/L) ± αketoglutarate (1 mmol/L) from 1 to 4 days.
Senescence and Apoptosis
Senescence-associated β-galactosidase (SA-β-gal) expressed in EOMA cells was analyzed according to the protocol described by Dimri et al.17 Stained cells were viewed and quantified under an inverted microscope at original magnification × 200. At least eight random fields were counted for each culture dish. In addition, quantification of apoptotic cells was performed using In Situ Cell Death Detection kit TMR red (Roche). The data presented were obtained by counting the number of apoptotic cells per 500 cells using fluorescence microscopy. In each experiment at least 15 to 20 randomly chosen fields were examined.
Statistical Analysis
All quantitative data were determined as the mean value ± SEM, unless otherwise noted. Statistical analyses of data were performed by one-way analysis of variance for multiple-group means or by Student’s t-test for comparisons between two group means. Statistical significance was set at the level of P < 0.05 or P < 0.001.
Results
Model Characterization
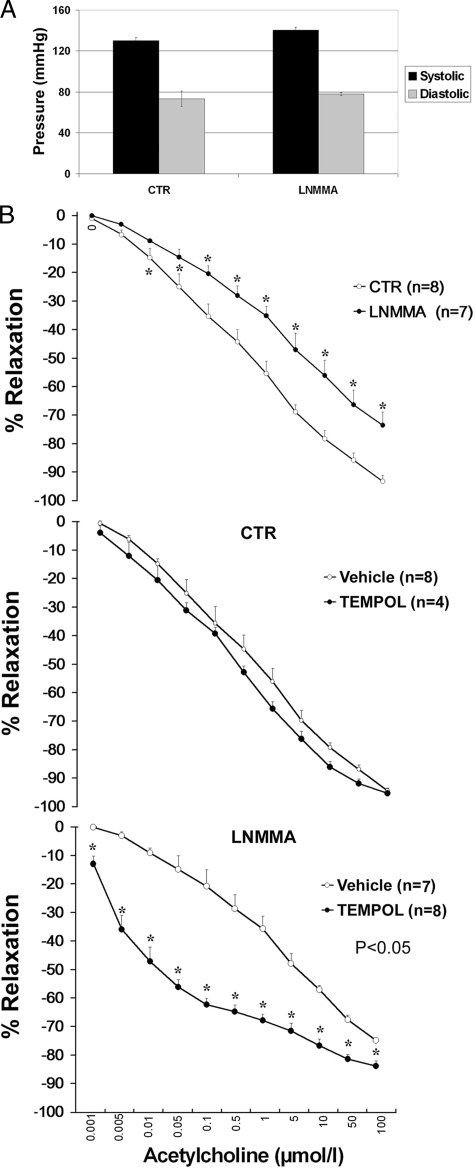
Mice fed L-NNMA were closely monitored for development of hypertension. The dose of 0.3 mg/ml L-NMMA in the drinking water caused no elevation in blood pressure (Figure 1A). This treatment resulted in no changes of weight and was not associated with proteinuria or elevation in plasma creatinine or abnormal glycemia (not shown), thus asserting the preservation of renal function. Screening for changes in cytokines and chemokines showed, however, the elevation of soluble ICAM-1 and VCAM-1, and MMP-9 (Table 1), all markers of endothelial activation, as well as granulocyte macrophage colony-stimulating factor (GM-CSF) and IL1α levels. Furthermore, acetylcholine-induced relaxation of aortic rings, a surrogate indicator of endothelial dysfunction, was modestly reduced in L-NMMA-treated mice (Figure 1B). Of note, exposure of the aortic rings to Tempol was without effect on relaxing responsiveness to acetylcholine in control mice, but greatly amplified the relaxing responsiveness of aortic rings from mice treated with L-NMMA (Figure 1B).
Figure 1.
Characterization of the model of mild chronic NOS inhibition in mice. A: Preservation of blood pressure control in L-NMMA-treated mice. L-NMMA was administered with the drinking water at concentration of 0.3 mg/ml. B: A modest defect in acetylcholine-induced vasorelaxation of aortic rings in L-NMMA-treated mice. Data represent a cumulative dose-response analysis of aortic relaxation (concentration of acetylcholine is shown in abscissa). *statistically significant differences from control.
