Abstract
Overexpression of the DEK gene is associated with multiple human cancers, but its specific roles as a putative oncogene are not well defined. DEK transcription was previously shown to be induced by the high-risk human papillomavirus (HPV) E7 oncogene via E2F and Rb pathways. Transient DEK overexpression was able to inhibit both senescence and apoptosis in cultured cells. In at least the latter case, this mechanism involved the destabilization of p53 and the decreased expression of p53 target genes. We show here that DEK overexpression disrupts the normal differentiation program in a manner that is independent of either p53 or cell death. DEK expression was distinctly repressed upon the differentiation of cultured primary human keratinocytes, and stable DEK overexpression caused epidermal thickening in an organotypic raft model system. The observed hyperplasia involved a delay in keratinocyte differentiation toward a more undifferentiated state, and expansion of the basal cell compartment was due to increased proliferation, but not apoptosis. These phenotypes were accompanied by elevated p63 expression in the absence of p53 destabilization. In further support of bona fide oncogenic DEK activities, we report here up-regulated DEK protein levels in both human papilloma virus-positive hyperplastic murine skin and a subset of human squamous cell carcinomas. We suggest that DEK up-regulation may contribute to carcinoma development at least in part through increased proliferation and retardation of differentiation.
The human DEK protein was originally identified as a fusion with the NUP214/CAN nucleoporin in a subset of patients with acute myeloid leukemia1 and was independently purified as a protein that modulates the topology of SV40 minichromosomes.2 DEK is abundantly expressed in proliferating cells, and a majority of the protein is bound to chromatin, whereas a small fraction is bound to RNA.3 The 43-kDa nuclear phosphoprotein is the only member of its family, and contains a conserved central SAP DNA binding domain with homology to SAF-A/B, acinus, and PIAS,4 and a second DNA binding motif within the C-terminus.5,6 DNA binding as well as DEK self-association can be regulated by C-terminal phosphorylation and N-terminal acetylation.7,8 Preferential DEK association with structured DNA templates and its ability to induce positive supercoils into circular DNA templates in vitro have led to the notion that DEK serves as an architectural protein.9,10,11,12 Multiple reports have implicated DEK in replication,2 positive and negative regulation of transcription13,14,15,16,17,18,19 as well as mRNA processing.20,21,22 How these activities translate into putative oncogenic DEK functions is presently unclear.
In agreement with the existence of such oncogenic DEK activities, we and others have described this molecule as an inhibitor of cell death and senescence in vitro. RNA-interference (i)-mediated depletion of DEK in primary human foreskin keratinocytes (HFKs) resulted in p53 dependent apoptosis,23 whereas the depletion of DEK in primary human fibroblasts was sufficient for senescence induction.24 DEK knockdown was also recently shown to render cells sensitive to genotoxic DNA damaging agents that activate poly(ADP-ribose)polymerase.25 Conversely, the overexpression of DEK partially inhibited HeLa cell senescence,26 as well as baseline apoptotic rates of primary HFKs.23 Transcriptional DEK up-regulation has been reported for many human cancers including bladder carcinoma,27 hepatocellular carcinoma,28,29 glioblastoma,30 melanoma,31 retinoblastoma,32 oral squamous cell carcinoma,33 advanced breast cancer,34 and acute myelogenous leukemia types that do not harbor the above translocation.35 We have described transcriptional DEK up-regulation in response to the high-risk, but not low-risk human papillomavirus (HPV) E7 oncogene through retinoblastoma pathways, suggesting that DEK may also be a marker, perhaps mediator, of HPV-associated cancers.26 Direct control of DEK expression by E2F family members together with the mapping of relevant binding sites within the promoter was subsequently reported by the Müller laboratory.36
Here we confirm the prediction that DEK protein levels are up-regulated in HPV16 E7 transgenic murine skin in vivo, and strongly induced in a subset of HPV positive human malignancies. By definition, aspiring cancer cells must acquire resistance to differentiation stimuli in addition to overcoming cell death. We show that human keratinocyte differentiation is accompanied by decreased DEK expression, and that the observed repression is important, since retroviral DEK overexpression in human keratinocytes resulted in hyperplasia in organotypic epithelial rafts. This was reflected by increased numbers of cell layers and proliferative expansion of the keratin 14- and p63-positive basal cell compartment, as well as by increased p63 and decreased loricrin expression in methylcellulose embedded keratinocytes. Importantly, p63 induction in the context of DEK overexpression appeared to occur in the absence of p53 regulation. Taken together, our data support a new, p53-independent role for DEK, which may mediate keratinocyte resistance to differentiation-associated cell cycle arrest and actively contribute to cancer development.
Materials and Methods
Cell Culture
Normal immortalized keratinocytes (NIKs)37 were maintained on irradiated murine J2 3T3 feeder fibroblasts in F media. F media is a mixture of three parts Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) to 1 part Ham’s F12 media (Invitrogen, Carlsbad, CA) supplemented with the following components: 5% fetal bovine serum, adenine (24 μg/ml), cholera toxin (8.4 ng/ml), epidermal growth factor (EGF; 10 ng/ml), hydrocortisone (2.4 μg/ml), insulin (5 μg/ml), 1% antibiotics, and 0.2% fungizone.38 Primary HFKs were prepared from human foreskins as described previously23 and maintained in Epilife Medium plus human keratinocyte growth supplement (Cascade Biologics, Portland, OR) with antibiotics.
Plasmids and Viral Constructs
The DEK open reading frame was amplified using the forward primer F- 5′-ATGTCCGCCTCGGCC-3′ and the reverse primer R- 5′- TCAAGAAATTAGCTCTTTTACAG-3′. The cDNA was cloned into the pGEM-Teasy vector (Promega, Madison, WI) according to the manufacturer’s instructions. The DEK cassette was sequenced, digested with NotI and cloned into a NotI digested FMEV type vector pSF91-I-enhanced green fluorescent protein (GFP) eGFP-PRE (R780) obtained from the Baum laboratory.39 Vector particles were generated in the Viral Vector Core facility at CCHMC and were pseudotyped with the feline endogenous virus (RD114) envelope protein.