Table 1.
Serum Levels of Adhesive Molecules, Cytokines, and Chemokines
| CTR (n = 5) | LNMMA (n = 5) | P value CTR versus LNMMA | NOS1 −/ − (n = 5) | P value CTR versus NOS1 −/ − | NOS3 −/ − (n = 5) | P value CTR versus NOS3 −/ − | |
|---|---|---|---|---|---|---|---|
| E-SELECTIN (ng/ml) | 91.14 ± 8.07 | 92.97 ± 7.48 | NS | 100.08 ± 11.61 | NS | 123.02 ± 11.49 | NS |
| MMP9 (ng/ml) | 257.73 ± 22.16 | 454.04 ± 79.95 | P < 0.05 | 91.09 ± 22.67 | P < 0.05 | 193.89 ± 10.64 | NS |
| ICAM-1 (ng/ml) | 12.98 ± 0.91 | 22.18 ± 3.29 | P < 0.05 | 19.68 ± 0.98 | P < 0.05 | 23.66 ± 2.45 | P < 0.05 |
| VCAM-1 (ng/ml) | 1011.61 ± 77.99 | 1319.68 ± 63.02 | P < 0.01 | 1245.41 ± 34.30 | P < 0.05 | 1304.63 ± 44.90 | P < 0.05 |
| MIP-1α (pg/ml) | 20.50 ± 2.56 | 22.11 ± 2.24 | NS | 43.26 ± 5.81 | P < 0.05 | 30.89 ± 7.56 | NS |
| GMCSF (pg/ml) | 6.12 ± 0.79 | 24.33 ± 3.14 | P < 0.01 | 7.80 ± 4.61 | NS | 57.06 ± 6.61 | P < 0.05 |
| MCP1 (pg/ml) | 50.74 ± 26.21 | 63.60 ± 34.10 | NS | 51.79 ± 47.46 | NS | 72.77 ± 4.90 | NS |
| KC (pg/ml) | 32.02 ± 4.83 | 25.94 ± 3.63 | NS | 39.25 ± 24.25 | NS | 37.89 ± 5.03 | NS |
| RANTES (pg/ml) | 10.34 ± 1.15 | 10.01 ± 2.32 | NS | 6.45 ± 0.47 | P < 0.05 | 18.68 ± 5.85 | NS |
| IFNγ (pg/ml) | 5.74 ± 2.53 | 7.25 ± 3.84 | NS | 14.39 ± 5.24 | NS | 36.89 ± 10.63 | P < 0.05 |
| IL1β (pg/ml) | 3.52 ± 0.19 | ND | NS | 9.04 ± 4.35 | NS | 4.42 ± 1.22 | NS |
| IL1α (pg/ml) | 49.78 ± 8.77 | 87.79 ± 7.43 | NS | 34.53 ± 16.53 | NS | 80.05 ± 16.72 | NS |
| GCSF (pg/ml) | 343.46 ± 51.49 | 156.81 ± 31.80 | NS | 58.80 ± 23.01 | P < 0.05 | 362.70 ± 99.40 | NS |
| IP10 (pg/ml) | 514.01 ± 144.67 | 899.51 ± 97.98 | NS | 660.01 ± 105.76 | NS | 1150.81 ± 303.92 | NS |
| IL-6 (pg/ml) | 10.17 ± 1.36 | 13.63 ± 2.04 | NS | 14.87 ± 9.53 | NS | 7.64 ± 2.04 | NS |
| IL-10 (pg/ml) | 79.88 ± 31.31 | 115.28 ± 45.24 | NS | 36.35 ± 13.72 | NS | 152.56 ± 17.73 | NS |
| TNF-α (pg/ml) | 7.727 ± 1.92 | 6.97 ± 1.18 | NS | 3.23 ± 0.23 | NS | 9.15 ± 1.37 | NS |
Each group was compared with control group (CTR), using Mann-Whitney-Wilcoxon test.
Maintenance of normotension and the apparent lack of any clinical manifestations of the mild eNOS inhibition together with the detectable abnormalities in soluble adhesion molecules and endothelium-dependent relaxation argued favorably that the used animal model achieved the goal of generating a preclinical phenotype of endothelial dysfunction. Indeed, previous studies demonstrated that changes in soluble adhesion molecules, sE-selectin, sICAM-1, and sVCAM-1, are reliable predictors of atherosclerosis in general population, thus representing early signs of atherogenesis and endothelial dysfunction.18 Observed effects of Tempol would imply that oxidative stress may play a role in this model of indolent endothelial dysfunction.
DIGE Analysis of Microvasculature
Microvascular trees (Figure 2) obtained from L-NMMA-treated and control animals were subjected to DIGE, as detailed in Methods. The overall number of detectable protein spots was 2200 in control and treated samples (Figure 2). Analysis of differentially expressed species revealed the presence of 14 prominent differentially expressed spots, each of which was further analyzed by in-gel trypsin digestion and matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy. A list of 13 non-redundant proteins identified with high level of confidence on the basis of amino acid sequencing of 6 to 21 peptides per digested spot is presented in Supplemental Table 1 (http://ajp.amjpathol.org). Intriguingly, five non-redundant proteins out of identified proteins were specific for mitochondria and all showed reduced abundance in L-NMMA-treated mice.
Figure 2.
Proteomic analysis of microvasculature obtained from control and L-NMMA-treated mice. A: A representative image of a vascular tree used for analyses. B,C: DIGE-resolved proteins and their abundance in control and experimental vessels respectively. Image scans of differentially expressed spots were performed using Typhoon TRIO (Amersham BioSciences, Piscataway, NJ) and subjected to in-gel analysis and cross-gel analysis using DeCyder software version 6.0 (Amersham BioSciences). The volume ratio change of the protein differential expression was obtained from in-gel DeCyder software analysis. Reported values are expressed as mean ± SD computed from a total of two analytical and one preparative 2D-DIGE.
Immunodetection of DIGE-Identified Proteins
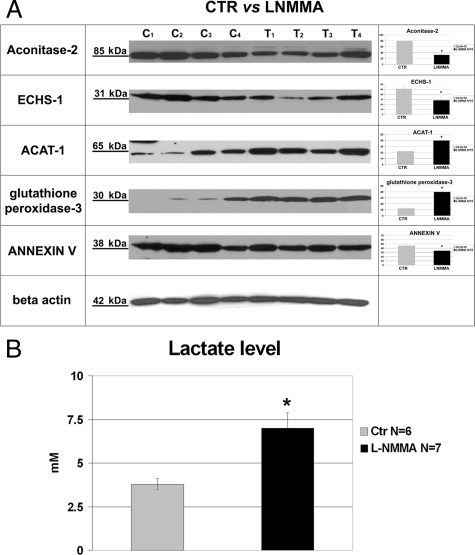
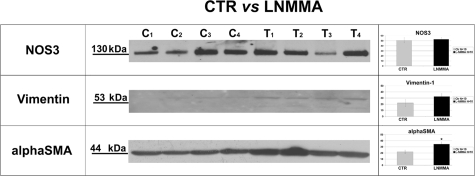
The above findings were validated in individual samples of isolated vascular trees by Western blotting. The expression of aconitase-2 and ECHS-1 was decreased, whereas the abundance of ACAT-1 and pyruvate carboxylase was increased in samples obtained from L-NMMA-treated animals, in accord with DIGE findings (Figure 3A); the expression of ATP-synthase was not detectably changed (not shown). These data confirmed differential expression of two mitochondrial proteins in microvasculature of mice with the mild endothelial dysfunction induced by subpressor doses of L-NMMA. Increased expression of extracellular glutathione peroxidase 3 and decrease in annexin V were confirmed using immunodetection (Figure 3A). We argued that functional consequences of aconitase-2 and ECHS-1 deficiency would result in the inhibition of the Krebs cycle and enhanced pyruvate shunting toward glycolytic pathway. Therefore, we measured lactate levels in the plasma of L-NMMA-treated mice. As shown in Figure 3B and consistent with this prediction, there was a significant elevation in the level of plasma lactate. The eNOS abundance in vascular trees obtained from L-NMMA-treated mice was not different from controls, anti-α-smooth muscle actin was elevated, and vimentin, used as an internal control for contamination with connective tissue, was undetectable (a faint band could be visualized only by pushing the exposure time to 2 hours) (Figure 4).