Retroviral Infections
NIKs were transduced with R780 or R780-DEK retroviral supernatant containing 8 μg/ml polybrene. The cells were washed with PBS after three hours and overlaid with fresh media. Cell pools were sorted for GFP expression using a fluorescence-activated cell sorter (VantageSEDiVa; Becton Dickinson, San Jose, CA). Cells were collected and maintained on feeder cells as above.
Western Blot Analyses
The cells were washed with PBS and whole cell lysates were harvested with RIPA buffer (1% Triton, 1% deoxycholate, 0.1% SDS, 0.16M NaCl, 10 mmol/L Tris pH 7.4, and 5 mmol/L EDTA) supplemented with a protease inhibitor cocktail (Pharmingen, San Diego, CA). Cancerous and normal human tissue samples were homogenized on ice in RIPA buffer. Protein concentrations were determined using Bradford Reagent (BioRad, Hercules, CA). Equal amounts of protein were boiled in SDS sample buffer and resolved by SDS-polyacrylamide electrophoresis. Proteins were transferred to a polyvinylidene difluoride membrane (BioRad, Hercules, CA) and probed with either the DEK monoclonal antibody (BD Biosciences, San Diego, CA), an involucrin monoclonal antibody (Sigma, St. Louis, MO), a p63 specific monoclonal antibody (Santa Cruz biotechnology, Santa Cruz, CA), a p53 (Ab-6) monoclonal antibody (Calbiochem, San Diego, CA), a cyclin A polyclonal antibody (Santa Cruz biotechnology, Santa Cruz, CA), or an actin specific monoclonal antibody, a generous gift from James Lessard. On the next day, the membrane was washed with TNET (10 mmol/L Tris, 2.5 mmol/L EDTA, 50 mmol/L NaCl, and 0.1% Tween 20) and secondary anti-mouse or anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham, Piscataway, NJ) were added for 30 minutes. Membranes were then washed and exposed to enhanced chemilluminescence reagents (Perkin Elmer, Boston, MA) and the protein bands were detected by autoradiography.
Analyses of Human Foreskin and Cancer Tissue
Human foreskins were collected in Dulbecco’s Modified Eagles Medium (DMEM) with antibiotics and were used for morphological and immunofluorescence analyses after fixation in 4% paraformaldehyde and paraffin embedding. Human normal or cancer tissue was obtained with informed consent and approval by the CCHMC Institutional Review Board (IRB protocol number 03-8–40x) through the Division of Gynecological Oncology at the University of Cincinnati, the Cooperative Human Tissue Network and the Gynecological Oncology Group at the Ohio State University. Normal tissue was derived from HPV unrelated hysterectomies. The samples were subjected for testing for the presence of 27 HPV types in the Division of Pathology using the Roche Linear Array (Roche Diagnostics, Indianapolis, IN). Snap frozen tissue was homogenized in RIPA buffer and used for immunoblot analysis as described above.
Mice
The K14E7 mice (line 2304, originally referred to as K14E6TTLE7 mice) used in this study were previously described.40 They express the HPV16 E7 gene from the human keratin 14 (K14) promoter, which drives expression of the transgene to stratified epithelia including the epidermis. The K14E7 mice were maintained in the hemizygous state on the inbred FVB/N genetic background. Tissues were harvested from K14E7 transgenic and nontransgenic FVB/N mice, fixed in formalin, paraffin-embedded, and thin (5 micron) sectioned. All use and handling of mice were performed in the American Association for Accreditation of Laboratory Animal Care-approved McArdle Laboratory Cancer Center Animal Care Facility according to an the Institutional Animal Care and Use Committee-approved protocol to P.F.L.
Organotypic Raft Cultures
A total of 1 × 106 sorted cells were plated for the generation of epithelial organotypic rafts as described.41 Briefly, dermal equivalents were generated from a collagen premix containing rat tail collagen (Upstate Biotechnology, Lake Placid, NY), antibiotics, F12 media, fetal bovine serum and sodium hydroxide, as well as 7.5 × 104 normal J2 3T3 mouse fibroblasts. NIKs infected with either R780-DEK or control vector were seeded on top of the gels. On day 4 after seeding, gels were lifted to the liquid air interface and allowed to stratify for eleven days. Bromodeoxyuridine was added to the media for 16 hours before harvest at a final concentration of 10 μmol/L. Rafts were harvested, fixed in 4% paraformaldehyde, and embedded in paraffin. Depending on the experiment, 5- to 8-μm sections were analyzed morphologically following H&E staining or for the detection of protein markers by immunofluorescence.
Methylcellulose Assays
R780-DEK or control NIKs were submerged in F media containing 1.5% methylcellulose, and allowed to differentiate for 24 hours. Cells were harvested from the methylcellulose by stringent washes with cold PBS. RNA was extracted from the cells for subsequent quantitation of relative transcript levels by real time reverse transcription (RT)-PCR (see below).
Immunofluorescence Microscopy
Raft sections were deparaffinized in xylene, and rehydrated in successive changes of ethanol and water. Antigen retrieval was performed by heating the sections in 10 mmol/L NaCitrate, pH 6.0, in a rice cooker for 20 minutes, and allowed to cool to room temperature. Sections were blocked with 5% donkey serum for 30 minutes at room temperature before the addition of primary antibody in dilution buffer (3% bovine serum albumen, 0.05% Tween 20, 0.5% Triton X-100, and 0.04% NaAzide in PBS) for one hour at 37°C. Dilutions were 1:20 for the monoclonal DEK antibody, 1:250 for the monoclonal E-cadherin antibody (BD biosciences, San Diego, CA), 1:2000 for the polyclonal K14 antibody (Covance, Denver, PA), 1:100 for the monoclonal p63 antibody (Santa Cruz biotechnology, Santa Cruz, CA), 1:100 for the monoclonal bromodeoxyuridine antibody (Zymed, Carlsbad, CA), and 1:200 for the monoclonal K10 antibody (Covance, Denver, PA). Raft sections were washed and incubated in secondary anti-mouse or anti-rabbit antibody conjugated to rhodamine or fluorescein isothiocyanate (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) for 30 minutes at 37°C. Sections were washed with PBS and counterstained with Vectashield mounting media containing 4,6-diamidino-2-phenylindole (DAPI; Vector laboratories, Burlingame, CA) and coverslipped. Immunofluorescence detection was via a Zeiss fluorescence microscope (Zeiss, Thornwood, NY), and images were captured using ×20 and ×40 magnification with an Axiovision camera (Lucas Microscope Service, Skokie, IL) driven by Axiovision software.