Figure 3.
Validation of the DIGE results: Western blot analysis of selected proteins. A: Representative Immunodetection of mitochondrial aconitase-2, ECHS-1, ACAT-1, glutathione peroxidase-3, and Annexin-V in individual microvascular trees of control (C1–C4) and L-NMMA-treated mice (T1–T4). Accordingly with 2D-DIGE, treated animals showed a reduced abundance of mitochondrial Aconitase-2 (80.8 ± 7.4 vs. 33.8 ± 6.1, controls versus L-NMMA treated respectively), ECHS-1 (51.9 ± 8.2 vs. 28.8 ± 6.2), and Annexin-V (47.2 ± 5.0 vs. 32.6 ± 5.9). Analysis showed an increased abundance of ACAT-1 (23.7 ± 5.0 vs. 39.4 ± 4.6) and Glutathione peroxidase 3 (12.2 ± 2.4 vs. 40.6 ± 4.4). Right-hand bar diagrams summarize densitometric analysis of individual proteins normalized for β-Actin abundance (N = 10 samples per group). Values are expressed in arbitrary units as means ± SD. *P < 0.05 vs. control. B: Lactate level in control and L-NMMA-treated mice. *P < 0.05 vs. control.
Figure 4.
Equal expression of eNOS and trace amounts of vimentin in microvasculature of control and L-NMMA-treated mice.
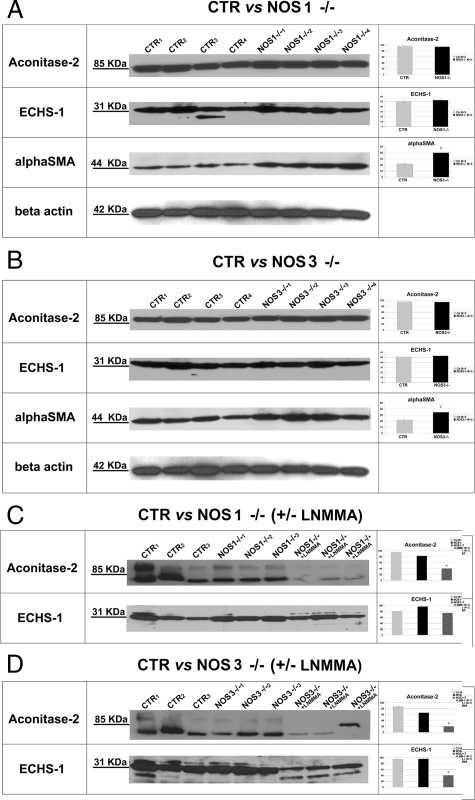
To test whether global eNOS or nNOS deficiency is accompanied by the equivalent changes in the proteome of the vascular wall, microvessels were similarly obtained from NOS-1 and NOS-3-deficient mice. Analysis of the expression of the above proteins showed no resemblance to the profile detected in vessels obtained from mice with chronic mild NOS inhibition (Figure 5). Therefore, we concluded that proteomic profile of our model of mild endothelial dysfunction had unique features not reproducible in the vasculature with the complete lack of constitutive NOS.
Figure 5.
A, B: Representative immunodetection of mitochondrial aconitase-2, ECHS-1, and anti-α-smooth muscle actin in microvascular trees obtained from NOS1−/− and NOS-3−/− mice. C, D: Immunodetection of Aconitase-2 and ECHS-1 in NOS1−/− and NOS3−/− mice ± L-NMMA treatment. Right-hand bar diagrams summarize densitometric analysis of individual proteins normalized for β-actin abundance. *P < 0.05 vs. control.
Mitochondrial Mass
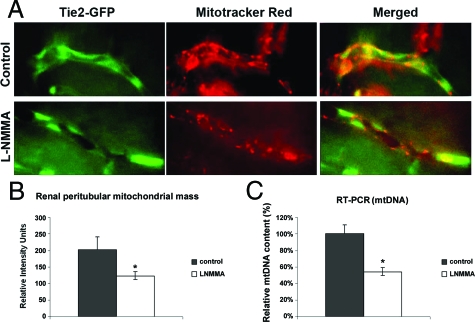
It has been demonstrated that NO plays a major role in mitochondrial biogenesis.19,20 To address the possibility that the results of our proteomic screen of blood vessels in L-NMMA-treated mice could be influenced by the depletion of mitochondrial mass, in vitro and in vivo studies were performed. Microvascular endothelial cells subjected to NOS inhibition showed decreased intensity of MitoTracker signal on fluorescence-activated cell sorter analysis (data not shown). To verify whether this also occurred in L-NMMA-treated mice, MitoTracker was loaded in vivo by infusion into the renal artery of Tie-2/GFP mice, as detailed in Methods. Co-localization of GFP and MitoTracker fluorescence was detected by intravital microscopy and fluorescence intensity of signals analyzed. As shown in Figure 6A, MitoTracker was readily detectable in endothelial cells. Quantitative analysis of fluorescence intensity showed that L-NMMA-treated animals exhibited lower fluorescence of MitoTracker in microvascular endothelia (Figure 6B). These findings were further confirmed by the analysis of mitochondrial DNA in the isolated vascular trees by quantitative RT-PCR (Figure 6C), which showed reduction of mtDNA in vessels obtained from L-NMMA-treated mice. Collectively, these data demonstrated reduction of mitochondrial mass in mice chronically treated with subpressor doses of L-NMMA.
Figure 6.
Intravital videomicroscopy of renal peritubular capillaries loaded with the MitoTracker in Tie-2/GFP mice. A: Consecutive images of GFP (left panels) and MitoTracker (middle panels) fluorescence, and the merged image (right panels). B: Quantitative pixel analysis of MitoTracker fluorescence in control and L-NMMA-treated mice.C: Quantitative RT-PCR for mitochondrial DNA. Data are expressed as percentage change, values are normalized to control mean. *P < 0.05.
Mitochondrial Oxidative Stress
It could not be excluded that in our model of mild endothelial dysfunction induced by low-dose L-NMMA, uncoupling of nitric oxide synthases and excessive generation of superoxide played a role. If that were the case, antioxidant treatment should ameliorate the existing defect in vasorelaxation. To test this prediction, acetylcholine-induced relaxation of aortic rings was performed in control and L-NMMA-treated mice with aortic samples being pre-treated with Tempol (Figure 1B). Inclusion of Tempol into the incubation medium did not affect acetylcholine-induced relaxation of control vessels, but significantly improved relaxation of aortic rings obtained from chronically L-NMMA-treated mice.
Functional Proof of Mitochondriopathy and Reliance on Glycolysis for Cell Survival: d-Glucose-to-Galactose Switch Assay
Having demonstrated reduced abundance of two key enzymes of oxidative phosphorylation, we sought to obtain functional confirmation of these findings. The ability to survive in culture media devoid of d-glucose, but instead containing galactose (thus disabling glycolysis as means of maintaining energy requirements) depends on the activity of the Krebs cycle. Hence, we used a glucose-to-galactose switch assay to test its activity. Glucose-to-galactose switch under these conditions resulted in a significant increase in the proportion of apoptotic and senescent cells. Addition of α-ketoglutarate, as an alternative fuel for the Krebs cycle bypassing aconitase-2 deficiency, salvaged endothelial cells from apoptotic death (Figure 7A,B). Collectively, these data, together with the in vivo demonstration of increased lactate levels, provide a functional confirmation of disabled oxidative phosphorylation in endothelial cells subjected to NOS inhibition and in mice chronically treated with non-pressor doses of L-NMMA.
Figure 7.