Quantitative RT-PCR
RNA was harvested in Trizol reagent (BRL, Bethesda, MD), treated with DNase and reverse transcribed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). Real time quantitative RT-PCR was performed using SYBR Green PCR Master Mix on an ABI 7300 system (Applied Biosystems, Foster City, CA). The following primer pairs were used for PCR at a concentration of 0.4 ng/ul each: c-Abl F-5′-TGCCCAGAGAAGGTCTATGAACT-3′, R-5′-AACATTGTTTCAAAGGCTTGGTG-3′, p63 F-5′-AGCAGCACCAGCACTTACTTCAGA-3′, R-5′-TGATAAGCTGGCTCACAGAAGGCA-3′, R-5′-TCTTGCTCAGGCAGTCCCTTTACA-3′, and loricrin F-5′-AGCGGCTGCATCATCAGTGG-3′, R-5′-TTCTGCTG-GGTCTGGTGGCAGAT-3′. All graphs represent mRNA levels relative to c-Abl expression.
Results
DEK Expression is Limited to the Basal Cell Layer in Normal Human Epithelium
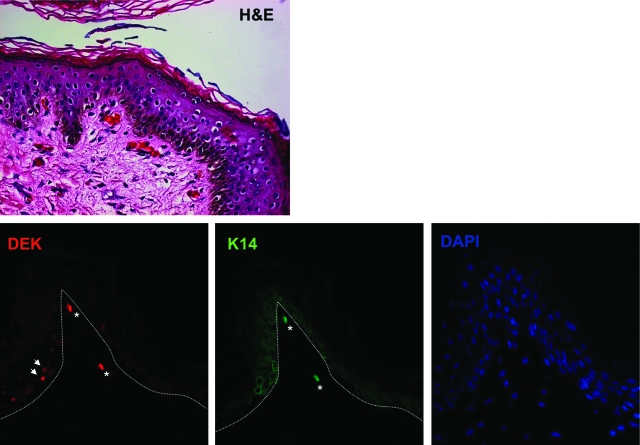
To determine normal DEK localization in human epithelium, human foreskin tissue was used for the immunofluorescence detection of DEK in comparison with the basal cell marker K14 (Figure 1). Epithelial morphology is depicted following H&E staining (top panel). The expression of both K14 and DEK was only observed in a subset of cells in the basal cell layer (bottom panel). Cells that were positive for a nuclear DEK signal are denoted with white arrows, autofluorescent red blood cells are denoted with an asterisk. These data suggested that DEK expression is limited to the proliferating, basal cell compartment in vivo, and that its expression is specifically inhibited during keratinocyte differentiation.
Figure 1.
DEK protein expression is restricted to the basal cell compartment in normal human epidermis. Human foreskin tissue was fixed, embedded and sectioned for analysis. Morphological appearance of human foreskin tissue following H&E staining is shown on the top. Immunofluorescence using DEK and K14 specific primary antibodies followed by rhodamine and fluorescein isothiocyanate-conjugated secondary antibodies respectively is shown in the bottom panels. The epidermal-dermal transition is indicated by the dotted line.White arrows indicate nuclear DEK staining, white asterisks indicate autofluorescent red blood cells.
Suppression of DEK Protein Expression Occurs during Primary Keratinocyte Differentiation
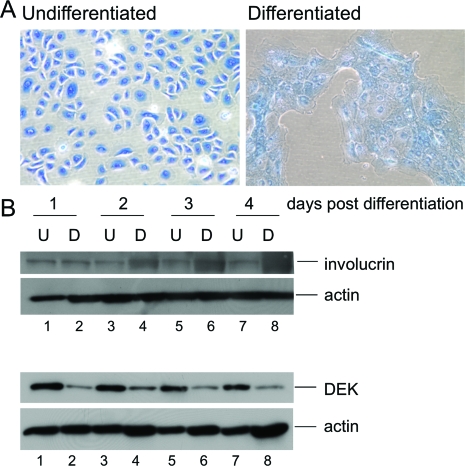
To determine whether DEK might be down-regulated during differentiation in vitro, primary monolayer HFKs were subjected to differentiation medium or left undifferentiated for the subsequent detection of DEK expression (Figure 2). Cultured primary keratinocytes exhibit many characteristics of basal epithelial cells in the presence of 0.06 mmol/L calcium, and differentiation can be induced by the addition of 1 mmol/L calcium. The underlying mechanism is not clearly understood, but is likely relevant to human skin, where a gradient of extracellular calcium concentrations from the basal to the most differentiated cell layers may serve as the primary differentiation signal.42 Loss of contact formation with the underlying substratum and a tightening of intercellular contacts via adherens junctions and desmosomes were reflected by the typical morphological changes (Figure 2A) and expression of the terminal differentiation marker involucrin (Figure 2B). Immunoblot analysis demonstrated that decreased DEK expression is indeed a consequence of differentiation as predicted from the above localization in the basal cell layer (Figure 2B).
Figure 2.
DEK suppression occurs during keratinocyte differentiation in vitro. A: Keratinocyte morphology following calcium addition. Primary human foreskin keratinocytes (HFKs) were either left untreated or subjected to 1 mmol/L CaCl2 and 10% fetal bovine serum to induce differentiation. Cells were fixed and stained with methylene blue. Images were captured 24 hours post treatment. B: Western blot analyses. HFKs were treated as above and total cell protein lysates were harvested between days 1 to 4 post-treatment. Lysates were subjected to Western blot analyses for the detection of DEK, involucrin and actin as a loading control.