Functional proof of mitochondriopathy and reliance on glycolysis for cell survival—d-glucose-to-galactose switch. A: The rate of apoptosis in endothelial cells cultured for 4 days in Dulbecco’s Modified Eagle’s Medium containing either d-glucose or galactose. B: The rate of senescence in endothelial cell after culture for 4 days in Dulbecco’s Modified Eagle’s Medium containing either d-glucose or galactose. *P < 0.05, **P < 0.01.
Discussion
The data presented herein establish a novel model of “subclinical” NOS inhibition detectable only through the targeted analysis of surrogate markers of endothelial dysfunction and identify inhibition of Krebs cycle and reduction in mitochondrial mass as its early manifestations. Specifically, a proteome-wide screen of microvasculature revealed a set of enzymes differentially expressed in animals treated with subpressor doses of L-NMMA. Admittedly, the detection of highly hydrophobic proteins using DIGE is difficult, but this system, nevertheless, allowed us to detect a reasonable number of proteins. Two mitochondrial enzymes, aconitase-2 and ECHS-1, were depleted in the microvasculature, affecting the Krebs cycle and leading to the accumulation of lactate. Notably, chronic L-NMMA treatment, akin to the elevation in ADMA levels, results in uncoupling of endothelial NOS resulting in enhanced generation of superoxide anion, rather than complete inhibition of its activity. Increased generation of superoxide anion appears to be the primary determinant of the impaired relaxing responsiveness to acetylcholine in the aorta of L-NMMA treated mice, since the impairment was mitigated in vessels exposed to the superoxide dismutase mimetic, Tempol.
There is substantial evidence linking mitochondrial biogenesis to NO bioavailability,19,20 which was partially confirmed in our model of mild endothelial dysfunction. Reduced mitochondrial mass could explain the reduction in abundance of aconitase-2 and ECHS-1. However, additional factors need to be invoked to explain the observed differences in the expression of these mitochondrial enzymes in the microvasculature of L-NMMA-treated and two constitutive NOS-deficient mice, NOS-1 and NOS-3 knockouts. Considering the uncoupling of eNOS and enhanced mitochondrial generation of reactive oxygen species as judged by the results of MitoSox fluorescence, as well as adaptive up-regulation of extracellular glutathione peroxidase-3 in the case of L-NMMA treatment, it is conceivable that the combination of oxidative stress and reduced NO bioavailability results in a unique proteomic profile: reduced mitochondrial mass and selective depletion of at least two key enzymes of the Krebs cycle, aconitase-2, and ECHS-1.
The decrease in aconitase-2 should slow down the Krebs cycle and therefore result in the reduced rate of citrate conversion to isocitrate and subsequently reduced rate of consumption of pyruvate. The excess pyruvate is converted to lactate by lactate dehydrogenase, as is the case in L-NMMA-treated mice. The structural predilection of iron-sulfur center in aconitase-2 to reactive oxygen species and peroxynitrite21 may in part explain its selective vulnerability. Prolonged exposure of mitochondria to oxidants results in disassembly of the [4Fe-4S]2+ cubane cluster, carbonylation, and degradation of the enzyme,22 potentially establishing the link between oxidant stress and enzyme inactivation, as detected during cardiac ischemia/reperfusion23 and eventual reduction in aconitase-2 abundance. Aconitase-2 has been previously found to be specifically targeted by oxidative damage during aging.24
In addition, inhibition of the Krebs cycle could be compounded by the depletion of ECHS-1. Enoyl-CoA hydratase-1 catalyzes the second step of β-oxidation in fatty acid metabolism, the process by which fatty acids in the form of acyl-CoA molecules are processed in mitochondria and/or peroxisomes to generate acetyl-CoA, the entry molecule for the Krebs cycle.