DEK Overexpression Causes Epithelial Hyperplasia
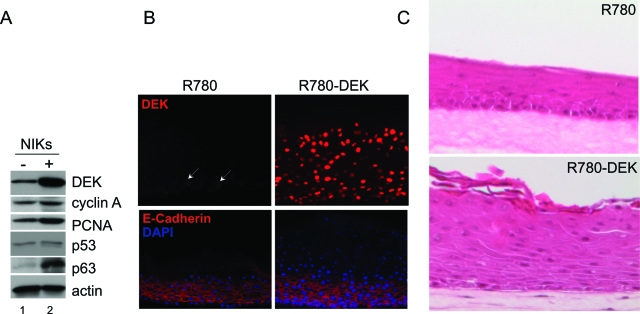
To determine whether the observed DEK repression was important for differentiation, we transduced NIKs with a retroviral DEK overexpression vector containing an EGFP cassette, and subsequently sorted for the EGFP expressing cell population. EGFP-expressing backbone vector transduced control cells were treated identically. NIKs are a spontaneously immortalized cell line, which has retained responsiveness to differentiation stimuli.43 Sorted, transduced DEK expressing versus control cells were subjected to organotypic epithelial raft culture. DEK overexpression in R780-DEK transduced cells was confirmed by Western blot analysis (Figure 3A, top panel), as well as by immunofluorescence detection in raft sections (Figure 3B). Levels of the cyclin A proliferation marker were perhaps slightly, albeit not necessarily reproducibly, up-regulated by DEK in monolayer cultures. Interestingly, DEK overexpression also did not affect the levels of p53 protein in the monolayer cultures (Figure 3A) or organotypic rafts (data not shown). This was in contrast to our previous results, where DEK expression in primary HFKs or HeLa cells repressed p53 protein levels and expression of p53 target genes such as p21CIP1 and bax.23 The lack of p53 repression in this system could be due to the use of NIKs, different culture conditions or DEK expression through retroviral rather than adenoviral delivery. However, we did observe a slight, but reproducible increase in the p53-related p63 protein in the DEK overexpressing NIKs (Figure 3A).
Figure 3.
DEK overexpression results in epithelial hyperplasia in organotypic rafts. A: Western blot analyses. NIKs infected with R780 empty vector (−) or DEK (+) were harvested for total cell protein lysates. Equal amounts of protein were subjected to Western blot analyses for the detection of DEK, Cyclin A, p53, p63 and actin. B: Immunofluorescence microscopy. Organotypic raft sections were generated as described in the Materials and Methods, and then incubated with DEK or E-cadherin antibody followed by rhodamine conjugated secondary antibody. Images were captured using an immunofluorescence microscope. C: H&E staining. Sections of NIKs rafts either overexpressing DEK or controls were stained with H&E and images were captured using light microscopy. White arrows denote DEK positive nuclei.
DEK expressing versus control NIKs were next used for the generation of organotypic epithelial rafts as described in the Materials and Methods. In agreement with its localization in normal human epithelium (Figure 1), DEK expression was weakly detectable in the basal cell layer of the R780 control rafts. However, DEK expression was absent in the suprabasal, differentiated compartment. In contrast, the R780-DEK transduced cells exhibited high levels of DEK expression throughout the raft (Figure 3B). E-cadherin expression was not affected in the DEK overexpressing rafts indicating normal cell-cell adhesion. However, E-cadherin positive cell layers were expanded upwards, as was the presence of nucleated cells detectable by DAPI staining (Figure 3B). Morphological examination of the rafts revealed a hyperplastic epithelial phenotype as a consequence of DEK overexpression (Figure 3C), a finding which supports a role for oncogenic DEK activities in human cancer. Similar phenotypes were observed in two other, independently derived, raft cultures (data not shown).
DEK Overexpression Inhibits the Epithelial Differentiation Program
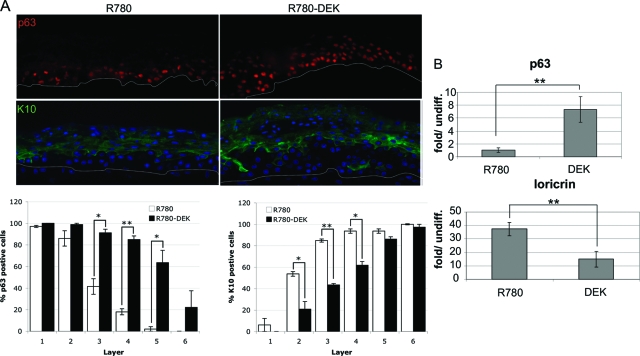
We hypothesized that the above hyperplastic phenotype may be due to the deregulation of keratinocyte differentiation. To investigate this possibility, we detected the differentiation marker K10, as well as the basal cell markers K14 and p63 in raft sections (Figure 4). Several isoforms exist within the p63 protein family. However, detection of p63 by the pan-p63 antibody reveals predominantly the ΔNp63 form in keratinocytes (reviewed in44). Isoform specific knockdown of ΔNp63 has recently verified the predominance of this growth stimulatory over its growth inhibitory TAp63 counterpart in this particular cell type.45 Keratin 10 (K10) is normally only expressed in suprabasal cells, whereas p63 is expressed in the basal cell layer in human epithelium. This was indeed observed in the control rafts (Figure 4A, left panels). However, in the presence of overexpressed DEK, an expansion of the basal at the expense of the differentiated cell compartment was reflected by the presence of additional p63-positive and fewer K10-positive cell layers (Figure 4A, right panels). These data suggest that under differentiation conditions, DEK activities expand the basal cell compartment containing epithelial stem and transit amplifying cells.
Figure 4.
DEK overexpression inhibits the normal differentiation program. A: Immunofluorescence microscopy. Raft sections were incubated with p63 or K10 primary antibodies followed by incubation with secondary rhodamine or fluorescein isothiocyanate-conjugated secondary antibody. Sections were counterstained with DAPI and images were captured using an immunofluorescence microscope. Positive cells were quantitated for each cell layer in two independent experiments and were averaged; SE is depicted. *P < 0.05, **P < 0.01. B: Quantitative RT-PCR. NIKs containing control or DEK expressing retrovirus were submerged in 1.5% methylcellulose to induce differentiation for 24 hours. Cells were harvested for RNA isolation and equal amounts of RNA were reverse transcribed and amplified quantitatively using SYBR green for detection. Levels of cDNA in each sample were calculated relative to the levels of c-abl, and fold mRNA levels are shown for differentiated relative to the respective undifferentiated cell populations. Real time RT-PCR data were derived from four independent experiments, SE is shown **P < 0.01.
To verify the observed DEK-mediated differentiation delay in a more quantitative manner, we exploited the known differentiation-dependent transcriptional regulation of p63 and the suprabasal marker loricrin46 (Figure 4B). R780-DEK and control NIKs were submerged in methylcellulose to initiate differentiation, and mRNA was harvested for real-time RT-PCR after 48 hours. In agreement with the above raft culture data, p63 expression was up-regulated by DEK overexpression in the differentiated cells. Conversely, loricrin expression was increased on differentiation in a manner that could be repressed by DEK (Figure 4B). Taken together, our data emphasize the importance of DEK repression during differentiation, and support DEK activities that delay differentiation and expand the basal cell compartment. Since neoplastic transformation is generally believed to originate within stem and perhaps transit amplifying cells in this compartment, such DEK activities by themselves could be envisioned as oncogenic in nature.
DEK Overexpression Increases Cell Proliferation
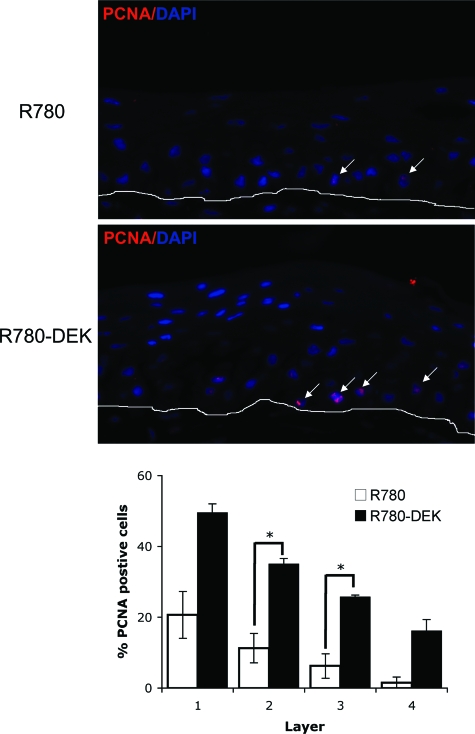
We next determined whether decreased rates of apoptosis or increased proliferation were responsible for the hyperplastic phenotype in DEK overexpressing epithelium (Figure 5). We did not observe differences in apoptosis between the control and DEK overexpressing rafts as determined by active caspase 3 and terminal deoxynucleotidyl transferase dUTP nick-end labeling staining (data not shown). However, statistically significant increases in the number of proliferating cell nuclear antigen (PCNA)-positive basal cells were found in DEK positive compared with control rafts (Figure 5). This suggests that DEK overexpression stimulates cellular proliferation within the basal as well as suprabasal cell compartment under differentiating conditions, and in a manner that is fundamentally different from its previously described cell death inhibitory functions.
Figure 5.
DEK overexpression results in increased basal cell proliferation. Immunofluorescence microscopy. R780-DEK transduced NIK raft sections compared with controls were incubated with PCNA monoclonal antibody followed by rhodamine conjugated secondary antibody. Rafts were counterstained with DAPI, images were captured and % PCNA positive cells were quantitated. Positive cells were quantitated from four images each per embedded sample per organotypic raft. Two independent rafts are represented. SE is depicted, *P < 0.05.
DEK Protein Levels Are Induced in Murine and Human Epithelial Cancers
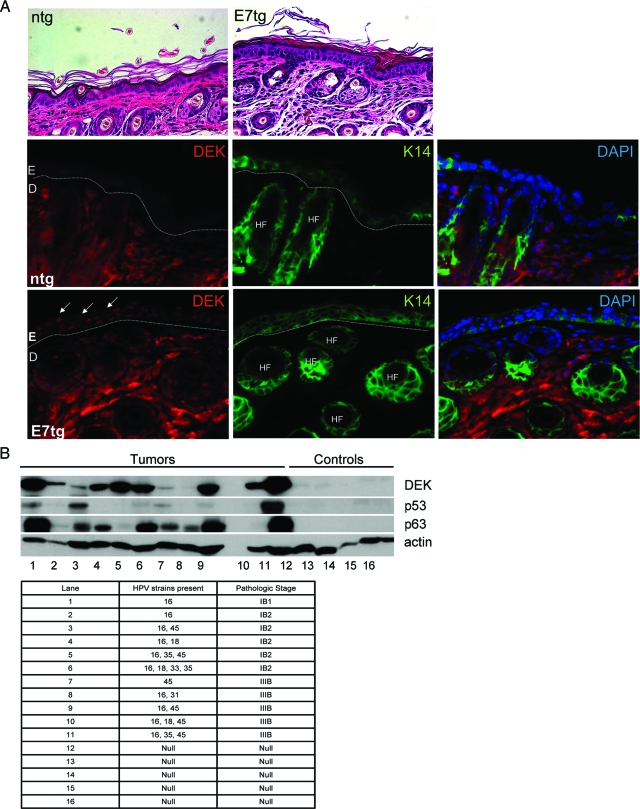
In vitro, DEK expression was induced by the high risk HPV E7 oncogene,26 and when overexpressed on its own, DEK was sufficient for causing hyperplasia (Figure 3). To determine whether DEK up-regulation may be a consequence of HPV16 E7 expression in vivo, murine skin sections from K14E7 transgenic versus nontransgenic mice were used for the detection of DEK by immunofluorescence. K14E7 mice are known to exhibit epithelial hyperplasia with expansion of the proliferating compartment in the skin.40 Morphological comparison and direct detection of the K14 positive basal compartment confirmed this phenotype (Figure 6A, top), and further demonstrated marked E7-dependent induction of nuclear DEK expression in the basal cell layers (Figure 6A, bottom panels).