Our recent studies provide evidence that tissue hypoxia does not develop in this model of subpressor dosing of L-NMMA.25 Hence, the observed switch to glycolytic metabolism in the face of normoxia is reminiscent of the Warburg effect, the phenomenon of preferential glycolysis in tumor cells.26 Most likely, selective depletion of aconitase-2 and ECHS-1 are responsible for the inhibition of Krebs cycle in our model, making it phenomenologically similar to the Warburg effect in cancer cells. In this context, the role of NO appears to be complex. Acute actions of NO result in “metabolic hypoxia” due to inhibition of mitochondrial electron transport.27 Chronic uncoupling of NOS, on the other hand, confers the same end-result, “metabolic hypoxia,” but through selective depletion of aconitase-2 and ECHS-1. Inhibition of the Krebs cycle may therefore augur the development of endothelial dysfunction. Moreover, it is possible to envisage that the chronic L-NMMA-induced mitochondriopathy may engender the accelerated development of atherosclerosis and insulin resistance3,28,29 further underscoring the pathogenetic significance of this early manifestation of endothelial dysfunction.
Supplementary Material
Footnotes
Address reprint requests to Michael S. Goligorsky MD, PhD, Departments of Medicine and Pharmacology, Renal Research Institute, Division of Nephrology, New York Medical College, Valhalla 10595, NY. E-mail: Michael_Goligorsky@nymc.edu.
Supported in part by NIH grants DK052783, DK45462, and DK054602 (M.S.G.).
References
- Koenig W. Cardiovascular biomarkers: added value with an integrated approach? Circulation. 2007;116:3–5. doi: 10.1161/CIRCULATIONAHA.107.707984. [DOI] [PubMed] [Google Scholar]
- Khot UN, Khot MB, Bajzer CT, Sapp SK, Ohman EM, Brener SJ, Ellis SG, Lincoff AM, Topol EJ. Prevalence of conventional risk factor in patient with coronary heart disease. JAMA. 2003;290:898–904. doi: 10.1001/jama.290.7.898. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation. 2006;113:2335–2362. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- Karra R, Vemullapalli S, Dong C, Herderick EE, Song X, Slosek K, Nevins JR, West M, Goldschmidt-Clermont PJ, Seo D. Molecular evidence for arterial repair in atherosclerosis. Proc Natl Acad Sci USA. 2005;102:16789–16794. doi: 10.1073/pnas.0507718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondjers C, He L, Takemoto M, Norlin J, Asker N, Hellstrom M, Lindahl P, Betsholtz C. Microarray analysis of blood microvessels from PDGF-B and PDGF-Rbeta mutant mice identifies novel markers for brain pericytes. FASEB J. 2006;20:1703–1705. doi: 10.1096/fj.05-4944fje. [DOI] [PubMed] [Google Scholar]
- King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A, Vailaya A, Kincaid R, Tsalenko A, Deng DX, Connolly A, Zhang P, Yang E, Watt C, Yakhini Z, Ben-Dor A, Adler A, Bruhn L, Tsao P, Quertermous T, Ashley EA. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- Bogdan C. Nitric oxide and the regulation of gene expression. Trends Cell Biol. 2001;11:66–75. doi: 10.1016/s0962-8924(00)01900-0. [DOI] [PubMed] [Google Scholar]
- Peterson TE, Poppa V, Ueba H, Wu A, Yan C, Berk BC. Opposing effects of reactive oxygen species and cholesterol on endothelial nitric oxide synthase and endothelial cell caveolae. Circ Res. 1999;85:29–37. doi: 10.1161/01.res.85.1.29. [DOI] [PubMed] [Google Scholar]
- O'Riordan E, Mendelev N, Patschan S, Patschan D, Eskander J, Cohen-Gould L, Chander P, Goligorsky MS. Chronic NOS inhibition actuates endothelial-mesenchymal transformation. Am J Physiol Heart Circ Physiol. 2007;292:H285–H294. doi: 10.1152/ajpheart.00560.2006. [DOI] [PubMed] [Google Scholar]
- Linder AE, Weber DS, Whitesall SE, D'Alecy LG, Webb RC. Altered vascular reactivity in mice made hypertensive by nitric oxide synthase inhibition. J Cardiovasc Pharmacol. 2005;46:438–444. doi: 10.1097/01.fjc.0000175879.14994.63. [DOI] [PubMed] [Google Scholar]
- Fuller AJ, Hauschild BC, Gonzalez-Villalobos R, Awayda MS, Imig JD, Inscho EW, Navar LG. Calcium and chloride channel activation by angiotensin II-AT1 receptors in preglomerular vascular smooth muscle cells. Am J Physiol Renal Physiol. 2005;289:F760–F767. doi: 10.1152/ajprenal.00422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura H, Parthasarathi K, Quadri S, Issekutz AC, Bhattacharya J. Mechano-oxidative coupling by mitochondria induces proinflammatory responses in lung venular capillaries. J Clin Invest. 2003;111:691–699. doi: 10.1172/JCI17271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JG, Fong J, Medeiros D, Finck B, Kelly D. Insulin-resistant heart exhibits a mitochondrial biogenesis response driven by the peroxisome proliferator-activated receptor-alpha/PGC-1alpha gene regulatory pathway. Circulation. 2007;115:909–917. doi: 10.1161/CIRCULATIONAHA.106.662296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungvari Z, Labinskyy N, Gupte S, Chander P, Edwards J, Csiszar A. Dysregulation of mitochondrial biogenesis in vascular endothelial and smooth muscle cells of aged rats. Am J Physiol Heart Circ Physiol. 2008;294:H2121–H2128. doi: 10.1152/ajpheart.00012.2008. [DOI] [PubMed] [Google Scholar]
- Obeso J, Weber J, Auerbach R. A hemangioendothelioma-derived cell line: its use as a model for the study of endothelial cell biology. Lab Invest. 1990;63:259–269. [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, Peacocket M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulaki I, Murray GD, Lee AJ, Rumley A, Lowe GD, Fowkes FG. C-reactive protein, interleukin-6, and soluble adhesion molecules as predictors of progressive peripheral atherosclerosis in the general population: Edinburgh Artery Study. Circulation. 2005;112:976–983. doi: 10.1161/CIRCULATIONAHA.104.513085. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Clementi E, Carruba MO, Moncada S. Defective mitochondrial biogenesis: a hallmark of the high cardiovascular risk in the metabolic syndrome? Circ Res. 2007;100:795–806. doi: 10.1161/01.RES.0000259591.97107.6c. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Carruba MO. Nitric oxide and mitochondrial biogenesis. J Cell Sci. 2006;119:2855–2862. doi: 10.1242/jcs.03062. [DOI] [PubMed] [Google Scholar]
- Tortora V, Quijano C, Freeman B, Radi R, Castro L. Mitochondrial aconitase reaction with nitric oxide. S-nitrosoglutathione, and peroxynitrite: mechanisms and relative contributions to aconitase inactivation. Free Radic Biol Med. 2007;42:1075–1088. doi: 10.1016/j.freeradbiomed.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Bulteau A, Ikeda-Saito M, Szweda L. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochem. 2003;42:14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- Bulteau A-L, Lundberg K, Ikeda-Saito M, Isaya G, Szweda L. Reversible redox-dependent modulation of mitochondrial aconitase and proteolytic activity during in vivo cardiac ischemia/reperfusion. Proc Natl Acad Sci USA. 2005;102:5987–5991. doi: 10.1073/pnas.0501519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L-J, Levine R, Sohal R. Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci USA. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoessel A, Paliege A, Theilig F, Addabbo F, Ratliff B, Waschke J, Patschan D, Goligorsky MS, Bachmann S. Indolent course of tubulointerstitial disease in a model of sub-pressor, low-dose nitric oxide synthase inhibition. Am J Physiol Renal Physiol. 2008;295:F717–F725. doi: 10.1152/ajprenal.00071.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Moncada S, Erusalimsky JD. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat Rev Mol Cell Biol. 2002;3:214–220. doi: 10.1038/nrm762. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Duchen MR. Endothelial mitochondria: contributing to vascular function and disease. Circ Res. 2007;100:1128–1141. doi: 10.1161/01.RES.0000261970.18328.1d. [DOI] [PubMed] [Google Scholar]
- Madamanchi NR, Runge MS. Mitochondrial dysfunction in atherosclerosis. Circ Res. 2007;100:460–473. doi: 10.1161/01.RES.0000258450.44413.96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.