Figure 6.
DEK protein levels are up-regulated in K14E7 transgenic mouse epithelium and in a subset of human HPV positive cancers. A: E7 transgenic versus control nontransgenic mouse skin sections were subjected to morphological examination following H&E staining. DEK and K14 detection was performed by immunofluorescence as described in the Materials and Methods, and the sections were counterstained with DAPI. E: epidermis, D: dermis, HF: hair follicle. B: Western blot analyses. HPV typed normal or cancerous human tissue samples were homogenized, lysed and equal amounts of total protein were subjected to Western blot analyses for the detection of DEK, p63, p53, and actin.
We next determined whether a similar up-regulation of DEK expression might be observed in human HPV positive tumors. To this end, immunoblot analysis was performed for five HPV negative normal human cervical tissue samples and eleven HPV positive cancer samples (Figure 6B). Expression of p63 was induced in 8 out of 11 samples to various extents, in line with the reported strong correlation between p63 up-regulation and squamous tumor morphology.47 In this limited sample set, DEK expression was markedly elevated in 10 out of 11 carcinomas. No correlation was observed between DEK and p53 expression as predicted from Figure 3A. In fact, some tumors expressed high levels of p53 in agreement with reports of up to 41% of high-grade squamous intraepithelial lesions overexpressing p53.48,49,50
A partial correlation between DEK and p63 expression was observed, in that eight tumor samples overexpressed both proteins. Such a correlation would be supported by our in vitro findings whereby DEK overexpression stimulated p63 expression at the level of mRNA and protein (Figures 3A and 4A). Conversely, RNA interference experiments demonstrate that DEK knockdown causes p63 repression (data submitted for publication). Based on our tumor data, however, if a link between p63 and DEK exists in vivo, is not expected to be absolute: tumor tissue in lane 8 exhibited p63, but not DEK overexpression, whereas tumor tissue in lanes 5 and 10 exhibited high DEK, but low or no p63 overexpression, respectively. Importantly, DEK expression was not associated with tumor stage, in agreement with a recent publication that reports increased DEK expression in the great majority of uterine cancers regardless of tumor stage.51 Taken together, these findings suggest that oncogenic DEK protein functions are reflected by the up-regulation of DEK protein levels in HPV positive murine and human tissue in vivo.
Discussion
Apart from bypassing cell death and senescence, epithelial transformation must also involve resistance to terminal differentiation stimuli that normally limit keratinocyte proliferation. Our data show that DEK protein expression was specifically repressed during keratinocyte differentiation and that the observed repression was important as DEK overexpression in organotypic raft cultures was sufficient for hyperplasia induction. Differentiation is known to be accompanied by Rb hypophosphorylation and cell cycle arrest.52 Based on the reported regulation of DEK transcription by Rb/E2F pathways,26,36 differentiation-associated DEK repression is therefore most likely mediated by members of the retinoblastoma protein family.
In contrast to our previous DEK overexpression studies in primary HFKs and HeLa cells, DEK did not affect cellular growth or death of NIKs under normal conditions in monolayer culture (data not shown). However, in line with a recent report from the Ferrando-May laboratory,25 DEK overexpression did protect from apoptosis when these same cells were challenged with the DNA damaging agent camptothecin (data not shown). A number of important differences could account for the lack of anti-apoptotic DEK activities in NIKs under baseline culture conditions: First, immortalized NIKs were used rather than primary HFKs. Immortality per se, however, is not sufficient to account for the observed differences, since DEK also repressed apoptosis in HeLa23 and U2OS cells (manuscript in preparation). Secondly, significant differences in cell culture conditions in previous compared with the above experiments could account for the different results. In particular, the NIKs were co-cultured with irradiated feeder cells. Such culture conditions are thought to represent less stressful conditions endowing primary cells with an extended cellular life span and delayed p16 expression.53,54 We have recently detected DNA damage induction in response to DEK depletion in a manner that lies upstream of p53 induction (manuscript in preparation, and25). The lack of p53 destabilization and apoptosis suppression in NIKs may be a reflection of a lower degree of culture stress including DNA damage in this system. Finally, DEK delivery was achieved stably from a retroviral expression vector in contrast to previous transient adenoviral delivery, with possible differential effects of the virus backbone and/or time-dependent cellular adaptation. Based on the lack of p53 protein repression in response to DEK in proliferating (Figure 3A) and differentiated (data not shown) cells, molecular mechanisms underlying the observed hyperplasia do not appear to involve the destabilization of p53. However, we cannot rule out at present the possible inhibition of p53 transcriptional activities. In fact, such inhibition could theoretically be achieved by some of the reported dominant negative functions of the ΔNp63 protein,55,56,57 since we observed p63 up-regulation in the presence of DEK. Incidentally, p63 is widely overexpressed in squamous cell carcinomas,44 further strengthening a correlation between DEK overexpression and human epithelial cancer development.
Our data demonstrate that DEK overexpression under differentiation conditions is sufficient for the induction of expanded proliferation in the basal cell compartment and resulting epithelial hyperplasia. DEK positive organotypic rafts contained more nuclei in the suprabasal portion of the raft (Figure 3B). The observed epidermal thickening correlated with an expansion of the K14 and p63-marked basal cell compartment (Figure 4A), and an increased number of K10 negative cell layers. DEK mediated repression of the differentiation marker loricrin was also observed in a methylcellulose differentiation system, and correlated with increased proliferation in the basal cell compartment. We propose that oncogenic DEK functions in differentiating epithelium are driven by increased proliferation in the basal cell compartment in a manner that is fundamentally different from the observed apoptosis repression in other cell systems. While loricrin is the only differentiation marker that was quantitatively assessed at the level of mRNA here, it is tempting to speculate that overexpression of DEK may suppress the expression of differentiation-associated genes more globally. If true, the regulation of differentiation genes might occur directly through known DEK transcriptional repressor functions15 and/or indirectly through p63.45 Taken together, our data represent the first line of evidence that DEK may exhibit pro-neoplastic activities. Its up-regulated expression in hyperplastic E7 positive mouse epidermis and in a subset of high and low stage HPV positive human tumors is suggestive of oncogenic DEK activities early in the development of human tumors. Based on Figure 6B, an absolute correlation between DEK and p63 expression does not exist in all squamous cell carcinomas in vivo despite the strong correlation between DEK and p63 expression in vitro. Alternatively, the lack of a stronger correlation between DEK and p63 protein levels in these studies may be due to our specific focus on the detection of Δp63, a single isoform of the up to twelve p63 isoforms that have been reported. The tumors analyzed in Figure 6B represent HPV-infected low and high stage tumors but unfortunately do not significantly differ with regard to tumor grade since all cases were diagnosed as moderately to poorly differentiated. Therefore our data does not allow an assessment of the correlation between DEK overexpression and tumor grade. However, Oncomine gene expression data were analyzed and supports elevated DEK expression as a function of tumor grade in head and neck cancers (see Supplemental Figure S1 at http://ajp.amjpathol.org). Interestingly, up to 30% of head and neck squamous cell carcinomas are associated with HPV infection, and it is therefore tempting to speculate that DEK induction in these cancers might function to suppress cellular differentiation. Future investigation of clinical and molecular differences between tumors expressing high versus those expressing baseline levels of DEK should allow for insights into its potential as a therapeutic marker and as a target in human cancer.
Supplementary Material
Acknowledgments
We thank Dr. Nilsa Ramirez and the Cooperative Human Tissue Network as well as Dr. Qualman and the Gynecological Oncology Group at the Ohio State University. We thank Dan Marmer and the Flow Cytometry and Cell Sorting Services at CCHMC, as well as Donna Diorio, Meredith Taylor, Mary Rolfes, and Pam Groen in the Department of Pathology for excellent technical assistance, and Gail Deutsch and James Wells for helpful discussions. We thank Han van der Loo and the CCHMC Viral Vector Core for retroviral production, and James Lessard for the monoclonal actin antibody.
Footnotes
Address reprint requests to Susanne Wells, Cincinnati Children’s Hospital Medical Center, Division of Hematology/Oncology, TCHRF Room S7.206, MLC 7013, 3333 Burnet Ave., Cincinnati, OH 45229, E-mail: Susanne.Wells@cchmc.org.
Supported by Public Health Service grants CA102357 and CA116316 from the National Cancer Institute to S.I.W., CA098428 to P.F.L., and by Public Health Service grant HL079193 to K.W.-B.T.M., Wise-Draper was supported by a training grant T32 CA59268 from the National Cancer Institute and by an Illick fellowship from the Albert J. Ryan Foundation.
References
- von Lindern M, Fornerod M, Soekarman N, van Baal S, Jaegle M, Hagemeijer A, Bootsma D, Grosveld G. Translocation t(6;9) in acute non-lymphocytic leukaemia results in the formation of a DEK-CAN fusion gene. Baillieres Clin Haematol. 1992;5:857–879. doi: 10.1016/s0950-3536(11)80049-1. [DOI] [PubMed] [Google Scholar]
- Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14:1308–1312. [PMC free article] [PubMed] [Google Scholar]
- Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276:26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Devany M, Kotharu NP, Matsuo H. Solution NMR structure of the C-terminal domain of the human protein DEK. Protein Sci. 2004;13:2252–2259. doi: 10.1110/ps.04797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, Cole PA, Markovitz DM. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280:31760–31767. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HG, Scholten I, Gruss C, Knippers R. The distribution of the DEK protein in mammalian chromatin. Biochem Biophys Res Commun. 2007;358:1008–1014. doi: 10.1016/j.bbrc.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277:24988–24994. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein–an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Campillos M, Garcia MA, Valdivieso F, Vazquez J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31:1571–1575. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Hilfinger JM, Markovitz DM. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J Biol Chem. 2001;276:25804–25812. doi: 10.1074/jbc.M006454200. [DOI] [PubMed] [Google Scholar]
- Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14:548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- Hu HG, Illges H, Gruss C, Knippers R. Distribution of the chromatin protein DEK distinguishes active and inactive CD21/CR2 gene in pre- and mature B lymphocytes. Int Immunol. 2005;17:789–796. doi: 10.1093/intimm/dxh261. [DOI] [PubMed] [Google Scholar]
- Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, Lee KS, Woo KM, Baek JH, Choo JK, Seo SB. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580:3217–3222. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- Sammons M, Wan SS, Vogel NL, Mientjes EJ, Grosveld G, Ashburner BP. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281:26802–26812. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, Nickerson JA, Awrey D, Greenblatt J, Grosveld G, Blencowe BJ. The acute myeloid leukemia-associated protein. DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150:309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol. 2007;81:2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Burkle A, Markovitz DM, Ferrando-May E. DEK is a poly(ADP-ribose)-acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Munger K, Wells SI. The human DEK proto-oncogene is a senescence inhibitor and an up-regulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan B, Williamson K. Molecular markers for predicting recurrence, progression and outcomes of bladder cancer (do the poster boys need new posters?). Curr Opin Urol. 2004;14:277–286. doi: 10.1097/00042307-200409000-00006. [DOI] [PubMed] [Google Scholar]
- Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita M, Tanaka K, Shuda M, Yamamoto M. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–4996. [PubMed] [Google Scholar]
- Lu ZL, Luo DZ, Wen JM. Expression and significance of tumor-related genes in HCC. World J Gastroenterol. 2005;11:3850–3854. doi: 10.3748/wjg.v11.i25.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroes RA, Jastrow A, McLone MG, Yamamoto H, Colley P, Kersey DS, Yong VW, Mkrdichian E, Cerullo L, Leestma J, Moskal JR. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156:191–198. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- Grottke C, Mantwill K, Dietel M, Schadendorf D, Lage H. Identification of differentially expressed genes in human melanoma cells with acquired resistance to various antineoplastic drugs. Int J Cancer. 2000;88:535–546. doi: 10.1002/1097-0215(20001115)88:4<535::aid-ijc4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Corson TW, Gallie BL. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer. 2007;46:617–634. doi: 10.1002/gcc.20457. [DOI] [PubMed] [Google Scholar]
- Nagpal JK, Das BR. Identification of differentially expressed genes in tobacco chewing-mediated oral cancer by differential display-polymerase chain reaction. Eur J Clin Invest. 2007;37:658–664. doi: 10.1111/j.1365-2362.2007.01841.x. [DOI] [PubMed] [Google Scholar]
- Abba MC, Sun H, Hawkins KA, Drake JA, Hu Y, Nunez MI, Gaddis S, Shi T, Horvath S, Sahin A, Aldaz CM. Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Mol Cancer Res. 2007;5:881–890. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas S, Nagy B, Elonen E, Aventin A, Larramendy ML, Sierra J, Ruutu T, Knuutila S. Aberrant expression of HOXA9. DEK, CBL and CSF1R in acute myeloid leukemia. Leuk Lymphoma. 2003;44:1935–1941. doi: 10.1080/1042819031000119299. [DOI] [PubMed] [Google Scholar]
- Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, Muller H. DEK expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5:1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- Allen-Hoffmann BL, Schlosser SJ, Ivarie CA, Sattler CA, Meisner LF, O'Connor SL. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line. NIKS J Invest Dermatol. 2000;114:444–455. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- Genther SM, Sterling S, Duensing S, Munger K, Sattler C, Lambert PF. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J Virol. 2003;77:2832–2842. doi: 10.1128/JVI.77.5.2832-2842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klump H, Schiedlmeier B, Vogt B, Ryan M, Ostertag W, Baum C. Retroviral vector-mediated expression of HoxB4 in hematopoietic cells using a novel coexpression strategy. Gene Ther. 2001;8:811–817. doi: 10.1038/sj.gt.3301447. [DOI] [PubMed] [Google Scholar]
- Herber R, Liem A, Pitot H, Lambert PF. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 1996;70:1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol. 1997;71:5161–5172. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP. Signal transduction pathways controlling the switch between keratinocyte growth and differentiation. Crit Rev Oral Biol Med. 1999;10:442–457. doi: 10.1177/10454411990100040201. [DOI] [PubMed] [Google Scholar]
- Lambert PF, Ozbun MA, Collins A, Holmgren S, Lee D, Nakahara T. Using an immortalized cell line to study the HPV life cycle in organotypic “raft” cultures. Methods Mol Med. 2005;119:141–155. doi: 10.1385/1-59259-982-6:141. [DOI] [PubMed] [Google Scholar]
- Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TY, Chen BF, Yang YC, Chen H, Wang Y, Cviko A, Quade BJ, Sun D, Yang A, McKeon FD, Crum CP. Histologic and immunophenotypic classification of cervical carcinomas by expression of the p53 homologue p63: a study of 250 cases. Hum Pathol. 2001;32:479–486. doi: 10.1053/hupa.2001.24324. [DOI] [PubMed] [Google Scholar]
- Bahnassy AA, Zekri AR, Saleh M, Lotayef M, Moneir M, Shawki O. The possible role of cell cycle regulators in multistep process of HPV-associated cervical carcinoma. BMC Clin Pathol. 2007;7:4. doi: 10.1186/1472-6890-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LW, Chao SL, Hwang JL, Chou YY. Down-regulation of p27 is associated with malignant transformation and aggressive phenotype of cervical neoplasms. Gynecol Oncol. 2002;85:524–528. doi: 10.1006/gyno.2002.6666. [DOI] [PubMed] [Google Scholar]
- Tsuda H, Hashiguchi Y, Nishimura S, Kawamura N, Inoue T, Yamamoto K. Relationship between HPV typing and abnormality of G1 cell cycle regulators in cervical neoplasm. Gynecol Oncol. 2003;91:476–485. doi: 10.1016/j.ygyno.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Wu Q, Li Z, Lin H, Han L, Liu S, Lin Z. DEK overexpression in uterine cervical cancers. Pathol Int. 2008;58:378–382. doi: 10.1111/j.1440-1827.2008.02239.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Hashimoto K, Yoshikawa K. Dephosphorylation of the retinoblastoma gene product induced by differentiation and its relevancy to growth inhibition in normal human keratinocytes. J Dermatol Sci. 1994;8:171–177. doi: 10.1016/0923-1811(94)90050-7. [DOI] [PubMed] [Google Scholar]
- Darbro BW, Lee KM, Nguyen NK, Domann FE, Klingelhutz AJ. Methylation of the p16(INK4a) promoter region in telomerase immortalized human keratinocytes co-cultured with feeder cells. Oncogene. 2006;25:7421–7433. doi: 10.1038/sj.onc.1209729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez RD, Morales CP, Herbert BS, Rohde JM, Passons C, Shay JW, Wright WE. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WM, Siu WY, Lau A, Poon RY. How many mutant p53 molecules are needed to inactivate a tetramer?. Mol Cell Biol. 2004;24:3536–3551. doi: 10.1128/MCB.24.8.3536-3551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helton ES, Zhang J, Chen X. The proline-rich domain in p63 is necessary for the transcriptional and apoptosis-inducing activities of TAp63. Oncogene. 2008;27:2843–2850. doi: 10.1038/sj.onc.1210948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.